Abstract
Genetic variation in contemporary South Asian populations follows a northwest to southeast decreasing cline of shared West Eurasian ancestry. A growing body of ancient DNA evidence is being used to build increasingly more realistic models of demographic changes in the last few thousand years. Through high-quality modern genomes, these models can be tested for gene and genome level deviations. Using local ancestry deconvolution and masking, we reconstructed population-specific surrogates of the two main ancestral components for more than 500 samples from 25 South Asian populations and showed our approach to be robust via coalescent simulations.
Our f3 and f4 statistics–based estimates reveal that the reconstructed haplotypes are good proxies for the source populations that admixed in the area and point to complex interpopulation relationships within the West Eurasian component, compatible with multiple waves of arrival, as opposed to a simpler one wave scenario. Our approach also provides reliable local haplotypes for future downstream analyses. As one such example, the local ancestry deconvolution in South Asians reveals opposite selective pressures on two pigmentation genes (SLC45A2 and SLC24A5) that are common or fixed in West Eurasians, suggesting post-admixture purifying and positive selection signals, respectively.
Keywords: ancestry deconvolution, South Asia, skin color, post-admixture selection
Introduction
Archeological and anthropological evidence points to a long-term human occupation of South Asia, dating the earliest anatomically modern humans dispersal in Indian subcontinent to at least 50 thousand years ago (ka) (Mellars et al. 2013; Bae et al. 2017). Genomic studies have described the South Asian genetic landscape as a composite of West Eurasian and East Asian exogenous components that mixed with the autochthonous South Asian groups in the last 10 ka (Reich et al. 2009; Chaubey et al. 2011; Metspalu et al. 2011; Moorjani et al. 2013; Basu et al. 2016; Lazaridis et al. 2016; Damgaard et al. 2018; Narasimhan et al. 2018; Pathak et al. 2018).
The two main components (i.e., autochthonous South Asian and West Eurasian) of Indian genetic variation form one of the deepest splits among non-African groups, which took place when South Asian populations separated from East Asian and Andamanese populations, shortly after having separated from West Eurasian populations (Mondal et al. 2016; Narasimhan et al. 2018). Thus, arguably these two components separated from each other for more than 40 ka (Jouganous et al. 2017; Terhorst et al. 2017). Methods estimating shared genetic drift based on allele frequency differences (Patterson et al. 2012) have been used to elucidate the relative proportions of these ancestry components within modern South Asian populations (Reich et al. 2009). Additionally, the increasing availability of ancient DNA sequences has significantly improved our understanding of the time frame as well as the exact genetic makeup of the populations involved in past demographic events (Damgaard et al. 2018; Narasimhan et al. 2018). However, due to low-coverage data in most cases, ancient DNA is inherently limited in providing phased haplotypes. Furthermore, the limited sample size of the ancient populations makes it difficult to provide accurate population-level allele frequency estimates. For instance, there seems to be a lack of unadmixed ancient samples from South Asia, which makes it difficult to assess ancestral components of modern populations in the region. Here, we propose a complementary method, similar to a previous study (Johnson et al. 2011), to harness the information embedded within the ancient DNA informed models, on good quality genomic data. This approach starts from genomes available from extant populations and maximizes the available information on the autochthonous “South Asian” and broader “West Eurasian” genetic components that can be observed in contemporary South Asian components, regardless of the number of admixture waves. We additionally introduce a new technique to create complete genomes from ancestry assigned genomic chunks. In this way, the modern genomes are seen as being arranged together from pieces of a jigsaw of ancient haplotypes that met and recombined in South Asia, during the last 10 ka. Following what has already been attempted for recently admixed populations (Pagani et al. 2012; Moreno-Estrada et al. 2013; Montinaro et al. 2017), one can extract these genomic regions and study them separately by combining existing and thoroughly tested methods. The benefit of this approach stems from our ability to make use of existing high-quality whole genomes, which can be deconvoluted to identify the genetic makeup of the West Eurasian and South Asian populations that admixed to form contemporary South Asians and that are now extinct in their unadmixed form. The real picture might be more complicated involving multiple components in modern South Asian genomes (Narasimhan et al. 2018). However, we only used source populations targeting these two major and deeply diverged components and only picked high probability regions for these two ancestral components during deconvolution, to minimize the effects of present complex structure.
Moreover, these two ancestral populations had undergone ∼40,000 years of independent evolution, including adaptation to different selective pressures. After their arrival in South Asia (∼5 ka) (Moorjani et al. 2013; Narasimhan et al. 2018), some West Eurasian alleles may have contributed positively or negatively to the fitness of local populations, hence leaving a signature of locus-specific admixture imbalance, which could be detected in modern South Asians.
We applied the proposed local ancestry approach to 25 Pakistani and Indian populations and focused on their West Eurasian (N) and South Asian (S) genomic ancestries, further exploring events of post-admixture recent adaptation as well as pinpointing the ancestral source of a set of phenotype-informative alleles. The retrieved genetic information provides the ideal substrate for subsequent analyses aimed to unveil the dynamics of the past 50 ka of human occupation in the area.
Results
Principal Component and Admixture Analyses
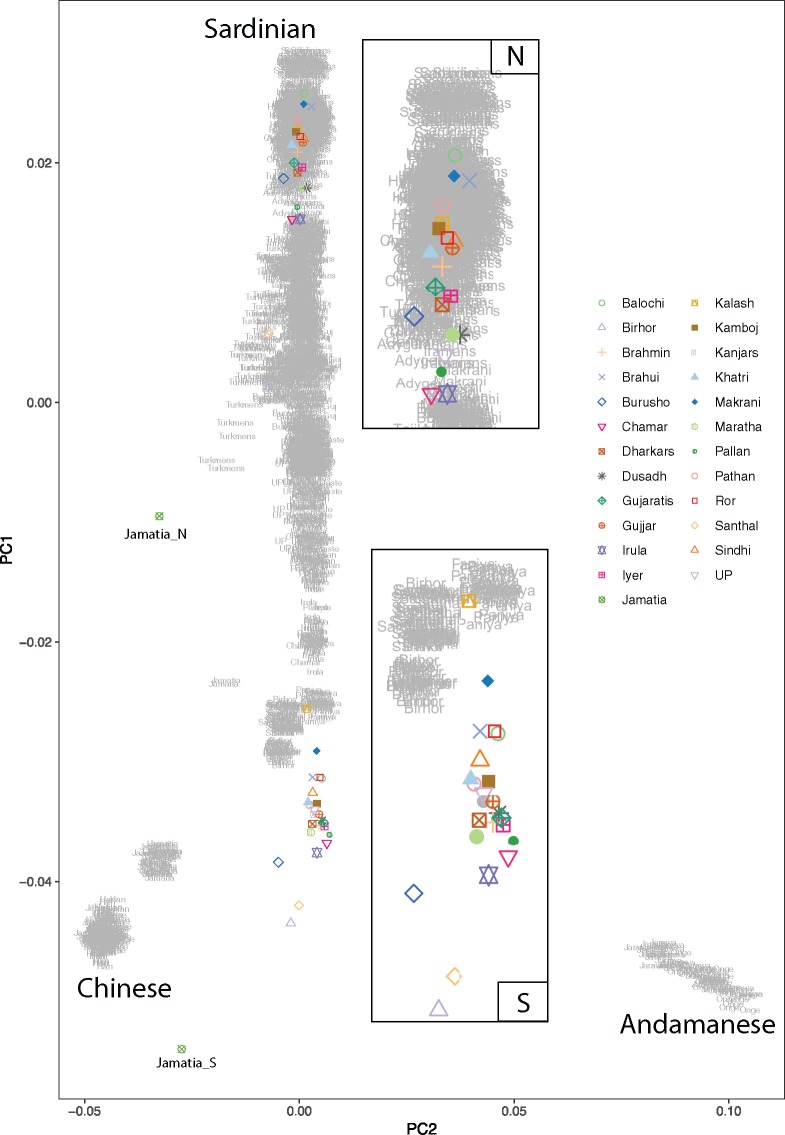
Using simulated data, we were able to show that our PCAdmix and masking approach is capable of effectively retrieving West Eurasian (N) and South Asian (S) haplotypes from admixed individuals, and that the Ancestral Random Breeder (ARB) individuals we created correctly cluster within the actual source populations (which were simulated to admix 5 ka to form India-like group) on Principal Component Analysis (PCA) (supplementary fig. 1, Supplementary Material online). We also observed that both masking and ARB approaches retrieve a good fraction of the original population heterozygosity (see Materials and Methods). Once applied on the actual single nucleotide polymorphism (SNP) array genotypes, our results show that reconstructed ARB_N samples occupy the area between Near East/Caucasus and Pakistan (close to Iranians and Lezgins), whereas reconstructed ARB_S tend to form two distinct clusters, one containing all the Indian Indo-European and Dravidian speaking populations, falling near Paniya and Irula, the other including Austroasiatic speaking populations (Birhor_S and Santhal_S) probably being attracted by the East Asian populations (fig. 1) . The separation of the N and S clusters can be observed both along the West/South Asia axis and Andaman/East Asia axis, correlated with the placement of modern populations. To control for potential artificial reduction of genetic diversity introduced by the ARB procedure at loci with decreased availability of S or N haplotypes for a given population, we reported the fraction of a given population genome where <5 S or N haplotypes are available (supplementary table 1, Supplementary Material online). Notably the scatter and positioning of ARB individuals on the PCA appears to be not correlated with such a fraction, since populations with reduced availability of S haplotypes (such as the Brahui_S, fig. 1) cluster together with other populations, such as Gujaratis_S, where either the higher fraction of S component or the higher number of available samples enable a better representation of the original S diversity (supplementary fig. 2, Supplementary Material online, see Materials and Methods). Masked samples (MASK_N and MASK_S) provided similar results, however their projected placement on PCA was wide due to missing information (introduced by the masking process itself), especially for MASK_S individuals (supplementary fig. 3, Supplementary Material online). Results were similar using Iranians, a modern population that is deemed to be closer to the West Eurasian groups that moved to South Asia, instead of French as a putative source for the N component (supplementary fig. 4a, Supplementary Material online), and more scattered when using North Indians and Onge as sources for N and S components (supplementary fig. 4b, Supplementary Material online).
Fig. 1.
PCA of ARBs (sources: French, Paniya-target: all South Asian populations) and modern populations. In order not to bias the PCA for an excess of South Asian samples, a single haploid ARB_N and ARB_S (colored) for each group was selected based on their proximity to medians of PC1 and PC2 values of each group obtained from initial PCA (supplementary fig. 19, Supplementary Material online, see Materials and Methods). Insets show a zoom on ARB_N and ARB_S samples, respectively.
Additionally, we projected ancient samples analyzed by Narasimhan et al. (2018) onto the PCA of our collection of modern populations and ARB individuals (supplementary fig. 5, Supplementary Material online). We did not observe substantial differentiation between ancient and modern samples from the South Asian region. Reconstructed ARB_N and MASK_N individuals were settled between ancient Steppe and Iran/Turan samples suggesting a successful extraction of the West Eurasian component via our approach.
ADMIXTURE analysis with a subset of the SNP array data set presents Gujarati population as a mixture of ARB_N and ARB_S components (supplementary fig. 6, Supplementary Material online). Additionally, Onge seems to have its own component which is not observed in Gujaratis, suggesting Paniya to be a better S source for the deconvolution method.
PCA results based on 1000 Genomes data attest to the appropriateness of our approach to sequence data as well (supplementary fig. 7, Supplementary Material online), though the lack of a Paniya-like population in the sequence-based data set and the putative higher N fraction present within Indian Telugu in the UK (ITU), the population we chose as South Asian source for the ancestry deconvolution, leads to a loss of precision, despite the absence of SNP ascertainment biases. We speculate that higher S ancestry estimation in the sequence data compared with the SNP array data (SNP array mean Gujaratis ancestry proportions: S = 0.20, N = 0.45, and Unknown = 0.35; sequence data mean Gujaratis ancestry proportions: S = 0.67, N = 0.12, and Unknown = 0.21) might indeed be due to the alleged presence of a sizeable N component within ITU, which causes Gujarati Indian in Houston, TX (GIH) individuals to yield a substantially greater S proportion (∼70%) compared with Gujaratis from SNP array, hence resulting in reduction of diversity and separate clustering of the ARB_N cluster on PCA plot.
Frequency-Based Analyses
To prevent ARB from introducing alterations to the original allele frequency spectrum, we used the MASK_S and MASK_N samples for a set of frequency-based analyses on SNP array data. Outgroup f3 results show, among the top scoring populations, different proxies for different MASK_N and MASK_S representatives (supplementary table 2, Supplementary Material online). Most of the reconstructed masked populations tend to have the highest scores when compared against their original unmasked population in both N and S groups. We repeated the analysis, now including ancient samples (Narasimhan et al. 2018) and sorted all the populations against MASK_N and MASK_S groups based on their mean rankings (supplementary fig. 8, Supplementary Material online). Highest ranking groups for MASK_S are other MASK_S followed by Southern Indian populations, Han and Onge. Highest ranking ancient groups are Saidu Sharif group from Pakistan (500–300 BCE) and Indus diaspora group from Iran (2550–2450 BCE), which reflects our PCA (supplementary fig. 5, Supplementary Material online). MASK_N top ranking groups are dominated by other N populations, interspersed with ancient samples such as Steppe_EMBA, which is also consistent with our PCA (supplementary fig. 5, Supplementary Material online). We note that the top two scoring ancient samples are reported to have very low SNP hits and therefore cannot be interpreted meaningfully.
We performed D-statistics as D(Gujaratis_N, French; Onge, Mbuti), D(Gujaratis_N, French; Han, Mbuti), and D(Irula_S, Onge; Steppe_EMBA, Mbuti) which resulted in Z = −0.827, 0.152, and 0.270, respectively and D(Irula_S, Onge; French, Mbuti) which resulted in Z = −2.357, indicating that these MASK_N and MASK_S are relatively clean deconvolutions. We additionally obtained D(Irula_S, Han; Onge, Mbuti) with Z = 0.268, D(Irula_S, Onge; Han, Mbuti) with Z = −0.068, D(Han, Onge; Irula_S, Mbuti) with Z = −0.348, which points to a trifurcation of S, Onge and Han as could also be noted in PCA (fig. 1). We followed up these findings by using qpAdm, which highlighted MASK_N populations as virtually free from any residual South Asian haplotypes, whereas MASK_S populations displayed a gradient of N residual ancestry (supplementary fig. 9a, Supplementary Material online). Such a difference in power of PCAdmix to obtain clean MASK_N and MASK_S samples may point to a likely pre-Neolithic or Neolithic wave (and hence finer graining) of N ancestry within the S haplotypes. Although this could have been related to the lack of power of PCAdmix approach, Irula_S being free of any N component, based on qpAdm and D analyses, eliminates this possibility (notably Irula_S can be seen as a better representative of the South Asian component, since Paniya, the population used as source, shows 25% of West Eurasian affinity in qpAdm). qpGraph analyses based on a representative set of MASK_N and MASK_S populations, however, showed that both masked groups can be represented as unadmixed West Eurasian or South Asian populations, respectively (supplementary fig. 9b–d, Supplementary Material online). More in details, the MASK_S populations can be modeled through a tree with only minor influence from Onge (supplementary fig. 9b, Supplementary Material online), which still maintains few f4 outliers, or through a slightly more complex tree (supplementary fig. 9c, Supplementary Material online) free from any outliers, in any case confirming the relatively homogeneous preadmixture S substrate (also referred to as AASI by Narasimhan et al. 2018) already highlighted by PCA. On the other hand, the treelike MASK_N model without admixture edges was significantly rejected (supplementary fig. 10a, Supplementary Material online) and the fit between the tree and data could be improved by invoking multiple admixture events (supplementary fig. 9c, Supplementary Material online), although none of our efforts managed to successfully eliminate all f4 outliers. Thus, the MASK_N groups cannot be described as a simple cluster of homogeneous populations, potentially pointing to multiple waves of arrival of West Eurasian components in the region. Importantly, the addition of an admixture edge from West Eurasian (Steppe_EMBA) into MASK_S or from South Asian (Onge) into MASK_N decreased the overall model fit of each qpGraph and increased the number of outlier f4 statistics, hence pointing to the lack of residual admixture in either of the masked populations. We further decomposed the reconstructed N ancestry with qpAdm, using various West Eurasian populations. Our results (supplementary fig. 10b, Supplementary Material online) confirm the heterogeneity within the N haplotypes already inferred with qpAdm and points to Armenia_MLBA as one of the putative major genetic contributor. As a proof of principle, we then moved on and constructed a simplified qpGraph where ancient populations already reported as a mixture of West Eurasian and South Asian ancestries (Indus Periphery and SGPT from Narasimhan et al. 2018) were described as either a combination of Steppe_EMBA and Onge frequencies (in absence of other South Asian genomes free from any West Eurasian component), or as our MASK_N and MASK_S genomes. Although the oversimplifications inherently present in the tested trees did not allow for appropriate removal of f2 and f4 outliers in any case, the overall final scores obtained when using S and N as admixture sources were up to one order of magnitude smaller than with their Onge/Steppe counterparts (supplementary fig. 11, Supplementary Material online).
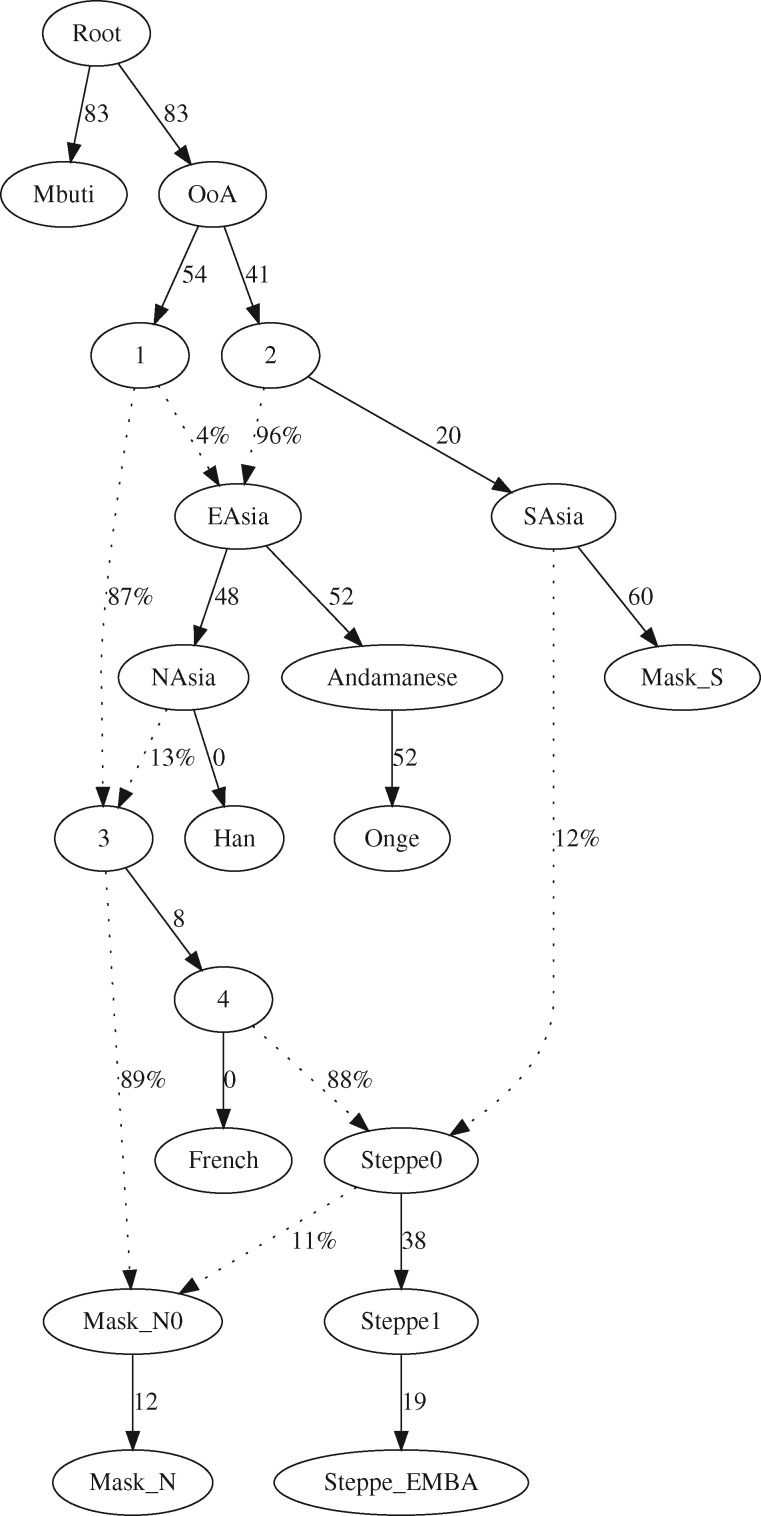
We followed up these preliminary explorations and included N (represented by Gujaratis_N) and S (represented by Irula_S) within a more comprehensive qpGraph aimed at yielding the broader picture of involved ancestries. The obtained qpGraph (fig. 2) has no f2 or f4 outliers and shows S to trifurcate from East Asian and Andamanese groups (here represented by Han Chinese and Onge). Notably all alternative trees invoking earlier splits of East Asians or Andamanese yielded higher final scores and f4 outliers, pointing to the slightly early separation of S as the best possible trifurcation. Also, alternative models reverting the directionality of admixture between precursors of S and of Steppe_EMBA increased the number of f4 outliers.
Fig. 2.
qpGraph model of N and S ancestries. The qpGraph reported here places the obtained N and S deconvoluted ancestries (represented here by Gujaratis_N and Irula_S, respectively) within the broader scenario of within Eurasia splits and subsequent admixtures. Alternative trees involving a different Onge/Han/S split order, or a reverse Steppe_EMBA – S admixture direction yielded f4 outliers and an overall poorer fit. Final score: 3,061.001, degrees of freedom: 2, no f2 outliers, no f4 outliers, worst f-stat: 1.747.
Phenotype-Informative SNPs
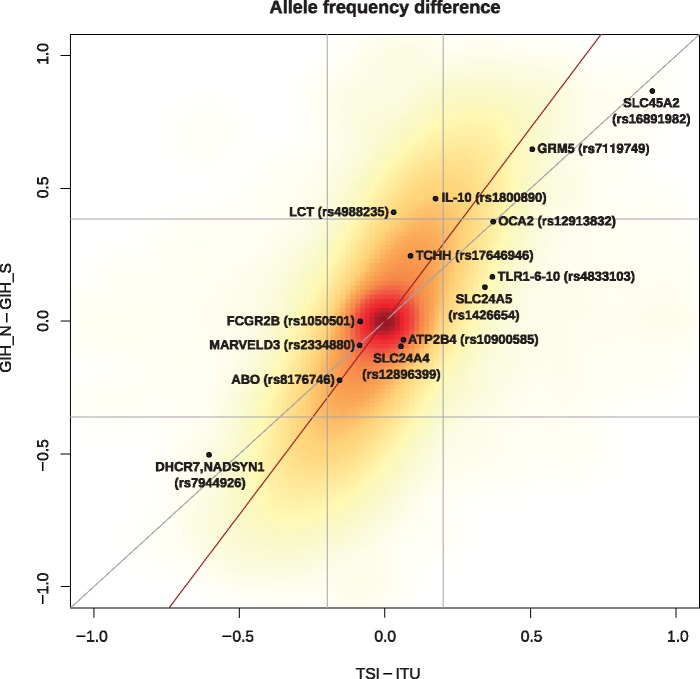
We compiled a list of phenotype-informative SNPs (Mathieson et al. 2015; Gelabert et al. 2017; van de Loosdrecht et al. 2018) and identified allele frequency differences between MASK_N and MASK_S genotypes (supplementary table 3, Supplementary Material online) to putatively link the presence of each SNP in South Asians to the arrival of the N component in the region, using the 1000 Genome GIH sequence data to maximize SNP coverage. Before interpreting these results, we calculated the genome-wide distribution of derived allele frequency differences between the source populations we used for the PCAdmix deconvolution on sequence data (Toscani in Italy [TSI] and ITU), and we calculated the same distribution between the GIH_N and GIH_S derived allele frequencies (fig. 3). These two distributions provided us with a reliability range, since moderate differentiation between the two PCAdmix sources (x axis, fig. 3) may influence the efficacy of the deconvolution process, whereas a small allelic differentiation between the GIH_S and GIH_N sequences (y axis, fig. 3) may point to a genuine similarity between the two components or to a poor PCAdmix performance for a specific region. We, therefore, identified the top and bottom 2.5% of each distribution and considered as reliable only information falling within the overlap of these highly differentiated ends of each axis (the area of the plot represented by the four peripheral square sections in fig. 3). By doing this, we were able to concentrate on SNPs which were differentiated between two source populations and which were also successfully assigned to the right masked counterparts (GIH_N or GIH_S). Highly positive values on the y axis suggest that the introduction of a given SNP in contemporary South Asians (here represented by GIH) can be conservatively attributed to the N ancestry (for cases such as SLC45A2 pigmentation allele in Europeans), whereas negative values reflect a high presence of a given allele within the S ancestry (for the case of DHCR7/NADSYN1). Other markers of interest, such as rs4988235 (associated with lactase persistence, directly upstream of LCT), rs12913832 in OCA2/HERC2, associated with pigmentation/eye color) and rs1800890 (in immune regulator IL-10) among others are shown as predominantly present within the N ancestry but do not meet our stringent acceptance criteria exemplified in figure 3. Notably, the SLC24A5 allele responsible for fair skin in West Eurasians falls within the unreliable area of the plot, appearing as weakly differentiated between the N and S ancestries. We followed up this result and explored the haplotype background of the SLC24A5 (rs1426654) variant using Haplostrips and discovered that both MASK_S and MASK_N haplotypes display the same “West Eurasian” haplotype (supplementary fig. 12, Supplementary Material online). This may point to a deeply shared ancestry between S and N at this locus or, more plausibly and as already shown by our reliability threshold (fig. 3), to inaccuracy in our PCAdmix ancestry deconvolution at this locus due to the high presence of this specific haplotype in both (West Eurasian and South Asian) source populations as a consequence of the high West Eurasian component in ITU. The absence of the rs1426654 “European” allele in other South Asian populations such as Onge and Birhor, as well as in any East Asian group seems to lend further support to the latter scenario. Our results point, regardless, to a high frequency of this haplotype in South Asia, reported as under selection in West Eurasia in the last 10 ka (Pickrell et al. 2009; Mathieson et al. 2015).
Fig. 3.
Derived frequency differences between source and deconvoluted populations. In the scatter plot, we report the difference in derived frequency between GIH_N and GIH_S against the same difference between the source populations TSI and ITU: all SNPs from 1000 Genomes sequences are included in the cloud. Phenotype-informative SNPs reported in supplementary table 3, Supplementary Material online, are provided as dots. The vertical and horizontal gray lines delimit the top and bottom 2.5% most extreme values in each axis independently. The oblique red and gray lines represent, respectively, the best fit linear regression and the diagonal.
Local Admixture Imbalance between Ancestral Components in South Asian Populations
Since their separation ∼40 ka, the N and S ancestry components (Mondal et al. 2016), are likely to have undergone adaptation to different West Eurasian and South Asian environments. The “N” alleles, once introduced into South Asia and exposed to the local environment could have had either beneficial or damaging effects on the resulting admixed populations. Given the time frame since the previous N/S admixture (∼5 ka), the interaction between the newly introduced N variants in the South Asian environment may have resulted in regional deviations from the expected admixture proportions, pushing the local N admixture fraction upward or downward depending on whether a given N haplotype had a positive or negative impact on the fitness of the admixed population. We looked for these local admixture fluctuations by calculating Ancestral Haplotype Frequency Difference (AHFD) on SNP array data for all 25 South Asian populations separately and tested whether these highly deviated regions are shared by different South Asian populations, suggesting the action of long-term (∼5 ky) selective forces and mitigating the confounding effect of population-specific random processes. Both our simulated data and real data failed normality tests for local single ancestry deviation (LSAD), local single ancestry deviation corrected (LSADC) as well as AHFD (data not shown). We suspect from our simulation that this is caused by a ceiling effect (as frequency of ancestry should be within 0 and 1). Thus, all the further results were converted to percentile as it is nonparametric. We found that AHFD is slightly more robust than the previously published methods from our simulation data (supplementary fig. 13, Supplementary Material online) in case of missing or unknown ancestry present in the data. In case of no missing data, they are interchangeable (see Materials and Methods).
For each ten SNPs region (∼130 kbp, close to expected length of ancestral regions given an admixture event happened ∼5 ka [Huerta-Sánchez et al. 2014]), we counted the number of populations displaying highly deviated (top 2.5%) N (represented by positive scores) and S (negative scores) admixture fractions. We used this summarized metric to construct a Manhattan plot (supplementary fig. 14, Supplementary Material online) that captures regions with shared biases of ancestry contribution in different populations (for more details see Materials and Methods). We then manually examined the most extreme 5/18569 regions for both the N and S component and annotated the genes they included (table 1) as representative of the underlying peaks, and reported all windows where at least 10 population showed significant admixture imbalance in supplementary table 4, Supplementary Material online.
Table 1.
Top Five Hits for N- and S-Related Selected Regions.
| Component | Position (Chr:Start–End) | Number of Populations with Significant Value | Genes (±50-kb Region) | Number of Ten SNPS Regions |
|---|---|---|---|---|
| N | 3:9,363,925–9,595,374 | 22 (percentile = 99.9949) | THUMPD3, SETD5 | 2 |
| 6:84,399,772–85,572,756 | 21 (percentile = 99.9814) | SNAP91, RIPPLY2, CYB5R4, MRAP2, CEP162, TBX18 | 2 | |
| 6:30,079,993–30,257,693 | 21 (percentile = 99.9814) | TRIM31, TRIM40, TRIM10, TRIM15, TRIM26, HLA-L | 1 | |
| 14:97,636,701–97,715,909 | 19 (percentile = 99.9383) | Intergenic | 1 | |
| 19:23,930,879–24,368,053 | 18 (percentile = 99.9195) | ZNF681, ZNF726, ZNF254 | 1 | |
| S | 5:33,944,217–34,032,014 | −21 (percentile = 0.0057) | RXFP3, SLC45A2, AMACR, C1QTNF3, ADAMTS12 | 2 |
| 20:652,097–694,894 | −16 (percentile = 0.038) | SRXN1, SCRT2, SLC52A3 | 1 | |
| 8:116,208,407–116,308,464 | −16 (percentile = 0.038) | Intergenic | 1 | |
| 9:12,276,668–12,460,256 | −15 (percentile = 0.0757) | Intergenic | 2 | |
| 8:54,578,044–55,071,319 | −14 (percentile = 0.1268) | ATP6V1H, RGS20, TCEA1, LYPLA1, MRPL15 | 1 |
Note.—Only populations with admixture imbalance score beyond 2.5 percentile were counted. Positive values represent N excess and negative values represent S excess.
One of the top five 10 SNP windows showing excess of N ancestry in most analyzed populations encompasses the SETD5 genic regions, potentially involved in regulation of insulin sensitivity (Palmer et al. 2015; Walford et al. 2016) (table 1). Zinc finger proteins (ZNF681, 726, 254) linked with colorectal cancer due to diet (Figueiredo et al. 2014) and HLA, a key immunity gene (Fairfax et al. 2012). The latter finding, although potentially important to elucidate the impact of the N/S admixture on the immune system of modern South Asians, maybe be due to balancing selection in ancient source populations or may as well be driven by ascertainment biases inherent to the SNP array data we used.
The top window showing excess of S ancestry (table 1) encompasses the SLC45A2 gene, reported to play an important role in skin and eye pigmentation in West Eurasians (Mathieson et al. 2015; Crawford et al. 2017; Martin et al. 2017), tanning (Visconti et al. 2018), and squamous cell carcinoma (Chahal et al. 2016) and already observed in our independent, SNP-based test (fig. 3).
Discussion
In this work, we demonstrate that haplotypes belonging to the broad N and S ancestries can still be retrieved from admixed South Asian contemporary groups even if the original source populations are no longer available, despite the long time since the gene flow event and even if the arrival of N in the area may have occurred through multiple waves or mediated by structured populations (Narasimhan et al. 2018). Nevertheless, we acknowledge that the process might introduce a slight bias toward the source populations as shown in our outgroup f3 tests where the source populations used are often present among the top populations after other masked N and S individuals. Given the comparable results obtained when using French or Iranians (a population deemed to be closer to the actual N component) as a West Eurasian source population during the ancestry deconvolution, we believe our choice of using French for the bulk of our analyses should mitigate the observed bias and allow for a reliable interpretation of the observed MASK_N and ARB_N components. Although providing somewhat similar results, using Onge as South Asian source and Pathan, a Northern South Asian population, as the West Eurasian source drastically reduces the fraction of confidently assigned genome and creates scattered MASK_S groups on PCA space (supplementary fig. 4b, Supplementary Material online), possibly due to the high genetic distance between Onge and S (fig. 2).
Despite the possible biases, our method provides substantial advantage as it yields full genomes for reconstructed population-specific surrogates of West Eurasian and South Asian source populations to be used for a wide range of analyses hence complementing the available low-coverage and low-quantity aDNA, which is currently not available for unadmixed South Asian ancient populations (also known as AASI). The S component we extracted, therefore, might be used as a proxy for “AASI” and seem to pinpoint its origin from a trifurcation involving S, East Asians and Andamanese (fig. 2). However, it is important to note that similarly straightforward demographic reconstructions may not be possible for the N component, as it is a mixture of all West Eurasian contributions to modern South Asian populations. We caution that our approach is specifically designed to disentangle West Eurasian from South Asian components and, hence, additional components (i.e., East Asian) documented for certain Austroasiatic groups may be masked as “unknown” or equally embedded within the N and S masked genomes.
Through qpGraph, we showed that the retrieved South Asian N haplotypes are comparable in their genetic composition to the mixture of West Eurasian groups initially defined by Reich et al. (2009), whereas the S component of most South Asian populations reconstructed here appears to be a good proxy for the AASI component described by Narasimhan et al. (2018), that is, a South Asian population with no detectable West Eurasian or East Asian ancestry. Overall, we believe the availability of authentic West Eurasian and South Asian haplotypes yielded by our approach, as opposed to inferred allele frequencies, is invaluable for downstream applications and we showed that our S and N genomes may serve as good genetic data to reconstruct modern and ancient admixture events.
From a functional viewpoint, allele frequency differences between N and S suggest that certain phenotypic traits in India might have been associated with different ancestry sources. The clearest example of extreme functional allele polarization by ancestry is represented by the SLC45A2 European allele (rs16891982-G) which is predominantly explained by the N ancestry. Other examples of alleles linked with the N ancestry are represented by the MCM6/LCT Lactase Persistence allele (rs4988235-A) which appear to have arrived in the region mediated by the N ancestry. Though it is important to note that our source populations for the sequence-based deconvolution method, which are ITU and TSI, have very similar frequencies for this allele, meaning that this position does not meet our stringent acceptance criteria.
We also show that the interaction between alleles that were highly polarized between the two ancestry sources that admixed in South Asia caused patterns of admixture imbalance across the majority of sampled groups, hence unlikely explainable by population specific random drift, and perhaps due to positive or negative environmental pressures. Interestingly, we report how loci that include genes involved with diabetes (SETD5), diet (ZNF) and the immune response (HLA) show West Eurasian (N) haplotypes to be significantly more represented compared with the South Asian (S) counterparts. This might be a stark contrast to what is expected, given the long-term history of local adaptation of S haplotypes in local environment. We speculate that the diet-related signal may be linked with post-Neolithic dietary shifts that might have followed the arrival of the West Eurasian component in the area, whereas the overrepresentation of West Eurasian HLA haplotypes might have some similarity, although at a different time scale, with what has happened in Native American populations after recent colonization likely caused by European borne epidemic (Lindo et al. 2016).
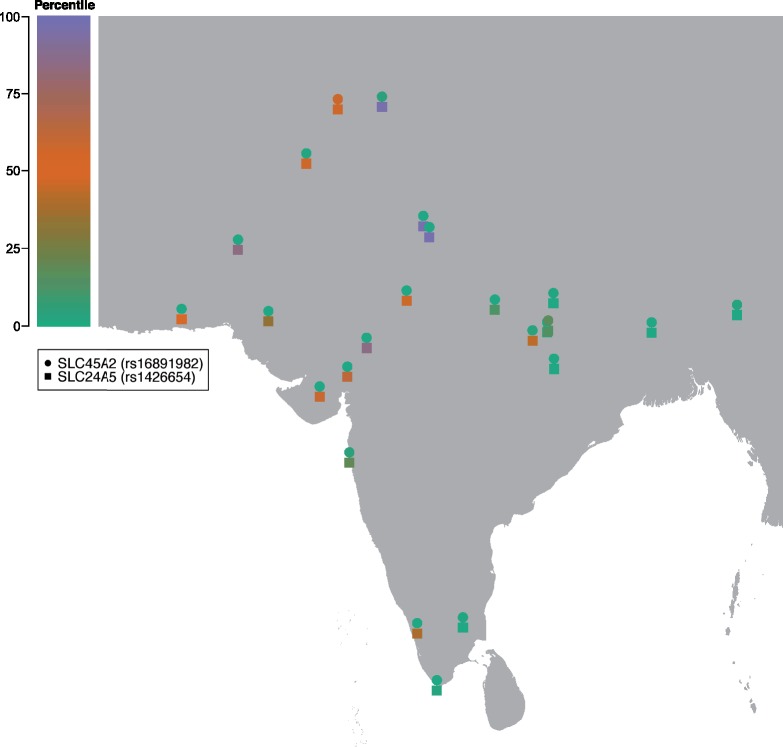
On the other hand, the top region for significant enrichment of South Asian ancestry includes the rs16891982-G allele of SLC45A2 gene (associated with light skin pigmentation in West Eurasians), suggesting purifying selection at this locus following admixture. From an allele frequency perspective (fig. 3 and supplementary table 3, Supplementary Material online, obtained from 1000 Genomes sequence data), we can clearly link the derived rs16891982-G allele with West Eurasian N haplotypes, but the overall abundance of these West Eurasian alleles is drastically reduced in 21 out of 25 South Asian populations analyzed here (fig. 4, dots, based on SNP array data). Such a strong negative pressure against a light pigmentation allele may be explained by the high ultraviolet (UV) radiation at South Asian latitudes and this result seems to be further corroborated by similar N ancestry deficiencies in TYRP1 and BNC2 genes for as many as 11 South Asian populations (supplementary table 4, Supplementary Material online). However, purifying selection against maladaptive light pigmentation alleles in high UV environment is not observed for all pigmentation alleles; in fact, the rs1426654-A allele of the SLC24A5 gene, which explains the highest proportion of skin pigmentation variation in individuals of European versus African ancestry (38%) (Lamason 2005), and Indians (27%) (Basu Mallick et al. 2013; Juyal et al. 2014), shows instead an increase of frequency in South Asians (assuming its absence in ancestral South Asians). Furthermore, this locus does not appear as an outlier for the reduction of N haplotypes in North and West South-Asian populations, despite our ancestry deconvolution is overestimating the A allele frequency in S populations (fig. 4, squares). Regardless of the accuracy of the PCAdmix ancestry deconvolution at this specific locus, as cautioned in figure 3, the West Eurasian rs1426654-A allele shows 95% frequency in 1000 Genomes GIH and is present at high frequency in most analyzed populations. Taken together, our results point to opposite pressures on some West Eurasian alleles involved in skin and eye pigmentation. On one hand, SLC45A2 seem to have undergone some selective pressure that removed most of West Eurasian alleles that arrived in the area after the admixture event. Conversely, the SLC24A5 (rs1426654-A) West Eurasian allele seems to have escaped such a negative pressure perhaps thanks to its apparent neutral role with respect to susceptibility to skin carcinoma caused by UV radiation (Gerstenblith et al. 2010; Chahal et al. 2016) or thanks to its beneficial effect through another yet to be understood phenotype.
Fig. 4.
Admixture imbalance (AHFD) percentile values of ten SNP windows including the rs1426654 (SLC24A5, squares) and rs16891982 (SLC45A2, dots) markers. The window including the SLC45A2 marker is in the bottom (green) percentile of most South Asian populations, hence showing a significant excess of the S haplotypes compared with the genome-wide average per population. The window including the SLC24A5 marker shows instead a more balanced pattern, showing moderate to high scores (orange to purple, hence toward an excess of N haplotypes) in West and North South Asia and orange to green (hence toward a prevalence of S haplotypes) in East and South Asia.
In conclusion, our approach allowed us to perform demographic and functional analyses on two major genetic components of South Asia, revealing both novel and confirmatory findings related to genetic structure of the region and providing population-size high-quality ancestral haplotypes (MASK and ARB) ready to be used for downstream applications, especially for analyses which are underpowered in presence of missing data, such as selection tests and haplotype-aware methods. We believe this approach can be extended and applied on other admixed populations where the source populations are no longer available, even in the absence of relevant ancient DNA.
Materials and Methods
Samples
We used SNP array data (initially total 1,286,187 SNPs) from various resources including a total of 565 samples from 28 populations from India and Pakistan (Li et al. 2008; Altshuler et al. 2010; Metspalu et al. 2011; Basu et al. 2016; Pathak et al. 2018) (supplementary table 5, Supplementary Material online), merged with 404 modern and 360 ancient reference samples from surrounding regions (Behar et al. 2010; Yunusbayev et al. 2012; Behar et al. 2013; Haak et al. 2015; Yunusbayev et al. 2015; Mörseburg et al. 2016; Narasimhan et al. 2018) (supplementary table 6, Supplementary Material online). SNP array data were filtered using PLINK 1.9 (Purcell et al. 2007; Chang et al. 2015) for minor allele frequency <0.01 for the whole sample of populations using the the –maf flag, missing call rates for variants <0.03 using the –geno flag and individuals <0.03 using the –mind flag. Additionally, to show the applicability of our methods to sequence data, we used 1000 Genomes phase III data (1000 Genomes Project Consortium et al. 2015). 1000 Genomes data were refined using VCFtools 0.1.14 (Danecek et al. 2011) filtering out SNPs with minor allele frequency <0.01 and multiallelic sites using maf and min-alleles 2 max-alleles 2, respectively.
Local Ancestry Deconvolution (PCAdmix)
PCAdmix uses PCA to infer the ancestry of fixed SNP windows of an admixed population with the help of given source populations (Brisbin et al. 2012). We set window size to ten SNPs after default LD (linkage disequilibrium) pruning (r2 > 0.8, based on a built-in window size), a value suggested by the software’s developers as the smallest size for reliable resolution and corresponding, on average, to 130 kbp in our data set. Additionally, we only used windows assigned to ancestral proxies with posterior probabilities (provided in the PCAdmix .fbk output) higher than 0.95. Based on simulations, thresholds of 0.90, 0.95, and 0.99 provided similar results (supplementary fig. 15, Supplementary Material online). For SNP array data, we used French and Paniya as proxies for West Eurasian (N) and South Asian (S) ancestry donors, respectively. Paniya were chosen for their low N component (Metspalu et al. 2011; Basu et al. 2016) and alleged similarity to the native S substrate, as opposed to Onge which may represent a deeper split on the South Asian population tree. On the other hand, French were preferred over more obvious West Eurasian populations (such as Iranian or Armenians or even Northern South Asians) to control for potential biases introduced by the ancestry deconvolution process such as artificial similarity of the deconvoluted haplotypes to the source population, or the loss of a percentage of the genome due to low differentiation between two parental proxy populations (supplementary table 1, Supplementary Material online), though we also present the PCA results with Iranians or Pathan (Northern South Asians) as a West Eurasian source, or Onge as South Asian source (supplementary fig. 4, Supplementary Material online). As good haplotypes are needed for the deconvolution process, only modern populations were considered. For 1000 Genomes data, we used GIH as the admixed population and TSI along with ITU as source populations for the N/S deconvolution, preferring ITU over other South or East Asian available populations to maximize the geographical and genetic affinity of the 1000 Genomes source with Paniya (for whom only SNP array data were available). French (for SNP array data) and Toscani (for 1000 Genomes Data), rather than other West Eurasian populations, were preferred to reduce the risk of any Eastern component presence in the source populations for N component, which would reduce the accuracy of deconvolution. Nevertheless, results were similar using Iranians (for SNP array data) as a putative source for the N component (supplementary fig. 4a, Supplementary Material online). The selected windows were masked based on their affiliation, yielding MASK_N and MASK_S haplotypes which we used for PCA, f3 and f4 tests, qpAdm and qpGraph via AdmixTools 4.1 as population based proxies of the N and the S ancestry component, respectively (Patterson et al. 2012). We therefore created MASK_N and MASK_S sets of individuals where, for each haploid individual, the haplotypes of the nonrelevant ancestry were masked out as missing data (e.g., haploid Individual N will carry only information for the sites identified as N by PCAdmix and will have the unknown and S sites masked out).
As PCAdmix requires phased haplotypes, we used SHAPEIT 2 (Delaneau et al. 2012) with default options for phasing all the modern samples together without using any reference. We used HapMap b37 genetic map (Altshuler et al. 2010) for SHAPEIT map input.
Constructing ARBs
For each analyzed population (n = 25), we generated a set of ARBs to limit issues related with PCA projection due to the reported shrinkage phenomenon (Haasl et al. 2013), which can be only partially mitigated in the case of samples with high missingness. ARBs were created for each N and S ancestry, by taking all the MASK_N or MASK_S individuals and replenishing the masked-out haplotypes by randomly picking (with replacement) a nonmasked haplotype from another donor within the same Pop_N or Pop_S population. This process hence created a number of ARB haploid individuals which feature the genetic makeup of the original individual used as a scaffold, and, where not available, a random set of haplotypes from a given ancestry, drawn from that individual’s population. The reconstructed ARB population can therefore be seen as a set of random breeders, to be considered as the best available proxy to the actual ancestry source within the studied population. Contrarily to the MASK_N and MASK_S samples, where allele frequencies are kept unaltered within each ancestral component, due to the potential introduction of artificial allele frequency shifts caused by the ARB-making process, the ARB_N and ARB_S samples, amicably dubbed “Frankensteins,” could not be used on frequency-based approaches such as f3/f4, qpAdm and qpGraph. Nevertheless, we show through simulation (see below) that ARB haploid individuals can be used reliably on PCA and ADMIXTURE, hence avoiding the introduction of any projection artifact. Notably the average size of each ten SNPs haplotype deconvoluted using PCAdmix is roughly 130 kbp, a length that is within the range of the expected ancestry chunks after an admixture event that took place between 100 and 200 generations ago (Huerta-Sánchez et al. 2014). Joining of ancestry chunks during the ARB construction procedure is, therefore, expected to have a limited impact on the creation of artificial switches within a given ancestral haplotype.
Alternative Ancestral Random Breeders (ARB2) and Rescued Heterozygosity Estimates
We also tried an alternative way of creating ARBs, working only on a small number of recipient ARB per population, by retaining only positions for which a minimum number of available donor haplotypes were available in a given ancestry status to minimize the donation of the same haplotype to multiple ARBs, and by maximizing the length of the donated haplotype and its affinity to the surrounding sequences of a given receiving ARB, to minimize the number of ancestry switches artificially introduced by the ARB making process. This alternative ARB approach (ARB2) yielded similar PCA results (supplementary fig. 16, Supplementary Material online) and increased the fraction of rescued heterozygosity at the cost of either reducing sample size or reducing SNP availability. We compared the reduction of expected average genomic heterozygosity between the two ARB making methods by applying N and S masks of 5 Gujarati individuals to five arbitrary Iranians (supplementary fig. 17, Supplementary Material online), showing MASK and ARB2 to perform better than the initial ARB approach at rescuing the expected average genomic heterozygosity (2pq).
Simulations
We simulated a model for four modern day populations using ms software (uchicago.app.box.com, March 1, 2018) (Hudson 2002), which is able to generate linked data, as proxies for European, Asian, African, and Indian-like groups (supplementary fig. 18, Supplementary Material online) to test the applicability of our PCAdmix approach which is fundamentally based on haplotypes. We used the following command line in 22 iterations (corresponding to chromosomes) with seeds (x) ranging from 1 to 22 to produce 10 mega base regions of 20 haplotypes for each population and sample 10 haplotypes from two ancient populations at 9 ka:
ms 80 1 -t 6493.248 -r 5861.96 10000000 -I 4 20 20 20 20 -n 1 8.977 -n 2 4.175 -n 3 2.104 -n 4 5.457 -g 1 267 -g 2 172.35 -g 4 235.5075 -m 1 2 1.889 -m 2 1 1.889 -m 1 3 0.216 -m 3 1 0.216 -m 2 3 0.496 -m 3 2 0.496 -es 0.0038236 4 0.70 -ej 0.007647201 4 2 -ej 0.007647201 5 1 -ej 0.034 1 2 -em 0.0956 2 3 7.124 -em 0.0956 3 2 7.124 -ej 0.0956 2 3 -en 0.239 3 1 -eA 0.006882481 4 10 -eA 0.006882481 5 10 -p 15 -seeds x x x
This yielded 511,540 total SNPs. Parameters for European, Asian, and African populations were selected from Jouganous et al. (2017). Arbitrary parameters within reasonable ranges relative to parameters for other simulated populations were used for the Indian-like group which was formed as an admixture of ancient European (N) and ancient Asian (S) groups (Ne = 61,520, Ne0 = 25,000, West Eurasia-N and South Asia-S population split = 10 ka, N/S admixture = 5 ka, N component = 70%, and S component = 30%). We performed filtering for minimum allele frequency (0.01) and population-based high linkage disequilibrium (0.8) for local ancestry deconvolution procedure. The simulated admixed genomes were then processed with PCAdmix using the simulated proxy sources and the resulting masked and ARB individuals were analyzed through PCA to see their placement relative to source and ancient samples. Median values for PC1 of each group were calculated with an initial PCA with all ARBs (supplementary fig. 19, Supplementary Material online). Subsequently, one haploid N individual and one haploid S individual with PC1 values closest to the corresponding medians for each ARB group were selected. We then used only these individuals for the second round of PCA. For all PCA with ARBs in the whole study, we used this approach (only one individual for each ARB group) to reduce excessive clustering related to the high number of ARBs. Additionally, ADMIXTURE runs were performed from K = 2 to K = 5 including ARB individuals with simulated modern and ancient samples to show that ARB components make up Indian-like group (supplementary fig. 20, Supplementary Material online).
PCA and Admixture Analyses
We performed PCA as an established initial screening method on the merged data set using smartpca (Patterson et al. 2006; Price et al. 2006), using the lsqproject option to project samples with missing genotypes. We used the ADMIXTURE (Alexander et al. 2009) software to perform unsupervised clustering of simulated and real data before and after ancestry deconvolution. We used R and ggplot2 package for visualization (Wickham 2017; R Development Core Team 2018).
Missingness and Principal Components
We analyzed the correlation between actual missingness (which corresponds to the windows not assigned to N or S ancestry given that our confidence fbk threshold is 0.95) and principal components (supplementary fig. 21, Supplementary Material online). Average missingness ranges between 0.22 and 0.40 for each population. We found out that there is a correlation between PC1 and missingness for both N and S sets which suggests a positive correlation between missingness and S proportion. This is likely to be related to the existence of West Eurasian (N) component in our S source population, Paniya, which results in lower confidence for ancestry assignment for certain windows during deconvolution. An alternative explanation may involve differential presence of N and S components within the fraction of genome labeled as “unassigned” by our deconvolution approach. We also compared the behavior of ARB and MASK populations on PCA space based on haplotype availability (supplementary fig. 2, Supplementary Material online). As expected, MASK populations cluster more loosely due to loss of SNP information. We used R and ggpubr package for the analyses (Kassambara 2018; R Development Core Team 2018).
Frequency-Based Allele-Sharing Analyses
We used Admixtools 4.1 to calculate outgroup f3 in the form f3(X, Y; Yoruba), X being MASK_N and MASK_S populations, Y being all other populations including masked ones. D-Statistics were performed with the same software using qpDstat to test for possible biases. With the same software, we also run the package qpAdm testing all of our South Asian populations and their MASK_N and MASK_S counterparts as a three-way mixture of West Eurasian (Armenia_MLBA) and East (Cambodians, as the best proxy for the documented Austroasiatic admixture event) and South (Onge) Asian groups and using the following populations as outgroups (Kankanaey, Karitiana, Mbuti, Papuan, Ust_Ishim, and Yoruba). We report here all combinations that yielded a qpWave P value > 0.05 and thus accepting the proposed mixture model. We also replicated the analyses using Han Chinese or Chukchis as an alternative proxy for the East Asian component and found no detectable differences between the already low amounts of East Asian components detected by the various runs of qpAdm. Once observed that no MASK_N population retained any South Asian component, we also tested each MASK_N as a three-ways combination of (Anatolia_N or Levant_N), EHG and Iran_N using a different set of outgroups (WHG, Han, Kankanaey, Karitiana, Mbuti, Papuan, Ust_Ishim, and Yoruba). We then used the information gathered from qpAdm to build a qpGraph model of our whole and MASK_N or MASK_S populations, taking four populations representative of the West/South Eurasia cline. Although qpGraph describing MASK_S specific relationships (supplementary fig. 9b and c, Supplementary Material online) and the broader South Asian picture (fig. 2) were successfully fitted to yield no f2 or f4 outliers with a positive number of degrees of freedom, MASK_N specific qpGraphs (supplementary fig. 9d, Supplementary Material online) were harder to model and, to the best of our efforts, always retained a number of f4 outliers.
Haplostrips
We used Haplostrips (Marnetto and Huerta-Sánchez 2017) to visualize the haplotype structure at SLC24A5 locus, using 1000 Genomes populations and MASK_N and MASK_S GIH sample groups. The plotted SNPs were filtered for a population-specific minor allele frequency >5%. See the application article for further details about the method.
Regions of N/S Admixture Imbalance
We painted every individual using ancestries detected by PCAdmix. Every individual is painted for three components:
N = for the region where the posterior probability is greater than 95% for N component.
S = for the region where the posterior probability is greater than 95% for S component.
Unknown = for the remaining regions where the posterior assignment probability inferred by PCAdmix is <95% certain of their ancestry.
After painting the individuals, we grouped them by population and counted N and S ancestry haplotypes for every locus per population. We were mainly interested in identifying regions where we observed deviations of ancestry which would suggest recent selection post admixture. Our approach is based on three step process for detecting highly deviated ancestral components in multiple Indian populations as follows:
(Step 1)
(Step 2)
(Step 3)
We defined the metric, AHFD where freq is the frequency (count/total number) of N and S haplotype, respectively. The total number count for the calculation of frequencies includes the Unknown ancestry, thus not biasing for regions where most of the haplotype is difficult to paint.
We simulated dummy local ancestry of a population to see the efficiency of AHFD over one parental ancestry deviation as was used previously (Jin et al. 2012; Schlebusch et al. 2012; Bhatia et al. 2014; Jeong et al. 2014; Busby et al. 2017; Patin et al. 2017; Pierron et al. 2018). For our simulations, we closely followed the Gujarati samples present in the Genotype data. We first simulated 0.69/0.31 admixture amount of N/S with standard deviation of 0.13 (with hard limit of around 0 and 1, which means any value lower or higher than that would be converted to 0 or 1 depending on the situation) for 100 individuals (or 200 haplotypes) and 18,569 windows using numpy.random.normal (numpy version 1.14.3) (Jones et al. 2007) to get a normal distribution. This produces a true known data set without any missing/unknown data. We then simulated a fraction of true data to be unknown, such fraction is normally distributed with mean 0.35, standard deviation 0.06, and a hard limit from 0 to 1. The unknown portion of data can come from either N or S with a probability generated by another normal distribution with mean 0.5 and standard deviation of 0.1. As the true ancestry haplotypes and unknown ancestry haplotypes are coming from independent normal distributions, there are some positions where unknown N or S haplotypes count are bigger than the true count of N or S haplotypes which cannot be possible in real cases. For those positions, we changed the missing N or S haplotypes count to the highest possibility (which is the true count of N or S haplotypes) and recalculated the total Unknown. We recalculated N and S after removing the missing data from their corresponding ancestry. Thus, these recalculated N, S, and Unknown closely resemble our real data set whose real admixture proportion is known from the previous step.
We then calculated AHFD alongside with previously published methods:
LSAD and LSADC is essentially the same in case of no missing/unknown data. Also in case of no unknown data, AHFD can be shown as
AHFD values are bounded from −1 to 1 because p is bounded by 0 to 1.
As we know the true admixture proportion for the simulated data (the known data set before Unknown calculation, see above), we used the true fraction of N ancestry in x axis and in the y axis we used LSAD (supplementary fig. 12a, Supplementary Material online), LSADC (supplementary fig. 12b, Supplementary Material online), and AHFD (supplementary fig. 12c, Supplementary Material online) after converting to percentile (for ease of comparison). We used scipy.stats.linregress (version 1.1.0) (Oliphant 2006) to calculate r_value and stderr.
After calculating AHFD for the 25 populations, we wanted to see if our highly deviated ancestral regions were present in multiple populations. Regions above the top 2.5 percentile of the AHFD (corresponding to those showing the most extreme excess of the N component) were each scored as +1, whereas those below the bottom 2.5 percentile (excess of the S component) were scored as −1. (Step 2). This conservative discretization step was preferable over simple percentile scaling of AHFD to overcome the “ceiling problem.” If the admixture proportion of N/S is highly deviated from 0.5, there would be more positions reaching the maximum on one side over the other side (for example; if the admixture proportion is 0.9/0.1, there would be many positions with AHFD score reaching maximum of +1, hence overpopulation the top 2.5 percentile, but very few if not zero on minimum of −1). Thus, AHFD scores are essentially admixture proportion specific. By choosing to retain the 2.5 percentile, we are essentially taking region which are necessarily <2.5% of the whole genome. This would make our approach more conservative, for example, if there are several positions (>2.5% of the whole data set) which reached the maximum, and therefore with the same value, none of them would be taken into consideration for step 3.
After applying these criteria to every population, we simply sum over all the results (step 3). A high positive value signifies the N component reached high frequency in multiple South Asian populations, potentially through the action of natural selection, whereas high negative value signifies the S component reached high frequency South Asian populations, again potentially through the action of natural selection. As empirical ranking score can be enriched for randomly drifted regions, we sum it over populations and concentrate upon regions which are outliers in multiple populations, thus pointing toward a consistent pattern across South Asia, which is less likely to be caused by random drift. We then converted the values to percentile score using Python Pandas (0.20.3) rank method with pct=True (McKinney 2010).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the European Union through the European Regional Development Fund (project number 2014-2020.4.01.16-0024, MOBTT53: L.P., D.M., B.Y.; project number 2014-2020.4.01.15-0012: M.Me.; project no. 2014-2020.4.01.16-0030: B.Y., M.Mo., F.M.); the European Union through Horizon 2020 grant number 810645: M.Me.; the Estonian Research Council grant PUT (PRG243): M.Me., L.P.; institutional research funding IUT (IUT24-1) of the Estonian Ministry of Education and Research: A.K.P., T.K.; the European Social Fund's Doctoral Studies 25 and Internationalisation Programme DoRa: A.K.P.
Author Contributions
B.Y.: designed the study, analyzed data, and wrote the manuscript; M.Mo.: analyzed data and wrote the manuscript; D.M.: analyzed data and revised the manuscript; A.K.P.: contributed to the interpretation of results and revised the manuscript; F.M.: contributed to the interpretation of results and revised the manuscript; I.G.R.: contributed to the interpretation of results and revised the manuscript; T.K.: analyzed data and wrote the manuscript; M.Me: contributed to the interpretation of results and wrote the manuscript; and L.P.: designed the study, analyzed data, and wrote the manuscript.
References
- 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. 2015. A global reference for human genetic variation. Nature 5267571: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 199: 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, et al. 2010. Integrating common and rare genetic variation in diverse human populations. Nature 4677311: 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae CJ, Douka K, Petraglia MD.. 2017. On the origin of modern humans: Asian perspectives. Science 3586368: pii: eaai9067. [DOI] [PubMed] [Google Scholar]
- Basu A, Sarkar-Roy N, Majumder PP.. 2016. Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc Natl Acad Sci U S A. 1136: 1594–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu Mallick C, Iliescu FM, Möls M, Hill S, Tamang R, Chaubey G, Goto R, Ho SYW, Gallego Romero I, Crivellaro F, et al. 2013. The light skin allele of SLC24A5 in South Asians and Europeans shares identity by descent. PLoS Genet. 911: e1003912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Metspalu M, Baran Y, Kopelman NM, Yunusbayev B, Gladstein A, Tzur S, Sahakyan H, Bahmanimehr A, Yepiskoposyan L, et al. 2013. No evidence from genome-wide data of a Khazar origin for the Ashkenazi Jews. Hum Biol. 856: 859–900. [DOI] [PubMed] [Google Scholar]
- Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, Rootsi S, Chaubey G, Kutuev I, Yudkovsky G, et al. 2010. The genome-wide structure of the Jewish people. Nature 4667303: 238–242. [DOI] [PubMed] [Google Scholar]
- Bhatia G, Tandon A, Patterson N, Aldrich MC, Ambrosone CB, Amos C, Bandera EV, Berndt SI, Bernstein L, Blot WJ, et al. 2014. Genome-wide scan of 29,141 African Americans finds no evidence of directional selection since admixture. Am J Hum Genet. 954: 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisbin A, Bryc K, Byrnes J, Zakharia F, Omberg L, Degenhardt J, Reynolds A, Ostrer H, Mezey JG, Bustamante CD.. 2012. PCAdmix: Principal Components-based Assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum Biol. 844: 343–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby G, Christ R, Band G, Leffler E, Le QS, Rockett K, Kwiatkowski D, Spencer C.. 2017. Inferring adaptive gene-flow in recent African history. bioRxiv doi:10.1101/205252. [Google Scholar]
- Chahal HS, Lin Y, Ransohoff KJ, Hinds DA, Wu W, Dai HJ, Qureshi AA, Li WQ, Kraft P, Tang JY, et al. 2016. Genome-wide association study identifies novel susceptibility loci for cutaneous squamous cell carcinoma. Nat Commun 7: 12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ.. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 41: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubey G, Metspalu M, Choi Y, Mägi R, Romero IG, Soares P, Van Oven M, Behar DM, Rootsi S, Hudjashov G, et al. 2011. Population genetic structure in Indian austroasiatic speakers: the role of landscape barriers and sex-specific admixture. Mol Biol Evol. 282: 1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NG, Kelly DE, Hansen MEB, Beltrame MH, Fan S, Bowman SL, Jewett E, Ranciaro A, Thompson S, Lo Y, et al. 2017. Loci associated with skin pigmentation identified in African populations. Science 3586365: eaan8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard PdB, Martiniano R, Kamm J, Moreno-Mayar JV, Kroonen G, Peyrot M, Barjamovic G, Rasmussen S, Zacho C, Baimukhanov N, et al. 2018. The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science 3606396. pii: eaar7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 2715: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF.. 2012. A linear complexity phasing method for thousands of genomes. Nat Methods 92: 179–181. [DOI] [PubMed] [Google Scholar]
- Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, Ellis P, Langford C, Vannberg FO, Knight JC.. 2012. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 445: 502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo JC, Hsu L, Hutter CM, Lin Y, Campbell PT, Baron JA, Berndt SI, Jiao S, Casey G, Fortini B, et al. 2014. Genome-wide diet-gene interaction analyses for risk of colorectal cancer. PLoS Genet. 104: e1004228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelabert P, Olalde I, De-DIos T, Civit S, Lalueza-Fox C.. 2017. Malaria was a weak selective force in ancient Europeans. Sci Rep. 71: 1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstenblith MR, Shi J, Landi MT.. 2010. Genome-wide association studies of pigmentation and skin cancer: a review and meta-analysis. Pigment Cell Melanoma Res. 235: 587–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, et al. 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 5227555: 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasl RJ, McCarty CA, Payseur BA.. 2013. Genetic ancestry inference using support vector machines, and the active emergence of a unique American population. Eur J Hum Genet. 215: 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. 2002. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 182: 337–338. [DOI] [PubMed] [Google Scholar]
- Huerta-Sánchez E, Jin X, Asan Bianba Z, Peter BM, Vinckenbosch N, Liang Y, Yi X, He M, Somel M, et al. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 5127513: 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, Alkorta-Aranburu G, Basnyat B, Neupane M, Witonsky DB, Pritchard JK, Beall CM, Di Rienzo A.. 2014. Admixture facilitates genetic adaptations to high altitude in Tibet. Nat Commun 5: 3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Xu S, Wang H, Yu Y, Shen Y, Wu B, Jin L.. 2012. Genome-wide detection of natural selection in African Americans pre- and post-admixture. Genome Res. 223: 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NA, Coram MA, Shriver MD, Romieu I, Barsh GS, London SJ, Tang H.. 2011. Ancestral components of admixed genomes in a Mexican cohort. PLoS Genet. 712: e1002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E, Oliphant T, Peterson P. 2007. SciPy: open source scientific tools for Python. Comput Sci Eng. 9: 90. [Google Scholar]
- Jouganous J, Long W, Ragsdale AP, Gravel S.. 2017. Inferring the joint demographic history of multiple populations: beyond the diffusion approximation. Genetics 2063: 1549–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juyal G, Mondal M, Luisi P, Laayouni H, Sood A, Midha V, Heutink P, Bertranpetit J, Thelma BK, Casals F.. 2014. Population and genomic lessons from genetic analysis of two Indian populations. Hum Genet. 13310: 1273–1287. [DOI] [PubMed] [Google Scholar]
- Kassambara A. 2018. ggpubr R package: ggplot2-based publication ready plots. R package version 0.1.8. https://CRAN.R-project.org/package=ggpubr [Google Scholar]
- Lamason RL. 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 3105755: 1782–1786. [DOI] [PubMed] [Google Scholar]
- Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, Fernandes D, Novak M, Gamarra B, Sirak K, et al. 2016. Genomic insights into the origin of farming in the ancient near east. Nature 5367617: 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 3195866: 1100–1104. [DOI] [PubMed] [Google Scholar]
- Lindo J, Huerta-Sánchez E, Nakagome S, Rasmussen M, Petzelt B, Mitchell J, Cybulski JS, Willerslev E, Degiorgio M, Malhi RS.. 2016. A time transect of exomes from a Native American population before and after European contact. Nat Commun. 7: 13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnetto D, Huerta-Sánchez E.. 2017. Haplostrips: revealing population structure through haplotype visualization. Methods Ecol Evol. 810: 1389–1392. [Google Scholar]
- Martin AR, Lin M, Granka JM, Myrick JW, Liu X, Sockell A, Atkinson EG, Werely CJ, Möller M, Sandhu MS, et al. 2017. An unexpectedly complex architecture for skin pigmentation in Africans. Cell 1716: 1340–1353.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, et al. 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 5287583: 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney W. 2010. Data structures for statistical computing in Python In: Proceedings of the 9th Python in Science Conference (SCIPY 2010), Austin, Texas. [Google Scholar]
- Mellars P, Gori KC, Carr M, Soares PA, Richards MB.. 2013. Genetic and archaeological perspectives on the initial modern human colonization of southern Asia. Proc Natl Acad Sci U S A. 11026: 10699–10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metspalu M, Romero IG, Yunusbayev B, Chaubey G, Mallick CB, Hudjashov G, Nelis M, Mägi R, Metspalu E, Remm M, et al. 2011. Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am J Hum Genet. 896: 731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal M, Casals F, Xu T, Dall’Olio GM, Pybus M, Netea MG, Comas D, Laayouni H, Li Q, Majumder PP, et al. 2016. Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation. Nat Genet. 489: 1066–1070. [DOI] [PubMed] [Google Scholar]
- Montinaro F, Busby GBJ, Gonzalez-Santos M, Oosthuitzen O, Oosthuitzen E, Anagnostou P, Destro-Bisol G, Pascali VL, Capelli C.. 2017. Complex ancient genetic structure and cultural transitions in Southern African populations. Genetics 2051: 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorjani P, Thangaraj K, Patterson N, Lipson M, Loh PR, Govindaraj P, Berger B, Reich D, Singh L.. 2013. Genetic evidence for recent population mixture in India. Am J Hum Genet. 933: 422–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Estrada A, Gravel S, Zakharia F, McCauley JL, Byrnes JK, Gignoux CR, Ortiz-Tello PA, Martínez RJ, Hedges DJ, Morris RW, et al. 2013. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 911: e1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörseburg A, Pagani L, Ricaut F, Yngvadottir B, Harney E, Castillo C, Hoogervorst T, Antao T, Kusuma P, Brucato N, et al. 2016. Multi-layered population structure in Island Southeast Asians. Eur J Hum Genet. 2411: 1605–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan VM, Patterson NJ, Moorjani P, Lazaridis I, Mark L, Mallick S, Rohland N, Bernardos R, Kim AM, Nakatsuka N, et al. 2018. The genomic formation of South and Central Asia. bioRxiv.:292581. doi:10.1101/292581.
- Oliphant TE. 2006. Guide to NumPy. USA: Trelgol Publishing.
- Pagani L, Kivisild T, Tarekegn A, Ekong R, Plaster C, Gallego Romero I, Ayub Q, Mehdi SQ, Thomas MG, Luiselli D, et al. 2012. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am J Hum Genet. 911: 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer ND, Goodarzi MO, Langefeld CD, Wang N, Guo X, Taylor KD, Fingerlin TE, Norris JM, Buchanan TA, Xiang AH, et al. 2015. Genetic variants associated with quantitative glucose homeostasis traits translate to type 2 diabetes in Mexican Americans: the GUARDIAN (Genetics underlying diabetes in Hispanics) consortium. Diabetes 645: 1853–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak AK, Kadian A, Kushniarevich A, Montinaro F, Mondal M, Ongaro L, Singh M, Kumar P, Rai N, Parik J, et al. 2018. The genetic ancestry of modern Indus Valley populations from Northwest India. Am J Hum Genet. 1036: 918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patin E, Lopez M, Grollemund R, Verdu P, Harmant C, Quach H, Laval G, Perry GH, Barreiro LB, Froment A, et al. 2017. Dispersals and genetic adaptation of Bantu-speaking populations in Africa and North America. Science 3566337: 543–546. [DOI] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D.. 2012. Ancient admixture in human history. Genetics 1923: 1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D.. 2006. Population structure and eigenanalysis. PLoS Genet. 212: 2074–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, et al. 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 195: 826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierron D, Heiske M, Razafindrazaka H, Pereda-Loth V, Sanchez J, Alva O, Arachiche A, Boland A, Olaso R, Deleuze JF, et al. 2018. Strong selection during the last millennium for African ancestry in the admixed population of Madagascar. Nat Commun. 9: 932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D.. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 388: 904–909. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 813: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a Language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/. [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L.. 2009. Reconstructing Indian population history. Nature 4617263: 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch CM, Skoglund P, Sjödin P, Gattepaille LM, Hernandez D, Jay F, Li S, De Jongh M, Singleton A, Blum MGB, et al. 2012. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science 3386105: 374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhorst J, Kamm JA, Song YS.. 2017. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet. 492: 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Loosdrecht M, Bouzouggar A, Humphrey L, Posth C, Barton N, Aximu-Petri A, Nickel B, Nagel S, Talbi EH, Hajraoui MA E, et al. 2018. Pleistocene North African genomes link near eastern and sub-Saharan African human populations. Science 3606388: 548–552. [DOI] [PubMed] [Google Scholar]
- Visconti A, Duffy DL, Liu F, Zhu G, Wu W, Chen Y, Hysi PG, Zeng C, Sanna M, Iles MM, et al. 2018. Genome-wide association study in 176,678 Europeans reveals genetic loci for tanning response to sun exposure. Nat Commun. 9: 1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walford GA, Gustafsson S, Rybin D, Stancáková A, Chen H, Liu CT, Hong J, Jensen RA, Rice K, Morris AP, et al. 2016. Genome-wide association study of the modified stumvoll insulin sensitivity index identifies BCL2 and FAM19A2 as novel insulin sensitivity loci. Diabetes 6510: 3200–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2017. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. [Google Scholar]
- Yunusbayev B, Metspalu M, Järve M, Kutuev I, Rootsi S, Metspalu E, Behar DM, Varendi K, Sahakyan H, Khusainova R, et al. 2012. The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Mol Biol Evol. 291: 359–365. [DOI] [PubMed] [Google Scholar]
- Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, Akhmetova V, Balanovska E, Balanovsky O, Turdikulova S, et al. 2015. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 114: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.