Abstract
Identifying genomic elements underlying phenotypic adaptations is an important problem in evolutionary biology. Comparative analyses learning from convergent evolution of traits are gaining momentum in accurately detecting such elements. We previously developed a method for predicting phenotypic associations of genetic elements by contrasting patterns of sequence evolution in species showing a phenotype with those that do not. Using this method, we successfully demonstrated convergent evolutionary rate shifts in genetic elements associated with two phenotypic adaptations, namely the independent subterranean and marine transitions of terrestrial mammalian lineages. Our original method calculates gene-specific rates of evolution on branches of phylogenetic trees using linear regression. These rates represent the extent of sequence divergence on a branch after removing the expected divergence on the branch due to background factors. The rates calculated using this regression analysis exhibit an important statistical limitation, namely heteroscedasticity. We observe that the rates on branches that are longer on average show higher variance, and describe how this problem adversely affects the confidence with which we can make inferences about rate shifts. Using a combination of data transformation and weighted regression, we have developed an updated method that corrects this heteroscedasticity in the rates. We additionally illustrate the improved performance offered by the updated method at robust detection of convergent rate shifts in phylogenetic trees of protein-coding genes across mammals, as well as using simulated tree data sets. Overall, we present an important extension to our evolutionary-rates-based method that performs more robustly and consistently at detecting convergent shifts in evolutionary rates.
Keywords: convergent evolution, evolutionary rates, molecular evolution
Introduction
Understanding the relationship between phenotype and genotype is a fundamental question in biological research. A mechanistic characterization of this relationship hinges on our ability to define how specific genetic elements contribute to biological processes at the molecular, cellular, and organismal level. High-throughput sequencing has enabled new experimental approaches that have uncovered a wealth of genetic elements with putative regulatory roles across tissues (ENCODE Project Consortium 2012; Andersson et al. 2014; Romanoski et al. 2015). However, identifying the precise biological functions of these elements remains a challenge. Even beyond noncoding elements, the precise biological roles of many protein-coding genes are still poorly understood, and many genes with statistical disease associations still lack a mechanistic explanation (Pennacchio et al. 2013; Radivojac et al. 2013; Sa et al. 2013; Shlyueva et al. 2014). While experimental validation for functional annotation remains challenging, there is considerable interest in developing new tools that can use existing data resources to further elucidate the function of genetic elements. These approaches have the potential to improve the diagnosis of disease susceptibility and the development of therapeutic interventions (Manolio et al. 2009; Esteller 2011).
Computational approaches learning from patterns of convergent phenotypic evolution across species provide a complementary approach to predict genotype–phenotype associations. The natural world is replete with examples of phenotypic convergence ranging from the independent evolution of flight in birds and mammals to diving in species that transitioned from a terrestrial to marine habitat to loss of complex phenotypes such as eyesight in animals colonizing the subterranean niche. Genome-scale studies aimed at identifying the genetic basis of phenotypic convergence take advantage of the growing availability of whole genome sequences for species across several orders, alongside the development of comparative methods to predict orthologous sequences (Eisen 1998; Pellegrini et al. 1999; Li et al. 2014). A common approach in such studies is to identify convergence at the molecular level, including substitutions at specific nucleotide or amino acid sites (Zhang and Kumar 1997; Parker et al. 2013; Stern 2013; Foote et al. 2015; Thomas and Hahn 2015; Zou and Zhang 2015). An alternative strategy to investigate the genetic basis of convergence is to search for convergent changes at the level of larger functional regions rather than specific nucleotide or amino acid sites. Sets of genes associated with a phenotype can respond to convergent changes in the selective pressure on the phenotype through nonidentical changes in the same gene, and as such, sites-based methods can fail to detect them. These limitations have encouraged researchers to search for convergent shifts in evolutionary rates of individual protein-coding genes and more recently conserved noncoding elements (Lartillot and Poujol 2011; Hiller et al. 2012; Chikina et al. 2016; Marcovitz et al. 2016; Prudent et al. 2016). An increased selective constraint can manifest as a slower evolutionary rate, whereas faster evolutionary rates can result from a release of constraint or from adaptation. Thus phenotypic associations for genetic elements can be predicted from correlated changes in their evolutionary rates on phylogenetic branches corresponding to the phenotypic change. Example approaches based on evolutionary rates include the Forward/Reverse Genomics methods that have identified protein-coding and noncoding genetic elements showing convergent regression in subterranean mammals and loss of limb-regulatory elements in snake lineages (Hiller et al. 2012; Marcovitz et al. 2016; Prudent et al. 2016; Roscito 2017).
We previously developed an evolutionary-rates-based method to identify genetic elements showing convergent shifts in evolutionary rates associated with two distinct phenotypic transitions (Chikina et al. 2016; Partha et al. 2017). Our original method calculates gene-specific evolutionary rates using a linear model, and gene-trait associations are inferred using correlations of these rates with the phenotype of interest. A genome-wide scan using this method to find protein-coding genes associated with the transition to the marine environment identified hundreds of genes that showed accelerated evolutionary rates on three marine mammal lineages (Chikina et al. 2016). These accelerated genes were significantly enriched for functional roles in pathways important for the marine adaptation including muscle physiology, sensory systems, and lipid metabolism. More recently, using our methods, we detected an excess of vision-specific genes as well as enhancers that showed convergent rate acceleration on the branches corresponding to four subterranean mammals (Partha et al. 2017). Genes showing convergent rate shifts associated with these two phenotypic transitions typically follow one of the following modes of change in the selective pressure—1) relaxation of constraint and 2) positive selection. Marine-accelerated and subterranean-accelerated genes identified in earlier scans were further probed using phylogenetic models of selective pressure to identify the underlying evolutionary process. In both cases, we found an excess of genes under relaxed constraint, as well as a smaller number of genes under positive selection. Overall, genome-scale efforts both from our group and others to find genetic elements responding to convergent changes in the selective pressures in their environment are gaining momentum in accurately describing precise genotype–phenotype associations.
Our original evolutionary-rates method has an important statistical limitation, namely strong mean–variance trends in the computed evolutionary rates. The distributions of branch lengths of gene trees in phylogenetic data sets are influenced by the choice of species, divergence from the most recent common ancestor, and species-specific properties, such as generation time, in addition to gene-specific constraints on the sequence evolution. These factors cause large differences in the average lengths as well as the variance of the branch lengths across the branches studied. In this article, we illustrate how this limitation can adversely impact the confidence with which we infer phenotypic associations for genetic elements, in particular making them sensitive to certain factors in phylogenomic analyses including choice of taxonomic groups and average rates of sequence divergence on phylogenetic branches showing the convergent phenotype. We demonstrate how introducing long branches in phylogenetic trees via the inclusion of distantly related species impacts the reliable estimation of evolutionary rates using gene trees across mammals, as well using a first-of-its-kind model for simulating gene trees. We present key improvements to our methods that address these limitations and overcome them. The next section New Approaches presents a detailed walk-through of our current approach to calculate relative evolutionary rates, the illustration of mean–variance trends (heteroscedasticity) in these rates, and our methodological updates that correct for the problem of heteroscedasticity in the rates. We subsequently demonstrate the improved reliability in relative rate calculations using our updated method, and, more importantly, in the robust detection of convergent rate shifts across a range of evolutionary scenarios in real and simulated phylogenetic data sets.
New Approaches
Original Relative-Evolutionary-Rates Method for Predicting Phenotypic Associations of Genetic Elements
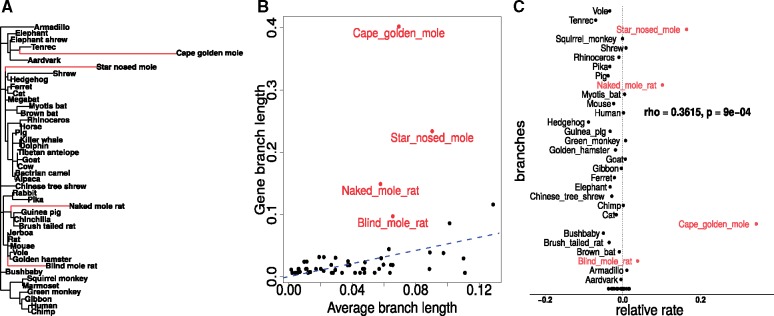
Our method infers genetic elements associated with a convergent phenotype of interest based on correlations between that phenotype and the rates of evolution of genetic elements. As input, the phenotype is encoded as a binary trait on a phylogenetic tree, and the evolution of each genetic element is similarly described by phylogenetic trees with the same fixed topology. Figure 1 provides an illustration of our method capturing the convergent acceleration of the Lens Intrinsic membrane 2 protein Lim2 on four subterranean mammal branches. We use maximum likelihood approaches to estimate the amount of sequence divergence of each genetic element on branches of the phylogenetic tree (Yang 2007). Using each tree’s branch lengths, we calculate the average tree across the individual trees reflecting the expected amount of divergence on each branch. Relative evolutionary rates (RERs) on individual trees are then calculated as the residuals of a linear regression analysis where the dependent variable corresponds to the branch lengths of individual trees, and the independent variable corresponds to branch lengths of the average tree. Thus the relative rates reflect the gene-specific rate of divergence in each branch, factoring out the expected divergence on the branch due to genome-wide effects (such as mutation rate and time since speciation). The relative rates method works downstream of estimating the trees, and hence considers protein-coding gene trees, noncoding genetic element trees, and simulated gene trees equivalently. For the sake of simplicity, we refer to the relative rates on the branches of each tree as the gene-specific relative rate; the term gene could in principle be referring to a protein-coding gene, noncoding genetic element, or a simulated tree depending on the data set being studied.
Fig. 1.
Predicting gene-trait associations using relative rates method. (A) Lens Intrinsic Membrane 2 (Lim2) protein-coding gene tree. Our phylogenetic data set is comprised of trees constructed from alignments of protein-coding genes in the mammalian genome across 59 species of placental mammals. (B) Relative rates on branches of phylogenetic trees are calculated using linear regression. (C) Gene-trait associations are identified using correlations of relative rates of the gene with binary trait of interest.
Estimating Mean–Variance Trends in Relative Rates
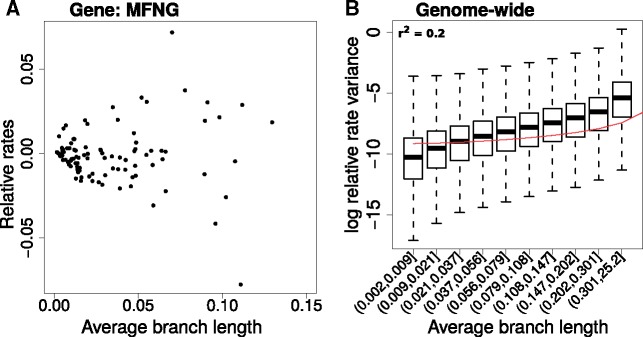
Our original method calculates the gene-specific rates by correcting for the genome-wide effects on branch lengths using linear regression. Consequently, the variance of the relative rates on individual branches strongly depends on the average length of the branch, illustrated here using an example protein-coding gene tree for MFNG, Manic Fringe Homolog Drosophila (fig. 2A). We see that longer branches have relative rates showing a higher variance, as can be inferred from the increasing spread of the relative rates. This pattern becomes clearer when we plot the genome-wide variance in relative rates for branches of different average lengths (fig. 2B). In statistical terms, the relative rates are heteroscedastic, meaning they show unequal variance across the range of values of the dependent variable, here the average branch length. The presence of a nonconstant mean–variance trend in the residuals stands in violation of one of the assumptions underlying linear regression, namely homoscedasticity, or constant variance of residuals with respect to the dependent variable. More importantly, we suspect that this heteroscedasticity of the relative rates adversely affects the confidence with which we can infer rate shifts on specific branches. For example, the presence of a mean–variance trend can increase the likelihood of observing higher relative rates on longer branches by chance, rather than due to gene-specific changes reflecting changes in selective pressure. A potential negative consequence could be a higher proportion of false positives while inferring convergent rate changes on such branches.
Fig. 2.
Heteroscedasticity in the relative rates computed using current method. (A) Relative rates on branches of Manic Fringe (MFNG) gene tree, calculated using original method. Points represent branches of the gene tree, with relative rates computed on the branches plotted against the genome-wide average length. Heteroscedasticity in the relative rates can be visualized as the increase in the variance of the relative rates with increasing average branch length. (B) Genome-wide mean–variance trends in relative rates. The logarithm of the relative rate variance within each bin is shown, where branches are binned based on their average lengths across all gene trees. Bin ranges were chosen to provide equal numbers of observations per bin. Higher variance in relative rates is observed with increasing branch lengths, and the extent of this heteroscedasticity is calculated using the “r-squared” of the quadratic model between the variables plotted.
Updated Method to Calculate Relative Rates
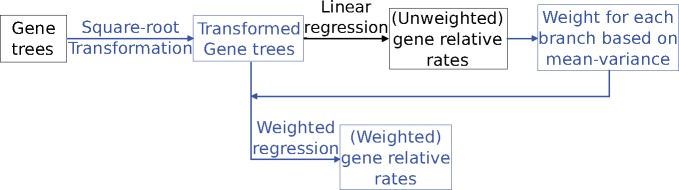
In this study, we present an approach relying on a combination of data transformation and weighted linear regression to calculate relative evolutionary rates that addresses the statistical limitations resulting from relative rates calculated using naive linear regression. The proposed method updates are based on the ideas presented in Law et al. (2014), who developed new linear modeling strategies to handle issues related to mean–variance relationship of log-counts in RNA-seq reads (Ritchie et al. 2015). We represent the branch lengths on individual gene trees as a matrix Y, where rows correspond to individual genes (g), and columns to the branches (b) on these trees. We first transform the branch length data using a square-root transformation (eq. 1).
| (1) |
Following the transformation, we perform a weighted regression analysis to calculate the relative evolutionary rates as follows: we calculate the average tree and perform a first-pass of linear regression using the transformed branch length matrix (eqs. 3 and 4).
| (2) |
where is the branch length for branch b in the average tree.
| (3) |
| (4) |
where are the coefficients of linear regression and R is the residuals matrix.
We then estimate the mean–variance trends in the residuals of the linear regression analysis by empirically fitting a locally weighted scatterplot smoothing (LOWESS) function capturing the relationship between the log of variance of the residuals and the branch lengths (eq. 5).
| (5) |
Subsequent to estimating this function, we assign each gene x branch observation a weight W based on the predicted value for the branch, obtained from the first pass linear regression (eq. 6).
| (6) |
For branches that are shorter on average, the variance in the residuals is smaller, thus resulting in a higher weight, and vice versa. Using the computed weights, we perform a weighted regression analysis between the individual branch length (dependent variable) and the average tree (independent variable). The weighted regression analysis attempts to remove the heteroscedasticity in the residuals by computing the residuals after minimizing the weighted sum of squared errors, as opposed to the raw sum of squared errors (eqs. 7 and 8).
| (7) |
| (8) |
| (9) |
where is the SD of the weighted residuals in branch b.
Subsequent to the weighted regression analysis, the weighted residuals (), are estimated by rescaling the regression residuals () with the weights, and the weighted residuals are additionally standardized to have unit variance within every branch across all genes (eq. 9). The weighted residuals () correspond to the weighted relative rate on branch b for gene g. The differences to the relative rate calculations introduced by the updated method result in changes to the scales of the relative rates computed. However, we note that this scale is arbitrary and the downstream gene-trait correlations for binary traits estimated using a Mann–Whitney test (see Materials and Methods) depend only on the ranks of the relative rates of each branch within any single gene tree. Figure 3 shows the workflows for computing relative evolutionary rates using the original and updated method.
Fig. 3.
Workflow for calculating relative evolutionary rates using the updated method. Black areas of the workflow represent steps implemented as part of current relative rates method, and blue/grey areas correspond to methodological updates.
Results
Improvements to Relative Evolutionary Rates Methods Mitigate Genome-Wide Mean–Variance Relationship
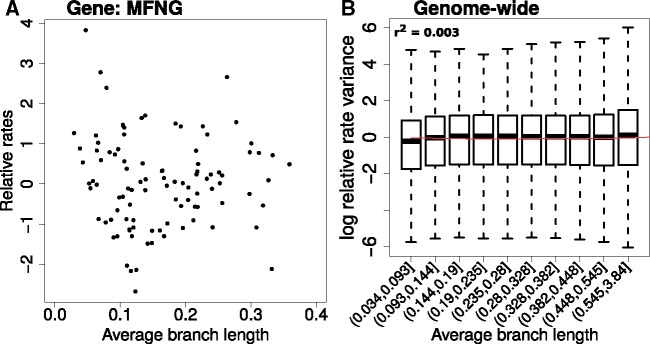
Our updated method to calculate relative rates using data transformation followed by weighted regression produces nearly homoscedastic relative rates that do not show a significant global mean–variance relationship. Figure 4A shows the relative rates computed for the MFNG protein-coding gene tree using the updated method. In comparison to the original method based on naive linear regression (fig. 2A), we observe that the updated method produces relative rates showing no apparent increase in the variance of relative rates on longer branches of the tree. Plotting the genome-wide mean–variance trends of the relative rates across all branches of all gene trees, we observe that the relative rates calculated from transformed-weighted residuals show nearly constant variance across branches of varying lengths (fig. 4B). We additionally checked the mean–variance relationships from intermediate steps in our method that can estimate relative rates, corresponding to two method variants which do not implement data transformation (linear-weighted regime) or a weighted regression (square-root unweighted regime) (supplementary fig. S1, Supplementary Material online). However, we find that the intermediate regimes, utilizing only one of the method updates (branch length transformation or weighted regression alone) are less effective at eliminating mean–variance trends. A combination of transformation and weighted regression steps works best at producing homoscedastic relative rates.
Fig. 4.
Updated method to calculate relative rates shows no apparent trends of heteroscedasticity. (A) Manic Fringe (MFNG) gene relative rates calculated using the updated method. In comparison to figure 2A, we do not observe an increase in the variance of relative rates of branches with increasing average branch length. (B) Genome-wide mean–variance trends for relative rates computed using the updated method show constant variance with increasing branch lengths. Contrasting the trends resulting from the application of original (fig. 2B) and updated method (fig. 4B), we observe that the updated method produces nearly homoscedastic relative rates. The extent of heteroscedasticity, computed as the “r-squared” of the quadratic model between the variables plotted, is nearly 100-fold lower with the updated method compared with original method.
Better Robustness to Inclusion of Distantly Related Species
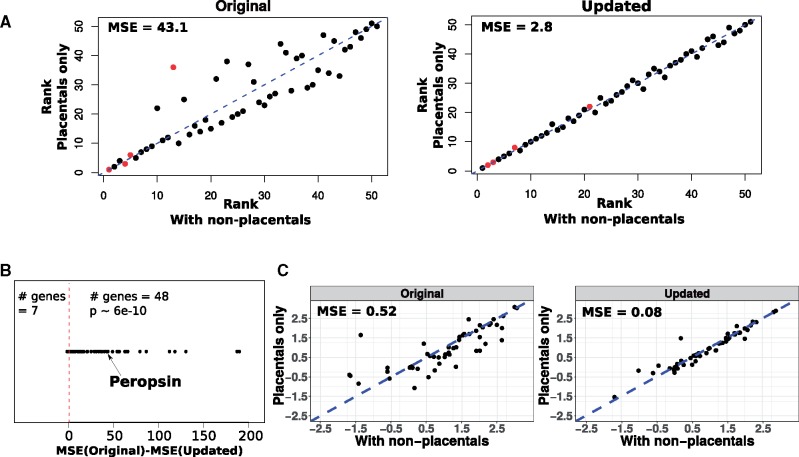
In earlier applications of our relative rates method to detect genetic elements convergently responding in subterranean mammals and marine mammals, respectively, we sampled alignments of placental mammal species to construct phylogenetic trees for each genetic element (Chikina et al. 2016; Partha et al. 2017). These alignments were derived from the placental mammal subset of the 100-way vertebrate alignments made publicly available by the UCSC genome browser (Casper et al. 2018). In addition to these placental mammals, the 100-way alignments include four other species of mammals, three marsupials—Opossum (monDom5), Wallaby (macEug2), Tasmanian Devil (sarHar1), and one monotreme—Platypus (ornAna1). Despite deep conservation of many genetic elements in these nonplacental mammals, human-and-mouse centered phylogenomic studies tend to exclude these species due to the introduction of long branches in the phylogenetic trees (Parker et al. 2013; Marcovitz et al. 2016; Prudent et al. 2016). For instance, in previous applications of our relative rates method, we deliberately excluded these nonplacental mammals since they produce wide variations in relative rates due to the introduction of long branches, which would adversely affect the confidence with which we make inferences of convergent rate acceleration in species exhibiting a convergent phenotype (Chikina et al. 2016; Partha et al. 2017). However, scanning for rate-trait associations across tree data sets with higher numbers of species would allow for more statistical power, and hence a relative rates method that can reliably include such distantly related species offers a clear advantage. To this end, we tested the robustness of our updated method to the inclusion of distantly related species at inferring convergent rate shifts. We chose two phylogenetic data sets 1. Genome-wide protein-coding gene alignments across 59 placental mammal species, and 2. across 63 mammals including four nonplacental mammals in addition to the placentals. An example demonstration of how our current method to calculate relative rates is sensitive to the inclusion of nonplacental mammals is illustrated in figure 5A. Using the Peropsin (RRH) gene for illustrative purposes, we show that the ranks of relative rates computed using the current method considerably vary upon the inclusion of nonplacental mammals. These changes in ranks are observed across many branches on the gene tree including one of the four subterranean branches (Cape golden mole). In comparison, the updated method displays a stronger concordance in the ranks of the computed relative rates (fig. 5A). Consequently, the subterranean acceleration scores for RRH computed using the updated method are more stable with the inclusion of nonplacental mammals (table 1).
Fig. 5.
Comparison of robustness of methods to inclusion of nonplacental mammals. (A) Relative rates of Peropsin (RRH) gene branches for trees with and without nonplacental mammals, using the original versus the updated method. The relative rate ranks of terminal lineage branches within the RRH tree are plotted with respect to the inclusion of nonplacental mammals. Red/grey points denote subterranean branches. Ranks of relative rates computed by the original method show wider variation with respect to the inclusion of nonplacental mammals, whereas updated methods reveal stronger concordance. Mean squared sum of residuals (MSE) were calculated based on a linear model between the ranks across the two tree data sets with slope coefficient equal to 1. (B) Updated method shows improved concordance in ranks of relative rates across trees with and without nonplacental mammals. For 48 out of 55 genes, rank concordance is better for relative rates computed using the updated method (MSE(original)-MSE(updated) > 0). For the other 7 genes, the two methods do not show strong differences in the MSE values, with the original method outperforming the updated by slight margins. (C) Updated method shows improved robustness to inclusion of nonplacental mammals at detecting subterranean acceleration of eye-specific genes. Individual points represent rate acceleration on subterranean branches for each of 55 eye-specific genes computed across two data sets using the two methods. Based on a linear model between the subterranean acceleration scores across the two tree data sets with slope coefficient equal to 1 we calculate the mean squared sum of residuals (MSE). An improved robustness (lower MSE) to the inclusion of nonplacental mammals is observed with the updated method.
Table 1.
Subterranean Acceleration Scores for Peropsin (RRH) Computed Using Two Methods, and across Two Data Sets.
| Data Set | Method |
|
|---|---|---|
| Original | Updated | |
| With nonplacentals | 2.70 (rho=0.31; P=0.002) | 2.1 (rho=0.27; P=0.008) |
| Placentals only | 1.38 (rho=0.21; P=0.041) | 2.0 (rho=0.26; P=0.01) |
Note.—In comparison to the original method, the updated method shows stronger consistency in the scores across the two tree data sets with and without the nonplacental mammals. The subterranean acceleration scores reflect the significance of convergent rate acceleration on the four subterranean branches.
We also performed a larger scale benchmarking of the robustness of our methods to the inclusion of nonplacental mammals across 55 genes showing eye-specific expression. These genes were identified based on mouse microarray expression data across 91 tissues (see Materials and Methods). We first compared the estimated concordance in ranks of relative rates computed using the original and updated method in trees including and excluding the nonplacental mammals. For each gene, we calculated concordance in ranks using the mean squared error of residuals of a linear model (see Materials and Methods), where lower MSE values reflect better robustness. We observed that for 48 (out of 55) eye-specific genes, the updated method shows improved concordance in the ranks of relative rates across the two sets of gene trees (fig. 5B). Using a pairwise Wilcoxon test, we compared the MSE values obtained using the original versus updated method, revealing a statistically significant (P ∼ 6e-10) decrease in MSE values obtained using the updated method.
For each of these eye-specific genes, we also calculated subterranean acceleration scores (see Materials and Methods) reflecting the convergent rate acceleration on the four subterranean branches independently in gene trees including and excluding the nonplacental mammals. Based on the relative rates calculated using each method, we compared the concordance of the subterranean acceleration scores across the two tree data sets. Ideally, we expect the scores produced by the methods to be highly consistent across the two data sets since the four nonplacental mammals are not subterranean, with only minor differences arising due to the inclusion of four additional background species. The results of the analysis revealed that the updated method produces superior concordance in the scores across the two tree data sets, reflecting its improved ability to handle the long branches introduced by the nonplacental mammals (fig. 5C).
Improved Power to Detect Convergent Rate Shifts in Simulated Trees
In order to compare the power of our methods to detect convergent rate shifts in branches across a range of evolutionary scenarios, we developed a model to simulate individual gene trees. Such a model allows us to rigorously examine method performance in relation to various parameters in phylogenetic data sets including number of foreground branches and length distribution of foreground branches, where foreground branches describe branches showing a convergent phenotype, while background branches do not. The limited availability of “ground truth” examples of convergently evolving genetic elements calls for the development of biologically realistic simulations of sequence evolution. Using our model to simulate trees (see Materials and Methods), we compared the power to detect rate shifts in relation to two factors: 1. Average lengths of foreground branches, in particular extreme foreground branches that are very short or very long on average. 2. Number of foreground branches. We investigated the performance of the updated method in detecting rate shifts in such extreme branches, assessing the power advantage resulting from calculating relative rates that do not suffer from a biased mean–variance relationship.
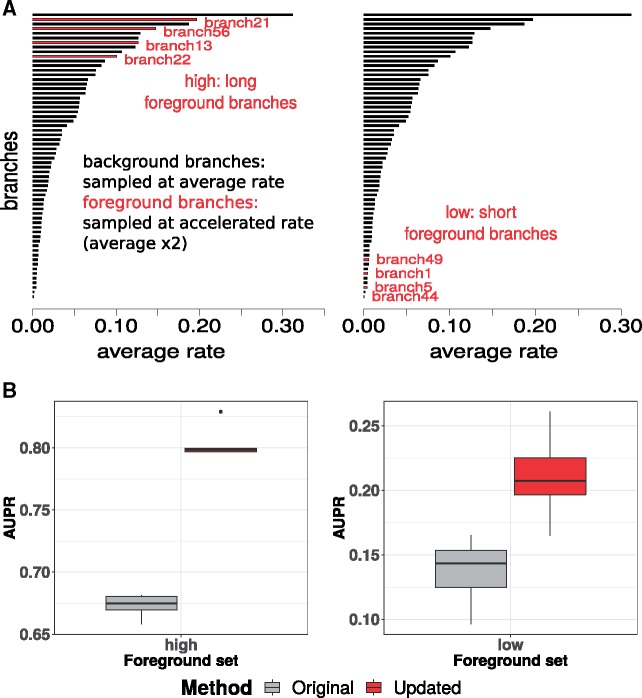
Our model to simulate phylogenetic trees allows for explicit control over choosing foreground branches showing convergent rate acceleration. We simulate “control” trees, where all branches are modeled to evolve at their respective average rates, and “positive” trees, where the chosen foreground branches are modeled to evolve at an accelerated rate. Initially, we chose a foreground rate multiplier value of 2, which corresponds to foreground branches in positive trees being sampled at twice their average rates (see Materials and Methods). We first compared the heteroscedasticity in the relative rates on the branches of the control trees calculated using the original and updated methods. Similar to the trends observed in mammalian gene trees (supplementary fig. S1, Supplementary Material online), we observed that the updated method outperformed the original method at producing homoscedastic relative rates (supplementary fig. S4, Supplementary Material online). We then calculated a foreground acceleration score for individual simulated trees, both control and positive. A more positive value of this score, calculated as a signed negative logarithm of the P value, reflects stronger convergent rate acceleration on the foreground branches (see Materials and Methods). Subsequent to estimating these scores, we evaluated the performance of the two methods, based on the power to distinguish the positive trees from control trees. In two independent simulation settings with foreground branches of long and short average lengths, we observed that the updated method offers more power to detect positive trees (fig. 6B and see supplementary fig. S5, Supplementary Material online, for precision-recall curves).
Fig. 6.
Comparison of method performance across simulated phylogenetic trees. (A) Branch length distributions for simulating phylogenetic trees with foreground branches labeled. Two independent simulations were performed with foreground branch sets comprised of long foreground branches (left panel) and short foreground branches (right panel), respectively. (B) Power to detect rate shift in foreground branches of simulated trees. Across five independent simulations of control trees and positive trees, we measured the area under the precision-recall curve (AUPR) to precisely detect positive trees using the foreground acceleration score. The AUPR distributions obtained using the updated method to calculate relative rates are significantly elevated compared with the original method across simulated scenarios involving foreground sets of long (left) and short branches (right), respectively.
We repeated the analyses with more conservative choices for modeling foreground acceleration using foreground rate multiplier values of 1.5 and 1.75 to ensure the improved power was robust to the choice of foreground rate multiplier (m). Consistent with the original analysis, the updated method was more powerful at precise detection of positive trees for all values of m (supplementary fig. S6, Supplementary Material online). We also observed that with increasing values of m, it becomes easier to detect positive trees (fig. 6B and supplementary fig. S6, Supplementary Material online) which is expected since the foreground branches will be longer for larger values of m. Our choices of foreground rate multiplier values in simulations (m = 1.5, 1.75, and 2) represent challenging scenarios for our method in comparison to foreground rate multiplier estimates observed in real data. For instance, our simulation choices are lower than the foreground rate multiplier estimates for genes showing strong relaxation of constraint in subterranean mammals, and more comparable to the estimates for genes under positive selection (see Materials and Methods and table 4). This proves the utility of our method at detecting genes showing rate acceleration due to positive selection, in addition to relaxation of constraint.
Table 4.
Mole Foreground Multiplier estimates for Genes Showing Strong Convergent Rate Acceleration on Mole Branches.
| Gene | Mole Foreground Rate Multiplier Estimate | Evolutionary Mode |
|---|---|---|
| LIM2 | 8.63 | Relaxed |
| CRYBB3 | 5.36 | Relaxed |
| CRYBB2 | 4.87 | Relaxed |
| CRYGC | 4.62 | Relaxed |
| CRYBA1 | 3.89 | Relaxed |
| GPR89B | 3.30 | Relaxed |
| KRTAP17-1 | 3.22 | Positive selection |
| GNAT1 | 2.66 | Relaxed |
| ROM1 | 2.58 | Relaxed |
| COL4A4 | 1.70 | Positive selection |
We also performed a control analysis using foreground acceleration scores computed using four length-matched control foreground branches that were not the true foreground, proving that the positive trees were not detected due to random chance (supplementary fig. S7, Supplementary Material online). Finally, in addition to the positive trees with foreground branches that were long or short, we compared the power to detect rate acceleration on foreground branches of intermediate length. Consistent with the findings in short/long foregrounds, we find a modest yet significant improvement offered by the updated method (supplementary fig. S8, Supplementary Material online). Overall, we find that our updated method to compute relative rates offers a significantly improved power to detect convergent rate shifts in simulated trees.
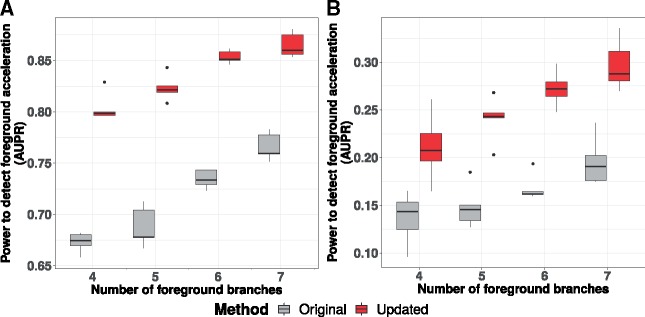
We then compared the power to detect rate shifts across varying numbers of foreground branches by simulating positive trees with seven foreground branches of long average lengths (supplementary fig. S9, Supplementary Material online). We subsequently generated positive trees with subsets of n branches (n ranging from 4 to 7) among these seven foreground branches (supplementary fig. S9, Supplementary Material online). Within each of these data sets, we calculated foreground acceleration scores for control and positive trees using each method independently. We observed that the updated method to calculate relative rates is consistently more powerful than the original method at precise detection of positive trees (fig. 7A). We repeated the analysis choosing seven foreground branches that were short on average rather than long (supplementary fig. S9, Supplementary Material online) and observed consistent gains in power using updated method to calculate relative rates (fig. 7B).
Fig. 7.
Improved power to detect foreground rate shifts using the updated method across different numbers of foreground branches. This analysis was performed across five independent simulations of control trees and positive trees with varying numbers of foreground branches (4–7). Within each simulation, we measured the area under the precision-recall curve (AUPR) to precisely detect positive trees using the foreground acceleration score. The AUPR distributions obtained using the updated method to calculate relative rates are consistently elevated compared with the original method across simulations with different numbers of foreground branches. These simulations were performed across two scenarios with different foreground branch sets consisting of short (A) and long branches (B), respectively.
Applying of our method to simulations with varying configurations of foreground branches also revealed that the power to detect foreground acceleration is higher for longer foreground branches. In other words, it is easier to detect rate acceleration on longer foreground branches compared with shorter ones (figs. 6A vs. B and 7A vs. B). In terms of sequence divergence, longer branches represent instances of higher sequence divergence or more changes, which are easier to detect as the method ranks the rates on branches relative to one another. The increased power to detect rate acceleration therefore becomes especially useful in convergent phenotypes involving short foreground branches, where the improvements are nearly 2-fold (fig. 7B).
Relative Rates-Based Inference Is Robust to Minor Uncertainties in Species Tree Topology
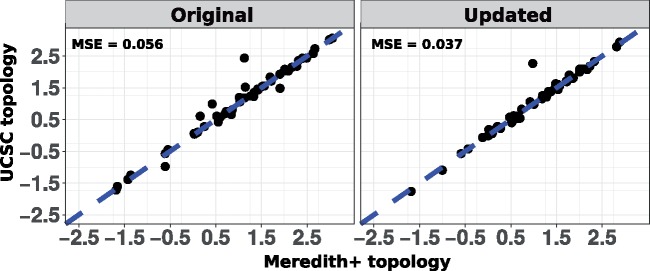
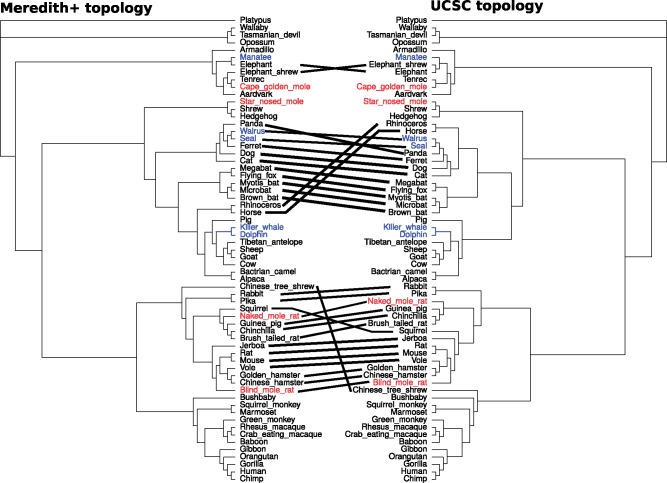
Our method relies on estimating sequence divergence on branches of phylogenetic trees with a fixed topology. Efforts to better resolve the phylogeny of extant mammals have resulted in continuous updates to the consensus species tree topology (Murphy et al. 2001, 2007). Topology trees commonly used in phylogenomic analyses of extant mammals include the UCSC genome browser’s 100-way tree, as well as the timetrees reported in Meredith et al. (2011) and Bininda-Emonds et al. (2007; Casper et al. 2018). Differences between these species tree topologies often involve entire clades, and the decision to choose a particular topology tree can potentially strongly influence the outcomes of phylogenetic analyses. Here, we benchmarked the robustness of our relative rates method to the choice of topology tree. We constructed protein-coding gene trees based on two different species tree topologies, namely the UCSC 100-way tree and our modified Meredith et al. (Meredith+) topology tree (see Materials and Methods). The Robinson–Foulds metric (calculated using the function RF.dist in the R package “phangorn”) between these two phylogenies is 22, reflecting differences in 22 partitions of species (Robinson and Foulds 1981; Schliep 2011). We observed that both the updated and original methods to calculate relative rates show robust signatures of subterranean rate acceleration for eye-specific genes with respect to the species tree topology used (fig. 8).
Fig. 8.
Comparison of robustness of methods to species tree topology. Both the original and updated relative rates methods are robust to choice of species tree topology used to construct individual gene trees. Points represent the strength of convergent subterranean acceleration for eye-specific genes whose trees were constructed using the Meredith+ topology (x-axis), and the UCSC topology (y-axis), respectively. Based on a linear model between the subterranean acceleration scores across the two tree data sets with slope coefficient equal to 1, we calculate the mean squared sum of residuals (MSE). We observed that the updated method offers a marginal improvement to the robustness, as reflected by a lower MSE value.
Comparison of Power to Detect Enriched Pathways Associated with Two Independent Convergent Phenotypes
Beyond examining individual genes, we further assessed our new method’s ability to detect pathway enrichments for genes under relaxation of constraint in subterranean mammals and marine mammals (see fig. 9 for respective foreground branches and supplementary fig. S10, Supplementary Material online, for average rates). Compared with our original method, the updated method detected more enriched Gene Ontology (GO) terms with accelerated evolutionary rates in subterranean mammals (table 2). Additionally, the fold enrichment for detected terms was significantly stronger with the updated method (supplementary fig. S10 and tables S1–S6, Supplementary Material online). On the other hand, the marine system showed mixed results. Both the updated and the original methods showed approximately equal power to detect enriched GO terms if we only consider the number of terms detected (table 3 and supplementary tables S7–S12, Supplementary Material online). However, when comparing the fold enrichment for detected terms, the original method was significantly better than the updated method (supplementary fig. S10, Supplementary Material online). These contrasting results from the subterranean data set versus the marine data set indicate the importance of tailoring the corrections we have developed to the data set of interest, as well as the importance of taking advantage of simulation-based power and robustness assessments to develop methods that are broadly applicable to many convergent phenotypes.
Fig. 9.
Cladograms describing relationships between 63 mammalian species used for constructing genome-wide maximum likelihood protein-coding gene trees. Final version of the tree we modified from the topology reported in Meredith et al. (2011) (left), and tree reported in UCSC genome browser (right) (Casper et al. 2018). Key differences between the placement of species are highlighted using black lines. Marine species are manatee, walrus, seal, killer whale, and dolphin. Subterranean species are cape golden mole, star nosed mole, naked mole-rat, and blind mole-rat.
Table 2.
Comparison of Number of Vision-Related Gene Ontology Terms Enriched in Top Subterranean-Accelerated Genes Discovered by the Original and Updated Methods.
| topN: Number of Top Accelerated Genes | # Subterranean-Accelerated GO Terms (FDR<0.05) |
|
|---|---|---|
| Original Method | Updated Method | |
| 20 | 2 | 9 |
| 100 | 11 | 28 |
| 200 | 16 | 32 |
Note.—Gene Ontology term enrichment analysis was performed individually on top subterranean accelerated genes discovered by each method. Across varying numbers of top target genes, genes discovered using the updated method were consistently enriched for higher numbers of vision-related GO terms.
Table 3.
Comparison of Number of Gene Ontology Terms Enriched in Top Marine-Accelerated Genes Discovered by the Original and Updated Methods.
| topN: Number of Top Accelerated Genes | # Marine-Accelerated GO Terms (FDR<0.05) |
|
|---|---|---|
| Original Method | Updated Method | |
| 50 | 16 | 10 |
| 100 | 27 | 31 |
| 200 | 59 | 59 |
Note.—Gene Ontology term enrichment analysis was performed individually on top marine-accelerated genes discovered by each method. Across varying numbers of top target genes, neither method showed a clear superiority over the other at detecting higher numbers of enriched terms. We chose the Top50 genes for the marine phenotype instead of Top20 as was the case with the subterranean analysis, since no terms were enriched across either method in the Top20 gene list.
Implementation and Availability
Our method is publicly available as an R package called RERconverge on GitHub at https://github.com/nclark-lab/RERconverge and described in (Kowalczyk et al. 2018). Also included are extensive vignette walkthroughs to guide users through the software. RERconverge requires as input gene trees with branch lengths that represent evolutionary rates and phenotype information. Functions within the package can take this input to calculate RERs, ancestral phenotype information, and statistical associations between phenotypes and evolutionary rates.
The RERconverge software is available for any platform, and its computational efficiency allows analysis of genome-scale data sets. When run on a set of 19,149 orthologous genes in 62 mammal species, the full analysis can be completed in 1 h and 18 min on a computer running Windows 10 with 16 Gb of RAM and an Intel Core i7-6500U 2.50GHz CPU (Kowalczyk et al. 2018). We hope that these capabilities will enable researchers worldwide to perform analyses to find relationships between genes and evolutionarily convergent traits.
Discussion
Our original evolutionary-rates-based method to detect genomic elements underlying convergent phenotypes has already proved to be a valuable technique to detect genes and enhancers associated with transitions to marine and subterranean habitats (Chikina et al. 2016; Partha et al. 2017). However, the original method suffered from reduced power to detect such genomic elements due to a heteroscedastic relationship between the mean and variance of branch lengths for a given branch across all gene trees, that is, branches that are longer on average have higher variance than branches that are shorter on average.
Here, we developed a method using a square-root transformation and a weighted regression based on the observed mean–variance relationship to correct for the heteroscedasticity. While our objective was to develop a method that robustly handles mean–variance trends in phylogenetic trees, we do not systematically investigate factors underlying this property. Previous genome-scale analyses in modern birds have showed evidence for base composition heterogeneity affecting variance of branch lengths in exon trees (Jarvis et al. 2014). However, in our phylogenetic data set of mammalian protein-coding genes, we found no evidence for base composition heterogeneity influencing sequence divergence at the gene level—we failed to detect any significant global trends between GC-content of our sequences and their raw branch lengths, relative rates computed using our original method, or from our new method (supplementary fig. S11, Supplementary Material online). Further comparative genomics analysis is required to better understand factors influencing branch length distribution patterns in phylogenetic trees.
We tested our new method on real and simulated phylogenies and observed improved robustness to wider ranges of branch lengths and increased ability to detect convergent evolutionary rate shifts. Our new method offers increased robustness to the inclusion of distantly related species with long branch lengths in our phylogeny, namely nonplacental mammals. When we compared results from an analysis using only placental mammals and an analysis that included nonplacental mammals using both our original and our updated methods, we found that our new method, unlike our original method, is unimpaired by the inclusion of nonplacental mammals. By improving our method’s robustness to inclusion of long branches, we increased the method’s applicability to a broader range of species and hence a broader range of convergent phenotypes. Additionally, our new method’s increased power could enable us to discover more convergently evolving genomic elements. One particular incentivizing example for these improvements is the recent efforts to sequence the northern marsupial mole, a completely blind mammal (Archer et al. 2011). When considering using subterranean species to find genes and enhancers associated with vision, the ability to include the nonplacental marsupial mole along with the other nonplacental mammals in our data set will allow for more power in a scan for vision-specific genetic elements showing convergent regression in the five blind mammals.
In addition to testing our method on real data, we also developed a simulation-based strategy to represent a “true positive” case of convergent evolution. Our simulations follow a similar approach to simulating RNA-seq counts where simulated rates are essentially capturing the number of substitutions that occur along a branch (Di et al. 2011). We showed that our new method demonstrates improved detection of rate shifts both when foreground species occupy long, high-variance branches and when foreground species occupy short, low-variance branches. This allows the method to detect convergent rate shifts given a variety of potential configurations of convergently evolving species. The types of simulations we developed are essential because relatively few concrete instances of sequence-level evolutionary convergence exist, so biologically accurate simulations of such evolution are essential to rigorously test methods that detect shifts in evolutionary rates. One simplification of our simulation method is that all species are present in all simulated trees, which is not the case in real genomic data because of genomic element gain and loss across species. However, maintaining constant species composition in our simulated trees should have little impact on our ability to compare our methods because we expect both to be equally impacted by species presence and absence. A second simplification is that we assume all convergently evolving species have the same phylogenetic relatedness, that is, each foreground branch is an independent instance of convergent evolutionary rates. We would like to be able to answer questions about our method’s power given more complex phylogenetic configurations. Developing methods to answer those types of questions will require a much higher degree of complexity in our simulations, but it will also allow us to determine which species to add to our genomic data sets to increase our power to find convergently evolving genomic features.
Our improved method has proved valuable for detecting genomic elements associated with two binary traits—subterranean-dwelling or not, and marine-dwelling or not—and we will extend our method for use in convergent continuous traits and nonbinary discrete traits. We will also assemble complementary analyses to assess the robustness and power of each method. By extending the scope of our method to nonbinary traits, we will expand the potential search-space of our method to a plethora of new convergent phenotypes. Our overarching goal is to develop an entire suite of methods that can utilize any conceivable phenotypes as inputs to accurately and robustly identify convergently evolving genomic elements.
Materials and Methods
Protein-Coding Gene Trees across 63 Mammalian Species
We downloaded the 100-species multiz amino acid alignments available at the UCSC genome browser, and retained only alignments with a minimum of ten species. We then pruned each alignment down to the species represented in figure 9 of the proteome-wide average tree. We added the blind mole rat ortholog of each gene based on the methods described in Partha et al. (2017). We estimated the branch lengths for each amino acid alignment using the aaml program from the package PAML (Yang 2007). We estimated these branch lengths on a tree topology modified from the timetree published in Meredith et al. We attempted to resolve conflicts between the topology inferred in Meredith et al. (2011) compared with that in Bininda-Emonds et al. (2007) based on a consensus of various studies employing a finer scale phylogenetic inference of the species involved. The differences between our final topology, which we call “Meredith+” topology and the Meredith et al. topology include setting the star-nosed mole as an outgroup to the hedgehog and shrew; cow as an outgroup to the Tibetan antelope, sheep and goat; and the ursid clade as an outgroup to mustelid and pinniped clades. For more details about the literature surveyed to resolve these differences, please refer to Meyer et al. (2018). The topology of our final “Meredith+” tree compared with the UCSC topology tree is reported in figure 9. In order to perform analyses benchmarking the method robustness to tree topology, we additionally generated the protein-coding gene trees based on the UCSC tree topology.
Genes Showing Eye-Specific Expression
We identified eye-specific gene sets using microarray expression data from 91 mouse tissues (Su et al. 2004). We identified genes specifically expressed in the following tissues of the eye—cornea, iris, lens, and retina (including retinal pigmented epithelium). These genes showed significant differential expression only in the tissue of interest compared with the other tissues at an alpha of 0.05 (t-test).
Calculating Concordance in Relative Rates Ranks across Data Sets with and without Nonplacental Mammals
To estimate the robustness of relative rates calculations to inclusion of nonplacental mammals, we calculate the concordance in relative rates ranks across two phylogenetic data sets with and without the nonplacental mammals, respectively. For each of the 55 eye-specific genes, we rank the extant branches in trees based on the ordering of relative rates independently in the two data sets. We then fit a linear model between the ranks across these two data sets, while forcing a slope coefficient of 1. We subsequently estimate the concordance in the ranks as the mean squared error of the residuals of this linear model. Lower MSE values reflect better concordance in the ranks, and thus superior robustness. We subsequently compare these MSE values for each eye-specific gene obtained using the original and updated methods to calculate relative rates. A positive value implies the updated method shows improved concordance in the ranks of relative rates, across data sets with and without the nonplacental mammals, respectively.
Simulating Phylogenetic Trees
Phylogenetic branch lengths have units of number of substitutions per site and thus can be thought of as normalized count data. However, we find that a Poisson distribution is unsuitable in this case as the real branch length data show considerable overdispersion, that is the variance is higher than the mean (supplementary fig. S2, Supplementary Material online). We thus model the branch lengths of the simulated trees using a negative binomial distribution, following ideas from studies simulating expression counts for RNAseq analysis (Robinson et al. 2009; Di et al. 2011; Law et al. 2014; Ritchie et al. 2015).
We simulated data sets of phylogenetic trees using the UCSC tree topology and branch lengths from the average proteome-wide tree across 19,149 mammalian protein-coding gene trees across 62 mammals. Supplementary figure S3, Supplementary Material online, describes the tree topology used for the simulations. We simulate the branch lengths (or rates) for every branch (j) on each tree (i) according to the following formula,
where Gamma is parametrized by mean and variance. Here, is a gene-specific scaling term, is the average rate of the corresponding branch so that is the expected rate on the i’th branch, and the simulated rate is drawn from a Gamma distribution with that mean. The composite Poisson-Gamma distribution is equivalent to the negative binomial distribution and thus in our simulation the mean variance relationship has a quadratic component, matching what we observe in real data (supplementary fig. S2, Supplementary Material online).
We simulate two classes of trees in every data set based on different input parameters. We simulate “control” trees, trees where the are simply the average rate on the branch j. These control trees do not show any explicit convergent rate shift on any of the branches. We additionally simulate “positive” trees showing convergent rate acceleration on foreground (fgd) branches by sampling at , only on these branches (m = 1.5, 1.75, or 2). Thus, the foreground branches in positive trees are effectively sampled at an accelerated rate compared with the foreground branches in control trees.
Estimating Foreground Rate Multiplier (m) for Genes Showing Convergent Rate Acceleration in Subterranean Mammals
We compared our choices for the foreground rate multiplier (m = 1.5, 1.75, or 2) in simulations to that observed in real data using branch lengths of ten genes showing strong convergent rate acceleration in the four subterranean mammals (moles). Of the 55 genes identified in Partha et al. as showing strongest convergent rate acceleration in the moles, we chose the top eight genes showing relaxation of constraint, and two genes undergoing positive selection on the four mole branches (supplementary table S5 in Partha et al. 2017). For each of these ten genes, we estimated the mole foreground rate multiplier as follows: we first fit a linear model between the gene branch lengths and the average branch lengths. Based on the predicted values for the mole branches from this linear model, we calculate the foreground rate multiplier for each mole branch by dividing the real mole branch length by their predicted value. The mole foreground rate multiplier estimate for each gene is subsequently calculated as the mean of the four individual foreground rate multipliers. Table 4 shows the mole foreground rate multiplier estimates for these ten genes.
Calculating Gene-Trait Correlations
The gene-trait correlations are computed under a Mann–Whitney U testing framework over the binary variable of foreground versus background branches. In the subterranean example, the four subterranean branches (fig. 1) are designated as foreground. We calculate a foreground acceleration score reflecting the strength of convergent rate acceleration on the foreground branches. The value is calculated as the negative logarithm of the P value of the Mann–Whitney test multiplied by the direction of the correlation as given by the sign of the rho statistic. A positive rho statistic indicates rate acceleration in the foreground species, and the negative logarithm of P value reflects the strength of the convergent rate shift. In simulated trees study, we generated trees for three sets of foreground branches with different branch length distributions—short, intermediate, and long as illustrated in figure 6 and supplementary figure S5, Supplementary Material online.
where rho and P are the correlation coefficient and statistical significance of the Mann–Whitney test for association between relative rates and binary trait.
Gene Ontology Term Enrichment Analysis
We performed functional enrichment analysis in target gene lists using the GOrilla tool (Eden et al. 2009). For each analysis, GO terms enriched in target gene lists were identified by comparing to a background gene list with all 19,149 genes used to construct gene trees.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dennis Kostka, as well as members of the Chikina and Clark labs for helpful discussion and feedback on earlier versions of this article. This work was supported by NIH R01 HG009299-01A1 to N.L.C. and M.C. A.K. was supported by NIH T32 training grant T32 EB009403 as part of the HHMI-NIBIB Interfaces Initiative.
References
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature. 5077493:455–461. doi:10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer M, Beck R, Gott M, Hand S, Godthelp H, Black K.. 2011. Australia’s first fossil marsupial mole (Notoryctemorphia) resolves controversies about their evolution and palaeoenvironmental origins. Proc R Soc B Biol Sci. 2781711:1498–1506. doi:10.1098/rspb.2010.1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, et al. 2018. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 46(D1):D762–D769. doi:10.1093/nar/gkx1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikina M, Robinson JD, Clark NL.. 2016. Hundreds of Genes Experienced Convergent Shifts in Selective Pressure in Marine Mammals. Mol Biol Evol. 339:2182–2192. doi:10.1093/molbev/msw112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Y, Schafer DW, Cumbie JS, Chang JH.. 2011. The NBP Negative Binomial Model for Assessing Differential Gene Expression from RNA-Seq. Stat Appl Genet Mol Biol. 101:1–28. doi:10.2202/1544-6115.1637 [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z.. 2009. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 101:48. doi:10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA. 1998. Phylogenomics: Improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Research. 83:163–167. doi:10.1101/gr.8.3.163 [DOI] [PubMed] [Google Scholar]
- Esteller M. 2011. Non-coding RNAs in human disease. Nature Reviews Genetics. 1212:861–874. doi:10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- Foote AD, Liu Y, Thomas GWC, Vinar T, Alföldi J, Deng J, Dugan S, Van Elk CE, Hunter ME, Joshi V, et al. 2015. Convergent evolution of the genomes of marine mammals. Nature Genetics. 473:272–275. doi:10.1038/ng.3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Schaar BT, Indjeian VB, Kingsley DM, Hagey LR, Bejerano G.. 2012. A “Forward Genomics” Approach Links Genotype to Phenotype using Independent Phenotypic Losses among Related Species. Cell Reports. 24:817–823. doi:10.1016/j.celrep.2012.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SYW, Faircloth BC, Nabholz B, Howard JT, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 3466215:1320–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A, et al. 2018. RERconverge: an R package for associating evolutionary rates with convergent traits. bioRxiv. Available at: http://biorxiv.org/content/early/2018/10/23/451138.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Poujol R.. 2011. A phylogenetic model for investigating correlated evolution of substitution rates and continuous phenotypic characters. Mol Biol Evol. 281:729–744. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK.. 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 152:R29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Calvo SE, Gutman R, Liu JS, Mootha VK.. 2014. Expansion of biological pathways based on evolutionary inference. Cell. 1581:213–225. doi:10.1016/j.cell.2014.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. 2009. Finding the missing heritability of complex diseases. Nature. 4617265:747–53. doi:10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcovitz A, Jia R, Bejerano G.. 2016. “reverse Genomics” predicts function of human conserved noncoding elements. Mol Biol Evol. 335:1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simão TLL, Stadler T, et al. 2011. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science. 3346055:521–524. doi:10.1126/science.1211028 [DOI] [PubMed] [Google Scholar]
- Meyer WK, Jamison J, Richter R, Woods SE, Partha R, Kowalczyk A, Kronk C, Chikina M, Bonde RK, Crocker DE, et al. 2018. Ancient convergent losses of Paraoxonase 1 yield potential risks for modern marine mammals. Science. 3616402:591–594. doi:10.1126/science.aap7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, O’Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, De Jong WW, et al. 2001. Resolution of the early placental mammal radiation using bayesian phylogenetics. Science. 2945550:2348–2351. doi:10.1126/science.1067179 [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W.. 2007. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Research. 174:413–421. doi:10.1101/gr.5918807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaf Bininda-Emonds, Cardillo Marcel, Jones Kate E., MacPhee Ross D. E., Beck Robin M. D., Grenyer Richard, Price Samantha A., Vos Rutger A., Gittleman JL, Purvis A.. 2007. The delayed rise of present-day mammals. Nature. 44629:507–512. [DOI] [PubMed] [Google Scholar]
- Parker J, Tsagkogeorga G, Cotton JA, Liu Y, Provero P, Stupka E, Rossiter SJ.. 2013. Genome-wide signatures of convergent evolution in echolocating mammals. Nature. 5027470:228–231. doi:10.1038/nature12511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partha R, Chauhan BK, Ferreira Z, Robinson JD, Lathrop K, Nischal KK, Chikina M, Clark NL.. 2017. Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. eLife. 6:e25884. doi:10.7554/elife.25884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO.. 1999. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proceedings of the National Academy of Sciences of the United States of America. 968:4285–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G.. 2013. Enhancers: Five essential questions. Nature Reviews Genetics. 144:288–295. doi:10.1038/nrg3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudent X, Parra G, Schwede P, Roscito JG, Hiller M.. 2016. Controlling for phylogenetic relatedness and evolutionary rates improves the discovery of associations between species’ phenotypic and genomic differences. Mol Biol Evol. 338:2135–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Project Consortium TE, Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M.. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature. 4897414:57–74. doi:10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Clark WT, Oron TR, Schnoes AM, Wittkop T, Sokolov A, Graim K, Funk C, Verspoor K, Ben-Hur A, et al. 2013. A large-scale evaluation of computational protein function prediction. Nature Methods. 103:221–227. doi:10.1038/nmeth.2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 437:e47. doi:10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DF, Foulds LR.. 1981. Comparison of phylogenetic trees. Mathematical Biosciences. 53(1–2):131–147. doi:10.1016/0025-5564(81)90043-2 [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2009. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 261:139–140. doi:10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanoski CE, Glass CK, Stunnenberg HG, Wilson L, Almouzni G.. 2015. Epigenomics: Roadmap for regulation. Nature. 5187539:314–316. doi:10.1038/518314a [DOI] [PubMed] [Google Scholar]
- Roscito JG, Sameith K, Parra G, Langer BE, Petzold A, Moebius C, Bickle M, Rodrigues MT, Hiller M.. 2018. Phenotype loss is associated with widespread divergence of the gene regulatory landscape in evolution. Nature Communications. 91. doi:10.1038/s41467-018-07122-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Y, Huarte M.. 2015. Long Non-Coding RNAs: Challenges for Diagnosis and Therapies. Nucleic Acid Therapeutics. 231:15–20. doi:10.1089/nat.2012.0414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KP. 2011. phangorn: Phylogenetic analysis in R. Bioinformatics. 274:592–593. doi:10.1093/bioinformatics/btq706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A.. 2014. Transcriptional enhancers: From properties to genome-wide predictions. Nature Reviews Genetics. 154:272–286. doi:10.1038/nrg3682 [DOI] [PubMed] [Google Scholar]
- Stern DL. 2013. The genetic causes of convergent evolution. Nat Rev Genet. 1411:751–764. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences. 10116:6062–6067. doi:10.1073/pnas.0400782101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GWC, Hahn MW.. 2015. Determining the null model for detecting adaptive convergence from genomic data: A case study using echolocating mammals. Molecular Biology and Evolution. 325:1232–1236. doi:10.1093/molbev/msv013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 248:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kumar S.. 1997. Detection of convergent and parallel evolution at the amino acid sequence level. Molecular Biology and Evolution. 145:527–536. doi:10.1093/oxfordjournals.molbev.a025789 [DOI] [PubMed] [Google Scholar]
- Zou Z, Zhang J.. 2015. No genome-wide protein sequence convergence for echolocation. Molecular Biology and Evolution. 325:1237–1241. doi:10.1093/molbev/msv014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.