Abstract
The sweet potato whitefly, Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) is a major agricultural pest that causes economic damages worldwide. In particular, B. tabaci MED (Mediterranean) has resulted in serious economic losses in tomato production of Korea. In this study, 1,145 B. tabaci MED females from 35 tomato greenhouses in different geographic regions were collected from 2016 to 2018 (17 populations in 2016, 13 in 2017, and five in 2018) and analyzed to investigate their population genetic structures using eight microsatellite markers. The average number of alleles per population (NA) ranged from 2.000 to 5.875, the expected heterozygosity (HE) ranged from 0.218 to 0.600, the observed heterozygosity (HO) ranged from 0.061 to 0.580, and the fixation index inbreeding coefficient (FIS) ranged from -0.391 to 0.872 over the three years of the study. Some significant correlation (p < 0.05) was present between genetic differentiations (FST) and geographical distance, and a comparatively high proportion of variation was found among the B. tabaci MED populations. The B. tabaci MED populations were divided into two well-differentiated genetic clusters within different geographic regions. Interestingly, its genetic structures converged into one genetic cluster during just one year. The reasons for this genetic change were speculated to arise from different fitness, insecticide resistance, and insect movement by human activities.
Introduction
The sweet potato whitefly, Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) is a major agricultural insect pest that is distributed worldwide. B. tabaci has an extremely broad host range [1] and causes serious damage to diverse host plant species. B. tabaci is also a vector for more than 100 pathogenic plant viruses [2], particularly known to be a vector for begomoviruses [3], and a major vector for tomato yellow leaf curl virus (TYLCV), one of the most devastating viruses in cultivated tomatoes in the world [4]. B. tabaci is a complex of 11 well-defined high-level groups consisting of at least 36 putative species identified based on mtCOI (mitochondrial cytochrome oxidase I) [5, 6]. These putative species are morphologically indistinguishable and differ in host range, virus transmission, fecundity, and insecticide resistance [7, 8]. Two major global putative species of B. tabaci, MEAM1 (Middle East-Asia Minor 1, formerly known as biotype B or B. argentifolii) and MED (Mediterranean, formerly known as biotype Q), are highly invasive and colonize large areas worldwide [9]. Three putative species (MEAM1, MED, and JpL (Lonicera japonica)) of the B. tabaci species complex are present in Korea. MEAM1 and MED were first detected in 1998 and 2004 [10, 11], respectively. JpL was first recorded in 2014 [12]. Currently, MED is predominant in most regions of the country, and MEAM1 and JpL are found only in a restricted region [12, 13].
Understanding the population genetic structure of a pest species is important for establishing pest management strategies [14]. Pest population structure assessments are helpful to reveal the origins and spread patterns of a target species [15], to delineate potential boundaries for their control [16], and to provide the statistical ability to differentiate between genetic groups [17], as well as to check whether they have mixed with other populations or not. When all population genetics information based on microsatellite markers is combined with environmental approaches, the construction of a powerful framework for managing B. tabaci is facilitated [18].
Over the past decades, various molecular genetics tools have considerably extended the boundary of population genetics [19]. Diverse DNA markers for insect genetics research (i.e., the amplified fragment length polymorphism (AFLP) marker, expressed sequence tags (EST), mitochondrial DNA (mtDNA), microsatellites, and random amplified polymorphic DNA (RAPD) [20]) have been identified and developed to determine the population genetic structure of a species. Among them, microsatellites are especially popular genetic markers because of their co-dominance, high abundant variation and polymorphism rates, multiple alleles, and quick allele detection by a wide variety of methods [21]. Microsatellite markers are also very effective tools in population genetic studies for insect species [22, 23]. Through molecular genetic diagnosis using population genetic analyses, effective control can be achieved in a short time at a low cost [24]. Different microsatellite markers were employed in several recent studies [25–29] to investigate the population genetic structure, genetic differentiation, genetic evolution, gene flow, and dispersal pattern of B. tabaci over relatively large geographic scales.
In this study, the population genetic structures and diversities of B. tabaci MED from tomato greenhouses were identified and their genetic relationships in Korea were examined.
Materials & methods
B. tabaci sampling
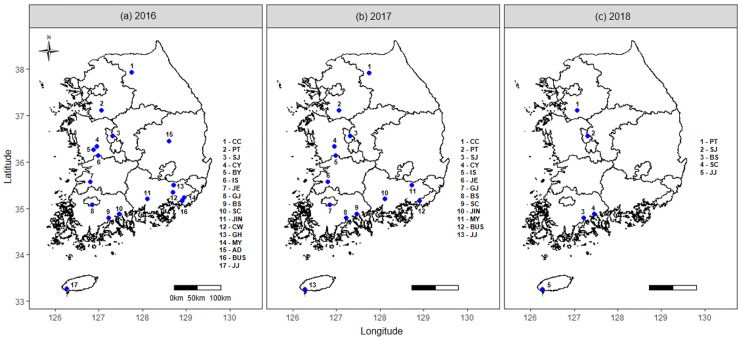
In total, 1,145 B. tabaci female adults were collected from 35 commercial tomato greenhouses in Korea using an aspirator during 2016–2018 (17 populations in 2016, 13 populations in 2017, and five populations in 2018) (Fig 1 and Table 1). The B. tabaci samples were collected from tomatoes plants at least 1 m apart to avoid the collection of full siblings in the greenhouses. All individual samples were preserved in 99.8% ethanol before DNA extraction.
Fig 1. B. tabaci MED sampling sites (see Table 1 for details) in Korea from (a) 2016, (b) 2017, and (c) 2018.
Table 1. Details of sampling information of B. tabaci MED in Korea.
| Sample site | Population | Collection date | GPS coordinates | Host plant | Sample size | GenBank accession No. |
|---|---|---|---|---|---|---|
| Seogwipo-si | 16’JJ | 2016-04-25 | 37°27′39.0″N,126°57′28.0″E | Tomato | 40 | HM802268 |
| 17’JJ | 2017-04-19 | 33°15′15.0″N,126°16′09.0″E | 20 | KY249477 | ||
| 18’JJ | 2018-10-10 | 33°15′15.0″N,126°16′09.0″E | 20 | KY249414 | ||
| Jinju-si * | 16’JIN | 2016-05-25 | 35°12′40.0″N,128°06′56.0″E | Tomato | 40 | EU386987 |
| 17’JIN | 2017-06-07 | 30 | EF694108 | |||
| Changwon-si | 16’CW | 2016-05-25 | 35°20′37.0″N,128°42′04.0″E | Tomato | 40 | KY468417 |
| Busan * | 16’BUS | 2016-05-25 | 35°10′18.0″N,128°54′56.0″E | Tomato | 40 | FJ375358 |
| 17’BUS | 2017-06-09 | 30 | HM597869 | |||
| Gimhae-si | 16’GH | 2016-05-26 | 35°14′06.0″N,128°57′42.0″E | Tomato | 40 | EU263626 |
| Miryang-si * | 16’MY | 2016-05-26 | 35°30′08.0″N,128°43′18.0″E | Tomato | 40 | EU760729 |
| 17’MY | 2017-06-08 | 30 | HM597849 | |||
| Jeongeup-si * | 16’JE | 2016-06-01 | 35°34′28.0″N,126°48′07.0″E | Tomato | 40 | EF667474 |
| 17’JE | 2017-06-20 | 30 | EU263630 | |||
| 18’JE | 2018-07-11 | 20 | KY249401 | |||
| Suncheon-si * | 16’SC | 2016-06-01 | 37°27′39.0″N,126°57′28.0″E | Tomato | 40 | MH357338 |
| 17’SC | 2017-06-19 | 30 | KY468420 | |||
| 18’SC | 2018-07-11 | 20 | HM597847 | |||
| Gwangju * | 16’GJ | 2016-06-02 | 35°04′31.0″N,126°51′11.0″E | Tomato | 40 | KY468410 |
| 17’GJ | 2017-06-20 | 30 | KY468415 | |||
| Boseong-gun * | 16’BS | 2016-06-02 | 34°47′33.0″N,127°13′15.0″E | Tomato | 40 | EU263629 |
| 17’BS | 2017-06-19 | 30 | HM597859 | |||
| Iksan-si | 16’IS | 2016-06-09 | 36°08'21.0"N,126°58'59.0"E | Tomato | 40 | HM597859 |
| 17’IS | 2017-06-20 | 36°08′20.0″N,126°58′55.0″E | 15 | EU427722 | ||
| Andong-si | 16’AD | 2016-06-09 | 36°27′23.0″N,128°36′11.0″E | Tomato | 40 | KP137475 |
| Buyeo-gun | 16’BY | 2016-06-30 | 36°15′60.0″N,126°52′49.0″E | Tomato | 40 | MH357340 |
| Cheongyang-gun * | 16’CY | 2016-06-30 | 36°20′21.0″N,126°57′18.0″E | Tomato | 40 | EU760736 |
| 17’CY | 2017-06-21 | 30 | KY249451 | |||
| Sejong-si | 16’SJ | 2016-06-30 | 36°34′11.6″N,127°19′02.8″E | Tomato | 40 | KY249434 |
| 17’SJ | 2017-06-19 | 36°34′19.0″N,127°18′40.0″E | 30 | MG565975 | ||
| 18’SJ | 2018-07-12 | 36°34′19.0″N,127°18′40.0″E | 20 | EU376987 | ||
| Chuncheon-si | 16’CC | 2016-07-29 | 37°56′02.9″N 127°44′57.7″E | Tomato | 40 | MH357339 |
| 17’CC | 2017-06-29 | 37°55′38.0″N 127°45′15.0″E | 30 | KY468408 | ||
| Pyeongtaek-si | 16’PT | 2016-08-05 | 37°07′20.0″N,127°03′29.0″E | Tomato | 40 | MH357340 |
| 17’PT | 2017-06-26 | 37°07′25.0″N,127°03′14.0″E | 30 | MH205752 | ||
| 18’PT | 2018-08-10 | 37°07′20.0″N,127°03′29.0″E | 20 | KY249438 |
*Same tomato greenhouse during two or three years
Molecular methods
DNA extraction
Genomic DNA (gDNA) extraction was performed using a Qiagen Gentra Puregene Tissue Kit (Qiagen, Gaithersburg, MD, USA) according to the manufacturer’s instructions. Since B. tabaci is a haplo-diploid species, producing male progeny from unfertilized eggs and female progeny from fertilized eggs [30], only adult females were used for the genetic analysis of each individuals. The extracted gDNA samples were finally stored at -20 °C until use. DNA quantification was performed with ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA).
Identification of B. tabaci putative species
Two individuals per population were randomly selected in order to identify the B. tabaci putative species. A fragment of the mtCOI gene was PCR-amplified using the primer pair C1-J-2195 (5’-TTGATTTTTTGGTCATCCAGAAGT-3’) and L2-N-3014 (5’-TCCAATGCACTAATCTGCCATATTA-3’) [31]. All PCR reactions were conducted using 1 μl forward primer (10 pmol/μl), 1 μl reverse primer (10 pmol/μl), and 2 μl template DNA in 20 μl reaction volumes consisting of 25 mM dNTPs, 10 mM Tris-HCl (pH 9), 30 mM KCl, 1.5 mM MgCl2, and 1 unit of Taq DNA polymerase using Accupower PCR PreMix (Bioneer, Seoul, Korea). The reaction conditions included an initial denaturation for 5 min at 94 °C, followed by 34 cycles of 1 min each at 94 °C, 1 min at 52 °C, and 1 min at 72 °C, with a final extension for 5 min at 72 °C [12]. PCR products were sent for sequencing to NICEM (Seoul, Korea). Putative species identification was based on direct sequence comparisons using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
PCR amplification of eight microsatellites
PCR primers were used to amplify microsatellite DNA loci 11, 53 [32], 68, 145, 177 [22], BT4, BT159 [33], and Bem23 [34] using the individual gDNA of B. tabaci MED as templates. PCR amplifications for the microsatellite primers and PCR reactions were performed as previously described [22]. A total of 1,145 individuals were genotyped using eight microsatellite loci distributed in two PCR multiplex sets. Two multiplex PCRs were performed for each individual at 10 pmol/μl (multiplex 1 loci: 11, 145, 177, BT4, and BT159; multiplex 2 loci: 53, 68, and Bem23). In order to analyze the length of the PCR products using a laser detection system, some of the forward and reverse primers were labeled with a fluorescent dye. The rTaq PCR kit (Takara Bio Inc., Kyoto, Japan) was used for these reactions. The total reaction volume was 10 μl, which contained 2.9 μl or 4.1 μl (multiplex 1: 2.9 μl, multiplex 2: 4.1 μl) distilled water, 1.0 μl 10X PCR buffer, 1.0 μl 2.5mM dNTP mixture, 0.2 μl of each primer, 0.1 μl of Taq polymerase, and 2.0 μl template DNA. The multiplex PCR products were analyzed using an ABI 3730xl (Applied Biosystems Inc., Foster, CA, USA). Allele size was detected using GENEMAPPER v.3.7 (Applied Biosystems Inc.). Multiplex 1 was amplified in PTC100 Thermocyclers (MJ Research, Waltham, MA, USA) as follows: 15 min at 94 °C, followed by 40 cycles for 30 s at 94 °C, 1 min 30 s at 57 °C, 1 min at 72 °C, ending with 30 min at 60 °C. Multiplex 2 was amplified as above except that the annealing temperature was increased from 57 to 60 °C. PCR was carried out as described by Dalmon et al. [22]. The 1 μl PCR product was diluted with 8.5 μl of Hi-Di formamide (Applied Biosystems Inc.) and 0.5 μl Genescan ROX-500 size standard (Applied Biosystems Inc.).
Analyses of genetic diversity
GENEPOP v.4.0 [35] and Micro-Checker v.2.2.3 [36, 37] were used to determine the microsatellite data for scoring errors, allelic dropouts, and null alleles. The estimated frequency of null alleles per loci for each population was calculated in FreeNa [38] using the expectation maximization (EM) algorithm [39]. Each of the 1,145 collected samples were used to test deviations from Hardy-Weinberg equilibrium (HWE) conditions, the number of alleles (NA), allele size range, and the observed (HO) and expected heterozygosities (HE), and the inbreeding coefficient (FIS) were computed using GenAlEx v.6.5 [40] and Microsatellite Toolkit [41].
Analysis of molecular variance (AMOVA)
AMOVA was performed using GenAlEx v.6.5. AMOVA was used to characterize genetic variation patterns and to estimate variance components. A two-part AMOVA analysis was conducted to check genetic divergence (FST) as a factor of variation among and within the populations. AMOVA computations were performed with 999 permutations to test for significance.
Analyses of genetic structure
The number of genetic clusters (K) was estimated in STRUCTURE v.2.3.2 with 60,000 Markov Chain Monte Carlo (MCMC) steps and a burn-in period of 600,000. The log-likelihood estimate was run for K = ranges from 1 to 10 with ten replicates each. They were used to determine the number of clusters based on a combination of the mean estimated Ln probability of the data [42] and the second-order rate of change in the log-probability of the data (ΔK) [43]. The Evanno method was then implemented in STRUCTURE HARVESTER Web v.0.6.93 [44].
Principal coordinate analysis (PCoA)
PCoA was conducted between multi-locus genotypes in all individuals. The codominant-genotypic option of GeneAlex v.6.5 was used to calculate the similarity genetic distance matrix [40]. The PCoA plot was based on factor scores along the two principal axes (axis 1 and 2) and enabled the visualization of population differences.
Discriminant analysis of principal components (DAPC)
DAPC was performed in the ‘adegenet’ package [45] of R software v.3.5.1 (R Development Core Team, 2018) to identify an optimal number of genetic clusters to describe the data. DAPC is a multivariate algorithm, similar to principal component analysis (PCA) that identifies genetic clusters and can be used as an efficient genetic clustering tool [46]. The number of clusters was identified based on Bayesian information criterion (BIC). If the value of BIC is positive and low, it is a suitable model. When the BIC value is negative, a high number is a suitable model.
Isolation by distance (IBD)
The Mantel test [47] was performed to assess isolation by distance. The relationship between pairwise geographic distance (Ln km) and pairwise genetic distance in terms of FST/(1-FST) with 1,000 random permutations was conducted using the GenAlEx v.6.5, GENEPOP v.4.0, and ‘ade4’ package [48] of R software v.3.5.1. The IBD graph was generated by using the R software v.3.5.1 with ‘ggpolt2’ package.
Bottleneck test
The BOTTLENECK v.1.2.02 [49] was used to detect the effect of a recent reduction in all population sizes. The possibility of bottleneck events in the 35 populations was examined using a one-tailed Wilcoxon signed-rank test under three mutation models, the infinite allele model (IAM), the two-phase model (TPM), and the stepwise mutation model (SMM) (parameters for TPM: variance = 30.0%, probability = 70.0%, 1,000 replications). The Wilcoxon signed-rank test has been shown to be effective and reliable when eight microsatellite loci are analyzed [49].
Pairwise comparisons of fixation index (FST)
To assess the level of genetic differentiation between the samples, pair-wise fixation index (FST) value estimates were computed using GENEPOP v.4.0. To correct for null alleles, pairwise estimators of FST values were calculated from each microsatellite dataset that potentially harbored null alleles using the excluding null alleles (ENA) correction method (FST-ENA) following 1,000 bootstrapping permutations over the loci. The ENA correction method was used to obtain unbiased pairwise FST values using FreeNA. To investigate the relationship between the genetic distance revealed by the FST values and geographic distance, an isolation-by-distance analysis was performed using a regression of FST/(1-FST) values against the logarithm of the geographical distance (km) between the populations. Significance of the correlation between the two data matrices was assessed using a Mantel test with 1,000 permutations. This was performed with the ISOLDE program implemented in GENEPOP v.4.0.
Results
Identification of the B. tabaci populations
All B. tabaci individuals collected were successfully sequenced and analyzed. Approximately 810 bp of the mtCOI gene was amplified from B. tabaci individuals by PCR. All populations identified belonged to the MED (Q1) species based on representative samples.
Genetic diversity
The values of the genetic diversity indexes for the Korea populations of B. tabaci MED are shown in Table 2. There were one to eight alleles per loci in the eight microsatellites and the estimated average frequency of null alleles ranged from 0.031 to 0.407 among the 35 populations. The average number of alleles per population (NA) ranged from 2.000 (17’JIN) to 5.875 (16’SJ). The expected heterozygosity (HE) ranged from 0.218 (16’JJ) to 0.600 (16’PT), whereas the observed heterozygosity (HO,) ranged from 0.061 (16’CW) to 0.580 (16’IS). The value of HE in each population was higher than the value of HO, except for 12 populations that showed negative values for FIS. The estimator of the fixation index inbreeding coefficient (FIS) ranged from -0.391 (17’CC) to 0.872 (16’CW). A positive value for FIS indicates the presence of heterozygotic deficiencies, whereas a negative value indicates the presence of homozygotic deficiencies. The analysis of genetic diversity for all different eight microsatellite loci of B. tabaci MED screened is given in S1 Table.
Table 2. Genetic diversity of the B. tabaci MED populations.
| Population | N | NA | HE | HO | FIS | Fnull |
|---|---|---|---|---|---|---|
| 16'JJ | 40 | 2.625 | 0.218 | 0.160 | 0.266 | 0.241 |
| 16'JIN | 40 | 5.500 | 0.423 | 0.274 | 0.353 | 0.217 |
| 16'CW | 40 | 3.500 | 0.480 | 0.061 | 0.872 | 0.393 |
| 16'BUS | 40 | 2.625 | 0.407 | 0.118 | 0.710 | 0.407 |
| 16'GH | 40 | 3.250 | 0.414 | 0.159 | 0.614 | 0.327 |
| 16'MY | 40 | 4.625 | 0.459 | 0.107 | 0.768 | 0.307 |
| 16'JE | 40 | 4.250 | 0.478 | 0.337 | 0.295 | 0.296 |
| 16'SC | 40 | 4.625 | 0.458 | 0.282 | 0.383 | 0.184 |
| 16'GJ | 40 | 2.875 | 0.462 | 0.231 | 0.499 | 0.284 |
| 16'BS | 40 | 4.750 | 0.521 | 0.187 | 0.642 | 0.292 |
| 16'IS | 40 | 5.750 | 0.549 | 0.580 | -0.057** | 0.174 |
| 16'AD | 40 | 5.125 | 0.486 | 0.272 | 0.440 | 0.284 |
| 16'BY | 40 | 3.000 | 0.256 | 0.136 | 0.466 | 0.231 |
| 16'CY | 40 | 2.875 | 0.391 | 0.180 | 0.540 | 0.031 |
| 16'SJ | 40 | 5.875 | 0.594 | 0.148 | 0.751 | 0.333 |
| 16'CC | 40 | 5.625 | 0.445 | 0.237 | 0.468 | 0.255 |
| 16'PT | 40 | 5.500 | 0.600 | 0.264 | 0.560 | 0.300 |
| 17'JJ | 20 | 3.000 | 0.369 | 0.391 | -0.058** | 0.268 |
| 17'JIN | 30 | 2.000 | 0.246 | 0.209 | 0.150 | 0.238 |
| 17'MY | 30 | 3.375 | 0.378 | 0.388 | -0.026** | 0.331 |
| 17'BUS | 30 | 3.250 | 0.406 | 0.304 | 0.251 | 0.329 |
| 17'SJ | 30 | 3.750 | 0.409 | 0.417 | -0.020** | 0.211 |
| 17'SC | 30 | 2.875 | 0.376 | 0.373 | 0.010* | 0.323 |
| 17'BS | 30 | 3.375 | 0.325 | 0.339 | -0.041** | 0.259 |
| 17'GJ | 30 | 3.000 | 0.443 | 0.425 | 0.041* | 0.382 |
| 17'JE | 30 | 3.375 | 0.394 | 0.499 | -0.265** | 0.308 |
| 17'IS | 15 | 3.000 | 0.379 | 0.426 | -0.123** | 0.272 |
| 17'CY | 30 | 3.000 | 0.356 | 0.299 | 0.160 | 0.406 |
| 17'PT | 30 | 3.250 | 0.424 | 0.513 | -0.211** | 0.240 |
| 17'CC | 30 | 2.625 | 0.387 | 0.539 | -0.391** | 0.343 |
| 18'SC | 20 | 2.500 | 0.368 | 0.413 | -0.122** | 0.303 |
| 18'BS | 20 | 2.875 | 0.302 | 0.319 | -0.054** | 0.273 |
| 18'SJ | 20 | 3.375 | 0.420 | 0.413 | 0.019* | 0.175 |
| 18'PT | 20 | 4.000 | 0.546 | 0.250 | 0.542 | 0.304 |
| 18'JJ | 20 | 2.375 | 0.286 | 0.350 | -0.225** | 0.292 |
N, number of individuals sampled; NA, Mean number of alleles per population; HE, Mean expected heterozygosity; HO, Mean observed heterozygosity; FIS, Mean fixation index inbreeding coefficient; and Fnull, average proportion of Homozygous for null allele. Significance FIS value is obtained after 1,000 permutation tests (*p < 0.05; **p < 0.01).
AMOVA
AMOVA among the 35 B. tabaci MED populations showed that 48.0% of the total genetic variation was accounted for by variation among the populations and 52.0% of the variation was accounted for by individual variation within the populations (Table 3). The AMOVA results revealed a relatively high proportion of variation among the populations.
Table 3. Analysis of molecular variance (AMOVA) for the 35 B. tabaci MED populations collected from different regions in Korea using eight microsatellite markers.
| Source of variation | Degrees of freedom | Sums of squares | Mean sums of squares | Estimated variance | % of variation | p-value |
|---|---|---|---|---|---|---|
| Among population | 34 | 5557.909 | 163.468 | 4.845 | 48.0% | 0.01 |
| Within population | 1110 | 5820.817 | 5.244 | 5.244 | 52.0% | |
| Total | 1144 | 11378.726 | 10.089 | 100% |
Significant at p < 0.01 (based on 999 permutations)
Genetic relationships and population structure analysis
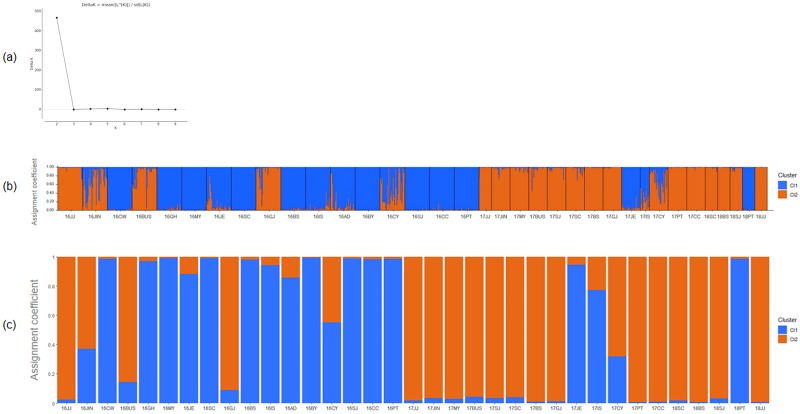
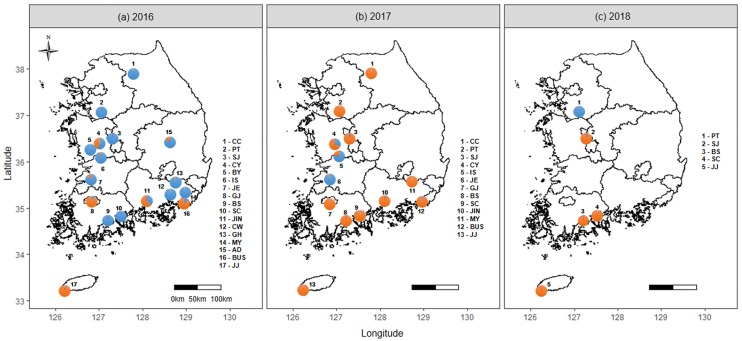
The genetic structure analysis of 35 B. tabaci MED populations using eight microsatellite marker genotypes revealed two dominant genetic clusters. The highest likelihood value was obtained for K = 2 (Fig 2a). The 16 populations (16’CC, 16’PT, 16’SJ, 16’BY, 16’CY, 16’IS, 16’JE, 16’BS, 16’SC, 16’CW, 16’GH, 16’MY, 16’AD, 17’IS, 17’JE, 18’PT) formed one cluster, and 19 populations (16’JIN, 16’GJ, 16’BUS, 16’JJ, 17’CC, 17’PT, 17’SJ, 17’CY, 17’GJ, 17’BS, 17’SC, 17’JIN, 17’MY, 17’BUS, 17’JJ, 18’SJ, 18’JE, 18’BS, 18’SC) formed the other cluster (Fig 2b and 2c). The populations of B. tabaci MED converged rapidly into one cluster (orange color) over time (Fig 3).
Fig 2. Scatter plots of ΔK = 2.
(a) The maximum value among the genotypes was 466.35 at ΔK = 2, using ΔK = m(|L“K|) / s[L(K)]. Bar plot of the population structure for B. tabaci from 35 populations in Korea (b) using STRUCTURE v.2.3.2 and (c) R software v.3.5.1. Each population is represented by a vertical line with different colors representing the probabilities assigned to each of the genetic clusters.
Fig 3. Bayesian clustering results from the structure for all samples (K = 2).
The geographical distribution of the population and the genetic structure of the B. tabaci MED in Korea revealed by STRUCTURE analysis in samples from (a) 2016, (b) 2017, and (c) 2018. Genetic changes were observed in six of the populations from 2016 to 2017. The maps were created by using the R software v.3.5.1.
PCoA of B. tabaci MED
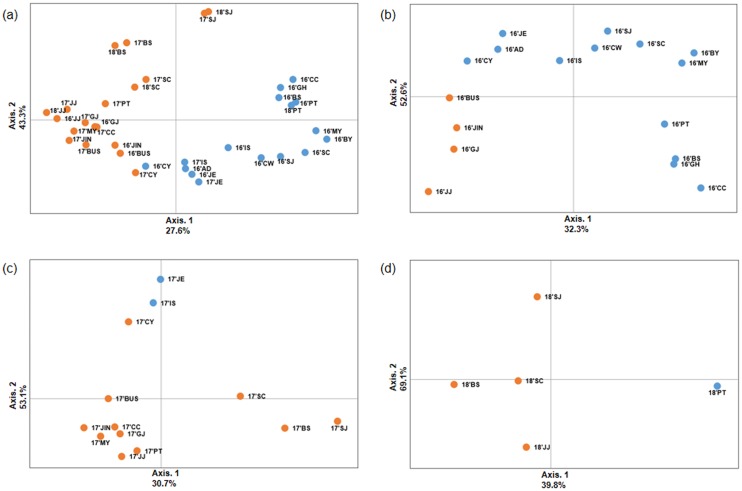
Principal component analysis of the 35 B. tabaci MED populations showed that the first principal components accounted for 27.6% of the total variation, followed by the second component, which accounted for 43.3% of the variation (Fig 4a). The first and second components of PCoA for each year are as follows: 32.3%, 52.6% for 2016 (Fig 4b), 30.7%, 53.1% for 2017 (Fig 4c), and 39.8%, 69.1% for 2018 (Fig 4d), respectively.
Fig 4. Principal component analysis (PCoA) plotting the relationships of 35 B. tabaci MED population samples.
Each axis indicates the percentage of the total variations. STRUCTURE with marked color is the same as one of Bayesian clustering (blue and orange colors) from (a) 2016–2018, (b) 2016, (c) 2017, and (d) 2018.
DAPC
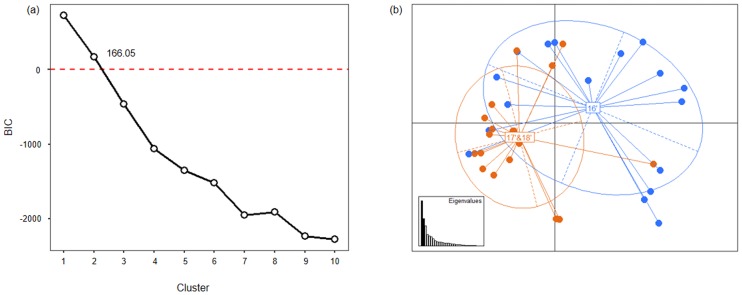
In DAPC, the elbow in the curve of BIC was at K = 2 using the find. cluster function of R software v.3.5.1 [50]. In this study, the value of BIC was found to be 166.05, which was positive and the smallest value (Fig 5a). The DAPC results showed that the populations of B. tabaci MED were split into two well-differentiated genetic clusters with low overlap between them. The first cluster contained populations from 2016 and the second cluster contained populations from 2017 and 2018 (Fig 5b). The DAPC results agreed with the STRUCTURE results.
Fig 5. Discriminant analysis of principal components (DAPC) analysis of 35 B. tabaci MED populations in Korea.
(a) The Bayesian information criteria (BIC) supported two distinct genetic clusters. (b) The eigenvalues of the analysis suggest that the first two components explained the maximum genetic structure of the dataset. Scatter-plot of the distribution of B. tabaci MED formed two genetic clusters (blue and orange colors).
IBD
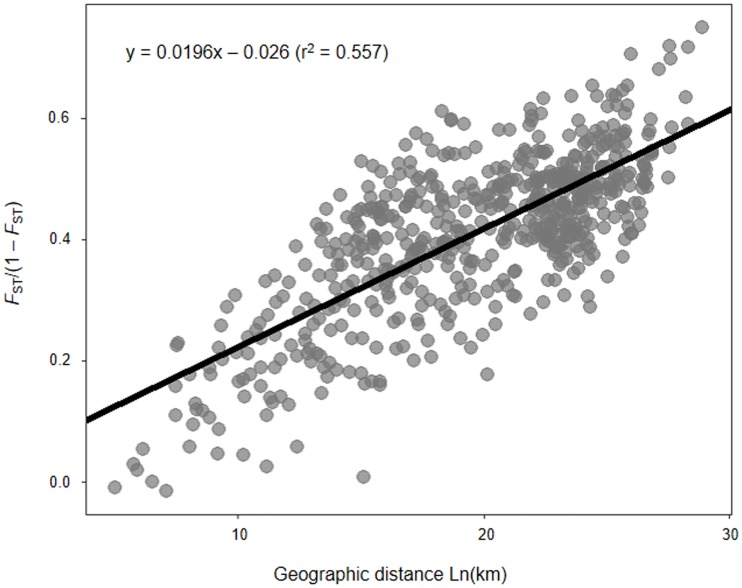
A significant correlation was detected between genetic and geographic distances in the B. tabaci MED populations based on the Mantel tests of IBD (r2 = 0.557; p = 0.01), indicating a pattern of isolation by distance (Fig 6). Multiple points in the scatterplot fit to the linear regression along the geographic distance range. This result indicates that gene flow between population increases with geographic distance. IBD analysis revealed that geographic distance had an effect on the population structure of the B. tabaci.
Fig 6. Relationship between genetic distance and the log of the geographical distance for B. tabaci MED.
The line represents the regression line and circles represent the logarithm transformation of distance (p = 0.01, 1,000 permutations).
Bottleneck test
The mode-shift analysis of bottleneck test, a signature of recent population reduction was found only for the 16’GJ and 18’PT populations (Table 4). Departure from mutation-drift equilibrium was observed in two populations, indicating that they remained relatively unstable in recent evolutionary history. Significant heterozygosity excess (Wilcoxon test p-values) was detected in eight populations under the IAM (16’CW, 16’BUS, 16’GJ, 16’CY, 17’BUS, 17’GJ, 17’CY, 18’PT) and two populations under the TPM (16’GJ, 17GJ), which accounted for 22.8% and 5.7% of the Korea populations (Table 4 bolded numbers), respectively. Under the SMM, however, significant heterozygosity excess was not detected in any population.
Table 4. Wilcoxon signed-rank tests for heterozygosity excess for the 35 B. tabaci MED populations.
| Population | WILCOXON Tests * (Heterozygosity Excess p-values) |
Mode-Shift | ||
|---|---|---|---|---|
| IAM | TPM | SMM | ||
| 16'JJ | 0.94531 | 0.97266 | 0.98047 | Normal |
| 16'JIN | 0.84375 | 0.99609 | 1.00000 | Normal |
| 16'CW | 0.03711 | 0.52734 | 0.72656 | Normal |
| 16'BUS | 0.01953 | 0.19147 | 0.52734 | Normal |
| 16'GH | 0.12500 | 0.37109 | 0.96289 | Normal |
| 16'MY | 0.67969 | 0.97266 | 0.99023 | Normal |
| 16'JE | 0.14844 | 0.59375 | 0.07813 | Normal |
| 16'SC | 0.52734 | 0.76953 | 0.99609 | Normal |
| 16'GJ | 0.00781 | 0.01172 | 0.05469 | Shifted mode |
| 16'BS | 0.32031 | 0.97266 | 0.99414 | Normal |
| 16'IS | 0.27344 | 0.76953 | 0.99023 | Normal |
| 16'AD | 0.62891 | 0.99414 | 1.00000 | Normal |
| 16'BY | 0.37109 | 0.97266 | 1.00000 | Normal |
| 16'CY | 0.01953 | 0.15625 | 0.52734 | Normal |
| 16'SJ | 0.32031 | 0.80859 | 0.98633 | Normal |
| 16'CC | 0.72656 | 0.97266 | 0.99609 | Normal |
| 16'PT | 0.12500 | 0.37109 | 0.84375 | Normal |
| 17'JJ | 0.42188 | 0.76953 | 0.84375 | Normal |
| 17'JIN | 0.28906 | 0.46875 | 0.65625 | Normal |
| 17'MY | 0.46875 | 0.76563 | 0.96094 | Normal |
| 17'BUS | 0.03906 | 0.65625 | 0.94531 | Normal |
| 17'SJ | 0.52734 | 0.67969 | 0.98047 | Normal |
| 17'SC | 0.32031 | 0.52734 | 0.76953 | Normal |
| 17'BS | 0.65625 | 0.96094 | 0.97266 | Normal |
| 17'GJ | 0.00781 | 0.01563 | 0.07813 | Normal |
| 17'JE | 0.40625 | 0.81250 | 0.94531 | Normal |
| 17'IS | 0.40625 | 0.65625 | 0.81250 | Normal |
| 17'CY | 0.03906 | 0.28906 | 0.94531 | Normal |
| 17'PT | 0.34375 | 0.46875 | 0.46875 | Normal |
| 17'CC | 0.15625 | 0.47266 | 0.76953 | Normal |
| 18'SC | 0.05469 | 0.23438 | 0.28906 | Normal |
| 18'BS | 0.57813 | 0.78125 | 0.96094 | Normal |
| 18'SJ | 0.23438 | 0.34375 | 0.65625 | Normal |
| 18'PT | 0.01953 | 0.18750 | 0.40625 | Shifted mode |
| 18'JJ | 0.34375 | 0.65625 | 0.65625 | Normal |
Infinite allele model (IAM), two-phase model (TPM), and stepwise mutation model (SMM) for detection of a recent population bottleneck event within each B. tabaci MED population.
*One-tailed Wilcoxon signed-rank test; Bolded numbers indicate they are significant at p < 0.05.
Pairwise comparisons of fixation index (FST)
The fixation index (FST) reflects the degree of genetic differentiation among the populations. FST is close to 0 when the genetic variation shows no difference in fixation among the populations. It is close to 1 when genetic differentiation is high. In this study, the FST values ranged from -0.0155 to 0.7501 and the ENA-corrected FST values ranged from -0.0139 to 0.7327 among the populations (Table 5). The highest FST value was detected between the 16’JJ and 16’BY populations (0.7327). The lowest FST value was found between the 17’SJ and 18’SJ populations (-0.0139). In practice, an FST value of 0.00–0.05 indicates low differentiation and FST values > 0.15 indicate a high level of differentiation. Negative FST values are allowed because correlations vary from -1 to +1 [51]. As a result, most B. tabaci in Korea showed high levels of differentiation.
Table 5. Pairwise FST values based on variation at eight microsatellite loci between the B. tabaci MED populations.
| 16'JJ | 16'JIN | 16'CW | 16'BUS | 16'GH | 16'MY | 16'JE | 16'SC | 16'GJ | 16'BS | 16'IS | 16'AD | 16'BY | 16'CY | 16'SJ | 16'CC | 16'PT | 17'JJ | |
| 16'JJ | 0.2894 | 0.5640 | 0.3427 | 0.5955 | 0.6336 | 0.5115 | 0.6201 | 0.2432 | 0.5249 | 0.5102 | 0.4856 | 0.7501 | 0.4542 | 0.5407 | 0.5687 | 0.5179 | 0.0949 | |
| 16'JIN | 0.2487 | 0.3983 | 0.1616 | 0.4852 | 0.4822 | 0.2634 | 0.4194 | 0.1808 | 0.4207 | 0.3661 | 0.2193 | 0.6080 | 0.2214 | 0.3667 | 0.4630 | 0.3748 | 0.2120 | |
| 16'CW | 0.5356 | 0.3746 | 0.3854 | 0.4038 | 0.3810 | 0.3126 | 0.3222 | 0.3871 | 0.3753 | 0.3226 | 0.2892 | 0.4910 | 0.3956 | 0.1769 | 0.4414 | 0.3712 | 0.4565 | |
| 16'BUS | 0.3232 | 0.1343 | 0.3635 | 0.4928 | 0.4916 | 0.2589 | 0.4466 | 0.2229 | 0.4599 | 0.3217 | 0.2005 | 0.5890 | 0.1597 | 0.3524 | 0.5230 | 0.4210 | 0.2364 | |
| 16'GH | 0.5750 | 0.4727 | 0.3767 | 0.4713 | 0.4588 | 0.4982 | 0.4018 | 0.4518 | 0.3322 | 0.4360 | 0.4740 | 0.5783 | 0.5415 | 0.4095 | 0.2679 | 0.3062 | 0.5041 | |
| 16'MY | 0.5957 | 0.4491 | 0.3105 | 0.4568 | 0.3863 | 0.4042 | 0.3066 | 0.5012 | 0.3076 | 0.4082 | 0.4239 | 0.4962 | 0.4622 | 0.3077 | 0.4060 | 0.3447 | 0.5371 | |
| 16'JE | 0.4903 | 0.2438 | 0.3042 | 0.2360 | 0.4778 | 0.3796 | 0.2899 | 0.3520 | 0.4515 | 0.3373 | 0.1653 | 0.5605 | 0.1794 | 0.2595 | 0.5084 | 0.3638 | 0.3963 | |
| 16'SC | 0.6078 | 0.4208 | 0.3201 | 0.4449 | 0.3631 | 0.2871 | 0.3052 | 0.4662 | 0.4038 | 0.3726 | 0.3251 | 0.5146 | 0.3628 | 0.2420 | 0.3949 | 0.2365 | 0.5267 | |
| 16'GJ | 0.2337 | 0.1458 | 0.3484 | 0.1597 | 0.4267 | 0.4396 | 0.3243 | 0.4479 | 0.4079 | 0.3568 | 0.2797 | 0.5975 | 0.3004 | 0.3999 | 0.4662 | 0.3857 | 0.1408 | |
| 16'BS | 0.5050 | 0.4093 | 0.3140 | 0.4370 | 0.2907 | 0.2504 | 0.4293 | 0.3864 | 0.3734 | 0.3923 | 0.4359 | 0.4639 | 0.4870 | 0.3374 | 0.2334 | 0.2769 | 0.4252 | |
| 16'IS | 0.4959 | 0.3545 | 0.2974 | 0.3289 | 0.4141 | 0.3624 | 0.3324 | 0.3668 | 0.3350 | 0.3629 | 0.3021 | 0.4615 | 0.3630 | 0.2219 | 0.4727 | 0.3570 | 0.3991 | |
| 16'AD | 0.4574 | 0.1882 | 0.2726 | 0.1810 | 0.4452 | 0.3766 | 0.1237 | 0.3191 | 0.2588 | 0.4001 | 0.2916 | 0.4904 | 0.1654 | 0.2954 | 0.4993 | 0.3674 | 0.3683 | |
| 16'BY | 0.7327 | 0.6041 | 0.4835 | 0.6019 | 0.5491 | 0.4690 | 0.5580 | 0.5133 | 0.5848 | 0.4452 | 0.4461 | 0.4874 | 0.5714 | 0.4263 | 0.5271 | 0.4362 | 0.6802 | |
| 16'CY | 0.4280 | 0.1899 | 0.3687 | 0.1489 | 0.5053 | 0.4182 | 0.1626 | 0.3460 | 0.2527 | 0.4528 | 0.3429 | 0.1610 | 0.5633 | 0.3328 | 0.5438 | 0.4008 | 0.3323 | |
| 16'SJ | 0.5188 | 0.3442 | 0.1298 | 0.3333 | 0.3651 | 0.2527 | 0.2440 | 0.2380 | 0.3489 | 0.2789 | 0.2008 | 0.2587 | 0.4266 | 0.2997 | 0.4219 | 0.2885 | 0.4372 | |
| 16'CC | 0.5446 | 0.4558 | 0.3950 | 0.5085 | 0.2155 | 0.3435 | 0.4867 | 0.3608 | 0.4410 | 0.2069 | 0.4456 | 0.4658 | 0.4969 | 0.5128 | 0.3771 | 0.2052 | 0.4913 | |
| 16'PT | 0.5045 | 0.3676 | 0.3051 | 0.3968 | 0.2395 | 0.2828 | 0.3490 | 0.2172 | 0.3495 | 0.2447 | 0.3328 | 0.3362 | 0.4351 | 0.3743 | 0.2344 | 0.1729 | 0.4150 | |
| 17'JJ | 0.1137 | 0.1934 | 0.4268 | 0.2116 | 0.4815 | 0.4971 | 0.3769 | 0.5198 | 0.1342 | 0.4088 | 0.3835 | 0.3419 | 0.6658 | 0.3078 | 0.4135 | 0.4781 | 0.4049 | |
| 17'JIN | 0.1919 | 0.1463 | 0.4985 | 0.2682 | 0.5684 | 0.5526 | 0.4082 | 0.5464 | 0.2399 | 0.4907 | 0.4580 | 0.3705 | 0.7034 | 0.2922 | 0.4741 | 0.5344 | 0.4787 | 0.1724 |
| 17'MY | 0.1117 | 0.1209 | 0.4169 | 0.1620 | 0.4798 | 0.4859 | 0.3333 | 0.4927 | 0.1089 | 0.4120 | 0.3816 | 0.3040 | 0.6444 | 0.2539 | 0.3960 | 0.4731 | 0.4040 | 0.0466 |
| 17'BUS | 0.2123 | 0.1147 | 0.3807 | 0.0491 | 0.4710 | 0.4581 | 0.2590 | 0.4534 | 0.1304 | 0.4159 | 0.3458 | 0.2388 | 0.6129 | 0.1434 | 0.3510 | 0.4811 | 0.3874 | 0.1098 |
| 17'SJ | 0.5808 | 0.4871 | 0.4695 | 0.4679 | 0.4718 | 0.4656 | 0.4873 | 0.4982 | 0.4380 | 0.4285 | 0.4474 | 0.4720 | 0.6113 | 0.4870 | 0.4215 | 0.4767 | 0.3945 | 0.4584 |
| 17'SC | 0.5147 | 0.3346 | 0.4464 | 0.3637 | 0.5174 | 0.4587 | 0.3982 | 0.4443 | 0.3429 | 0.4546 | 0.3993 | 0.3630 | 0.6230 | 0.3265 | 0.3838 | 0.5090 | 0.3893 | 0.3870 |
| 17'BS | 0.5535 | 0.4179 | 0.4811 | 0.4274 | 0.5289 | 0.5364 | 0.4585 | 0.5515 | 0.3894 | 0.5132 | 0.4804 | 0.4277 | 0.6750 | 0.4395 | 0.4627 | 0.5422 | 0.4618 | 0.4104 |
| 17'GJ | 0.2570 | 0.1775 | 0.3932 | 0.1874 | 0.4730 | 0.4779 | 0.3466 | 0.4923 | 0.0413 | 0.4109 | 0.3746 | 0.2914 | 0.6128 | 0.2695 | 0.3881 | 0.4743 | 0.3875 | 0.1429 |
| 17'JE | 0.5528 | 0.3073 | 0.3220 | 0.3023 | 0.5127 | 0.4031 | 0.0499 | 0.3069 | 0.3693 | 0.4648 | 0.3589 | 0.1696 | 0.5906 | 0.2140 | 0.2874 | 0.5226 | 0.3958 | 0.4405 |
| 17'IS | 0.5543 | 0.3683 | 0.3920 | 0.3368 | 0.5073 | 0.4588 | 0.3790 | 0.4408 | 0.3433 | 0.4486 | 0.1003 | 0.3096 | 0.5353 | 0.3189 | 0.2951 | 0.5181 | 0.3958 | 0.4252 |
| 17'CY | 0.4606 | 0.2164 | 0.3974 | 0.1638 | 0.5385 | 0.4563 | 0.1890 | 0.3979 | 0.2645 | 0.4869 | 0.3560 | 0.1587 | 0.5969 | 0.0245 | 0.3400 | 0.5521 | 0.4166 | 0.3233 |
| 17'PT | 0.3562 | 0.2511 | 0.4435 | 0.3172 | 0.4814 | 0.4811 | 0.4080 | 0.4963 | 0.1767 | 0.4039 | 0.4019 | 0.3760 | 0.6363 | 0.3570 | 0.4040 | 0.4634 | 0.3905 | 0.2466 |
| 17'CC | 0.2495 | 0.1793 | 0.4270 | 0.2110 | 0.4729 | 0.4679 | 0.3580 | 0.4688 | 0.1806 | 0.4003 | 0.3679 | 0.3349 | 0.6273 | 0.2505 | 0.3824 | 0.4672 | 0.3999 | 0.1486 |
| 18'SC | 0.5227 | 0.3110 | 0.4460 | 0.3442 | 0.5241 | 0.4650 | 0.3734 | 0.4413 | 0.3369 | 0.4622 | 0.3888 | 0.3383 | 0.6383 | 0.3066 | 0.3724 | 0.5184 | 0.3861 | 0.3809 |
| 18'BS | 0.5666 | 0.4143 | 0.4869 | 0.4249 | 0.5462 | 0.5491 | 0.4596 | 0.5573 | 0.3838 | 0.5249 | 0.4759 | 0.4287 | 0.6976 | 0.4369 | 0.4611 | 0.5563 | 0.4671 | 0.4145 |
| 18'SJ | 0.5997 | 0.4883 | 0.4632 | 0.4744 | 0.4720 | 0.4572 | 0.4824 | 0.4931 | 0.4375 | 0.4195 | 0.4376 | 0.4663 | 0.6166 | 0.4884 | 0.4114 | 0.4694 | 0.3840 | 0.4664 |
| 18'PT | 0.5779 | 0.4134 | 0.3525 | 0.4479 | 0.2800 | 0.3409 | 0.3892 | 0.2336 | 0.3949 | 0.3150 | 0.3621 | 0.3687 | 0.5085 | 0.4221 | 0.2693 | 0.2308 | 0.0098 | 0.4621 |
| 18'JJ | 0.0441 | 0.2128 | 0.4835 | 0.2585 | 0.5312 | 0.5446 | 0.4286 | 0.5592 | 0.1658 | 0.4604 | 0.4280 | 0.3947 | 0.7042 | 0.3508 | 0.4610 | 0.5147 | 0.4497 | 0.0181 |
Significant values (p < 0.05) for pairwise FST are in bold.
Discussion
This study is the first comprehensive genetic structure analysis of B. tabaci MED (Q1) populations in Korea using eight microsatellite loci. The Korean populations of tomato B. tabaci MED appeared to be classified into two genetic clusters based on STRUCTURE and DAPC analyses, and their genetic structure converged rapidly into one genetic cluster. This phenomenon was reported previously by Dinsdale et al. [52] in Australia. They reported that the genetic cluster of B. tabaci rapidly changed even in a period of just four months. The results of this study and those by Dinsdale et al. [52], suggested that one out of the two B. tabaci MED genetic clusters in Korea might become the dominant species in the future.
This phenomenon could be caused by different fitness between the two B. tabaci MED genetic clusters in Korea. Although the two B. tabaci MED genetic clusters might have been mixed when they were first introduced in new areas, one genetic cluster would become dominant if there is fitness difference between them. Fitness difference between two genetic clusters could result from different susceptibilities to insecticides. The use of various insecticides, such as neonicotinoids, organophosphates, and carbamates, has been the main control method for B. tabaci MED in Korea. Extensive use of these insecticides has rapidly resulted in high levels of insecticide resistance in B. tabaci MED populations [53]. The two genetic clusters of B. tabaci MED might have different potentials for developing resistance to different insecticides. This differentiation was partially supported by changing the frequencies and diversity caused by chemical control [54, 55]. Results of the current study also showed low genotype frequencies and diversities, and limited founder or bottleneck effects.
However, the speed of this genetic cluster change in Korea could differ by areas. For example, the Jeju populations showed one genetic cluster of B. tabaci MED and this trend was maintained during the past three years. However, in the Pyeongtaek area, the genetic cluster of B. tabaci MED changed every year. The differences in the speed of genetic cluster change could be caused by human-related factors because B. tabaci has a low dispersal ability over long distances [56]. In the case of Jeju, the B. tabaci MED populations should not have been affected by other populations because almost all growers produce tomato seedlings themselves and Jeju is isolated because it is an island. On the other hand, the Pyeongtaek tomato growers have purchased tomato seedlings from different nurseries every year. Moreover, the city of Pyeongtaek has one of the most active agricultural trades of all Korean cities. Whitefly populations are generally affected by human activities, such as the movement of infested plants from nurseries, material shipments, and commercial trading, rather than by active flight [54, 57]. Thus, the populations in areas with high human activities and diverse nursery routes (i.e., the Pyeongtaek populations) might show accelerated genetic cluster changes compared to populations in isolated areas with limited nursery routes (i.e., the Jeju populations).
The information on the genetic characteristics of B. tabaci in areas where it usually occurs should be useful for efficient management of B. tabaci [58–60]. The genetic structure information gathered from the long-term and large-scale field analysis in this study facilitates a better understanding of the population dynamics of B. tabaci MED as an invasive pest in Korea. Thus, the results of this study could be a valuable foundation to develop efficient management strategies for B. tabaci MED in Korea. However, further studies are needed to clearly find the fitness differences between the two B. tabaci MED genetic clusters in Korea.
Supporting information
aNumber of alleles. bExpected heterozygosity. cObserved heterozygosity. dMean fixation index inbreeding coefficient. eAverage proportion of homozygous for null allele.
(XLSX)
Data Availability
The data is contained within the manuscript and Supporting Information files.
Funding Statement
This research was funded by grants from the Rural Development Administration (project no. PJ01194804) and by the Brain Korea 21 Plus project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Perring TM. The Bemisia tabaci species complex. Crop Protection. 2001;20(9):725–37. [Google Scholar]
- 2.Simón B, Cenis JL, Demichelis S, Rapisarda C, Caciagli P, Bosco D. Survey of Bemisia tabaci (Hemiptera: Aleyrodidae) biotypes in Italy with the description of a new biotype (T) from Euphorbia characias. Bull Entomol Res. 2003;93(3):259–64. 10.1079/BER2003233 [DOI] [PubMed] [Google Scholar]
- 3.Khan MS, Ji SH, Chun SC. Begomoviruses and Their Emerging Threats in South Korea: A Review. Plant Pathology Journal. 2012;28(2):123–36. [Google Scholar]
- 4.Li M, Hu J, Xu F-C, Liu S-S. Transmission of Tomato Yellow Leaf Curl Virus by two invasive biotypes and a Chinese indigenous biotype of the whitefly Bemisia tabaci. International Journal of Pest Management. 2010;56(3):275–80. [Google Scholar]
- 5.De Barro PJ, Liu SS, Boykin LM, Dinsdale AB. Bemisia tabaci: a statement of species status. Annu Rev Entomol. 2011;56:1–19. 10.1146/annurev-ento-112408-085504 [DOI] [PubMed] [Google Scholar]
- 6.Boykin LM, Armstrong KF, Kubatko L, De Barro P. Species delimitation and global biosecurity. Evolutionary Bioinformatics. 2012;8:EBO. S8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinsdale A, Cook L, Riginos C, Buckley YM, De Barro P. Refined Global Analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) Mitochondrial Cytochrome Oxidase I to Identify Species Level Genetic Boundaries. Annals of the Entomological Society of America. 2010;103(2):196–208. [Google Scholar]
- 8.Wang P, Ruan YM, Liu SS. Crossing experiments and behavioral observations reveal reproductive incompatibility among three putative species of the whitefly Bemisia tabaci. Insect Science. 2010;17(6):508–16. [Google Scholar]
- 9.Hu J, De Barro P, Zhao H, Wang J, Nardi F, Liu SS. An extensive field survey combined with a phylogenetic analysis reveals rapid and widespread invasion of two alien whiteflies in China. PLoS One. 2011;6(1):e16061 10.1371/journal.pone.0016061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee M-l, Ahn S-B, Cho W-S. Morphological Characteristics of Bemisia tabaci (Gennadius) (Homoptera: Aleyrodidae) and Discrimination of Their Biotypes in Korea by DNA Markers. Korean Journal of Applied Entomology 2000;39(1):5–12. [Google Scholar]
- 11.Lee M-H, Kang S-Y, Lee S-Y, Lee H-S, Choi J-Y, Lee G-S, et al. Occurrence of the B- and Q-biotypes of Bemisia tabaci in Korea. Korean Journal of Applied Entomology. 2005;44(3):169–75. [Google Scholar]
- 12.Lee W, Lee S-M, Kim C-S, Choi H-S, Akimoto S-I, Lee K-Y, et al. Three species of the Bemisia tabaci (Hemiptera: Aleyrodidae) complex in the Republic of Korea; detection by an extensive field survey combined with a phylogenetic analysis. Florida entomologist. 2014;97(1):155–61. [Google Scholar]
- 13.Lee W, Kim C-S, Lee K-Y, Lee G-S. The JpL species of the Bemisia tabaci complex in Korea: Detection by an extensive field survey and analysis of COI sequence variability. Journal of Asia-Pacific Entomology. 2016;19(1):23–9. [Google Scholar]
- 14.Kim H, Hoelmer KA, Lee S. Population genetics of the soybean aphid in North America and East Asia: test for introduction between native and introduced populations. Biological Invasions. 2016;19(2):597–614. [Google Scholar]
- 15.Kim KS, Cano-Ríos P, Sappington TW. Using genetic markers and population assignment techniques to infer origin of boll weevils (Coleoptera: Curculionidae) unexpectedly captured near an eradication zone in Mexico. Environmental entomology. 2006;35(4):813–26. [Google Scholar]
- 16.Streito JC, Clouet C, Hamdi F, Gauthier N. Population genetic structure of the biological control agent Macrolophus pygmaeus in Mediterranean agroecosystems. Insect science. 2017;24(5):859–76. 10.1111/1744-7917.12370 [DOI] [PubMed] [Google Scholar]
- 17.Hedrick PW. Conservation genetics: where are we now? Trends Ecol Evol. 2001;16(11):629–36. [Google Scholar]
- 18.Ben Abdelkrim A, Hattab T, Fakhfakh H, Belkadhi MS, Gorsane F. A landscape genetic analysis of important agricultural pest species in Tunisia: The whitefly Bemisia tabaci. PLoS One. 2017;12(10):e0185724 10.1371/journal.pone.0185724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sunnucks P. Efficient genetic markers for population biology. Trends Ecol Evol. 2000;15(5):199–203. [DOI] [PubMed] [Google Scholar]
- 20.Behura SK. Molecular marker systems in insects: current trends and future avenues. Molecular ecology. 2006;15(11):3087–113. 10.1111/j.1365-294X.2006.03014.x [DOI] [PubMed] [Google Scholar]
- 21.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5(6):435–45. 10.1038/nrg1348 [DOI] [PubMed] [Google Scholar]
- 22.Dalmon A, Halkett F, Granier M, Delatte H, Peterschmitt M. Genetic structure of the invasive pest Bemisia tabaci: evidence of limited but persistent genetic differentiation in glasshouse populations. Heredity (Edinb). 2008;100(3):316–25. [DOI] [PubMed] [Google Scholar]
- 23.Boopathi T, Mohankumar S, Karuppuchamy P, Kalyanasundaram M, Ravi M, Preetha B, et al. Genetic Evidence for Diversity of Spiralling Whitefly, Aleurodicus dispersus (Hemiptera: Aleyrodidae) Populations in India. Florida Entomologist. 2014;97(3):1115–22. [Google Scholar]
- 24.Oliveira EJ, Padua JG, Zucchi MI, Vencovsky R, Vieira MLC. Origin, evolution and genome distribution of microsatellites. Genetics and Molecular Biology. 2006;29(2):294–307. [Google Scholar]
- 25.Chu D, Gao CS, De Barro P, Wan FH, Zhang YJ. Investigation of the genetic diversity of an invasive whitefly (Bemisia tabaci) in China using both mitochondrial and nuclear DNA markers. Bull Entomol Res. 2011;101(4):467–75. 10.1017/S0007485311000022 [DOI] [PubMed] [Google Scholar]
- 26.Dickey AM, Osborne LS, Shatters RG, Hall PM, McKenzie CL. Population Genetics of Invasive Bemisia tabaci (Hemiptera: Aleyrodidae) Cryptic Species in the United States Based on Microsatellite Markers. Journal of Economic Entomology. 2013;106(3):1355–64. 10.1603/ec12512 [DOI] [PubMed] [Google Scholar]
- 27.Tahiri A, Halkett F, Granier M, Gueguen G, Peterschmitt M. Evidence of gene flow between sympatric populations of the Middle East-Asia Minor 1 and Mediterranean putative species of Bemisia tabaci. Ecology and Evolution. 2013;3(8):2619–33. [Google Scholar]
- 28.Díaz F, Endersby NM, Hoffmann AA. Genetic structure of the whitefly Bemisia tabaci populations in Colombia following a recent invasion. Insect Sci. 2015;22(4):483–94. 10.1111/1744-7917.12129 [DOI] [PubMed] [Google Scholar]
- 29.Mouton L, Gnankiné O, Henri H, Terraz G, Ketoh G, Martin T, et al. Detection of genetically isolated entities within the Mediterranean species of Bemisia tabaci: new insights into the systematics of this worldwide pest. Pest Manag Sci. 2015;71(3):452–8. 10.1002/ps.3834 [DOI] [PubMed] [Google Scholar]
- 30.Liu S-s, Colvin J, De Barro PJ. Species Concepts as Applied to the Whitefly Bemisia tabaci Systematics: How Many Species Are There? Journal of Integrative Agriculture. 2012;11(2):176–86. [Google Scholar]
- 31.Frohlich DR, Torres-Jerez I, Bedford ID, Markham PG, Brown JK. A phylogeographical analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Molecular Ecology. 1999;8(10):1683–91. [DOI] [PubMed] [Google Scholar]
- 32.Delatte H, Reynaud B, Granier M, Thornary L, Lett JM, Goldbach R, et al. A new silverleaf-inducing biotype Ms of Bemisia tabaci (Hemiptera: Aleyrodidae) indigenous to the islands of the south-west Indian Ocean. Bulletin of Entomological Research. 2005;95(1):29–35. [DOI] [PubMed] [Google Scholar]
- 33.Tsagkarakou A, Roditakis N. Isolation and characterization of microsatellite loci in Bemisia tabaci (Hemiptera: Aleyrodidae). Molecular Ecology Notes. 2003;3(2):196–8. [Google Scholar]
- 34.De Barro PJ, Scott KD, Graham GC, Lange CL, Schutze MK. Isolation and characterization of microsatellite loci in Bemisia tabaci. Molecular Ecology Notes. 2003;3(1):40–3. [Google Scholar]
- 35.Raymond M, Rousset F. GENEPOP (VERSION-1.2)—Population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86(3):248–9. [Google Scholar]
- 36.Brookfield JFY. A simple new method for estimating null allele frequency from heterozygote deficiency. Molecular Ecology. 1996;5(3):453–5. [DOI] [PubMed] [Google Scholar]
- 37.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4(3):535–8. [Google Scholar]
- 38.Chapuis MP, Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol. 2007;24(3):621–31. 10.1093/molbev/msl191 [DOI] [PubMed] [Google Scholar]
- 39.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society: Series B (Methodological). 1977;39(1):1–22. [Google Scholar]
- 40.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28(19):2537–9. 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park S. The Excel microsatellite toolkit. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. 2001.
- 42.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67(1):170–81. 10.1086/302959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14(8):2611–20. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- 44.Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2011;4(2):359–61. [Google Scholar]
- 45.Jombart T, Ahmed I. adegenet 1.3–1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27(21):3070–1. 10.1093/bioinformatics/btr521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. Bmc Genetics. 2010;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer research. 1967;27(2 Part 1):209–20. [PubMed] [Google Scholar]
- 48.Chessel D, Dufour AB, Thioulouse J. The ade4 package-I-One-table methods. R news. 2004;4(1):5–10. [Google Scholar]
- 49.Piry S, Luikart G, Cornuet JM. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity. 1999;90(4):502–3. [Google Scholar]
- 50.Deperi SI, Tagliotti ME, Bedogni MC, Manrique-Carpintero NC, Coombs J, Zhang R, et al. Discriminant analysis of principal components and pedigree assessment of genetic diversity and population structure in a tetraploid potato panel using SNPs. PloS one. 2018;13(3):e0194398 10.1371/journal.pone.0194398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wright S. The genetical structure of populations. Annals of eugenics. 1949;15(1):323–54. [DOI] [PubMed] [Google Scholar]
- 52.Dinsdale A, Schellhorn NA, De Barro P, Buckley YM, Riginos C. Rapid genetic turnover in populations of the insect pest Bemisia tabaci Middle East: Asia Minor 1 in an agricultural landscape. Bull Entomol Res. 2012;102(5):539–49. 10.1017/S0007485312000077 [DOI] [PubMed] [Google Scholar]
- 53.Lee Y-S, Lee S-Y, Park E-C, Kim J-H, Kim G-H. Comparative toxicities of pyriproxyfen and thiamethoxam against the sweetpotato whitefly, Bemisia tabaci (Homoptera: Aleyrodidae). Journal of Asia-Pacific Entomology. 2002;5(1):117–22. [Google Scholar]
- 54.Chu D, Guo D, Tao Y, Jiang D, Li J, Zhang Y. Evidence for rapid spatiotemporal changes in genetic structure of an alien whitefly during initial invasion. Scientific reports. 2014;4:4396 10.1038/srep04396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gauthier N, Clouet C, Perrakis A, Kapantaidaki D, Peterschmitt M, Tsagkarakou A. Genetic structure of Bemisia tabaci Med populations from home-range countries, inferred by nuclear and cytoplasmic markers: impact on the distribution of the insecticide resistance genes. Pest management science. 2014;70(10):1477–91. 10.1002/ps.3733 [DOI] [PubMed] [Google Scholar]
- 56.Byrne DN. Migration and dispersal by the sweet potato whitefly, Bemisia tabaci. Agricultural and forest meteorology. 1999;97(4):309–16. [Google Scholar]
- 57.Hadjistylli M, Roderick GK, Brown JK. Global population structure of a worldwide pest and virus vector: Genetic diversity and population history of the Bemisia tabaci sibling species group. PloS one. 2016;11(11):e0165105 10.1371/journal.pone.0165105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karsten M, van Vuuren BJ, Barnaud A, Terblanche JS. Population genetics of Ceratitis capitata in South Africa: implications for dispersal and pest management. PLoS One. 2013;8(1):e54281 10.1371/journal.pone.0054281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cullingham CI, Roe AD, Sperling FA, Coltman DW. Phylogeographic insights into an irruptive pest outbreak. Ecology and evolution. 2012;2(5):908–19. 10.1002/ece3.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rollins LA, Woolnough AP, Sherwin WB. Population genetic tools for pest management: a review. Wildlife Research. 2006;33(4):251–61. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
aNumber of alleles. bExpected heterozygosity. cObserved heterozygosity. dMean fixation index inbreeding coefficient. eAverage proportion of homozygous for null allele.
(XLSX)
Data Availability Statement
The data is contained within the manuscript and Supporting Information files.