Summary
Broadly neutralizing monoclonal antibodies protect against HIV-1 infection in animal models, suggesting that a vaccine that elicits them in humans would be effective. However, it has not yet been possible to elicit adequate serologic responses by vaccination. To activate B-cells expressing precursors of broadly neutralizing antibodies within polyclonal repertoires, we developed a new immunogen, RC1, which facilitates recognition of the V3-glycan patch on HIV-1 envelope while concealing non-conserved immunodominant regions by addition of glycans and/or multimerization on virus-like particles. Mouse, rabbit and rhesus macaque immunizations with RC1 elicited serologic responses targeting the V3-glycan patch. Antibody cloning and cryo-electron microscopy structures of antibody-envelope complexes confirmed that RC1 immunization expands clones of B-cells carrying anti-V3-glycan patch antibodies that resemble precursors of human broadly neutralizing antibodies. Thus, RC1 may be a suitable priming immunogen for sequential vaccination strategies to elicit V3-glycan antibodies in the context of polyclonal repertoires.
Introduction
Single cell antibody cloning from HIV-1–infected human donors revealed that broadly neutralizing antibodies (bNAbs) have undergone unusually extensive somatic mutation1–4. Moreover, the high degree of somatic mutations is essential for binding to the native HIV-1 envelope spike (Env) and for bNAb neutralizing activity5. The accumulation of large numbers of mutations suggests that bNAbs evolve in response to iterative rounds of somatic hypermutation and selection in germinal centers (GCs)6. Studies in humans revealed that this occurs in response to viral escape variants arising from antibody pressure4. Together these observations suggest that vaccination to elicit bNAbs requires a series of sequential immunogens starting with an immunogen that induces the 7expansion of B-lymphocytes expressing appropriate germline precursors8.
Sequential immunization to shepherd bNAb development was demonstrated in genetically-modified mice that carry inferred germline (iGL) precursors of human bNAbs8,9. However, the priming immunogens used to initiate the response failed to activate and expand B-cells expressing inferred bNAb precursors in animals with polyclonal antibody repertoires. Thus, a goal of HIV-1 vaccine development has been to design immunogens that recruit B-cells expressing bNAb precursors into GC reactions in animals with polyclonal repertoires.
The germline targeting approach to immunogen design focuses on producing immunogens that bind with high affinity to specific bNAb precursors, the rationale being that B-cell recruitment to GCs is in part dependent on receptor affinity for antigen10–14. However, this methodology effectively limits the repertoire of recruited B-cells qualitatively and quantitatively. Moreover, it fails to account for the findings that each GC accommodates different founder B-cells with a wide range of affinities and that GC entry is limited by competition and not absolute affinity7,10.
Here we describe RC1, an immunogen designed to recruit and expand diverse V3-glycan specific B-cells by improving accessibility of the V3-glycan patch epitope, which includes a group of high-mannose and complex-type N-glycans surrounding V3 (gp120 residues N133, N137, N156, N295, N301, N332, N339, N385 and N392)15. bNAbs targeting this site, including PGT12116, 10–107417, and BG1818, reach through these glycans using elongated CDRH3 loops and portions of CDRL1 and CDRL3 to contact the highly-conserved GDIR motif (G324-D325-I326-R327) at the base of V319. Here we show that RC1 activates and expands a diverse group of B-cells expressing antibodies that resemble human V3-glycan patch bNAb precursors in mice, rabbits and rhesus macaques.
Results
RC1 facilitates antibody binding to the V3-glycan patch
RC1 was designed using 11MUTB20, a modified native-like Env trimer (SOSIP.664) derived from clade A/E BG505 Env21, as a template. Compared to BG505, 11MUTB includes substitutions in V1 and lacks potential N-linked glycosylation sites (PNGSs) at N133 and N13720 (Fig. 1a). We reasoned that removal of the N156 PNGS (N156Q) to create RC1 would facilitate recognition of the V3-glycan patch by increasing accessibility of V1 residues that interact with V3-glycan bNAbs22,23. Consistent with this idea, the absence of the N156 PNGS enhances neutralization by PGT121 and 10–1074, whereas the absence of other glycans; e.g., N301 or N137, reduces neutralization (Extended Data Fig. 1a). In addition, we hypothesized that removal of the N156 glycan, which includes negatively-charged terminal sialic acids22,24, would produce a more electrostatically-neutral Env surface that could facilitate the binding of the largely neutral precursor of PGT121 and 10–1074 (iGL PGT121/10–1074)25.
Fig. 1. |. Characterization of the RC1 immunogen.
a, N-glycans (colored spheres) and GDIR motif (red surfaces) mapped onto BG505 (PDB 5T3Z) (N137 glycan from PDB 5FYL) in the top-down orientation. b, Side-views of structures of BG505 and RC1 complexed with 10–1074 (glycan atoms are colored spheres). Middle: superimposition of the boxed regions with protein in cartoon representations (dark and light purple: 10–1074 VH,VL, red: GDIR, wheat: other portions of RC1, grey: BG505, orange spheres: N156 glycan). Regions of V1 showing displacement (gp120 residues 139–140) are indicated by dots and an arrow. c, SPR data for iGL PGT121/10–1074 binding to Env trimers. N.B. = no binding above background. Representative plot from 3 independent experiments.
To characterize RC1, we compared its antigenic properties to BG505 (Extended Data Fig. 1b) and solved a 4.0 Å single-particle cryo-EM structure of RC1 complexed with 10–1074, comparing it to a BG505–10-1074 structure22 (Fig. 1b; Extended Data Fig. 2; Extended Data Table 1). Both structures showed three 10–1074 Fabs bound to the V3-glycan patch epitopes of a closed Env trimer (Fig. 1b). Compared with BG505, the V1 loop in RC1 included more ordered residues and was shifted towards the 10–1074 CDRH3, allowing for increased interactions between RC1 and 10–1074 (Fig. 1b).
Despite V1 structural changes resulting from deletion of the N156 glycan (Fig. 1b), iGL PGT121/10–107417 bound RC1 and 11MUTB with similar affinities (KDs~50μM) (Fig. 1c), and priming immunizations with RC1 and 11MUTB elicited comparable V3-glycan-specific serologic responses in knock-in (KI) mice carrying genes encoding the iGL PGT121/10–10749 (Fig. 2a–c). Thus, RC1 exhibited structural changes resulting from N156 glycan deletion that did not affect its affinity for iGL PGT121/10–1074.
Fig. 2. |. Wild-type mouse immunization with RC1 elicits V3-glycan patch antibodies.
a, Immunization protocol. b,d,f,h, Representative ELISAs showing serum binding to indicated immunogens. Controls include naïve serum (red), purified PGT121 (green) and iGL-PGT121/10–1074 (black). b, iGL PGT121 KI mice9. d, f, h. Wild-type mice. c,e, Area under the curve (AUC) for ELISAs in b and d, respectively, but combined results from 2 experiments using 3 or 4 mice each. Each dot represents serum from one mouse. f, Binding to RC1 and RC1-glycanKO. g, AUC for RC1 vs RC1-glycan KO ELISAs from 7 experiments with 2 or 3 mice immunized with RC1. i, Ratio of the AUC for RC1 vs RC1-glycan KO ELISAs for wild-type mice immunized with RC1(7 experiments) or RC1–4fill (5 experiments). j, Pie charts show clonal expansion of RC1 binding germinal center B-cells. Colored slices are proportional to the number of clonal relatives. White indicates single IgVH sequences. The number of heavy chains analyzed is indicated in the center. k, IgH nucleotide mutations from naïve and RC1 immunized mice in j. l, ELISA binding of representative mAbs from RC1-immunized mice to RC1 and RC1-glycanKO. m, ELISA binding of Ab275MUR and Ab276MUR to indicated Env proteins. Unpaired t-test in c, e, i. Data in c, g, i, k are mean.
RC1 elicits V3-glycan patch antibodies in wild-type mice
To determine whether RC1 can activate B-cells carrying V3-glycan patch-specific antibodies in wild-type mice, we immunized C57BL/6J mice once with RC1 or 11MUTB (Fig.2a). 11MUTB did not produce a measurable serologic response, but RC1-immunized mice showed reproducible anti-V3-glycan patch-specific serologic responses as shown by ELISA comparing binding to RC1 and a mutant RC1 (RC1-glycanKO) that lacks two V3 PNGSs (N301 and N332) critical for human V3-glycan patch bNAbs (Fig. 2d–g; Extended Data Table 2). Moreover, serum from the RC1-immunized mice cross-reacted with 11MUTB but not with the more native-like 10MUT20 or with BG505 (Extended Data Fig. 3a). The improved immunogenicity of the V3-glycan patch epitope of RC1 results from specific removal of the N156 glycan from 11MUTB because removal of a nearby glycan at N301 that is also part of the glycan patch (11MUTBΔ301) (Extended Data Table 2) failed to induce detectable serologic responses (Fig. 2h). We conclude that, unlike 11MUTB and 11MUTBΔ301, RC1 elicits V3-glycan-specific serologic responses in wild-type mice.
To reduce antibody responses to off-target epitopes26–29 and further focus responses on the V3-glycan patch, we produced an RC1 variant, RC1–4fill, by introducing PNGSs to add glycans to gp120 positions 230, 241, 289 and 344 (Extended Data Fig. 4). Compared with RC1, RC1–4fill elicited serologic responses that were more focused on the V3-glycan patch in wild-type mice (Fig. 2i). We conclude that RC1–4fill focuses antibody responses to the V3-glycan patch.
Clonal expansion of V3-glycan patch specific B-cells in wild-type mice
To further characterize humoral responses elicited by RC1 and RC1–4fill, we sequenced antibody genes from single GC B-cells that bound RC1 but not RC1-glycanKO (Extended Data Fig. 3b). All RC1- and RC1–4fill–immunized mice analyzed showed expansion of GC B-cell clones (Fig. 2a, j). The expanded clones predominantly expressed heavy chain V genes VH5–6, VH9–3 and VH2–9, and light chain genes VK3–4 and VK14–111 (Fig. 2j; Supplementary Tables 1,2; Extended Data Table 4). The CDRH3 sequences in expanded clones showed similarities to human V3-glycan patch bNAbs such as Tyr-rich or RxY motifs and longer-than-average CDRH3s (Supplementary Table 1; Extended Data Table 4). Consistent with a single immunization, the VH genes of the expanded clones had an average of 3.2 nucleotide mutations (Fig. 2k; Supplementary Table 1).
We mapped the target sites of these antibodies by ELISAs against RC1 and RC1 mutant proteins. A diverse group of monoclonal antibodies (mAbs) showed V3-glycan patch-specific binding (Fig. 2l). Further characterization of two mAbs showed that they bind the V3-glycan patch of RC1 (Ab275MUR KD~30nM) in a GDIR- and N301-glycan dependent manner, and Ab275MUR and Ab276MUR both retained binding to 11MUTB (Ab275MUR KD~230nM), demonstrating accommodation of the N156 glycan, but did not bind BG505 or a peptide that covers the crown of the V3 loop (Fig. 2m; Extended Data Table 2; Extended Data Fig. 3c,d). Acquired mutations were essential for binding because RC1 did not bind to the Ab276MUR reverted iGL (Extended Data Fig. 3e). Consistent with a single immunization, neither Ab275MUR nor Ab276MUR showed detectable neutralizing activity against a panel of tier 1B and tier 2 HIV-1 isolates in TZM-bl assays. We conclude that RC1 and RC1–4fill expand mouse B-cell clones expressing antibodies that target the V3-glycan patch.
VLP-RC1-4fill elicits V3-glycan patch antibodies in rabbits and rhesus macaques
To enhance potential avidity effects and limit exposure of off-target epitopes at the Env base, we multimerized RC1–4fill on virus-like particles (VLPs) using the SpyTag-SpyCatcher system30,31 (Fig. 3a,b). VLPs were used to prime rabbits and rhesus macaques. Single immunizations of 4 rabbits and 16 macaques with VLP-RC1–4fill elicited serologic responses that were partially specific for the V3-glycan patch in all animals (Fig. 3c–f and Extended Data Figure 5a). Serum from macaques primed with VLP-RC1–4fill showed sequentially reduced binding to the more native-like immunogens 11MUTB and 10MUT20 (Extended Data Fig. 5b) and no neutralizing activity against a small panel of HIV-1 isolates that included fully-glycosylated tier 2 and glycan-deleted viruses (Extended Data Table 3). Thus, VLP-RC1–4fill elicited robust serologic responses that mapped in part to the V3-glycan patch in rabbits and rhesus macaques.
Fig. 3. |. Macaque immunization with RC1–4fill VLPs elicits anti-V3-glycan patch antibodies that resemble iGLs of bNAbs.
a, Model of VLP-RC1–4fill: RC1–4fill (wheat and pink), SpyTag (gold), SpyCatcher (cyan), and bacteriophage A P205 (green). b, Negative-stain EM comparing VLPs (top) and VLP-RC1 (bottom). Arrows point to the VLP surface (black) and to RC1 (red). Representative image from three independent experiments. c,d, Immunization protocols for rabbits (c) and NHPs (d). e,f, AUC for ELISAs with serum from 4 rabbits (e) and 8 NHPs (f) primed with VLP-RC1–4fill to RC1 (black) and RC1-glycanKO (grey). g,h, Flow cytometry shows frequency of GC B-cells that bind to RC1 but not to RC1-glycanKO (g) representative, (h) 4 naïve and 4 immunized NHPs. i, Pie charts showing clonal expansion of RC1 binding GC B-cells (see legend for Fig.2j). j, IgVH mutations for the sequences in clones in I (Supplementary Table 3). k, Logo plots compare CDRL3 of iGL-PGT121/10–1074 and all IgL from GC B-cells from i. l, Fraction of CDRL3 sequences from i that show a DSS-like motif. Unpaired t-test in h, l. Data in h, j, l are mean.
To further characterize responses elicited by VLP-RC1–4fill in macaques, we purified draining lymph node GC B-cells that bound RC1 but not RC1-glycanKO by flow cytometry (RC1+RC1-glycanKO-). Whereas RC1+ cells were absent from GCs of naïve macaques, RC1+RC1-glycanKO- GC B-cells were found at an average frequency of 0.4% of GC B-cells in the lymph nodes in the 4 macaques analyzed (Fig. 3g,h).
Antibody cloning from 4 immunized macaques revealed expanded B-cell clones that used a variety of VH genes, as found for human V3-glycan patch bNAbs32, with an average of 5.6 nucleotide somatic mutations (Fig. 3i,j; Supplementary Table 3). Most characterized human V3-glycan patch bNAbs contain a lambda light chain18,33. Analysis of lambda genes revealed that macaque RC1-binding cells preferentially used gene segments VL132 (91% nt sequence identity to VL2–8 germline gene segments in PGT125–128 and PGT130–131) and VL124 (94% identity to the VL3–21 germline in PGT121–123/10–1074) (Fig. 3k). 86% of the lambda light chains had CDRL3s that included a DSS motif present in the iGLs of PGT121–123 and 10–1074/PGT12417 (Fig. 3l; Supplementary Table 4). This motif mutates to DSR in the mature bNAbs, which is critical for PGT121 neutralization activity34. Thus, there is congruence between the sequence of human V3-glycan patch bNAb precursors and the antibodies expressed by macaque B-cell clones elicited by priming with VLP-RC1–4fill.
We expressed 38 macaque GC antibodies with CDRL3s that resembled the CDRL3s of iGL V3-glycan patch bNAbs (Supplementary Table 5). The CDRL3s of 33 of 38 antibodies contained a DSS motif and a Q at position 89 (QxxDSS motif), also found in the CDRL3s of the PGT121–3, 10–1074, PGT124 and BG18 iGLs (Extended Data Table 5)17,18. Five of 38 antibodies contained a SYAG motif, which is present in the CDRL3s of the PGT125–7, PGT128, PGT130 and PGT131 iGLs (Extended Data Table 5). Thirty of 33 QxxDSS motif-containing antibodies and 2 of 5 SYAG motif-containing antibodies bound to the V3-glycan patch epitope, as determined by ELISA using RC1 and RC1-glycanKO with additional mutations in the GDIR sequence (RC1-glycanKO-GAIA) (Fig 4a; Supplementary Table 5). In addition, the CDRL1 of all 38 macaque antibodies contained a NIG-like motif present in the iGL PGT121/10–1074 antibody (33 NIG, 4 DIG, 1 NLG) (Supplementary Table 5). The CDRH3 lengths of the 38 antibodies were relatively long (11–21 residues; average=15.5) (Fig. 4b). Longer CDRH3s were enriched in Tyr and/or Phe residues, similar to the long CDRH3s found in human V3-glycan patch bNAbs16–18 (Supplementary Table 5). The antibody VH and VL genes included an average of 4.9 and 3.3 nucleotide mutations, respectively (Fig. 4c). Consistent with recruitment of antibodies with a range of affinities to GCs and their subsequent affinity maturation7,10, the iGL versions of the macaque antibodies showed lower affinity for RC1 than their mutated counterparts, ranging from levels that were below quantification to μM KDs (Extended Data Fig. 5c). Similarity between the macaque antibodies and the iGL PGT121/10–1074 was corroborated by ELISAs using an anti-idiotypic antibody specific for iGL PGT121/10–1074 (Extended Data Fig. 5d). iGL-reverted versions of 5 of 11 macaque antibodies were recognized by the anti-iGL PGT121/10–1074 antibody (Extended Data Fig. 5d).
Fig. 4. |. Monoclonal antibodies from macaques bind to the V3-glycan patch.
a, ELISA binding of representative macaque mAbs to RC1 and RC1-glycanKO-GAIA. b, CDRH3 length of 32 V3-glycan patch specific mAbs. c, Nucleotide mutations in IgVH and IgVL of 32 V3-glycan patch-specific mAbs. d,e, AUC for ELISA binding of mAbs to indicated proteins. Each AUC value corresponds to one ELISA curve. Data in b and c are mean.
To further characterize the target site of the macaque antibodies, we performed ELISAs against additional proteins: RC1-glycanKO, RC1-GAIA, RC1-glycanKO-GAIA, 11MUTBΔ301, RC1Δ301, RC1Δ332, 11MUTB20 and BG505 (Fig. 4d,e; Extended Data Table 2). The ELISAs suggested four distinct RC1-binding patterns among antibodies that contained a CDRL3 QxxDSS motif (Fig. 4d) and another pattern among antibodies containing a SYAG motif (Fig. 4e). Whereas all antibodies were glycan-dependent as determined by no binding to RC1-glycanKO, they differed in binding to 11MUTB or 10MUT and dependence on GDIR and N301, N332, and N156 glycans (Fig. 4a,d,e). Although none of the antibodies recognized BG505, Ab933NHP, Ab936NHP, and Ab1170NHP bound 11MUTB, indicating that they can accommodate the N156 glycan (Fig. 4d,e). Consistent with no binding to BG505, none exhibited neutralizing activity, and removal of the N133, N137, and N156 glycans did not render the BG505/T332N and JRCSF.JB viruses sensitive to neutralization (Extended Data Table 3b,c), suggesting that the lack of neutralization is not due to clashes with those glycans. We conclude that macaque immunization with VLP-RC1–4fill elicits V3-glycan patch-specific antibodies that resemble precursors of human bNAbs targeting this site.
Cryo-EM structures of mouse and macaque antibodies in complex with RC1
We determined structures of one mouse and two macaque Fabs complexed with RC1 using single-particle cryo-electron microscopy. Ab275MUR (4.4Å resolution) and Ab874NHP (3.9Å) (derived from the same clone as Ab876NHP) bound similarly to each other, consistent with their 69% VH domain amino acid sequence identity, whereas Ab897NHP (4.4Å) (related by 48% and 54% VH sequence identity to Ab275MUR and Ab874NHP, respectively) adopted a distinct angle of approach (Fig. 5a; Extended Data Figure 5e).
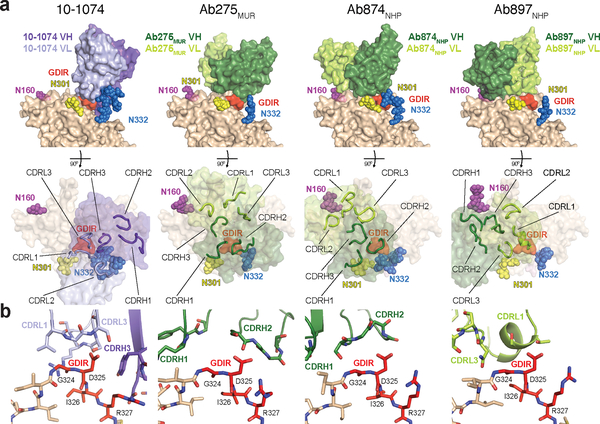
Fig. 5. |. Structures of 10–1074 and elicited antibodies bound to RC1.
a, Top: VH-VL domains of 10–1074 and elicited antibodies bound to one protomer of RC1 (GDIR residues are red; glycans are colored spheres). Bottom: Antibody combining sites (CDRs shown as loops) mapped onto gp120 (glycans as colored spheres; GDIR in red). b, Comparisons of interactions of GDIR motif with 10–1074 and elicited antibodies (colors as in panel b).
All three Fabs in the RC1 complexes bound to the V3-glycan patch epitope and contacted the GDIR motif, but with different orientations and footprints from each other and from V3-glycan patch bNAbs (Fig. 5a; Extended Data Figure 5e). 10–1074 contacts the conserved GDIR motif using CDRH3, CDRL1, and CDRL335 (Fig. 1c, 5a), Ab874NHP and Ab275MUR made GDIR contacts using their CDRH2s, whereas Ab897NHP utilized CDRL1 and CDRL3 (Fig. 5a,b). In addition, Ab874NHP and Ab897NHP contain the conserved CDRL3 QxxDSS motif, which make contacts with conserved regions of the V3-glycan epitope in mature bNAbs22,36. Similar to mature V3-glycan patch bNAbs, Ab897NHP contains a substitution (S93N) within the QxxDSS motif that enables contacts with gp120GDIR and also uses its conserved CDRL1 NIG motif to contact the V1 loop (Extended Data Figure 5f).
Ab275MUR and Ab874NHP also interacted with the N332 glycan, consistent with mature V3-glycan bNAbs (Fig 5a; Extended Data Figure 5e). However, unlike 10–1074, which interacts with the N332 glycan via its CDRL1, FRWL3, CDRH2, and CDRH335, Ab275MUR made contacts using CDRH2, while Ab874NHP engaged the N332 glycan with CDRH2 and FRWH3. We did not observe N332 glycan interactions in the Ab897NHP-RC1 structure. Despite reduced binding of Ab275MUR, Ab876NHP (same clone as Ab874NHP), and Ab897NHP to RC1Δ301 (Fig. 2l), none of these Fabs showed interactions with the N301 glycan in our EM structures, suggesting either glycan heterogeneity obscures this interaction and/or conformational heterogeneity in a V3-glycan patch lacking this glycan diminishes binding37. We conclude that RC1 elicits V3-glycan patch-targeting antibodies with distinct binding modes in animals with polyclonal antibody repertoires.
Conclusions
HIV-1 bNAbs develop in infected humans by sequential rounds of somatic mutation in response to a rapidly-evolving pathogen4. Vaccination with a series of related antigens can reproduce this progression of events in genetically-engineered mice that carry supraphysiological numbers of B-lymphocytes expressing the iGL precursors of bNAbs9. An important goal of HIV-1 vaccine design is to develop immunogens that initiate this response in organisms with polyclonal immune systems and then reproduce these responses in humans.
HIV-1 immunogen design has focused upon increasing the affinity of candidate immunogens for specific iGL bNAb precursors with the objective of recruiting a specific group of rare precursors into the GC1. This approach typically fails to account for increases in apparent affinity produced by interactions between multimerized antigen and clusters of bivalent antigen receptors on the surface of a B-cell. Moreover, GC entry is primarily limited by competition7,10,11,14. Thus, the importance of affinity is relative, as evidenced by the observation that B-cells bearing low affinity receptors are frequently found in GCs under physiological conditions10,38, and by our finding that iGL precursors of macaque antibodies elicited by RC1 showed relatively low affinity for the immunogen.
The principles employed to produce RC1 did not take affinity for a germline B-cell receptor into account. Instead, RC1 was designed to increase the number of bNAb progenitors that compete for GC entry by making the antigenic target site more available and facilitating binding to electrostatically-neutral iGL precursors25. In addition, VLP-RC1–4fill incorporates the idea that masking competing off-target epitopes26,29 by addition of glycans27 and tethering the bottom of the trimer to a VLP minimizes competition for GC entry.
RC1 differs from other HIV-1 vaccine candidates in that it induces B-cells expressing antibodies against a targeted epitope to undergo clonal expansion in GCs in animals with a fully polyclonal B-cell repertoire. In macaques, these B-cells express antibodies that show sequence and structural similarities to iGL precursors of bNAbs targeting the V3-glycan patch. Like the precursors of human bNAbs, they do not bind to wild-type Env or neutralize HIV-15. Importantly, biochemical and structural results showed that antibodies with distinct mechanisms of targeting the V3-glycan patch were elicited by RC1, increasing the probability that one or more might develop breadth and potency after boosting9. Thus, VLP-RC1–4fill is a suitable candidate immunogen for further evaluation in sequential vaccination strategies to elicit V3-glycan bNAbs.
Methods
Envelope proteins
Env trimers were expressed as soluble native-like gp140 trimers21. The newly-engineered Env SOSIP trimers, RC1, RC1–4fill, RC1-Avitag, RC1-SpyTag, RC1-glycanKO, RC1-glycanKO–Avitag, RC1-glycanKO-GAIA and RC1-GAIA, BG50521, and the BG505 variants 11MUTB, 10MUT, 7MUT, 5MUT20 were cloned in the pPPPI4 expression vector using synthetic gene fragments (Integrated DNA technologies (IDT)). The glycan variants RC1Δ301, RC1Δ332, and 11MUTBΔ301 were produced by site-directed-mutagenesis (QuikChange Lightning Multi-site directed mutagenesis kit, Agilent Technologies). Specific modifications of each protein are listed in Extended Data Table 2.
Soluble Env trimers were expressed by transient transfection in HEK293–6E cells (National Research Council of Canada) or Expi293 cells (Life Technologies) and purified from cell supernatants by 2G12 or NIH45–46 immunoaffinity chromatography and size exclusion chromatography (SEC) as previously described39. Proteins were stored at 4˚C in 20 mM Tris pH 8.0, and 150 mM sodium chloride (TBS buffer). SpyTagged immunogens were buffer exchanged into 20 mM sodium phosphate pH 7.5, 150 mM NaCl.
VLP production and conjugation
A C-terminal SpyTag sequence (13 residues) was added to RC1–4fill to form an irreversible isopeptide bond to SpyCatcher protein31. We produced and purified SpyCatcher-AP20540 VLPs as described30 and separated conjugated VLPs from free Env trimers by SEC on a Superdex 200 column. Conjugation of Env trimers was verified by negative-stain EM and/or SDS-PAGE (Fig.3; Extended Data Fig. 4), and immunogen concentrations were estimated by comparing to known amounts of free immunogen run on the same SDS-PAGE gel. Conjugated and unconjugated VLPs were compared by negative-stain EM on a FEI Tecnai T12 transmission electron microscope at 120 keV using a Gatan Ultrascan 2k x 2k CCD detector.
Mass spectrometry
The glycosylation profiles of RC1 and RC1–4fill were determined as previously described [REF]. Briefly, samples were denatured with Lys-C (Promega), Arg-C (Promega), Glu-C (Promega), and chymotrypsin (Promega). Following digestion, the samples were deglycosylated by Endo-H (Promega) and PNGaseF (Glyko®, Prozyme) in the presence of 18O-water (Cambridge Isotope Laboratories). The resulting peptides were separated on an Acclaim PepMap RSLC C18 column (75 μm x 15 cm) and analyzed using an Orbitrap Fusion™ Lumos™ Tribrid™ mass spectrometer (Thermo Fisher Scientific) with a 240-min linear gradient consisting of 1–100% solvent B over 180 min at a flow rate of 200 nL/min. Full MS scans were acquired using the Fusion instrument software (v2.0, Thermo Fisher Scientific), and the resulting spectra were analyzed and filtered using SEQUEST (Proteome Discoverer 1.4, Thermo Fisher Scientific) and ProteoIQ (v2.7, Premier Biosoft). Site occupancy was calculated using spectral counts assigned to the 18O-Asp-containing (PNGaseF-cleaved) and/or HexNAc-modified (EndoH-cleaved) peptides and their unmodified counterparts.
Animals
Mice carrying the iGL IgH and IgL human PGT121 and 10–1074 bNAbs (GLHL121 knock-in mice) were previously described9. 6–8-week-old C57BL/6J male mice from The Jackson laboratory were used for immunizations. All animal procedures were performed in accordance to protocols approved by the Rockefeller University IACUC. Male and female GLHL121 knock-in mice or C57BL/6J wild-type mice were equally distributed in groups and immunized intraperitoneally with 10μg of soluble SOSIP Envelope trimer in Ribi adjuvant (Sigma) (1:1).
Six-month-old New Zealand White rabbits (Covance) were used for immunizations. Rabbits were immunized subcutaneously with ~22 μg of RC1–4fill SOSIP Env trimer conjugated to VLP (VLP-RC1–4fill) in an ISCOMs-like saponin adjuvant (see below). Serum samples were collected from mice and rabbits on weeks 0 and 2 after immunization. All procedures in rabbits were approved by the Denver PA IACUC Committee.
Sixteen rhesus macaques (Macaca mulatta) of Indian genetic origin, 2 to 4 years of age, were housed and cared for in accordance with Guide for Care and Use of Laboratory Animals Report no. NIH 82–53 (Department of Health and Human Services, Bethesda, Maryland, 1985) in a biosafety level 2 NIH facility. All animal procedures and experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of NIAID, NIH.
Macaques were immunized subcutaneously in the medial inner forelegs and hind legs (total of 4 sites/animal) with ~200μg (Experiment 1; Fig. 3f ) or 100μg (Experiment 2; Extended Data Figure 5a) of RC1–4fill SOSIP trimer conjugated to VLP (RC1–4fill VLP) adjuvanted in IscoMPLA. Blood and lymph node biopsies were obtained from naïve macaques and from the immunized macaques 3 weeks after immunization.
Adjuvant synthesis
ISCOM-like saponin adjuvant was prepared as described41. Final adjuvant concentration was determined by cholesterol quantification (Sigma MAK043).
ELISA
ELISAs with SOSIP Env trimers 11MUTB, RC1, 11MUTBΔ301, RC1Δ301, RC1-GAIA, RC1-glycan-knockout (RC1-glycanKO), RC1-glycanKO-GAIA, RC1Δ332, BG505, 10MUT, 7MUT, 5MUT or the V3 loop-consensus C peptide (KGKGKGKGKGCTRPNNNTRKSIRIGPGQTFYATGDIIGDIRQAHC) were performed as described9. Serum samples were assayed at a 1:100 or 1:30 starting dilution and seven additional 3-fold serial dilutions. Mouse and human IgGs or human Fabs were evaluated at concentrations specified in the Results.
Alternatively, 96-well plates were directly coated with 50 μl of a solution of Fab at 20μg/ml in 1xPBS overnight at 4ºC, washed, and blocked as above and incubated in 50μl of a solution of RC1 or RC1-glycanKO-GAIA at 2μg/ml in blocking buffer for 1 h at RT. Plates were washed as above and developed using a chimeric version (human Fabs and mouse Fc) of the CD4-binding site bNAb 3BNC6042 at 3-fold serial dilutions starting at 5μg/ml followed by anti-mouse IgG secondary antibody conjugated to HRP (Jackson ImmunoResearch #115–035-071).
For anti-idiotype ELISAs, 96-well plates were coated with 50μl of a solution of IgG at 10μg/ml in 1xPBS overnight at 4ºC, washed and blocked as above and incubated with biotinylated anti-iGL PGT121 idiotypic antibody. Plates were developed with streptavidin conjugated to HRP.
Flow cytometry and single B-cell sorting
Single cell suspensions were obtained from the draining lymph nodes and spleens of immunized mice, and mature B-cells were isolated by negative selection using anti-CD43 magnetic beads (MACS) following the manufacturer’s instructions.
Frozen PBMCs or cells from lymph node biopsies obtained from the naïve and immunized macaques were thawed and washed in RPMI medium 1640 (1x) (Gibco #11875–093). Mouse or macaque cells were incubated with 100 μl of FACS buffer (PBS 1x with 2% fetal bovine serum and 1mM EDTA) with mouse (BD Biosciences #553142) or human (BD Biosciences #564219) Fc Block, respectively, at a 1:500 dilution for 30 min on ice.
RC1 and RC1-glycanKO (RC1+RC1 glycanKO-) tetramers were prepared by incubating 5 μg of Avitagged and biotinylated RC1 (RC1-AviBio) or Avitagged and biotinylated RC1-glycanKO (RC1-glycanKO AviBio) with fluorophore-conjugated streptavidin at a 1:200 dilution in 1xPBS for 30 min on ice.
RC1+RC1-glycanKO- mouse B-cells were isolated using RC1-AviBio conjugated to streptavidin BV711 (BD Biosciences, #563262) and RC1-glycanKO AviBio conjugated to streptavidin PE (BD Biosciences, #554061) as baits. RC1+RC1-glycan KO- macaque B-cells were isolated using: RC1-AviBio conjugated with streptavidin PE and streptavidin AF647 and RC1-glycanKO AviBio conjugated with streptavidin BV605 (BD Biosciences, #563260). Tetramers were mixed with the human or mouse antibody cocktails indicated below to a final concentration of 5μg/ml each.
Mouse cells were stained with anti-CD4 APC-eFluor780 (Invitrogen, #47–0042-82), anti-CD8 APC-eFluor780 (Invitrogen, #47–0081-82), anti-F4/80 APC-eFluor780 (Invitrogen, #47–4801-82), anti-NK1.1 APC-eFluor780 (Invitrogen, #47–5941-82), anti-CD11b APC-eFluor780 (eBioscience #47–0112-82), anti-CD11c APC-eFluor780 (eBioscience #47–0114-82),anti-Gr-1 APC-eFluor780 (Invitrogen, #47–5931-82), anti-B220 APC (Biolegend, #103212), anti-GL7 FITC (BD Biosciences #553666) and anti-CD95 BV421 (BD Biosciences #562633) at 1:200 dilution and the live/dead marker Zombie NIR (Biolegend, #77184) at a 1:400 dilution in FACS buffer. Macaque cells were stained with anti-CD16 APC-eFluor780 (Invitrogen, #47–0168-41), anti-CD8a APC-eFluor780 (Invitrogen, #47–0086-42), anti-CD3 APC-eFluor780 (Invitrogen, #47–0037-41), anti-CD14 APC-eFluor780 (eBiosciences, #47–0149-41), anti-CD20 PeCy7 (BD, #335793), anti-CD38 FITC (Stem Cell technologies, #60131FI), anti-IgG BV421 (BD Biosciences, #562581), anti-IgM PerCP-Cy5.5 (BD Biosciences, #561285) at a 1:200 dilution and the live/dead marker Zombie NIR at a 1:400 dilution in FACS buffer.
Zombie NIR-/CD4-/CD8-/F4/80-/NK1.1-/CD11b-/CD11c-/B220+/GL7+/CD95+/RC1+/RC1-glycanKO- single cells were isolated from the mouse cell homogenates and Zombie NIR-/CD16-/CD8a-/CD3-/CD14-/CD20+/CD38+/IgG+/−/double RC1+RC1-glycanKO- single cells were isolated from the macaque cell homogenates using a FACS Aria III (Becton Dickinson).
Single cells were sorted into individual wells of a 96-well plate containing 5 μl of lysis buffer (TCL buffer (Qiagen #1031576) with 1% of 2-β-mercaptoethanol). Plates were immediately frozen on dry ice and stored at −80ºC.
Antibody sequencing and cloning
Single cell RNA was purified using magnetic beads (RNAClean XP, #A63987 Beckman Coulter). RNA was eluted from the magnetic beads with 11μl of a solution containing (14.5 ng/μl of random primers (Invitrogen, #48190–011), 0.5% of tergitol, (Type NP-40, 70% in H2O, Sigma-Aldrich, #NP40S-100ML), 0.6U/μl of RNAse inhibitor (Promega #N2615) in nuclease free water (Qiagen), and incubated at 65ºC for 3 min. cDNA was synthesized by reverse transcription (SuperScript® III Reverse Transcriptase, Invitrogen, #18080–044, 10’000U)43. cDNA was stored at −80ºC or used for antibody gene amplification by nested Polymerase chain reaction (PCR) after addition of 10 μl of nuclease-free water.
Mouse and macaque antibody genes were cloned43 using the primers in Supplementary Table 6. PCR protocols: (annealing (ºC)/ elongation (sec)/ number of cycles): 1st PCR (IgG IgH and Igλ): 46/55/50; 2nd PCR (IgG IgH and Igλ): 50/55/50.
iGL macaque IgGs and Fabs were produced by reverting all nucleotide mutations in the V(D)J antibody genes to their corresponding iGL sequences while conserving the N-nucleotides from the V(D)J junctions found in the mutated antibodies.
Antibody production and purification
Igs were purified from 200μl of mouse or macaque serum using Ab Spin Trap Protein G Sepharose columns (GE Healthcare, #28–4083-47). Ig-containing fractions were buffer exchanged with PBS by overnight dialysis at 4ºC (dialysis cassettes 20000 MWCO Thermo Scientific, #66005).
For structural studies, mouse IgGs and macaque His6-tagged Fabs were expressed by transient transfection in HEK293–6E or Expi293 cells and purified from cell supernatants using protein A or G (GE Healthcare) (for IgGs) or Ni-NTA (GE Healthcare) or Ni Sepharose 6 Fast Flow (GE Healthcare) (for Fabs) chromatography and SEC44. Mouse Fab was obtained by digesting IgG at 1–5 mg ml−1 with ficin (Sigma). Fab was purified by protein G (GE Healthcare) and SEC chromatography45, followed by monoQ 5/50 (GE Healthcare) ion exchange chromatography. The common iGL of the PGT121 and 10–1074 bNAbs17 was expressed as a His6-tagged Fab.
In vitro neutralization assay
TZM-bl assays were performed as described46. In brief, neutralization activity was calculated as a function of the reduction in Tat-induced luciferase expression in the TZM-bl reporter cell line after a single round of virus infection with Env-pseudoviruses.
SPR and Octet binding studies
SPR experiments were performed using a T200 (Biacore). For measuring the affinity for PGT121/10–1074 iGL Fab, Protein A was immobilized on a CM5 chip by primary amine chemistry (Biacore manual) and 200 nM 8ANC195G52K5 IgG or a non-HIV Env-binding IgG (mG053) was injected as described44. 1μM human Fc was injected to block remaining protein A sites. After capturing 10 μM RC1, 11MUTB, or 10MUT, a concentration series of PGT121/10–1074 iGL Fab (4-fold dilutions from a top concentration of 160 μM for 10MUT, and 2-fold dilutions from a top concentration of 150 μM for 11MUTB and RC1) was injected, and binding reactions were allowed to reach equilibrium. KDs were derived by nonlinear regression analysis of plots of Req (equilibrium binding response) versus the log of the injected protein concentration, and the data were fit to a 1:1 binding model47. For measuring the affinity of Ab275MUR, a concentration series of Fab was injected over immobilized RC1 or 11mutB (4-fold dilutions from a top concentration of 50 μM). KDs were calculated from the on/off rates (ka/kd), which were derived using a 1:1 binding model from seven concentrations of Ab275MUR Fab (3.125μM to 0.763nM). Flow cells were regenerated with 1 M guanidine HCl and/or 10 mM glycine pH 2.0 at a flow rate of 90 μl/min44.
OCTET experiments were performed using the OCTET Red96 system to determine affinities of iGL and mutated macaque Fabs for RC1. Biotinylated RC1-Avitag was immobilized on high precision streptavidin (SAX) biosensors (FORTÉBIO) using a solution of biotinylated-RC1-Avitag at 400nM in dilution buffer (FORTÉBIO). Four serial dilutions of each macaque Fab, one irrelevant Fab, and 3BNC60 Fab were prepared in dilution buffer (FORTÉBIO). The binding experiment was performed at 30ºC using the following protocol: Baseline 1 (60 secs)/ Load RC1 (300 secs)/ Baseline 2 (200 secs)/ Fab association (300 secs)/ Fab dissociation (600 secs). Analysis was performed using OCTET software Data Analysisi HT 10.0 (FORTÉBIO).
Cryo-EM Sample and Grid Preparation
RC1 complexed with 10–1074 was prepared by incubating purified RC1 with 10–1074 Fab and a CD4-binding site (CD4bs) Fab at a 1:3:3 molar ratio (gp140 protomer:10–1074 Fab:CD4bs Fab) overnight at room temperature. The RC1-Fab complex was isolated by SEC in TBS (20 mM Tris pH 8.0, 100 mM NaCl) using a Superdex-200 Increase 10/300 column (GE Healthcare). RC1–mouse/macaque Fab complexes were prepared by incubating purified RC1 with a mouse or macaque Fab and with 8ANC195 Fab42 at a 1:1.3:1.3 molar ratio as above and used without SEC purification. RC1-Fab complexes were diluted to 0.75–1.4 mg/ml in TBS, and 3μl was added to Quantifoil R1.2/1.3 300 mesh copper grids (Electron Microscopy Services) that had been freshly glow-discharged using a PELCO easiGlow (Ted Pella). Samples were vitrified in 100% liquid ethane using a Mark IV Vitrobot (Thermo Fisher). Sample preparation conditions are summarized in Extended Data Table 1.
Cryo-EM Data Collection
RC1–Fab complexes were collected on a 200 kV Thermo Fisher Talos Arctica electron microscope using EPU automated image acquisition software48. Movies were collected on a Falcon 3EC direct electron detector (Thermo Fisher) operating in counting mode at a nominal magnification of 73,000x (1.436 Å/pix) using a defocus range of −1.4μm to −3.0μm or −0.8μm to −2.5 μm. Data for the RC1–10–1074 complex were collected across two separate sessions and combined during data processing. Microscope conditions are summarized in Extended Data Table 1.
Cryo-EM Data Processing
Movies were motion corrected and dose-weighted using the MotionCor249 frame alignment program in RELION-350. Dose-weighted summed images were used for CTF determination using Gctf51, and reference-free particle picking from each micrograph was achieved using Laplacian-of-Gaussian filtering in RELION-350. Unbinned extracted particles were imported into cryoSPARC v252 and subjected to reference-free 2D-classification using a 240Å circular mask. Particles from the best 2D classes were selected for heterogeneous ab initio model generation (two models). The best model exhibited C3 symmetry and was used as an initial model for homogenous 3D auto-refinement in cryoSPARC v252. Resolutions were estimated using the Gold Standard Fourier shell correlation (FSC=0.143)53, and maps were auto-sharpened in cryoSPARC52. For interpreting N-linked glycans, maps were generated with overall B-factors ranging from −150 to −400 Å2 to improve local features and map connectivity54. See Extended Data Fig. 2 and Extended Data Table 1.
Model Building
Initial coordinates were generated by docking reference models into the maps using UCSF Chimera55. For the RC1–10–1074 complex, BG505 Env, 10–1074 Fab, and 8ANC131 Fab (PDBs 53TZ and 4RWY) were docked into the density maps. For RC1 complexes with mouse or macaque Fabs, BG505 Env, PGT121/10–1074 iGL, and 8ANC195 (PDBs 5CEZ, 4FQQ, 5CJX) coordinates were docked into density maps. Initial models were refined into EM maps using rigid body refinement55. Models were built using Fab and RC1 sequences following iterative rounds of real-space refinement in Coot and Phenix56,57. (Models of only the RC1–10–1074 and RC1–mouse/macaque Fab portions of the complexes are shown in structure figures and deposited as coordinates in the EMDB and PDB.) Coordinates for glycans were added as Man9 and then trimmed to fit the maps at σ=5. Model validation was done using MolProbity58 and Privateer59. Superposition calculations and molecular representations were generated with PyMOL (Version 1.5.0.4 Schrodinger, LLC), UCSF Chimera55, and ResMap60.
Analysis
MacVector 15.5.3 was used for sequence analysis and graphs were created using R language. Flow cytometry data was processed using FlowJo 10.5.0. GraphPad Prism 7 was used for data analysis. Ig gene sequence AB1 files were converted to FASTQ format using biopython package. FASTQ files were trimmed by quality using cutadapt v1.18 software. Igblast v1.9.0 was used for VDJ assignment and clone analysis was performed using Change-O software v0.3.7. For macaques, a custom VDJ database was created using previously reported Ig gene sequences61.
Quantification and statistical analysis
Statistical information including n, mean and statistical significance values are indicated in the text or the figure legends. GraphPad Prism 7 was used for statistical analysis by unpaired T-Test. Data were considered statistically significant at *p≤0.05, **p≤0.01, ***p≤0.001 and ****p≤0.0001.
Extended Data
Extended Data Fig. 1. RC1 characterization.
a, Comparison of geometric mean IC50 values for V3-glycan patch bNAbs (10–1074 and PGT121) and a N156 glycan-dependent V1V2 bNAb (BG1) evaluated against HIV-1 strains either containing or not containing a PNGS at the indicated positions (number of HIV-1 strains indicated in the parentheses). IC50values > 50 μg/mL set to 50 μg/mL for geometric mean calculations. Whereas V3-glycan patch bNAbs show enhanced neutralization upon removal of the N156 glycan, removal of nearby glycans (N137, N301) diminished or had little effect on neutralization. b, Graphs of ELISA data showing binding of different classes of bNAbs to RC1, RC1–4fill, and BG505. bNAbs were evaluated at 5μg/ml and seven additional 3-fold dilutions.n=2. RC1 and RC1-fill show similar binding patterns for V3-glycan patch bNAbs, CD4-binding site bNAbs (CD4bs), gp120-gp41 interface bNAbs, but not to BG1, a V1V2 bNAb that interacts with the N156 glycan (see panel a).
Extended Data Fig. 2. Cryo-EM data collection and processing for RC1 complexes.
a, 10–1074–RC1. b, Ab275MUR–RC1. c, Ab874NHP–RC1. d, Ab897NHP–RC1. A representative micrograph, selected 2D class averages, orientation distribution summary, GSFSC Resolution plot, local resolution (calculated using ResMap), and representative density maps contoured at 7σ for a gp41 helix and antibody CDRH3 are shown for each.
Extended Data Fig. 3. Antibody responses in wild-type mice.
a, ELISA cross-reactivity of serum from RC1-immunized wild-type mice to 11MUTB. ELISA graphs show the binding of the serum from wild-type mice primed with RC1 to RC1, 11MUTB, 10MUT and BG505. The binding of the human bNAbs 10–1074 (green) and 3BNC117 (red) was evaluated at 5μg/ml as a control. n=2. b, FACS plots showing the gating strategy to isolate single RC1+RC1-glycanKO- GC B-cells (DUMP- (CD4-, CD8-, F4/80-, NK1.1-, CD11b-, CD11c-, Gr-1-) B220+ CD95+ GL7+ RC1-glycanKO- RC1+ ) from the spleen and draining lymph nodes of wild-type mice primed with RC1 or RC1–4fill. c, ELISA graphs showing binding of the mouse antibodies Ab275MUR and Ab276MUR to a V3 loop-Consensus C peptide (see Methods). Human antibodies 3869 and 3074 were used as positive controls. Antibodies were evaluated at 30μg/ml. n=2. d, Representative sensograms from two independent SPR binding experiments of Ab275MUR Fab injected over immobilized RC1 (panel b) or 11MUTB (panel c). Experimental binding curves (red) are overlaid with predicted curves (black) derived from a 1:1 binding model. Representative plot from 3 independent experiments. e, ELISAs showing the binding of Ab276MUR and its iGL version (Ab276MUR-GL) to RC1 (left) and RC1-glycanKO (right) at 30μg/ml. The human monoclonal antibodies PGT121 (green) and 3BNC60 (red) were used as controls at 5μg/ml.
Extended Data Fig. 4. Characterization of RC1, RC1–4fill, and VLPs.
a, SEC profiles of RC1 and RC1–4fill showing a larger apparent hydrodynamic radius for RC1–4fill compared with RC1, consistent with addition of extra glycans at the introduced PNGSs. b, ELISAs showing comparable binding of PGT122 Fab to RC1 and RC1–4fill. c, Glycan site occupancy for each PNGS in RC1 and RC1–4fill determined by mass spectrometry. d, SDS-PAGE analysis for RC1 and RC1–4fill under non-reducing (NR), reducing (R), and PNGaseF-treated (PNG) conditions. e, SDS-PAGE analysis for VLP, SpyTagged RC1–4Fill, and VLP–RC1–4fill under non-reducing (NR) and reducing (R) conditions.
Extended Data Fig. 5. Characterization of antibody responses in macaques.
a, ELISA binding of serum from macaques primed with RC1–4fill VLPs. Bar graph shows the ELISA binding of the serum from 8 macaques primed with RC1–4fill VLPs and PGT121 to RC1 (black) and RC1-glycanKO (gray). b, ELISA binding of serum from macaques primed with RC1–4fill VLPs. Bar graph shows the ELISA binding of the serum from 8 macaques primed with RC1–4fill VLP and one naïve macaque to RC1 (black) and the sequentially less modified Env proteins 11MUTB (gray) and 10MUT (white). The human bNAbs PGT121 and 3BNC60 were used as controls at 5μg/ml, and the serum was evaluated at a 1:100 dilution and seven additional 3-fold serial dilutions. c, Table showing the affinities (KD) for RC1 of different macaque antibodies isolated after a prime with VLP-RC1–4fill, and the corresponding iGL-reverted antibodies as determined by bio-layer interferometry (OCTET). d, ELISA binding of an anti-idiotypic antibody that recognizes the iGL PGT121/10–1074 to mAbs isolated from macaques primed with -RC1–4fill VLPs. The iGL PGT121/10–1074, two chimeric antibodies comprising the mutated HC and iGL LC of PGT121 (PGT121HCMT-LCGL) or the iGL HC and the mutated LC of PGT121 (PGT121 HCGL-LCMT) and different iGL bNAbs were used as controls. Results in a, b, and d are shown as area under the ELISA curve (AUC). e, Comparison of binding mode between the vaccine-elicited antibodies (Ab275MUR, Ab874NHP, and Ab897NHP) and the V3-glycan patch bNAbs 10–1074, PGT128, and PGT135. RC1 trimer is shown in gray from above and all Fabs are modeled onto the same trimer. For clarity, only one Fab per trimer is shown. f, Interactions between Ab897NHP conserved light chain motifs and RC1 gp120. DNS motif in CDRL3 (lime); gp120 GDIR (red); NIG motif in CDRL1 (pink); gp120 V1 loop (teal). Each AUC value corresponds to one ELISA curve.
Extended Data Table 1.
Cryo-EM data collection, refinement, and validation statistics.
| RC1–10–1074 (EMDB-xxxx) (PDB xxxx) | RC1–Ab275MUR (EMDB-xxxx) (PDB xxxx) | RC1–Ab874NHP (EMDB-xxxx) (PDB xxxx) | RC1–Ab897NHP (EMDB-xxxx) (PDB xxxx) | |
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 73,000× | 73,000× | 73,000× | 73,000× |
| Voltage (kV) | 200 | 200 | 200 | 200 |
| Electron exposure (e–/Å2) | 39.1 | 40 | 40 | 40 |
| Defocus range (μm) | 1 – 3.4 | 0.8 – 2.5 | 0.8 – 2.5 | 0.8 – 2.5 |
| Pixel size (Å) | 1.436 | 1.436 | 1.436 | 1.436 |
| Symmetry imposed | C3 | C3 | C3 | C3 |
| Initial particle images (no.) | 145,907 | 172,558 | 188,004 | 314,471 |
| Final particle images (no.) | 122,013 | 49,308 | 86,564 | 158,954 |
| Map resolution (Å) | 4.05 | 4.39 | 3.90 | 4.43 |
| FSC threshold - 0.143 | ||||
| Map resolution range (Å) | 3.6 – 6 | 4.2 – 7 | 3.5 – 6.5 | 4.2 – 7 |
| Refinement | ||||
| Initial model used (PDB code) | ab initio | ab initio | ab initio | ab initio |
| Model resolution (Å) | 4.0 | 4.3 | 3.8 | 4.3 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Model resolution range (Å) | 3.8 – 4.2 | 3.8 – 4.4 | 3.6 – 4.0 | 4.0 – 4.5 |
| Map sharpeningB factor (Å2) | −192.6 | −252.4 | −230.0 | −322.1 |
| Model composition | ||||
| Non-hydrogen atoms | 19,947 | 19,932 | 20,010 | 19,623 |
| Protein residues | 2,418 | 2,430 | 2,421 | 2,400 |
| Ligands | BMA:6 NAG:48 | BMA:6 NAG:54 | BMA:9 NAG:66 | BMA:6 NAG:48 |
| MAN:18 | MAN:6 | MAN:12 | MAN:18 | |
| B factors (Å2) | ||||
| Protein | 169.5 | 118.0 | 72.1 | 155.1 |
| Ligand | 177.2 | 203.2 | 88.6 | 210.7 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.007 | 0.008 | 0.012 | 0.009 |
| Bond angles (°) | 1.045 | 1.219 | 1.378 | 1.205 |
| Validation | ||||
| MolProbity score | 2.10 | 2.22 | 2.17 | 2.29 |
| Clashscore | 12.87 | 11.06 | 9.60 | 12.87 |
| Poor rotamers (%) | 0.47 | 0.57 | 0.29 | 0.57 |
| Ramachandran plot | ||||
| Favored (%) | 92.3 | 84.8 | 84.7 | 84.7 |
| Allowed (%) | 7.7 | 15.0 | 15.2 | 15.3 |
| Disallowed (%) | 0 | 0.1 | 0.1 | 0 |
Extended Data Table 2. HIV-1 Envelope-based proteins.
Table summarizes the modifications of all the Envelope proteins used in this study.
| Protein | PNGS |
Other modifications | |
|---|---|---|---|
| Deleted | Added | ||
| RC1 | 133, 137, 156 | - | V134Y, T135A, I138L, T139L, D140S, D141N, T320F, Q328M, T415V, MD39* |
| RC1–4FILL | 133, 137, 156 | 230, 241, 289, 344 | V134Y, T135A, I138L, T139L, D140S, D141N, T320F, Q328M, T415V, MD39* |
| RC1-glycanKO | 133, 137, 156, 301, 332 | - | V134Y, T135A, I138L, T139L, D140S, D141N, T320F, Q328M, T415V, H330A, MD39* |
| RC1-glycanKO-GAIA | 133, 137, 156, 301, 332 | - | V134Y, T135A, I138L, T139L, D140S, D141N, T320F, Q328M, T415V, GDIR/GAIA, H330A, MD39* |
| RC1-GAIA | 133, 137, 156 | - | V134Y, T135A, I138L, T139L, D140S, D141N, T320F, Q328M, T415V, GDIR/GAIA, MD39* |
| 11MUTBΔ301 | 133, 137, 301 | - | V134Y, T135A, I138L, T139L, D140S, D141N, T320F, Q328M, T415V, MD39* |
| RC1Δ301 | 133, 137, 156, 301 | - | V134Y, T135A, I138L, T139L, D140S, D141N, T320F, Q328M, T415V, MD39* |
| RC1Δ332 | 133, 137, 156, 332 | V134Y, T135A, I138L, T139L, D140S, D141N, T320F, Q328M, T415V, MD39* | |
| 11MUTB | 133, 137 | - | V134Y, T135A, I138L, T139L, D140S, D141N, T320F, Q328M, T415V, MD39* |
| 10MUT | 133, 137 | - | V134Y, T135A, N136P, N137F, I138L, T139I, D140N, T320F, Q328M, MD39* |
| 7MUT | 133, 137 | - | V134Y, T135A, N136P, N137F, I138L, T139I, D140N, MD39* |
| 5MUT | - | - | V134Y, N136P, I138L, D140N, MD39* |
| BG505 | - | - | MD39* |
| RC1–4fill VLP | 133, 137, 156 | 230, 241, 289, 344 | V134Y, T135A, I138L, T139L, D140S, D141N, T320F, Q328M, T415V, Spytag, MD39* |
| RC1-Avitag | 133, 137, 156 | - | V134Y, T135A, I138L, T139L, D140S, D141N, T320F, Q328M, T415V, Avitag, MD39* |
| RC1-glycanKO-Avitag | 133, 137, 156, 301, 332 | - | V134Y, T135A, I138L, T139L, D140S, D141N, T320F, Q328M, T415V, H330A, Avitag, MD39* |
Extended Data Table 3. Results of neutralization assays in TZM-bl cells.
a, Results of neutralization assays in TZM-bl cells using the serum of 8 macaques (NHP) primed with VLP-RC1–4fill and the serum from a naive macaque at 1:25 dilution against 2 tier 2 HIV-1 isolates. b, Results of neutralization assays in TZM-bl cells using the serum from 8 macaques primed with VLP-RC1–4fill at 1:50 dilution against the fully glycosylated and glycan deleted JRCSF.JB and BG505/T332N HIV-1 isolates. c, Results of neutralization assays in TZM-bl cells using mAbs isolated from macaques primed with VLP-RC1–4fill at a 1:20 dilution of a 1.5 mg/ml (Ab988NHP), 1mg/ml (Ab893NHP, Ab876NHP, Ab936NHP) or 0.5 mg/ml (Ab935NHP, Ab934NHP, Ab1170NHP) antibody solution. Values are the serum dilution at which relative luminescence units (RLUs) were reduced 50% compared to virus control wells (no test sample).
| a | |||||||||
|
ID50 in TZM-bl cells |
|||||||||
| Sample | BG505ΔCT/T332N | Du156.12 | |||||||
| NHP 1 | <25 | <25 | |||||||
| NHP 2 | <25 | <25 | |||||||
| NHP 3 | <25 | <25 | |||||||
| NHP 4 | <25 | <25 | |||||||
| NHP 5 | <25 | <25 | |||||||
| NHP 6 | <25 | <25 | |||||||
| NHP 7 | <25 | <25 | |||||||
| NHP 8 | <25 | <25 | |||||||
| NAÏVE NHP | <25 | <25 | |||||||
| PGT121 | 0.03 | <0.02 | |||||||
| b | |||||||||
|
ID50 in TZM-bl cells |
|||||||||
| Virus ID | NHP 1 | NHP 2 | NHP 3 | NHP 4 | NHP 5 | NHP 6 | NHP 7 | NHP 8 | NAÏVE NHP |
| JRCSF.JB WT | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 |
| JRCSF_N156A | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 |
| JRCSF_N133A_N137A | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 |
| JRCSF_N133A_N137A_N156A | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 |
| JRCSF_N332A | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 |
| BG505/T332N | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 |
| BG505/T332N_N156A | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 |
| BG505/T332N_N133A_N137A | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | 99 |
| MuLV | <50 | <50 | 78 | <50 | <50 | <50 | <50 | <50 | <50 |
| c | |||||||||
|
ID50 in TZM-bl cells |
|||||||||
| Virus ID | Ab893NHP | Ab876NHP | Ab935NHP | Ab934NHP | Ab936NHP | Ab988NHP | Ab1170NHP | ||
| JRCSF.JB WT | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| JRCSF_N156A | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| JRCSF_N133A_N137A | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| JRCSF_N133A_N137A_N156A | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| JRCSF_N332A | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| BG505/T332N | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| BG505/T332N_N156A | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| BG505/T332N_N133A_N137A | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| MuLV | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
Extended Data Table 4. Mouse monoclonal antibodies.
Monoclonal antibodies isolated from wild-type mice immunized with RC1 or RC1–4fill. All the RC1 binding antibodies targeted the V3-glycan patch epitope of RC1 except antibody 341 (marked with *) that shows similar binding to RC1 and RC1-glycanKO.
| ANTIBODY | MOUSE | IMM. | VH | CDRH3 | LENGTH (AA) | VK | CDRL3 | LENGTH (AA) | RC1 BINDING |
|---|---|---|---|---|---|---|---|---|---|
| 271 | 1 | RC1 | IGHV5–6*01 | ARHSRTGTGAMDY | 13 | IGKV3–4*01 | QQSNEDPPWT | 10 | YES |
| 340 | 2 | RC1 | IGHV1–81*01 | ARPYYYGSSPYFDY | 14 | IGKV4–57*01 | QQRSSYPPT | 9 | NO |
| 341 | 2 | RC1 | IGHV5–17*01 | ARSIVPDY | 8 | IGKV14–100*01 | VQYVQFPLT | 9 | YES* |
| 343 | 2 | RC1 | IGHV5–6*01 | ASLYGNAFDY | 10 | IGKV3–4*01 | QQSNEDPFT | 9 | YES |
| 344 | 2 | RC1 | IGHV9–3*01 | ASGGNYFDY | 9 | IGKV14–111*01 | LQYDEFPPFT | 10 | YES |
| 346 | 2 | RC1 | IGHV5–6*01 | ARHVGDHAMDY | 11 | IGKV3–4*01 | QQSNEDPFT | 9 | YES |
| 347 | 2 | RC1 | IGHV1–81*01 | ARPYYYGSSPNFDY | 14 | IGKV3–4*01 | QQSNEDPWT | 9 | NO |
| 351 | 3 | RC1 | IGHV9–3*01 | GTGKNYFDH | 9 | IGKV14–111*01 | LQYDEFPYT | 9 | YES |
| 352 | 3 | RC1 | IGHV5–6*01 | ATNYGAWFPY | 10 | IGKV3–4*01 | QQSNEDPYT | 9 | YES |
| 274 | 4 | RC1 | IGHV5–6*01 | ARHGITTVGVAMDY | 14 | IGKV3–4*01 | QQSNEDPWT | 9 | YES |
| 275 | 4 | RC1 | IGHV5–6*01 | ARHGITTVGVAMDY | 14 | IGKV3–4*01 | QQSNEDPYT | 9 | YES |
| 276 | 6 | RC1–4 | IGHV5–6*01 | ARHGRLTGTGAMDY | 14 | IGKV3–4*01 | QQSNEDPPWT | 10 | YES |
| 278 | 6 | RC1–4 | IGHV5–6*01 | ARHGRLTGTGAMDY | 14 | IGKV3–4*01 | HQSNEDPPWT | 10 | YES |
| 280 | 6 | RC1–4 | IGHV5–6*01 | ARHGHYYGSSYGMDY | 15 | IGKV3–4*01 | QQSNEDPPWT | 10 | YES |
| 294 | 6 | RC1–4 | IGHV2–9*01 | ANIPKDRLCYG | 11 | IGKV3–4*01 | QQSNEDPWT | 9 | YES |
| 348 | NS | RC1 | IGHV1–62–2*01 | ARHEGNYLYAMDY | 13 | IGKV4–62*01 | QQCSGYPLT | 9 | YES |
| 349 | NS | RC1 | IGHV1–7*01 | ARPPFITVVANYFDY | 15 | IGKV10–94*01 | QQYSKLPWT | 9 | YES |
Extended Data Table 5. Ig genes and CDRL3 amino acid sequences of V3-glycan patch bNAbs.
Table shows the amino acid sequence of the mature and iGL CDRL3s of several V3-glycan patch human bNAbs.
| bNAb | VH | VL | CDRL3 (MT) | CDRL3 (iGL) |
|---|---|---|---|---|
| PGT121 | 4–59 | L3–21 | HIWDSRVPTKWV | QVWDSSSDHPWV |
| PGT122 | 4–59 | L3–21 | HIWDSRRPTNWV | QVWDSSSDHPWV |
| PGT123 | 4–59 | L3–21 | HIYDARGGTNWV | QVWDSSSDHPWV |
| 10–1074 | 4–59 | L3–21 | HMWDSRSGFSWS | QVWDSSSDHPWV |
| PGT124 | 4–59 | L3–21 | MWDSRSGFSWS | QVWDSSSDHPWV |
| BG18 | 4–4 | L3–25 | QSSDTSDSYKM | |
| PGT125 | 4–39 | L2–8 | GSLVGNWDVI | SSYAGSNXXX |
| PGT126 | 4–39 | L2–8 | SSLVGNWDVI | SSYAGSNXXX |
| PGT127 | 4–39 | L2–8 | SSLVGNWDVI | SSYAGSNXXX |
| PGT128 | 4–39 | L2–8 | GSLVGNWDVI | SSYAGSNXXX |
| PGT130 | 4–39 | L2–8 | SSLFGRWDVV | SSYAGSNXXX |
| PGT131 | 4–39 | L2–8 | SSLSGRWDIV | SSYAGSNXXX |
| DH270.6 | 1–2 | L2–23 | SFGGSATVV | SYAGSSTVI |
Supplementary Material
Acknowledgements
We thank members of the Bjorkman, Martin and Nussenzweig laboratories for discussions, Thomas Eisenreich and Steven Tittley (Rockefeller) for animal husbandry, Kristie Gordon (Rockefeller) for flow cytometry, Susan Zolla-Pazner (Mount Sinai) for providing the V3-Consensus C peptide and anti-V3 monoclonal antibodies, Mark Howarth (Oxford) for providing plasmids and advice for VLP expression and purification, and Andrey Malyutin (Caltech) for help with cryo-EM data collection. Cryo-EM was done in the Beckman Institute Resource Center for Transmission Electron Microscopy at Caltech. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) Grant HIVRAD P01 AI100148 (to P.J.B. and M.C.N.), NIH Grant P50 GM082545-06 (P.J.B.), NIH Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) 1UM1 AI100663-01 (M.C.N.) the Intramural Research Program of the NIAID (M.M), the National Center for Biomedical Glycomics P41GM103490 (L.W.), Gates CAVD grant OPP1146996 (M.S.S. and D.C.M.), NIH/NIAID P01 AI138212 (L.S., A.T.M. and MCN), and the Robertson Fund of the Rockefeller University (M.C.N.). Additional support included an NSF GRFP (M.E.A.), an EMBO fellowship (J.M.), the HHMI Hanna Gray Fellowship and the Postdoctoral Enrichment Program from the Burroughs Welcome Fund (C.O.B.). M.C.N. and D.J.I. are HHMI investigators.
Footnotes
Author information
Reprints and permissions information is available at www.nature.com/reprints.
There are patents on 3BNC117 and 10–1074 on which M.C.N. and P.J.B. are inventors. M.C.N. is a member of the Scientific Advisory Boards of Celldex and Frontier Biosciences.
Readers are welcome to comment on the online version of the paper.
Data availability
Data availability statement
The atomic models and cryo-EM density maps generated during the current study have been deposited in the Protein Data Bank and Electron Microscopy Data Bank with accession numbers XXXX and EMD-XXX (RC1–10–1074), XXXX and EMD-XXXX (RC1–Ab275MUR), XXXX and EMD-XXXX (RC1–Ab874NHP), and XXXX and EMD-XXXX (RC1–Ab897NHP). Sequence data sets generated and analysed during the current study are available from the corresponding author on reasonable request.
Extended data is available for this paper.
Supplementary Information is available in the online version of the paper at www.nature.com/nature.
References
- 1.McCoy LE & Burton DR Identification and specificity of broadly neutralizing antibodies against HIV. Immunol Rev 275, 11–20, doi: 10.1111/imr.12484 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.West AP Jr. et al. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 156, 633–648, doi: 10.1016/j.cell.2014.01.052 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwong PD & Mascola JR HIV-1 Vaccines Based on Antibody Identification, B Cell Ontogeny, and Epitope Structure. Immunity 48, 855–871, doi: 10.1016/j.immuni.2018.04.029 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Bonsignori M et al. Antibody-virus co-evolution in HIV infection: paths for HIV vaccine development. Immunol Rev 275, 145–160, doi: 10.1111/imr.12509 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein F et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126–138, doi: 10.1016/j.cell.2013.03.018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Victora GD & Nussenzweig MC Germinal centers. Annu Rev Immunol 30, 429–457, doi: 10.1146/annurev-immunol-020711-075032 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Schwickert TA et al. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J Exp Med 208, 1243–1252, doi: 10.1084/jem.20102477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escolano A, Dosenovic P & Nussenzweig MC Progress toward active or passive HIV-1 vaccination. J Exp Med 214, 3–16, doi: 10.1084/jem.20161765 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escolano A et al. Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell 166, 1445–1458 e1412, doi: 10.1016/j.cell.2016.07.030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tas JM et al. Visualizing antibody affinity maturation in germinal centers. Science 351, 1048–1054, doi: 10.1126/science.aad3439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dal Porto JM, Haberman AM, Shlomchik MJ & Kelsoe G Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J Immunol 161, 5373–5381 (1998). [PubMed] [Google Scholar]
- 12.Abbott RK et al. Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48, 133–146 e136, doi: 10.1016/j.immuni.2017.11.023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dosenovic P et al. Anti-HIV-1 B cell responses are dependent on B cell precursor frequency and antigen-binding affinity. Proc Natl Acad Sci U S A 115, 4743–4748, doi: 10.1073/pnas.1803457115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shih TA, Meffre E, Roederer M & Nussenzweig MC Role of BCR affinity in T cell dependent antibody responses in vivo. Nat Immunol 3, 570–575, doi: 10.1038/ni803 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Kong L et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 20, 796–803, doi: 10.1038/nsmb.2594 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker LM et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470, doi:nature10373 [pii] 10.1038/nature10373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mouquet H et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109, E3268–3277, doi: 10.1073/pnas.1217207109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freund NT et al. Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci Transl Med 9, doi: 10.1126/scitranslmed.aal2144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sok D et al. A Prominent Site of Antibody Vulnerability on HIV Envelope Incorporates a Motif Associated with CCR5 Binding and Its Camouflaging Glycans. Immunity 45, 31–45, doi: 10.1016/j.immuni.2016.06.026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steichen JM et al. HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies. Immunity 45, 483–496, doi: 10.1016/j.immuni.2016.08.016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders RW et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9, e1003618, doi: 10.1371/journal.ppat.1003618 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gristick HB et al. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat Struct Mol Biol, doi: 10.1038/nsmb.3291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garces F et al. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell 159, 69–79, doi: 10.1016/j.cell.2014.09.009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrabi R et al. Glycans Function as Anchors for Antibodies and Help Drive HIV Broadly Neutralizing Antibody Development. Immunity 47, 1004, doi: 10.1016/j.immuni.2017.10.012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scharf L et al. Structural basis for germline antibody recognition of HIV-1 immunogens. Elife 5, doi: 10.7554/eLife.13783 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCoy LE et al. Holes in the Glycan Shield of the Native HIV Envelope Are a Target of Trimer-Elicited Neutralizing Antibodies. Cell Rep 16, 2327–2338, doi: 10.1016/j.celrep.2016.07.074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan H et al. Glycan Masking Focuses Immune Responses to the HIV-1 CD4-Binding Site and Enhances Elicitation of VRC01-Class Precursor Antibodies. Immunity 49, 301–311 e305, doi: 10.1016/j.immuni.2018.07.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrity RR et al. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J Immunol 159, 279–289 (1997). [PubMed] [Google Scholar]
- 29.Klasse PJ et al. Epitopes for neutralizing antibodies induced by HIV-1 envelope glycoprotein BG505 SOSIP trimers in rabbits and macaques. PLoS Pathog 14, e1006913, doi: 10.1371/journal.ppat.1006913 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brune KD et al. Plug-and-Display: decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci Rep 6, 19234, doi: 10.1038/srep19234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zakeri B et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci U S A 109, E690–697, doi: 10.1073/pnas.1115485109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longo NS et al. Multiple Antibody Lineages in One Donor Target the Glycan-V3 Supersite of the HIV-1 Envelope Glycoprotein and Display a Preference for Quaternary Binding. J Virol 90, 10574–10586, doi: 10.1128/JVI.01012-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burton DR & Hangartner L Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu Rev Immunol 34, 635–659, doi: 10.1146/annurev-immunol-041015-055515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sok D et al. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog 9, e1003754, doi: 10.1371/journal.ppat.1003754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gristick HB et al. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat Struct Mol Biol 23, 906–915, doi: 10.1038/nsmb.3291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JH, de Val N, Lyumkis D & Ward AB Model Building and Refinement of a Natively Glycosylated HIV-1 Env Protein by High-Resolution Cryoelectron Microscopy. Structure 23, 1943–1951, doi: 10.1016/j.str.2015.07.020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zolla-Pazner S et al. Structure/Function Studies Involving the V3 Region of the HIV-1 Envelope Delineate Multiple Factors That Affect Neutralization Sensitivity. J Virol 90, 636–649, doi: 10.1128/JVI.01645-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murugan R et al. Clonal selection drives protective memory B cell responses in controlled human malaria infection. Sci Immunol 3, doi: 10.1126/sciimmunol.aap8029 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Wang H et al. Asymmetric recognition of HIV-1 Envelope trimer by V1V2 loop-targeting antibodies. Elife 6, doi: 10.7554/eLife.27389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tissot AC et al. Versatile virus-like particle carrier for epitope based vaccines. PLoS One 5, e9809, doi: 10.1371/journal.pone.0009809 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lövgren-Bengtsson K & Morein B in Vaccine Adjuvants: Preparation Methods and Research Protocols (ed O’Hagan D) 239–258 (Humana Press, 2000). [Google Scholar]
- 42.Scheid JF et al. Sequence and Structural Convergence of Broad and Potent HIV Antibodies That Mimic CD4 Binding. Science 333, 1633–1637, doi: 10.1126/science.1207227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Boehmer L et al. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat Protoc 11, 1908–1923, doi: 10.1038/nprot.2016.102 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Scharf L et al. Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env. Cell 162, 1379–1390, doi: 10.1016/j.cell.2015.08.035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diskin R, Marcovecchio PM & Bjorkman PJ Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat Struct Mol Biol 17, 608–613, doi:nsmb.1796 [pii] 10.1038/nsmb.1796 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montefiori DC Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 485, 395–405, doi: 10.1007/978-1-59745-170-3_26 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Vaughn DE & Bjorkman PJ High-affinity binding of the neonatal Fc receptor to its IgG ligand requires receptor immobilization. Biochemistry 36, 9374–9380 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Tan YZ, Cheng A, Potter CS & Carragher B Automated data collection in single particle electron microscopy. Microscopy (Oxf) 65, 43–56, doi: 10.1093/jmicro/dfv369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, doi: 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang K Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12, doi: 10.1016/j.jsb.2015.11.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296, doi: 10.1038/nmeth.4169 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Scheres SH & Chen S Prevention of overfitting in cryo-EM structure determination. Nat Methods 9, 853–854, doi: 10.1038/nmeth.2115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terwilliger TC, Sobolev OV, Afonine PV & Adams PD Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr D Struct Biol 74, 545–559, doi: 10.1107/S2059798318004655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goddard TD, Huang CC & Ferrin TE Visualizing density maps with UCSF Chimera. J Struct Biol 157, 281–287 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221, doi:S0907444909052925 [pii] 10.1107/S0907444909052925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501, doi:S0907444910007493 [pii] 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21, doi: 10.1107/S0907444909042073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agirre J et al. Privateer: software for the conformational validation of carbohydrate structures. Nat Struct Mol Biol 22, 833–834, doi: 10.1038/nsmb.3115 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65, doi: 10.1038/nmeth.2727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corcoran MM et al. Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat Commun 7, 13642, doi: 10.1038/ncomms13642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.