Abstract
Background and Objectives
Human amniotic fluid-derived mesenchymal stem cells (AF-MSCs) may be a valuable source for cardiovascular tissue engineering and cell therapy. The aim of this study is to verify angiotensin II and transforming growth factor-beta 1 (TGF-β1) as potential cardiomyogenic differentiation inducers of AF-MSCs.
Methods and Results
AF-MSCs were obtained from amniocentesis samples from second-trimester pregnant women, isolated and characterized by the expression of cell surface markers (CD44, CD90, CD105 positive; CD34 negative) and pluripotency genes (OCT4, SOX2, NANOG, REX1). Cardiomyogenic differentiation was induced using different concentrations of angiotensin II and TGF-β1. Successful initiation of differentiation was confirmed by alterations in cell morphology, upregulation of cardiac genes-markers NKX2-5, TBX5, GATA4, MYH6, TNNT2, DES and main cardiac ion channels genes (sodium, calcium, potassium) as determined by RT-qPCR. Western blot and immunofluorescence analysis revealed the increased expression of Connexin43, the main component of gap junctions, and Nkx2.5, the early cardiac transcription factor. Induced AF-MSCs switched their phenotype towards more energetic and started utilizing oxidative phosphorylation more than glycolysis for energy production as assessed using Agilent Seahorse XF analyzer. The immune analysis of chromatin-modifying enzymes DNMT1, HDAC1/2 and Polycomb repressive complex 1 and 2 (PRC1/2) proteins BMI1, EZH2 and SUZ12 as well as of modified histones H3 and H4 indicated global chromatin remodeling during the induced differentiation.
Conclusions
Angiotensin II and TGF-β1 are efficient cardiomyogenic inducers of human AF-MSCs; they initiate alterations at the gene and protein expression, metabolic and epigenetic levels in stem cells leading towards cardiomyocyte-like phenotype formation.
Keywords: Myocytes, Cardiac, Amniotic fluid, Cell differentiation, Chromatin, Histones
Introduction
Cardiovascular diseases are among the leading causes of morbidity and mortality in the whole world nowadays. Myocardial infarction when the heart muscle tissue is damaged due to the impaired supply of blood and oxygen is one of them. The traditional treatment (pharmacological, interventional or operative therapies) is very expensive and usually ineffective, mostly because of the irreversible loss of functional cardiomyocytes and their very low regenerative capability. Thus cardiovascular tissue engineering and cell therapy for cardiac diseases have been extensively studied in recent years. Many different types of stem cells, from embryonic to bone marrow or induced pluripotent (1), were tested for this application and one of the alternative sources of stem cells could be human amniotic fluid-derived mesenchymal stem cells (AF-MSCs). These stem cells are obtained from pregnant women during amniocentesis procedure and their main features are the expression of mesenchymal cell surface markers and the ability to differentiate into osteogenic, adipogenic and other lineages (2). AF-MSCs were also demonstrated to have the potential to differentiate into cardiomyocyte-like cells using DNA methyltransferases or p53 inhibitors, such as decitabine, zebularine, pifithrin-α (3, 4) or other chemical agents, for example, hyaluronic, butyric or retinoic acid (5).
However, little is known on the use of biomolecules, such as angiotensin II and transforming growth factor-β1 (TGF-β1), as cardiomyogenic differentiation inducers of human AF-MSCs and underlying mechanisms. Angiotensin II is one of the components of the renin-angiotensin system and this signaling pathway plays a pivotal role in stem cells and cardiovascular system (6). It was also shown that angiotensin II is able to induce cardiomyogenic differentiation of rat bone marrow mesenchymal stem cells (7) as well as to act synergistically with VEGF-A to enhance the differentiation of bone marrow-derived mesenchymal stem cells into endothelial cells (8). TGF-β1 is a member of cytokines family that participates in many biological processes including cardiovascular development and regulation of renal renin-angiotensin system (9). It has also been assessed in the induction of cardiac commitment, but not the full maturation, of bone marrow stem cells (10). What is important, in vitro it was as effective as widely used chemical compound 5-azacytidine and even more effective than other biomolecules, for example, IGF-1, bFGF, dynorphin B, insulin or oxytocin (11). In contrast to angiotensin II, TGF-β1 was also investigated in vivo by intramyocardial transplantation of differentiated rat bone marrow mesenchymal stem cells into the injured rat heart (12). Also, it was shown that TGF-β1 had an effect on the contractile function of adult ventricular rat cardiomyocytes in vitro and in vivo by modulating laminin receptor 37/67 dependent regulation of cardiac performance signaling pathway (13). However, to our best knowledge, most of the studies investigated the effects of these biomolecules at the gene and protein levels but few looked into other pivotal processes, such as metabolic or epigenetic alterations.
In this study, the potential of angiotensin II and TGF-β1 to induce cardiomyogenic differentiation of human AF-MSCs was evaluated in vitro. We detected phenotypical alterations and upregulation of specific cardiac genes-markers (NKX2-5, TBX5, GATA4, MYH6, TNNT2 and DES) as well as cardiac ion channels genes (SCN5A, CACNA1D, KCNJ12, KCND3, HCN2). In addition, enhanced levels of Connexin43 protein, the main component of gap junctions, and Nkx2.5, the early cardiac transcription factor, were observed indicating successful initiation of differentiation with both agents. Moreover, for the first time, cellular energetics, metabolic potential and the relative expression of genes, associated with metabolism regulation, were assessed demonstrating functional changes in induced to differentiate cells. Also, epigenetic markers, such as chromatin remodeling proteins and histone modifications keeping the active state of chromatin, were investigated. Thus the present work extends the potential use of angiotensin II and TGF-β1 as cardiomyogenic differentiation inducers in vitro and expands the knowledge of human amniotic fluid-derived mesenchymal stem cells functionality at the initial stages of induced differentiation.
Materials and Methods
Human amniotic fluid mesenchymal stem cells isolation and cultivation
Amniotic fluid mesenchymal stem cells were isolated from amniocentesis samples from second-trimester amniotic fluid obtained from healthy women who needed prenatal diagnostics but no genetic abnormalities were detected (protocols approved by the Ethics Committee of Biomedical Research of Vilnius District, No 158200-123-428-122). A two-stage protocol was used as previously described (2). Selected and isolated cells were maintained in the growth medium AmnioMAX-C100 basal with Amnio-MAX-C100 supplement (Gibco, Thermo Fisher Scientific, NY, USA), 100 U/ml penicillin and 100 μg/ml streptomycin.
Flow cytometry analysis
Cell surface markers of AF-MSCs were detected as described by Glemžaitė and Navakauskienė (14) using the following antibodies: CD44 (Invitrogen, Thermo Fisher Scientific, CA, USA), CD34 (Miltenyi Biotec, Bergisch Gladbach, Germany) and CD90 (Molecular Probes, Thermo Fisher Scientific, OR, USA) – FITC conjugated mouse anti-human, mouse anti-human CD105, labeled with PE (Invitrogen, Thermo Fisher Scientific, CA, USA) and mouse IgG2A-FITC (Miltenyi Biotec, Bergisch Gladbach, Germany), IgG1-FITC, IgG2b-FITC (Invitrogen, Thermo Fisher Scientific, CA, USA) or IgG1-PE (Molecular Probes, Thermo Fisher Scientific, OR, USA) as isotype controls. The expression of cell surface markers was measured using BD FACSCantoTM II flow cytometer and BD FACSDIVATM software (BD Biosciences, CA, USA).
Differentiation assay
Cardiomyogenic differentiation of AF-MSCs was induced using decitabine (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany), two concentrations of angiotensin II (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany) and two concentrations of TGF-β1 (R&D Systems, MN, USA). Differentiation conditions are provided in Table 1. Differentiation time – 12 days, chosen as the minimum time required to achieve the evidence of initiated differentiation according to our previous publication (4). Differentiation of each cell population was performed in 3 replicates and undifferentiated cells were used as a control.
Table 1.
Differentiation inducers and conditions
| Inducer | Working concentration | Induction time | Differentiation medium | Reference for the applied concentration |
|---|---|---|---|---|
| Decitabine | 10 μM | 24 h, then medium without inducer every 3 days | BMa+horse serum | 3, 4 |
| Angiotensin II | 0.1 μM | The whole differentiation period, every 3 days | BM | 7 |
| Angiotensin II | 1 μM | The whole differentiation period, every 3 days | BM | 19 |
| TGF-β1 | 5 ng/ml | The whole differentiation period, every 3 days | BM | 10, 13 |
| TGF-β1 | 10 ng/ml | The whole differentiation period, every 3 days | BM | 11 |
BMa: basal differentiation medium; DMEM-low glucose, 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco, Thermo Fisher Scientific, NY, USA).
RNA isolation and RT-qPCR
Total RNA from undifferentiated and differentiated cells was isolated using TRIzol® reagent (Thermo Fisher Scientific, CA, USA). For the gene expression analysis, cDNA synthesis was carried out using “Maxima First Strand cDNA Synthesis Kit for RT-qPCR” (Thermo Fisher Scientific, Vilnius, Lithuania). RT-qPCR was performed with Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific, Vilnius, Lithuania) and Rotor-Gene 6000 system (QIAGEN Instruments AG, Hombrechtikon, Switzerland). GAPDH gene was used for normalization of the mRNA amount and ΔΔCt method (compared to undifferentiated control) was applied to calculate the relative gene expression. The list of primers (Metabion International AG, Planegg-Steinkirchen, Germany) is provided in the Supplementary Methods, Supplementary Table 1.
Total protein isolation and Western blot analysis
Total proteins from AF-MSCs were isolated as described previously (14). Adult mouse heart was washed with PBS, homogenized in liquid nitrogen and lysed using 2X SDS lysis buffer. Proteins from cells and mouse heart were fractionated in 7.5~15% gradient SDS/PAGE gel, transferred onto the PVDF membrane and target proteins were detected using antibodies against DNMT1, HDAC1, HDAC2 (Santa Cruz Biotechnology, Texas, USA), SUZ12 (Cell Signaling Technology, MA, USA), BMI1, H3K4me3, H3K9Ac, H4hyperAc (Millipore, MA, USA), EZH2, Connexin43 (Thermo Scientific, IL, USA) and Nkx2.5 (GeneTex, CA, USA). Antibodies against GAPDH (Abcam, Cambridge, UK) were used as a protein loading control. Secondary anti-mouse, anti-rabbit and anti-goat antibodies (DAKO, Glostrup, Denmark), conjugated with horseradish peroxidase, were used and enhanced chemiluminescence was detected using ClarityTM Western ECL Substrate and ChemiDocTM XRS+ system with Image LabTM Software (Bio-Rad Laboratories, CA, USA). Semiquantitative analysis of blots was performed using Image J software (NIH, MD, USA), the relative density of each band was normalized to the GAPDH loading control.
Immunofluorescence
AF-MSCs were seeded on the coverslips, fixed using 4% paraformaldehyde solution in 1X PBS for 15 min RT, washed with 1X PBS and permeabilized using 10% Triton X-100/PBS for 20 min RT. After washing with PBS, cells were blocked with 1% BSA/10% goat serum/PBS for 30 min at 37°C. Detection of Connexin43: the coverslips were incubated with primary rabbit against Connexin43 and secondary goat anti-rabbit IgG (H+L) Highly Cross-Adsorbed, Alexa Fluor-488 antibodies (Thermo Fisher Scientific, IL, USA) for 1 hour at 37°C each, in a humid chamber. F-actin was labeled with Alexa Fluor-594 Phalloidin (Thermo Fisher Scientific, IL, USA) for 30 min RT in a humid chamber. Detection of Nkx2.5: the coverslips were incubated with primary rabbit against Nkx2.5 and secondary goat anti-rabbit IgG (H+L) Highly Cross-Adsorbed, Alexa Fluor-594 antibodies (Thermo Fisher Scientific, IL, USA) for 1 hour at 37°C each, in a humid chamber. F-actin was labeled with Alexa Fluor-488 Phalloidin (Thermo Fisher Scientific, IL, USA) for 30 min RT in a humid chamber. After each step coverslips were washed with 1% BSA in PBS. 300 nM DAPI (Invitrogen, OR, USA) was used for staining the nuclei, 10 min RT. Coverslips were mounted using Dako Fluorescent Mounting Medium (Agilent Technologies, CA, USA) and labeled samples were analyzed using Zeiss Axio Observer (Oberkochen, Germany) fluorescent microscope, the 63 X objective with immersion oil and Zen BLUE software.
Quantitative evaluation of Nkx2.5 nuclear localization
Custom image processing and analysis tools were employed to perform a quantitative evaluation of the Alexa Fluor-594-labelled nuclear-localized Nkx2.5 protein. Nuclear quantity (NQ) of Nkx2.5 was computed as a number of dots detected in the nucleus region of the immunofluorescent image. Required tools have been implemented in MatlabTM environment (The MathWorks, Natick, MA, USA). To obtain NQs, it was necessary to segment DAPI images (find borders of the nuclei) and sum up all the dots detected in Alexa Fluor-594 image that belong to the nucleus region. Image segmentation to acquire nuclei regions included image manipulation steps like Gaussian image smoothing for noise reduction and thresholding. Other minor processing steps were used to ensure the solidity of nuclei regions. Symmetrical feature detection and Laplacian of Gaussian (LoG) filtering were performed to enhance visible dots in Alexa Fluor-594 image. Symmetrical feature detector generates a map of second order symmetries using Johansson method (15), and LoG filter rejects non-spot (concave) symmetrical areas. Detection of local maxima of dots produces a binary image with marked centers of fluorescent dots. Previously detected nuclei areas in DAPI images were used as regions of interest (ROIs) for calculations of the total fluorescent dots count, expressed as NQ:
where (x, y) represents pixel coordinates, and is a binary image, which contains only centers of fluorescent dots marked as single white pixels.
Wilcoxon rank sum test was used to test the hypothesis on the equality of medians of two samples. Statistical significance of changes between samples was presented over Tukey-style box plots.
Extracellular flux analysis
Energetic profile of differentiated and control cells was determined using Seahorse XFp Extracellular Flux Analyzer and Cell Energy Phenotype Test Kit (Agilent Technologies, CA, USA). Mitochondrial respiration was measured using Cell Mito Stress Test Kit. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured simultaneously, firstly without inhibitors of the electron transfer chain (oligomycin, FCCP, rotenone and antimycin A) – the baseline, and then after the addition of the above-mentioned inhibitors. After the measurements, cells were collected and lysed using RIPA buffer (150 mM NaCl, 10 mM EDTA, pH 8.0, 10 mM Tris, pH 7.4, 0.1% SDS, 1% deoxycholate, 1% NP-40 in PBS, pH 7.6). Total protein concentrations were measured using DC Protein Assay (BioRad Laboratories, CA, USA) and spectrophotometer Infinite M200 Pro (Tecan, Switzerland). In all calculations, OCR and ECAR values were normalized to the total amount of protein in each well and expressed per μg of protein. Cell energy phenotype, OCR/ECAR ratio, determined from the normalized OCR and normalized ECAR, and the metabolic potential, calculated as the percentage increase of stressed OCR over baseline OCR and stressed ECAR over baseline ECAR, were evaluated from the Cell Energy Phenotype Test data. Cell Mito Stress Test was used to obtain maximal respiration (the difference between OCR after FCCP injection and non-mitochondrial OCR, expressed as fold induction over baseline), spare respiratory capacity (the ratio between maximal respiration and basal respiration, where basal respiration reflects only mitochondrial respiration, expressed in percent) and ATP production (OCR difference before and after the addition of oligomycin). More details are provided in the Supplementary Methods.
Statistical analysis
All experiments were performed in triplicates, data were expressed as mean values with SDs. One-way ANOVA with Tukey post-hoc test in GraphPad Prism software (CA, USA) was used for statistical analysis.
Results
Characterization of human AF-MSCs
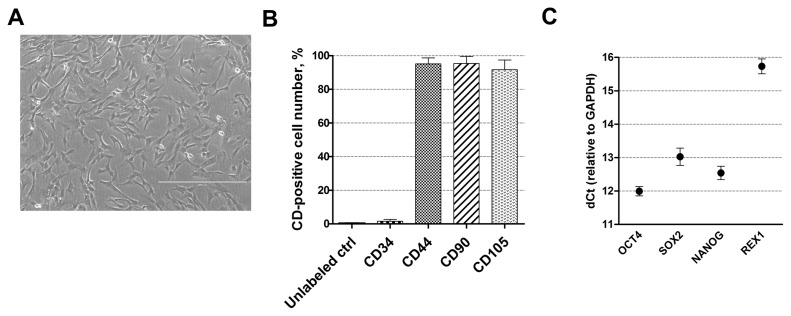
For cardiomyogenic differentiation induction human amniotic fluid-derived mesenchymal stem cells, obtained from the amniotic fluid of the second-trimester pregnancy, were used. These stem cells have typical spindle-shaped morphology (Fig. 1A) and express high levels (over 95%) of mesenchymal cell surface markers, such as CD44 (cell adhesion molecule), CD90 (Thy-1, thymocyte antigen-1) and CD105 (endoglin) but do not express (less than 1%) hematopoietic cells marker CD34 (Fig. 1B) as detected by flow cytometry. They are also positive for pluripotency genes-markers, such as OCT4, SOX2, NANOG, and REX1 (Fig. 1C) as determined using RT-qPCR.
Fig. 1.
Characterization of human AF-MSCs. (A) The typical spindle-shaped morphology of human amniotic fluid-derived mesenchymal stem cells, cultivated in cell culture. Scale bar=400 μm. (B) The expression of cell surface markers CD44, CD90, CD105 and CD34 as detected by flow cytometry. Unlabeled ctrl–non-labeled, undifferentiated control cells. Results are presented as mean±SD (n=3). (C) The relative expression of pluripotency genes-markers, such as OCT4, SOX2, NANOG and REX1 as determined by RT-qPCR. Data, relative to GAPDH, are presented as mean±SD (n=3).
Assessment of alterations during the induced cardiomyogenic differentiation
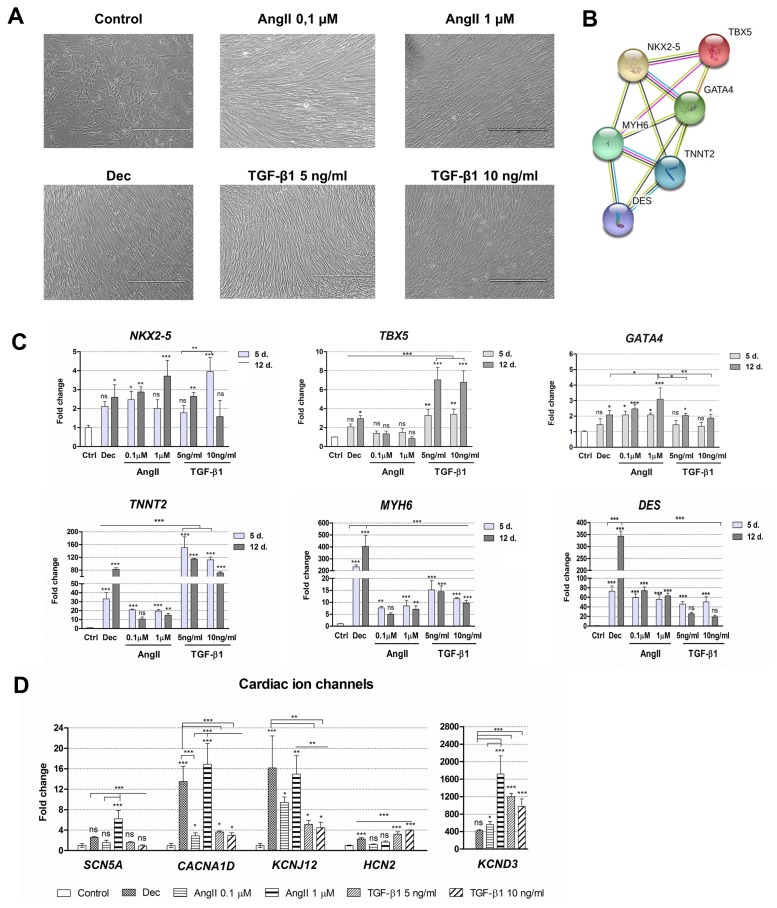
Cardiomyogenic differentiation was induced with two concentrations of angiotensin II and two concentrations of TGF-β1. Decitabine was used as a positive control because it was previously demonstrated as the potential differentiation inducer (4). All these agents caused morphological alterations in induced cells (Fig. 2A), cells became elongated and grew in a tight fibrous monolayer forming junctions between adjacent cells. No differences in morphology were detected between different concentrations of angiotensin II and TGF-β1. Then, induced differentiation was evaluated at the gene expression level. Firstly, the expression of three early transcription factors (TF) – NKX2-5, TBX5 and GATA4 – regulating the expression of structural and functional genes of cardiomyocytes was assessed. As shown in Fig. 2B and Supplementary tables, obtained from STRING database, NKX2-5 interacts with other TFs TBX5 and GATA4 forming the network of transcription factors that cooperate with MYH6 (α-myosin heavy chain), TNNT2 (cardiac Troponin T) and DES (Desmin) in the early cardiac phenotype induction, heart morphogenesis and development. The results revealed that the relative gene expression of NKX2-5 as well as of GATA4 was upregulated in AF-MSCs induced with all agents while the expression of TBX5 increased significantly only in decitabine and TGF-β1 differentiated cells (Fig. 2C). Angiotensin II caused a very slight upregulation of TBX5. Next, the relative expression of several structural genes of cardiomyocytes considered as cardiac genes-markers, i.e. TNNT2, MYH6 and DES was determined. These genes were strongly upregulated after the initiation of differentiation with all agents (Fig. 2C). Decitabine caused the highest upregulation of MYH6 and DES while TNNT2 increased the most in TGF-β1 treated cells. Both concentrations of angiotensin II and TGF-β1 increased the expression of MYH6 and DES to the comparable level. Furthermore, several cardiac ion channels were observed at the gene expression level: SCN5A – sodium voltage-gated channel α-subunit 5, CACNA1D – L-type calcium channel, KCNJ12 – the ATP-sensitive inward rectifier potassium channel, KCND3 – the transient outward potassium channel and HCN2 – the hyperpolarization-activated cyclic nucleotide-gated channel (Fig. 2D). The data indicated that decitabine, angiotensin II and TGF-β1 upregulated the expression of assessed ion channels genes to a different extent: angiotensin II (especially 1 μM concentration) and decitabine caused the highest increase in the expression while TGF-β1 had a smaller effect (5 ng/ml of TGF-β1 was slightly more effective than 10 ng/ml).
Fig. 2.
Alterations in morphology and gene expression patterns during decitabine, angiotensin II and TGF-β1 induced cardiac differentiation. (A) Assessment of the cell morphology 12 days after the cardiac differentiation initiation. Dec: decitabine, AngII: angiotensin II, TGF-β1: transforming growth factor-beta 1. Scale bar=400 μm. (B) The connections between proteins that are considered as cardiomyogenic differentiation markers. NKX2-5, TBX5 and GATA4 are the early transcription markers, initiating the expression of structural proteins of cardiomyocytes, such as MYH6 (α-myosin heavy chain), TNNT2 (cardiac troponin T) and DES (Desmin). The data was obtained from the STRING database. (C) The relative expression of the main cardiac : genes-markers, such as NKX2-5, TBX5, GATA4, MYH6, TNNT2 and DES at the days 5 (5 d.) and 12 (12 d.) of induced differentiation. Ctrl–non-differentiated control cells. (D) The relative expression of cardiac ion channels genes at the day 12 of differentiation: SCN5A (sodium voltage-gated channel α-subunit 5), CACNA1D (L-type calcium channel), KCNJ12 and KCND3 (voltage-gated potassium) and HCN2 (hyperpolarization-activated cyclic nucleotide-gated channel). The relative gene expression was determined by RT-qPCR, normalized to GAPDH and presented as a fold change over undifferentiated control. The data are presented as mean±SD (n=3), p≤0.05 (*), p≤0.01 (**), p≤0.001 (***), ns: non-significant.
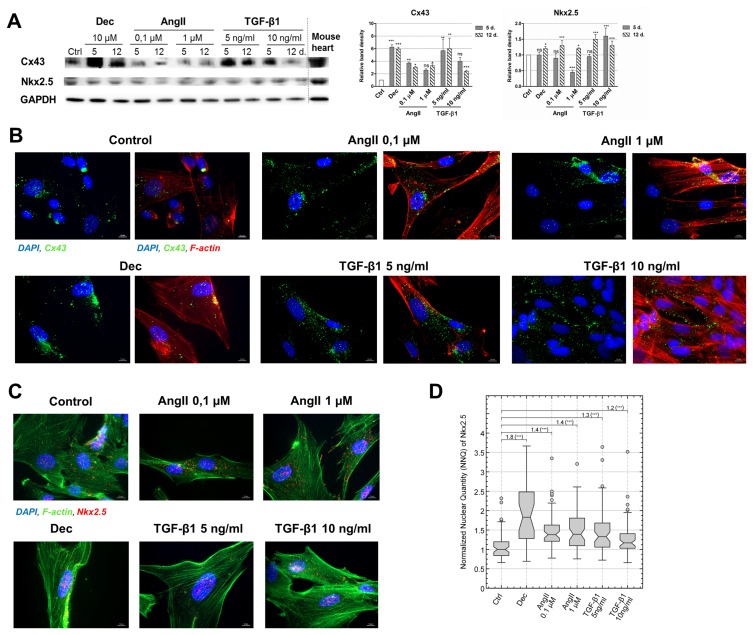
In addition to the alterations at the gene expression level, two proteins, vital for the development of the cardiomyocyte-like phenotype were investigated, i.e. Connexin43 (Cx43) – the main component of gap junctions and Nkx2.5 – the early cardiac transcription factor. As the Western blot data uncovered, the levels of Cx43 increased in all induced AF-MSCs, especially in decitabine and 5 ng/ml TGF-β1 treated cells (Fig. 3A). Concomitant with that very high levels of Cx43 were detected in the adult mouse heart lysate. Immunofluorescence data complemented Western blot results and revealed that Cx43 started to migrate towards the boundaries of the cells and accumulate at the peripheral of the differentiated cells where it could form functional gap junctions channels (Fig. 3B). The levels of Nkx2.5 also increased in all induced to differentiation stem cells, to the rather similar extent, however, the upregulation was slightly lesser than Cx43 (Fig. 3A). High levels of Nkx2.5 in the mouse heart revealed its importance for the cardiac tissue. Immunofluorescence of Nkx2.5 supplemented Western blot data and exhibited that it was mainly located in the nuclei (Fig. 3C). Concomitant with that, quantitative evaluation of Nkx2.5 localization in the nuclei from the immunofluorescence images demonstrated higher nuclear levels of this cardiac transcription factor in angiotensin II and TGF-β1 treated AF-MSCs compared to undifferentiated control (Fig. 3D).
Fig. 3.
Evaluation of Connexin43 (Cx43) and Nkx2.5 levels in induced AF-MSCs. (A) The levels of Cx43, the main gap junction protein, and Nkx2.5, an early cardiac transcription factor, as determined by Western blot in non-differentiated cells (Ctrl) and differentiated cells at the days 5 and 12. As a positive control mouse heart lysate was used. The relative density of each band was measured using ImageJ software (NIH, USA), normalized to the GAPDH loading control and presented as a fold difference over control. The data are presented as mean±SD (n=3), p≤0.05 (*), p≤0.01 (**), p≤0.001 (***), ns: non-significant. The blots represent one of three independent experiments showing similar results. (B) The distribution of Cx43 in control and induced cells at the 12 day of differentiation as obtained using the immunofluorescence assay. The data from each differentiation sample is presented in two images: the left panel–two channels view (blue nuclei and green Cx43) and the right panel–the same image in three channels view (blue nuclei, green Cx43 and red F-actin). Samples were observed using Zeiss Axio Observer fluorescence microscope, 63X magnification with immersion oil, scale bar=10 μm. (C) The immunofluorescence data of Nkx2.5 protein in control and induced cells at the 12 day of differentiation. Nuclei stained blue, Nkx2.5 – red and F-actin – green. Samples were observed using Zeiss Axio Observer fluorescence microscope, 63X magnification with immersion oil, scale bar=10 μm. (D) Quantitative evaluation of Nkx2.5 nuclear localization at the day 12 of differentiation as calculated from the Nkx2.5 immunofluorescence images. The nuclear quantity of Nkx2.5 was normalized to the median value of undifferentiated Control (Ctrl) and presented as normalized nuclear quantity (NNQ). All NNQs were graphed as Tukey-style box plots with sample size n=66, 31, 57, 58, 64 and 60. Relative change of medians and the statistical significance of changes are presented above the box plots. Wilcoxon rank sum test was used for statistical analysis, p≤0.001 (***).
Evaluation of metabolic changes during the induced differentiation
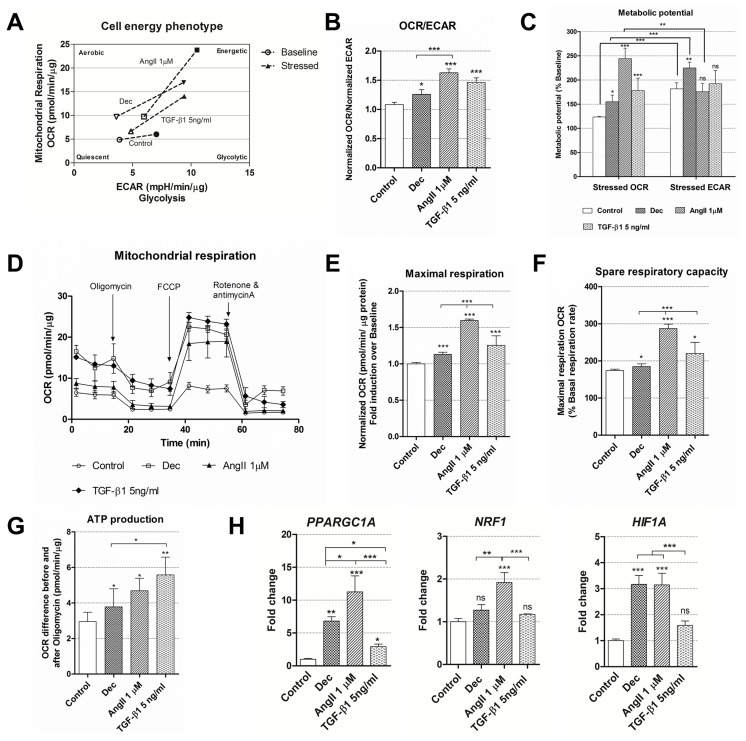
After the initiation of differentiation was confirmed at the gene and protein expression level, cellular energetics and mitochondrial function were observed using Seahorse extracellular flux analyzer. For the assessment of these parameters, cells were induced using decitabine, 1 μM of angiotensin II and 5 ng/ml of TGF-β1 as these differentiation conditions caused the highest upregulation rate of specific genes and proteins (Fig. 2 and 3). Firstly, the rates of oxygen consumption (OCR) and extracellular acidification (ECAR) were measured simultaneously under basal and induced stressed conditions and the switch in cell energy phenotype towards more energetic was obtained in all induced cells while undifferentiated control cells remained almost at the same level (Fig. 4A). In addition, OCR/ECAR ratio at the basal conditions was higher in the differentiated cells compared to control cells (Fig. 4B) indicating that treated cells started using Oxidative phosphorylation more than glycolysis for the energy production. Under induced stress conditions, mimicking the maximal energy demand for the cell, angiotensin II-treated cells had significantly higher OCR compared to untreated cells and it was higher than ECAR at the same conditions (Fig. 4C), meaning that these AF-MSCs relied more on the mitochondrial respiration. On the other hand, decitabine differentiated cells possessed higher stressed ECAR than OCR as well as undifferentiated cells suggesting that they may utilize glycolysis more. Interestingly, TGF-β1 treated cells had higher stressed OCR than control cells but the difference between OCR and ECAR under induced stress conditions was insignificant demonstrating the use of both energetic pathways. In association with the upregulated oxidative phosphorylation, mitochondrial respiration was analyzed in control and cells, induced to differentiation, measuring OCR after the addition of electron transfer chain inhibitors (Fig. 4D). It revealed that maximal respiration and its derivative spare respiratory capacity, indicating the capability of the cell to respond to an energetic demand, were increased in AF-MSCs differentiated with all agents, especially in angiotensin II-treated cells (Fig. 4E, 4F) in correlation with cell energy phenotype data. ATP production also increased in differentiated cells (Fig. 4G). Changes in cell metabolism induced changes at the transcriptional level too and the relative expression of several genes, related to metabolism, was measured (Fig. 4H). Angiotensin II upregulated the expression of PPARGC1A (peroxisome proliferator-activated receptor gamma coactivator 1 alpha), NRF1 (nuclear respiratory factor 1) and HIF1A (hypoxia-inducible factor 1-alpha) the most, decitabine – to the less extent and TGF-β1 only caused a significant upregulation of PPARGC1A.
Fig. 4.
Cell energy phenotype and metabolic alterations during cardiac differentiation of AF-MSCs. (A) Cell energy diagram showing normalized oxygen consumption rate (OCR) and normalized extracellular acidification rate (ECAR) of control and differentiated cells at the basal (the hollow points) and stressed conditions (colored points). (B) The ratio of normalized OCR to normalized ECAR in control and induced AF-MSCs. (C) Metabolic potential (in percent) of undifferentiated and induced AF-MSCs was calculated from stressed OCR/ECAR over Baseline OCR/ECAR. (D) Mitochondrial respiration expressed as normalized OCR of control and differentiated cells after the addition of ETC inhibitors oligomycin, FCCP, rotenone and antimycin A. The time in the x-axis represents time points when each measurement was done. All calculations of mitochondrial function were done referring to these measurements. (E) Maximal respiration, calculated as normalized OCR fold induction over baseline. (F) Spare respiratory capacity, a derivative from Maximal respiration, was expressed in percent for the control and induced cells. (G) ATP production was calculated as the OCR difference between the baseline and after the addition of oligomycin. (H) The relative expression of genes, related to the cell metabolism and respiration: PPARGC1A (Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 Alpha), NRF1 (nuclear respiratory factor 1) and HIF1A (hypoxia-inducible factor 1-alpha), in control and differentiated cells as determined using RT-qPCR. The data were normalized to GAPDH and presented as a fold change over undifferentiated control. The data in A~G was obtained using Seahorse XFp Extracellular Flux Analyzer (Agilent, USA), “Cell energy phenotype test” (parts A~C) or “Cell Mito Stress Test” kits (parts D~G) and cells differentiated for 12 days. Results are presented as mean±SD. p≤0.05 (*), p≤0.01 (**), p≤0.001 (***), ns: non-significant.
Analysis of epigenetic changes during induced differentiation
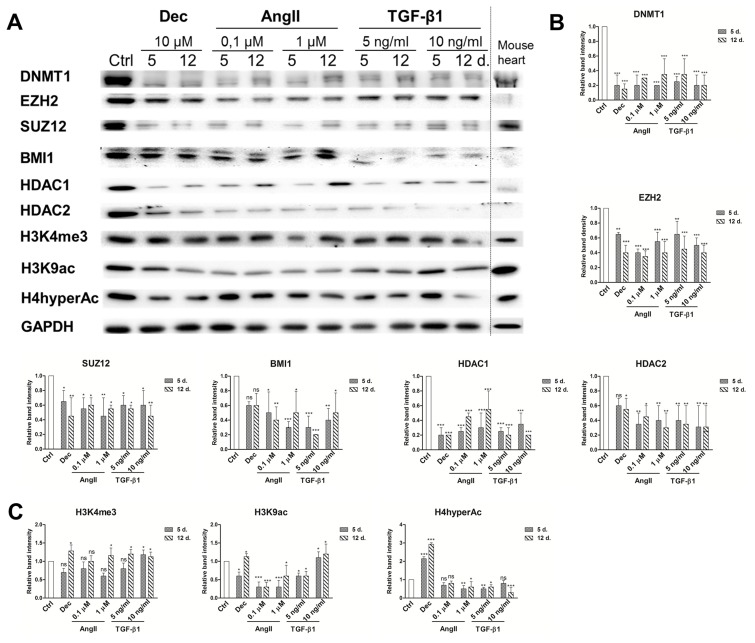
Global epigenetic changes of chromatin modifying proteins and histone modifications were evaluated using immunoanalysis in the cells induced to differentiate with decitabine and both concentrations of angiotensin II and TGF-β1 (Fig. 5A). The obtained results exhibited the downregulation of all studied chromatin remodeling proteins (Fig. 5B). The highest decrease was observed in DNA methyltransferase 1 (DNMT1) levels during differentiation compared to undifferentiated control. In addition, the levels of Polycomb repressive complex 2 components EZH2 and SUZ12 as well as Polycomb repressive complex 1 component BMI1, were also diminished in induced cells. Histone deacetylases 1 and 2 were also downregulated in the treated cells. Decitabine, angiotensin II and TGF-β1 caused very similar changes in chromatin compaction proteins levels. These results are in accordance with mouse heart lysate control exhibiting small levels of chromatin remodeling proteins. On the other hand, variations in histone modifications, associated with transcriptionally active chromatin state, were higher among differently induced cells (Fig. 5C). Histone H3 lysine 4 trimethylation (H3K4me3) was slightly upregulated in all treated cells. In contrast, only decitabine and TGF-β1 increased the levels of H3K9 acetylation and H4 hyper-acetylation was upregulated only in decitabine induced cells, while other agents downregulated the levels of these modified histones. On the other hand, the levels of these histone modifications in the mouse heart were high.
Fig. 5.
Epigenetic alterations during induced cardiomyogenic differentiation. (A) Total proteins were extracted from control cells (Ctrl) and cells, induced using angiotensin II and TGF-β1 at the days 5 and 12 of differentiation. Alterations in the expression levels of Polycomb repressive complex 2 (PRC2) proteins EZH2 and SUZ12, Polycomb repressive complex 1 (PRC1) component BMI1, DNA and histones modifying proteins DNMT1, HDAC1 and HDAC2 as well as in histone modifications, considered as the markers of active chromatin state (H3K4me3, H3K9Ac and H4hyperAc) are presented as determined by Western blot. Mouse heart lysate was used as a positive control. The blots represent one of three independent experiments showing similar results. (B) Relative band density of chromatin remodeling proteins and (C) histone modifications, as measured using ImageJ software (NIH, USA) and normalized to the GAPDH loading control. The data are presented as mean±SD (n=3), p≤0.05 (*), p≤0.01 (**), p≤0.001 (***), ns: non-significant.
Discussion
Nowadays the field of cell therapy for the treatment of cardiovascular diseases is developing very fast as there is a high demand for new and effective application of stem cells strategies. One of the possible sources of stem cells is human amniotic fluid-derived stem cells having cardiomyogenic capacity in vitro. In this work, differentiation was induced using two biomolecules, angiotensin II and TGF-β1, which have a wide pleiotropic effect in different cells. They induce various signaling pathways in cardiac and renal cells, brain, adipocytes and even immune system (16). It was demonstrated that under specific differentiation conditions, these biomolecules can induce multilineage differentiation potential of embryonic or mesenchymal stem cells into insulin-producing cells, keratinocytes, chondrocytes, smooth muscle cells, osteocytes, neurocytes, immune system cells Th1 or even play a significant role in the wound closure (17). However, both AngII and TGF-β1 inhibit differentiation into adipocytes or skeletal muscle cells (17).
In this study, these two biological compounds initiated alterations in human AF-MSCs leading towards the cardiac-like phenotype formation, in contrast to Gaafar and co-authors study where adipose tissue-derived MSCs were used (18). In agreement with previously published studies (7, 11, 12, 19, 20), our results demonstrate that angiotensin II and TGF-β1, as well as decitabine, that was used as a positive control, together with morphological changes, upregulated the expression of the main cardiac genes-markers: early transcription factors NKX2-5, TBX5 and GATA4 as well as structural genes of cardiomyocytes, such as MYH6, TNNT2 and DES. Also, the increased expression of cardiac ion channels (sodium, calcium, potassium) genes observed with decitabine, angiotensin II and TGF-β1, to a greater or smaller extent, suggests of governed cell fate to become functional cardiac myocytes upon maturation in vitro or in vivo (21). It was previously reported that angiotensin II and TGF-β1 may enhance the contractibility of mouse bone marrow MSCs (19); but, similarly to our results, no changes in electrophysiological characteristics were detected after differentiation induction. In addition to that, the enhancement of Connexin43 protein was inspected in our induced to differentiate AF-MSCs. These results confirmed the association between Cx43, angiotensin II and TGF-β1 described by Cao et al. (22). Connexin43 is the main component of gap junctions that mediate electrical and chemical coupling between adjacent cardiomyocytes (23). Despite the fact that Cx43 localization in the induced AF-MSCs was more unspecific, diffusive in the whole cell, the tendency of its accumulation at the boundaries of the cells could be detected. This indicates the morphological basis of the intercalated discs formation where Cx43 forms gap junctions and maintains electrical activity necessary for myocardial function. Moreover, the higher nuclear localization of Nkx2.5 protein in AF-MSCs treated with angiotensin II and TGF-β1 compared to control cells is strongly related to Nkx2.5 function upon induced differentiation (24): it activates the transcription of various genes participating in the cardiac atrium and ventricle morphogenesis, development, cardiac muscle cell differentiation and heart development (Gene Ontology pathways, STRING database).
To our knowledge, we were the first to demonstrate cellular energetics and metabolic alterations after AF-MSCs treatment with decitabine, angiotensin II and TGF-β1. We indicated that undifferentiated MSCs may produce ATP either via a tricarboxylic acid cycle or anaerobic glycolysis, consistent with Capasso et al. results (25). Meanwhile differentiated cells or specialized progenitors start utilizing mitochondrial oxidative phosphorylation more (26). Here we obtained that decitabine, angiotensin II and TGF-β1 shifted AF-MSCs phenotype towards more energetic and increased metabolic potential. They also upregulated maximal respiration, spare respiratory capacity and ATP production indicating the enhanced energetic needs and metabolic activity. Previous research showed that cardiomyocytes, differentiated from human induced pluripotent stem cells in vitro, mostly applied oxidative phosphorylation to meet the increased energy demand (27, 28). In addition to that, PPARGC1A, NRF1 and HIF1A genes, associated with metabolism regulation, were upregulated in differentiated cells. The most important – PPARGC1A – is enriched in tissues with high oxidative activity, like heart, and is induced in the states of enhanced energy demand (29) as well as stimulates mitochondrial proliferation and activates nuclear respiratory factors 1 and 2 (NRF1/2) (30). NRF1 plays the main role in cardiac mitochondrial biogenesis and respiration (31). What is more, HIF1A, a transcriptional regulator of the adaptive response to hypoxia, was linked to cardiac differentiation promotion in embryonic stem cells (32). Considering these extracellular flux analysis results together with the enhanced expression of cardiac and metabolic genes and proteins, the slight metabolic switch in angiotensin II and TGF-β1 induced cells can be considered as the indicator of successful commitment towards cardiomyocytes forming the basis for further maturation. Depending on the cellular and molecular environment, TGF-β1 can also generate mature cells, for example, functional cardiomyocytes from cardiomyocytes progenitors (33). Also, several studies have successfully applied TGF-β1 induced bone marrow MSCs to rodent infarcted myocardium where they assisted in the myocardial regeneration (12, 20) suggesting that all the features of mature cardiomyocytes probably are not necessary for successful clinical application. This suggests that AF-MSCs, pre-treated with angiotensin II or TGF-β1, may also have potential clinical utility, for example, for transplantation into the myocardium, subsequent maturation and regeneration of the damaged tissue. As stem cells from amniotic fluid are considered non-immunogenic and do not cause immune response (34), they could be tested not only in autologous but also in allogeneic transplantation having wider clinical applications.
Epigenetic changes observed in this study involved chromatin remodeling proteins and histone modifications. DNA methyltransferases together with histone deacetylases and other chromatin modifying proteins regulate chromatin organization during stem cells differentiation (35). As our results revealed, the levels of DNMT1, as well as the levels of HDAC1 and HDAC2, decreased during the induced differentiation with decitabine, angiotensin II and TGF-β1. These results are in accord with Liu et al. (36) and Montgomery et al. (37) who suggested that HDAC1 inhibition induced cardiomyogenic differentiation and that both HDAC1 and HDAC2 controlled the expression of cardiac genes-markers, such as TNNT2 or MYH6. Also, the detected downregulation of PRC2 components EZH2 and SUZ12 that was observed in our study could be associated with the promotion of differentiation towards cardiomyogenic lineage (38). In addition, PRC1 protein BMI1 is very significant in cardiac programming as it was exhibited that reducing levels of BMI1 significantly enhanced the efficiency of fibroblasts conversion into contractile induced cardiomyocytes (39). As our results indicated, BMI1 levels were diminished after the induction of differentiation with both agents, consistent with the gene and protein expression pattern as well as metabolic changes. Moreover, reduced BMI1 expression corresponded with the increased levels of active histone modification H3K4me3 at cardiogenic loci (39) and similar global effect of increased H3K4me3 was observed in our study. Interestingly, the global levels of acetylated histones H3 and H4, also associated with the transcriptional activity, increased only in decitabine and TGF-β1 treated AF-MSCs despite the global decrease in HDAC1/2 levels. As Wang and colleagues demonstrated (40), promoters of cardiac-specific genes (MYH6, TNNT2) were enriched in acetylated H3 and H4 and higher levels of these modifications correlate with stronger cardiomyogenic capacity. Regarding this, the relative expression of cardiac-specific genes was higher in decitabine than angiotensin II or TGF-β1 induced cells in our study possibly having an effect on the cardiomyogenic capacity. On the other hand, the global level of acetylation may not reflect the actual epigenetic status of cardiomyogenesis-related genes as decitabine and used biomolecules have considerably different nature and mechanisms of action.
In conclusion, angiotensin II and TGF-β1 are able to induce alterations in gene and protein expression patterns in human AF-MSCs as well as metabolic and epigenetic landscape leading towards the formation of cardiomyocyte-like cells in vitro. Comparing the used differentiation conditions, 1 μM of angiotensin II had higher differentiation induction efficiency than 0.1 μM and 5 ng/ml of TGF-β1 – higher than 10 ng/ml. Overall, in our study, angiotensin II seemed to induce cardiac differentiation more efficiently than TGF-β1, however, not as effective as decitabine. The obtained phenotype is not fully mature and the generated cells can be considered as committed progenitors having the basis of becoming functional cardiomyocytes under relevant conditions in vitro or in vivo. All in all, our results complement the knowledge of processes happening at the onset of induced cardiomyogenic differentiation of AF-MSCs. Future research is needed to evaluate the potential use of these induced AF-MSCs in cell therapy for the treatment of cardiovascular diseases.
Supplementary Information
Acknowledgments
This work was supported by the Research Council of Lithuania (Project No. MIP-57/2015). We thank Natalija Krasovskaja (Vilnius University, Faculty of Medicine) for providing samples of human amniotic fluid.
Footnotes
Potential Conflict of Interest
The authors have no conflicting financial interest.
Supplementary Materials
Supplementary data including supplementary methods and tables can be found with this article online at http://pdf.medrang.co.kr/paper/pdf/IJSC/IJSC-12-s18126.pdf.
References
- 1.Bagno L, Hatzistergos KE, Balkan W, Hare JM. Mesenchymal stem cell-based therapy for cardiovascular disease: progress and challenges. Mol Ther. 2018;26:1610–1623. doi: 10.1016/j.ymthe.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savickiene J, Treigyte G, Baronaite S, Valiuliene G, Kaupinis A, Valius M, Arlauskiene A, Navakauskiene R. Human amniotic fluid mesenchymal stem cells from second-and third-trimester amniocentesis: differentiation potential, molecular signature, and proteome analysis. Stem Cells Int. 2015;2015 doi: 10.1155/2015/319238. 319238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan X, Delo DM, Atala A, Soker S. In vitro cardiomyogenic potential of human amniotic fluid stem cells. J Tissue Eng Regen Med. 2011;5:220–228. doi: 10.1002/term.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gasiūnienė M, Zubova A, Utkus A, Navakauskienė R. Epigenetic and metabolic alterations in human amniotic fluid stem cells induced to cardiomyogenic differentiation by DNA methyltransferases and p53 inhibitors. J Cell Biochem. 2018 doi: 10.1002/jcb.28092. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 5.Maioli M, Contini G, Santaniello S, Bandiera P, Pigliaru G, Sanna R, Rinaldi S, Delitala AP, Montella A, Bagella L, Ventura C. Amniotic fluid stem cells morph into a cardiovascular lineage: analysis of a chemically induced cardiac and vascular commitment. Drug Des Devel Ther. 2013;7:1063–1073. doi: 10.2147/DDDT.S44706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmadian E, Jafari S, Yari Khosroushahi A. Role of angiotensin II in stem cell therapy of cardiac disease. J Renin Angiotensin Aldosterone Syst. 2015;16:702–711. doi: 10.1177/1470320315621225. [DOI] [PubMed] [Google Scholar]
- 7.Xing Y, Lv A, Wang L, Yan X. The combination of angiotensin II and 5-azacytidine promotes cardiomyocyte differentiation of rat bone marrow mesenchymal stem cells. Mol Cell Biochem. 2012;360:279–287. doi: 10.1007/s11010-011-1067-z. [DOI] [PubMed] [Google Scholar]
- 8.Ikhapoh IA, Pelham CJ, Agrawal DK. Synergistic effect of angiotensin II on vascular endothelial growth factor-A-mediated differentiation of bone marrow-derived mesenchymal stem cells into endothelial cells. Stem Cell Res Ther. 2015;6:4. doi: 10.1186/scrt538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azhar M, Schultz Jel J, Grupp I, Dorn GW, 2nd, Meneton P, Molin DG, Gittenberger-de Groot AC, Doetschman T. Transforming growth factor beta in cardiovascular development and function. Cytokine Growth Factor Rev. 2003;14:391–407. doi: 10.1016/S1359-6101(03)00044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohanty S, Bose S, Jain KG, Bhargava B, Airan B. TGFβ1 contributes to cardiomyogenic-like differentiation of human bone marrow mesenchymal stem cells. Int J Cardiol. 2013;163:93–99. doi: 10.1016/j.ijcard.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Abdel-Latif A, Zuba-Surma EK, Case J, Tiwari S, Hunt G, Ranjan S, Vincent RJ, Srour EF, Bolli R, Dawn B. TGF-beta1 enhances cardiomyogenic differentiation of skeletal muscle-derived adult primitive cells. Basic Res Cardiol. 2008;103:514–524. doi: 10.1007/s00395-008-0729-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv Y, Liu B, Wang HP, Zhang L. Intramyocardial implantation of differentiated rat bone marrow mesenchymal stem cells enhanced by TGF-β1 improves cardiac function in heart failure rats. Braz J Med Biol Res. 2016;49:e5273. doi: 10.1590/1414-431x20165273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wenzel S, Henning K, Habbig A, Forst S, Schreckenberg R, Heger J, Maxeiner H, Schlüter KD. TGF-beta1 improves cardiac performance via up-regulation of laminin receptor 37/67 in adult ventricular cardiomyocytes. Basic Res Cardiol. 2010;105:621–629. doi: 10.1007/s00395-010-0108-1. [DOI] [PubMed] [Google Scholar]
- 14.Glemžaitė M, Navakauskienė R. Osteogenic differentiation of human amniotic fluid mesenchymal stem cells is determined by epigenetic changes. Stem Cells Int. 2016;2016 doi: 10.1155/2016/6465307. 6465307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson B. Low level operations and learning in computer vision [PhD dissertation] Linköping: Linköping University; 2004. [Google Scholar]
- 16.Forrester SJ, Booz GW, Sigmund CD, Coffman TM, Kawai T, Rizzo V, Scalia R, Eguchi S. Angiotensin II signal transduction: an update on mechanisms of physiology and pathophysiology. Physiol Rev. 2018;98:1627–1738. doi: 10.1152/physrev.00038.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang MK, Sun HQ, Xiang YC, Jiang F, Su YP, Zou ZM. Different roles of TGF-β in the multi-lineage differentiation of stem cells. World J Stem Cells. 2012;4:28–34. doi: 10.4252/wjsc.v4.i5.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaafar T, Shawky S, Attia W, Hamza H, El Hawary R. The role of angiotensin II in cardiomyogenic differentiation of human adipose tissue-derived mesenchymal stem cells. Comp Clin Pathol. 2015;24:879–885. doi: 10.1007/s00580-014-2001-z. [DOI] [Google Scholar]
- 19.Liu BW, Lü AL, Hou J, Huang W, Hou H, Hou ZL, DAJ, Ai SY. Electrophysiological characteristics of cardiomyocyte-like cells from rat bone marrow derived mesenchymal stem cells by four inductors. Chin Med J (Engl) 2013;126:3528–3533. [PubMed] [Google Scholar]
- 20.Li TS, Hayashi M, Ito H, Furutani A, Murata T, Matsuzaki M, Hamano K. Regeneration of infarcted myocardium by intramyocardial implantation of ex vivo transforming growth factor-beta-preprogrammed bone marrow stem cells. Circulation. 2005;111:2438–2445. doi: 10.1161/01.CIR.0000167553.49133.81. [DOI] [PubMed] [Google Scholar]
- 21.Honda M, Kiyokawa J, Tabo M, Inoue T. Electrophysiological characterization of cardiomyocytes derived from human induced pluripotent stem cells. J Pharmacol Sci. 2011;117:149–159. doi: 10.1254/jphs.11038FP. [DOI] [PubMed] [Google Scholar]
- 22.Cao L, Chen Y, Lu L, Liu Y, Wang Y, Fan J, Yin Y. Angiotensin II upregulates fibroblast-myofibroblast transition through Cx43-dependent CaMKII and TGF-β1 signaling in neonatal rat cardiac fibroblasts. Acta Biochim Biophys Sin. 2018;50:843–852. doi: 10.1093/abbs/gmy090. [DOI] [PubMed] [Google Scholar]
- 23.Boengler K, Schulz R, Heusch G. Connexin 43 signalling and cardioprotection. Heart. 2006;92:1724–1727. doi: 10.1136/hrt.2005.066878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armiñán A, Gandía C, Bartual M, García-Verdugo JM, Lledó E, Mirabet V, Llop M, Barea J, Montero JA, Sepúlveda P. Cardiac differentiation is driven by NKX2.5 and GATA4 nuclear translocation in tissue-specific mesenchymal stem cells. Stem Cells Dev. 2009;18:907–918. doi: 10.1089/scd.2008.0292. [DOI] [PubMed] [Google Scholar]
- 25.Capasso S, Alessio N, Squillaro T, Di Bernardo G, Melone MA, Cipollaro M, Peluso G, Galderisi U. Changes in autophagy, proteasome activity and metabolism to determine a specific signature for acute and chronic senescent mesenchymal stromal cells. Oncotarget. 2015;6:39457–39468. doi: 10.18632/oncotarget.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11:596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rana P, Anson B, Engle S, Will Y. Characterization of human-induced pluripotent stem cell-derived cardiomyocytes: bioenergetics and utilization in safety screening. Toxicol Sci. 2012;130:117–131. doi: 10.1093/toxsci/kfs233. [DOI] [PubMed] [Google Scholar]
- 28.Denning C, Borgdorff V, Crutchley J, Firth KS, George V, Kalra S, Kondrashov A, Hoang MD, Mosqueira D, Patel A, Prodanov L, Rajamohan D, Skarnes WC, Smith JG, Young LE. Cardiomyocytes from human pluripotent stem cells: from laboratory curiosity to industrial biomedical platform. Biochim Biophys Acta. 2016;1863:1728–1748. doi: 10.1016/j.bbamcr.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ventura-Clapier R, Garnier A, Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc Res. 2008;79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- 30.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 31.Xia Y, Buja LM, Scarpulla RC, McMillin JB. Electrical stimulation of neonatal cardiomyocytes results in the sequential activation of nuclear genes governing mitochondrial proliferation and differentiation. Proc Natl Acad Sci U S A. 1997;94:11399–11404. doi: 10.1073/pnas.94.21.11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng KM, Lee YK, Chan YC, Lai WH, Fung ML, Li RA, Siu CW, Tse HF. Exogenous expression of HIF-1 alpha promotes cardiac differentiation of embryonic stem cells. J Mol Cell Cardiol. 2010;48:1129–1137. doi: 10.1016/j.yjmcc.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Goumans MJ, de Boer TP, Smits AM, van Laake LW, van Vliet P, Metz CH, Korfage TH, Kats KP, Hochstenbach R, Pasterkamp G, Verhaar MC, van der Heyden MA, de Kleijn D, Mummery CL, van Veen TA, Sluijter JP, Doevendans PA. TGF-beta1 induces efficient differentiation of human cardiomyocyte progenitor cells into functional cardiomyocytes in vitro. Stem Cell Res. 2007;1:138–149. doi: 10.1016/j.scr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 34.De Coppi P, Bartsch G, Jr, Siddiqui MM, Xu T, Santos CC, Perin L, Mostoslavsky G, Serre AC, Snyder EY, Yoo JJ, Furth ME, Soker S, Atala A. Isolation of amniotic stem cell lines with potential for therapy. Nat Biotechnol. 2007;25:100–106. doi: 10.1038/nbt1274. [DOI] [PubMed] [Google Scholar]
- 35.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Liu Z, Li T, Liu Y, Jia Z, Li Y, Zhang C, Chen P, Ma K, Affara N, Zhou C. WNT signaling promotes Nkx2.5 expression and early cardiomyogenesis via downregulation of Hdac1. Biochim Biophys Acta. 2009;1793:300–311. doi: 10.1016/j.bbamcr.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coppola A, Romito A, Borel C, Gehrig C, Gagnebin M, Falconnet E, Izzo A, Altucci L, Banfi S, Antonarakis SE, Minchiotti G, Cobellis G. Cardiomyogenesis is controlled by the miR-99a/let-7c cluster and epigenetic modifications. Stem Cell Res. 2014;12:323–337. doi: 10.1016/j.scr.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Y, Wang L, Vaseghi HR, Liu Z, Lu R, Alimohamadi S, Yin C, Fu JD, Wang GG, Liu J, Qian L. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell. 2016;18:382–395. doi: 10.1016/j.stem.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M, Yu Q, Wang L, Gu H. Distinct patterns of histone modifications at cardiac-specific gene promoters between cardiac stem cells and mesenchymal stem cells. Am J Physiol Cell Physiol. 2013;304:C1080–C1090. doi: 10.1152/ajpcell.00359.2012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.