Abstract
Peroxisomal disorders are genetically heterogeneous metabolic disorders associated with a deficit of very long chain fatty acid β-oxidation that commonly manifest as early-onset neurodegeneration. Brain microvascular endothelial dysfunction with increased permeability to monocytes has been described in X-linked adrenoleukodystrophy, one of the most common peroxisomal disorders caused by mutations of the ATP binding cassette subfamily D member 1 (ABCD1) gene. The present study demonstrated that dysregulation of sirtuin 1 (SIRT1) in human brain microvascular endothelial cells (HBMECs) mediates changes in adhesion molecules and tight-junction protein expression, as well as increased adhesion to monocytes associated with peroxisomal dysfunction due to ABCD1 or hydroxysteroid 17-β dehydrogenase 4 silencing. Furthermore, enhancement of the function of SIRT1 by resve-ratrol attenuated this molecular and functional dysregulation of HBMECs via modulation of the nuclear factor-κB and Krüppel-like factor 4 signaling pathways.
Keywords: peroxisomal disorders, X-linked adrenoleukodystrophy, multifunctional protein 2, human brain microvascular endothelial cells, sirtuin1, resveratrol
Introduction
Peroxisomes are ubiquitous organelles in eukaryotic cells with multiple metabolic functions (1,2). The three established functions of peroxisomes are α-oxidation (release of one carbon of a 3-methyl-branched or 2-hydroxyfatty acid), β-oxidation (release of two carbons of the backbone of diverse substrates) and ether lipid synthesis. Almost all diseases caused by peroxisome dysfunction are manifested by a variety of neurological abnormalities, underscoring the importance of peroxisomes in the nervous system.
X-linked adrenoleukodystrophy (X-ALD), the most frequent peroxisomal disorder, is caused by inactivity of a transmembrane ATP binding cassette (ABC) transporter, which imports very long chain fatty acid (VLCFA)-CoA into the peroxisome for β-oxidation (3). Up to 60% of male patients develop a catastrophic form of cerebral inflammatory demyelination, cerebral ALD (CALD), characterized by increased blood-brain barrier (BBB) permeability to monocytes with marked perivascular microglial/monocyte activation at the lesion edge (4-7).
Multifunctional protein 2 (MFP2) deficiency, another peroxisomal disorder, is caused by mutations in the hydroxysteroid 17-β dehydrogenase 4 (HSD17B4) gene, which encodes D-bifunctional protein, the central enzyme in the second step of the peroxisomal β-oxidation pathway that is essential for the shortening of branched-chain and straight-chain compounds (8). Defects of MFP2 are associated with a spectrum of neurological disorders, encompassing developmental and degenerative pathologies, and marked monocytic activation and neuroinflammation have been demonstrated in a mouse model lacking MFP2 (9,10). While the pathogenetic mechanisms underlying cerebral degeneration following peroxisomal dysfunction remain largely elusive, previous studies on X-ALD suggest that abnormal BBB function precedes inflammatory demyelination (11,12). As the key component of the BBB, brain microvascular endothelial cells (BMECs) strictly control the flow of substances into and out of the brain, as well as the entry of leukocytes through regulation of adhesion molecules and intercellular tight-junction protein expression (13-15). Abnormal microvascular function in peroxisomal disorders has been previously demonstrated in CALD: i) Ex vivo histopathology demonstrated distorted microvascular permeability beyond the edge of the demyelinating lesions (12) and ii) in vitro experiments indicated that lack of ABC subfamily D member 1 (ABCD1) in human BMECs causes dysregulation of adhesion molecules and tight-junction proteins, leading to an increase in permeability to leukocytes (12). However, how loss of peroxisomal function secondary to ABCD1 or MFP2 deficiency leads to endothelial dysfunction has remained largely elusive.
As the most conserved mammalian sirtuin (SIRT) family NAD+ dependent protein deacetylase, SIRT1 has been demonstrated to be the key metabolic sensor across different tissue types, regulating numerous transcriptional factors and co-factors involved in systemic metabolic homeostasis, including peroxisomal disorders (16,17). Endothelial cell dysfunction is markedly associated with activation of nuclear factor (NF)-κB signaling, a pathway that is strictly regulated by SIRT1 (11,12,18). Reduced SIRT1 expression has been observed in numerous abnormalities of vascular endothelial function, including diabetic vasculopathy, whereas improvement of SIRT1 activity by either overexpression or replacement treatment markedly ameliorated the dysfunction and had significant beneficial effects (19-21). Given the master regulator role of SIRT1 in energy metabolism homeostasis and its protective role in endothelial cells (22,23), understanding whether SIRT1 is also able to modulate endothelial dysfunction in peroxisomal disorders may lead to novel therapeutic and less toxic interventions.
In the present study, an in vitro model of the human BBB was utilized to investigate the effects of ABCD1 and MFP2 deficiency on the BME and assess whether modulation of SIRT1 levels can ameliorate the endothelial dysfunction and normalize its interactions with monocytes.
Materials and methods
Cell cultures
Primary human brain microvascular endothelial cells (HBMECs) from CSC systems were purchased from ScienCell Research Laboratories, Inc. They were maintained in EGM-2 Endothelial Cell Growth Medium-2 Bullel kit (Lonza Group, Ltd.) on a Collagen type 1 (Corning, Inc.)-coated 10 cm diameter plate in a 37°C humidified atmosphere of 95% air and 5% CO2 in incubator and used for experiments at 80% confluency.
THP-1 was purchased from ZQ Cell Research and cultured in RPMI1640 (GE Healthcare) medium supplemented with 0.05 mM β-Mercaptoethanol (Sigma-Aldrich; Merck KGaA), 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin streptomycin (Thermo Fisher Scientific, Inc.). Cells were grown in a humidified atmosphere of 5% CO2 at 37°C and used for experiments at >80% confluency.
Chemical treatment
Resveratrol was purchased from Sigma-Aldrich; Merck KGaA and a 50 mM storage solution in DMSO was prepared. Per experimental needs, cells were treated with 20 µM Resveratrol for 48 h before further analysis. Sirtinol (Sigma-Aldrich; Merck KGaA) was made into 20 mM storage solution by DMSO and diluted into a 20 µM working solution for cell treatment. For tumor necrosis factor (TNF)-α treatment, cells were treated with 10 ng/ml TNF-α (eBioscience; Thermo Fisher Scientific, Inc.) for 6 h before different assays.
ABCD1, HSD17B4 and Krüppel-like factor 4 (KLF4) gene silencing
SiGENOME Human ABCD1 (cat. no. M009605010005) small interfering (si)RNA-SMARTpool and siGENOME non-targeting siRNA (cat. no. D0012100105) were purchased from GE Healthcare Dharmacon, Inc. HSD17B4siRNA-Homo-427/990/1870 and KLF4 siRNA were purchased from Shanghai GenePharma Co., Ltd. Silencing was performed according to the protocol provided by the manufacturer. Briefly, 1.5-2×105 HBMECs were seeded in 6-well plates for 24 h. siRNA (5 µM) dissolved in serum free medium was mixed with DharmaFECT transfection reagent (GE Healthcare Dharmacon, Inc.) and incubated for 20 min at room temperature. Cells were then replaced with fresh medium containing 25 nM siRNA and then cultured for 48 h for further analysis.
Reverse transcription quantitative-PCR
Total RNA was extracted with Qiagen RNeasy kit. First-strand cDNA was synthesized (25°C, 10 min; 42°C, 30 min; 85°C, 5 min) using Hiscript QRT SuperMix (Thermo Fisher Scientific, Inc.) in 20 µl reaction. Thermocycling conditions were 95°C for 10 min and 40 PCR cycles (95°C, 20 sec; 55°C, 30 sec; 72°C, 20 sec). After the amplification reaction, the temperature was 95°C for 5 sec and 65°C for 60 sec. PCRs were performed using SYBR master mix (Thermo Fisher Scientific, Inc.) and the data were analyzed by calculating the ∆Cq value (24). The primers used were listed as below: Claudin 5 forward: 5′-CTC TGC TGG TTC GCC AAC AT-3′, and Claudin 5 reverse: 5′-CAG CTC GTA CTT CTG CGA CA-3′; β-actin forward: 5′-CGA GCA CAG AGC CTC GCC TTT GCC-3′ and β-actin reverse: 5′-TGT CGA CGA CGA GCG CGG CGA TAT-3′.
Western blotting protein analysis
HBMEC protein was extracted with radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate and 0.1% SDS). After lysis on an ice for 0.5 h, samples were collected and centrifuged at 4°C at 20,817 g for 0.5 h. The supernatant was transferred into fresh tubes and protein concentration was determined by the bicinchoninic acid (BCA) method. Nuclear Protein was extracted using Nucleoprotein Extraction kit from Beyotime Institute of Biotechnology according to the manufacturer's protocol. Briefly, cells were lysed with RIPA buffer on ice for 0.5 h, which were collected and centrifuged at 4°C at 20,817 × g for 0.5 h. The supernatant was removed. After adding 100 µl buffer C, the precipitation was centrifuged at 4°C at 20,817 × g for 0.5 h. The supernatant was transferred into fresh tubes for next step.
Equal amounts of protein (60 µg/lane) were separated by 10% SDS-PAGE and transferred onto nitrocellulose filter membrane (EMD Millipore). Membranes were blocked with 5% non-fat milk in phosphate-buffered saline containing 0.05% Tween-20 at room temperature for 2 h and probed with antibodies at 4°C for 14 h including Anti-ABCD1 (1:1,000; OriGene Technologies, Inc.; cat. no. TA803208), and anti-hydroxysteroid (17B4) dehydrogenase 4 (1:2,000; Abcam; cat. no. ab128565), anti-SIRT1 (1:1,000; Santa Cruz Biotechnology, Inc.; cat. no. sc15404), anti-intracellular adhesion molecular (ICAM1; 1:1,000; Santa Cruz Biotechnology, Inc.; cat. no. sc7891), anti-vascular cellular adhesion molecule (VCAM1; 1:1,000; Santa Cruz Biotechnology, Inc.; cat. no. sc13160), anti-Claudin 5 (1:2,000; Abcam; cat. no. ab15106), anti-KLF4 (1:2,000; Proteintech; cat. no. 11880), anti-phosphorylated NF-κB inhibitor (p-IκBα; 1:2,000; Abcam; cat. no. ab133462), anti-NF-κB (1:2,000; Abcam; cat. no. ab16502) and anti-Acetyl-Lysine (1:2,000; ABclonal; cat. no. A2391). Anti-β-actin (1:5,000; Santa Cruz Biotechnology, Inc.; cat. no. sc47778) was used as a protein loading control while anti-Lamin b (1:5,000; Santa Cruz Biotechnology, Inc. cat. no. sc56143) was elected as nuclear protein loading control. Membranes were developed with Odyssey Infrared laser scanning image-forming system after being washed and incubated with appropriate secondary antibodies (1:2,000; horseradish peroxidase, Goat anti mouse or Goat anti rabbit, Vicmed Company; cat. no. C30409) at room temperature for 2 h.
Immunoprecipitation (IP)
The protein lysates, which obtained as aforementioned, were incubated with 1 µg IgG (Beyotime Institute of Biotechnology) and 20 µl resuspended Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Inc.) for 30 min at 4°C, then centrifugation was used to remove the non-specific protein adhering to protein A/G at 4°C at 2,500 × g for 5 min. After collecting and measuring the concentration of protein by the bicinchoninic acid method, the supernatants were made up to a total amount of 500 µl with IP buffer. The corresponding primary antibody with 5 µl was added to the samples including anti-KLF4 (ProteinTech Group, Inc.; cat. no. 11880), or anti-SIRT1 (Santa Cruz Biotechnology, Inc.; cat. no. sc-15404) or IgG (Beyotime Institute of Biotechnology) which as a contrast, followed by incubation overnight at 4°C. Subsequently, the samples were incubated with 100 µl Protein A/G PLUS-Agarose at 4°C for 4 h. Finally, the result was produced by western blot analysis.
Human monocyte-endothelial adhesion assay
For the adhesion assay, HBMECs were plated in 6-well plates at a density of 5×104 cells/well until 80-90% confluent, with certain groups treated with tumor necrosis factor (TNF)-α (10 ng/ml) for 6 h. Furthermore, 1×105 THP-1 cells/well were incubated with 1 µl Calcein AM (Sigma-Aldrich; Merck KGaA; 4 mM) at a dilution of 1:1,000 for 1 h at 37°C. Subsequently, the Calcein AM-labeled THP-1 cells were seeded onto the endothelial monolayer at a density of 1×105 cells/well and incubated with RPMI-1640 medium plus 10% FBS for 1 h at 37°C. The cells were washed with PBS and fixed in 4% paraformaldehyde at room temperature for 20 min prior to imaging. Images were captured in five randomly selected microscopic fields at a ×10 magnification using an inverted fluorescence microscope (TH4-200; Olympus Corp.) and fluorescence was quantified using ImageJ software (Version no. 1.4.3.67, National Institutes of Health).
Determination of the NADP/NADPH ratio
The Amplite™ Fluorimetric NADP/NADPH Ratio Assay kit was purchased from AAT Bioquest. The assay was performed according to the manufacturer's protocol. In brief, HBMECs were plated at a density of 1×105 cells/well in 6-well plates and allowed to attach for 24 h. After removing the medium, 100 µl NADP/NADPH lysis buffer (AAT Bioquest, Inc.) was added to each well, followed by incubation at room temperature for 15 min. The supernatant was collected by centrifugation for subsequent analysis at room temperature at 2,000 × g for 10 min. The fluorescence intensity was measured using a microplate reader (Gene Company, Ltd.) with excitation and emission wavelengths of 530 and 590 nm, respectively.
Malondialdehyde (MDA) and superoxide dismutase (SOD) activity detection
The MDA Assay kit and the SOD assay kit were purchased from NJJC Institute of Bioengineering. First, HBMECs were plated at a density of 1×105 cells/well in 6-well plates and allowed to attach for 24 h, followed by treatment with or without TNF-α (10 ng/ml) for 6 h. Next, the cells were collected and the corresponding reagents were added according to the manufacturer's protocols. The absorbance was measured using a microplate reader (Gene Company, Ltd.) at 550 nm for SOD and 532 nm for MDA, respectively.
Statistical analysis
All the results in this study were analyzed by SPSS 16.0 statistical software; GraphPad Prism 6.0 software (GraphPad Software, Inc.) was used for the generation of graphs. Values are expressed as the mean ± standard deviation. Statistical analyses were performed using one-way ANOVA with Tukey multiple comparison test to determine statistical significance between the groups. P<0.05 was considered to indicate a statistically significance difference.
Results
ABCD1 or HSD17B4 silencing in HBMECs leads to reduction of SIRT1 levels and alteration in adhesion molecules and tight-junction proteins
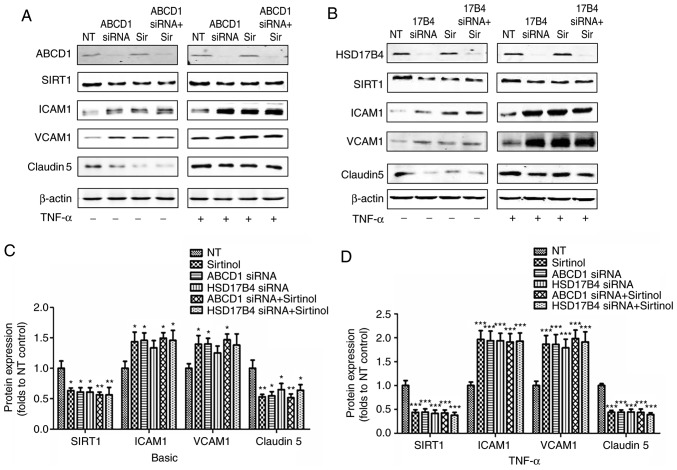
In the first set of experiments, the direct impact of ABCD1 and HSD17B4 deficiency in HBMECs was investigated. Several key protein markers involved in endothelial interactions and permeability to leukocytes, including tight-junction proteins and adhesion molecules, as well as changes in SIRT1 expression levels prior to and after activation with TNF-α (10 ng/ml), were measured. As presented in Fig. 1, silencing of ABCD1 led to a significant 35% reduction in Claudin 5 (P<0.05), a 38% reduction in SIRT1 (P<0.05) as well as a 41% increase in ICAM1 (P<0.05) and a 33% increase in VCAM1 (P<0.05), while stimulation with TNF-α lead to further decreases in Claudin 5 (53%, P<0.001) and SIRT1 (47%, P<0.001), as well as a 2 and 2.8-fold increase in ICAM1 (P<0.001) and VCAM1 (P<0.001), respectively. Similar results were observed following HSD17B4 silencing, which led to a ~25% reduction in Claudin 5 (P<0.05), a 36% increase in ICAM1 and a ~25% increase in VCAM1, accompanied by a 39% reduction in SIRT1 (P<0.05) at the basal level without TNF-α stimulation. Upon TNF-α stimulation, a 57% reduction of Claudin 5 (P<0.001), a 58% reduction of SIRT1 (P<0.001), a 2.1-fold increase of ICAM1 (P<0.001) and a 2.2-fold increase of VCAM1 (P<0.001) were observed. Of note, inhibition of SIRT1 by treatment with Sirtinol (20 µmol/l) led to comparable alterations of the expression of adhesion molecules and Claudin 5 prior to and after endothelial activation (39/58% reduction in Claudin 5; 43/94% increase in ICAM1 and 36/300% increase of VCAM1 expression prior to/after stimulation with TNF-α, respectively). A increased higher expression of ICAM1 and VCAM1 and decreased Claudin 5 levels were detected in the groups with ABCD1 silencing plus Sirtinol and HSD17B4 silencing plus Sirtinol, but this was not statistically significant (Fig. 1).
Figure 1.
ABCD1 or HSD17B4 silencing leads to molecular biomarker alteration and SIRT1 downregulation in HBMECs; an effect mimicked by SIRT1 inhibition using sirtinol. Representative western blot images as well as intensity quantification showing SIRT1, tight-junction protein (Claudin 5) and adhesion molecule (ICAM1 and VCAM1) changes in (A) ABCD1 or (B) HSD17B4 silenced HBMECs with or without sirtinol (20 µM) treatment at (C) basal or (D) TNF-α (10 ng/ml) treated condition. Data are the mean ± standard deviation of three different experiments. *P<0.05, **P<0.01 and ***P<0.001 vs. NT control group. NT, non-targeting; ABCD, ATP binding cassette subfamily D member 1; HBMECs, human brain microvascular endothelial cells; TNF, tumor necrosis factor; ICAM, intracellular cellular adhesion molecule; VCAM, vascular cellular adhesion molecule; NT, non-targeting.
SIRT1 activation by resveratrol restores protein changes in HBMECs after ABCD1 or HSD17B4 silencing
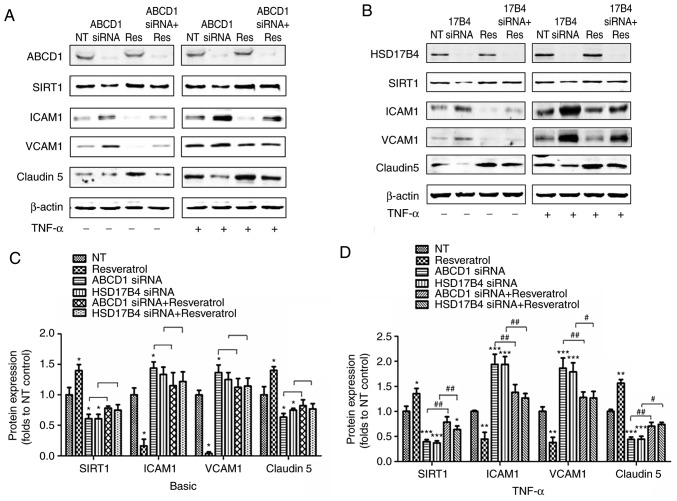
It was then investigated whether increasing SIRT1 function using the SIRT1 activator resveratrol (20 µmol/l) attenuates the protein alterations associated with peroxisomal dysfunction caused by ABCD1 or HSD17B4 silencing. The targeting association was demonstrated by significant increases in SIRT1 protein levels after resveratrol treatment at a baseline and following TNF-α treatment (P<0.05). Resveratrol also partially normalized SIRT1 levels after ABCD1 and HSD17B4 silencing, particularly when HBMECs were activated with TNF-α (P<0.01). HBMECs treated with resveratrol exhibited a reduction of ICAM1 and VCAM1 and increased Claudin 5 expression under basal conditions (P<0.05) and following TNF-α treatment (P<0.01). Of note, resveratrol treatment of HBMECs with silencing of ABCD1 or HSD17B4 attenuated the overexpression of ICAM1 and VCAM1 and significantly increased Claudin 5 expression after activation with TNF-α (Fig. 2).
Figure 2.
SIRT1 activation by resveratrol restores protein biomarker changes in HBMECs after ABCD1 or HSD17B4 silencing. Representative western blot images as well as intensity quantification showing SIRT1, tight-junction protein (Claudin 5) and adhesion molecule (ICAM1 and VCAM1) changes in (A) ABCD1 or (B) HSD17B4 silenced HBMECs with or without resveratrol (20 µM) treatment at (C) basal or (D) TNF-α (10 ng/ml) treated condition. Data are presented as the mean ± standard deviation of three different experiments. *P<0.05, **P<0.01 and ***P<0.001 vs. NT control group. #P<0.05 and ##P<0.01. NT, non-targeting; ABCD, ATP binding cassette subfamily D member 1; HBMECs, human brain microvascular endothelial cells; TNF, tumor necrosis factor; ICAM, intracellular cellular adhesion molecule; VCAM, vascular cellular adhesion molecule; si, small interfering; SIRT1, sirtuin1; NT, non-targeting.
SIRT1 expression levels in HBMECs modulate endothelial-monocyte interactions
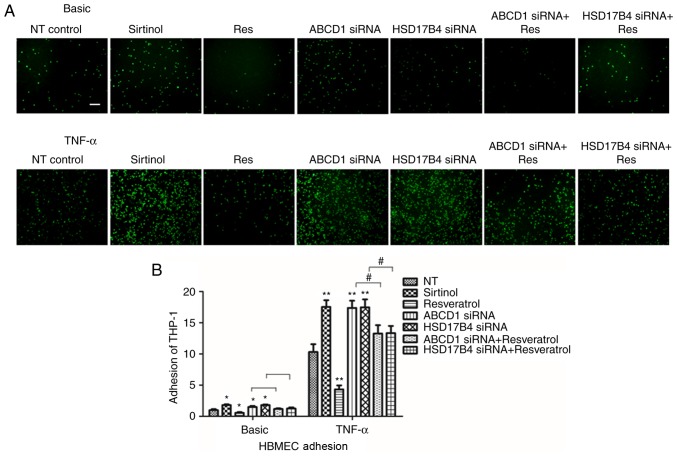
After determining the changes of molecular biomarkers, the functional consequences of low SIRT1 levels in HBMECs achieved by inhibition with Sirtinol or silencing of ABCD1 or HSD17B4. As previously described, a significant increase in adhesion of THP-1 cells was identified after ABCD1 silencing under basal conditions (P<0.05) and following TNF-α treatment (P<0.01; Fig. 3). Similarly, a significant increase in adhesion was observed after HSD17B4 silencing and Sirtinol treatment under basal conditions (P<0.05) and upon stimulation with TNF-α (P<0.01; Fig. 3).
Figure 3.
SIRT1 expression levels in HBMECs modulate endothelial-monocyte interactions. (A) Representative images showing adherence of fluorescent THP-1 cells to endothelial monolayers at different treatment condition. (B) Quantification of calcein AM labelled THP-1 cells that adhere to HBMECs at different conditions with or without TNF-α (10 ng/ml) treatment. Data are presented as the mean ± standard deviation of three different experiments. *P<0.05 and **P<0.01 vs. NT control group. #P<0.05. Scale bar: 10 µm. HBMECs, human brain microvascular endothelial cells; TNF, tumor necrosis factor; si, small interfering; SIRT1, sirtuin1.
To further assess whether restoration of SIRT1 levels in ABCD1- and HSD17B4-silenced cells is able to attenuate their increased adhesion to monocytes that had been observed, silenced HBMECs were treated with resveratrol. First, resveratrol treatment alone significantly reduced monocyte adhesion to 48 and 41% under basal conditions (P<0.05) and following TNF-α treatment (P<0.01), respectively. Furthermore, in ABCD1-silenced HBMECs, treatment with resveratrol markedly inhibited monocyte adhesion by 32 and 41% under basal and TNF-α treatment conditions, respectively (P<0.05). Similarly, in HSD17B4-silenced HBMECs, treatment with resveratrol also markedly inhibited monocyte adhesion by 31 and 40% at basal and TNF-α treatment conditions, respectively (P<0.05), suggesting functional correction by resveratrol treatment (Fig. 3).
Disruption of NADP/NADPH and increased oxidative stress in HBMECs after ABCD1 or HSD17B4 silencing
As a NAD+-dependent protein deacetylase and the key metabolic sensor, SIRT1 tightly regulates numerous cellular activities involved in systemic metabolic homeostasis (16). In the present study, it was revealed that the NADP/NADPH ratio in HBMECs was significantly increased after ABCD1 or HSD17B4 silencing (P<0.01; Table I), suggesting a disturbance of metabolic homeostasis. By measuring oxidative stress makers MDA and SOD, it was unveiled that ABCD1 and HSD17B4 silencing led to significantly increased MDA levels and decreased SOD levels in HBMECs, suggesting increased oxidative stress. This effect was mimicked by sirtinol treatment, while resveratrol treatment significantly reduced the oxidative stress under basal and TNF-α treatment conditions (P<0.01; Table II).
Table I.
NADP/NADPH ratio in human brain microvascular endothelial cells after ABCD1 and HSD17B4 silencing.
| NT | ABCD1 SIRNA | HSD17B4 SIRNA | |
|---|---|---|---|
| NADP/NADPH | 0.95±0.20 | 1.40±0.28A | 1.41±0.22A |
P<0.01 vs. the non-targeting group. ABCD, ATP binding cassette subfamily D member 1; si, small interfering.
Table II.
MDA and SOD levels in HBMECs at different treatment conditions.
| Basic
|
TNF-α
|
|||
|---|---|---|---|---|
| MDA (nmol/ml) | SOD (U/ml) | MDA (nmol/ml) | SOD (U/ml) | |
| Control | 0.75±0.12 | 64.08±4.38 | 1.54±0.27 | 103.41±5.88 |
| ABCD1 siRNA | 1.12±0.23a | 33.38±5.27a | 3.24±0.29a | 71.72±4.75a |
| Sirtinol | 1.15±0.28a | 34.05±4.75a | 3.03±0.39a | 72.05±8.18a |
| ABCD1 siRNA+sirtinol | 1.23±0.32a | 34.39±4.35a | 3.31±0.40a | 72.42±4.78a |
| Resveratrol | 0.39±0.14a | 83.73±5.57a | 0.60±0.34a | 125.93±5.51a |
| ABCD1 siRNA+resveratrol | 0.94±0.20a,b | 45.48±6.40a,b | 2.23±0.31a,b | 83.43±6.65a,b |
| Control | 0.73±0.18 | 63.62±5.62 | 1.51±0.36 | 101.75±6.33 |
| 17B4 siRNA | 1.13±0.31a | 35.82±6.49a | 3.28±0.39a | 68.72±9.57a |
| Sirtinol | 1.16±0.34a | 34.38±5.12a | 3.21±0.48a | 67.95±7.53a |
| 17B4 siRNA+sirtinol | 1.27±0.31a | 33.71±5.90a | 3.41±0.45a | 70.38±8.39a |
| Resveratrol | 0.40±0.18a | 83.89±6.59a | 0.47±0.56a | 126.93±4.76a |
| 17B4 siRNA+resveratrol | 0.89±0.21a,b | 44.23±5.86a,b | 2.13±0.73a,b | 81.86±8.26a,b |
TNF, tumor necrosis factor; si, small interfering; MDA, malondialdehyde; SOD, superoxide dismutase; ABCD, ATP binding cassette subfamily D member 1.
P<0.01 vs. the respective control group:
P<0.01 vs. the respective siRNA group. n=9. SOD, superoxide dismutase; MDA, malondialdehyde; si, small interfering; TNF, tumor necrosis factor.
NF-κB and Krüppel-like factor 4 (KLF4) mediate SIRT1- dependent transcriptional regulation of ICAM1 and Claudin 5 in HBMECs following ABCD1 or HSD17B4 silencing
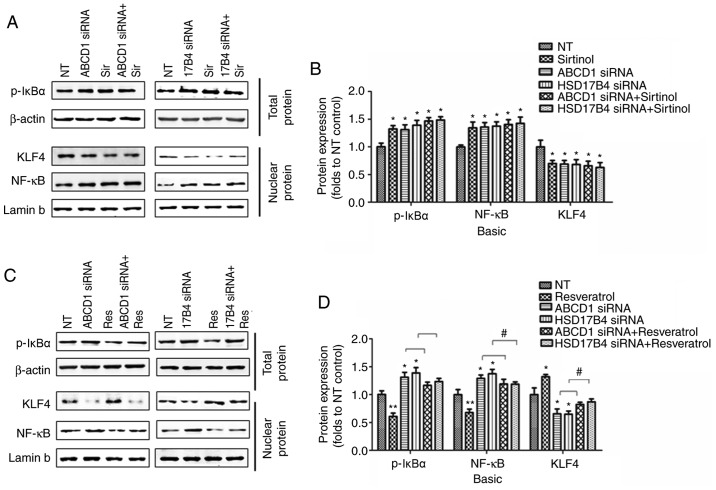
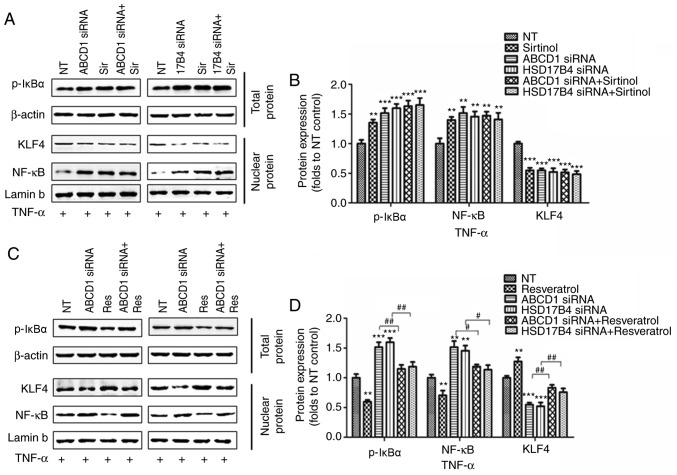
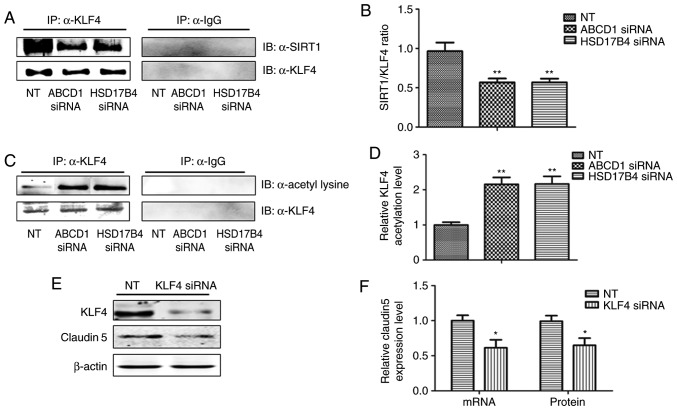
In order to investigate the molecular mechanisms that mediate the effect of SIRT1 upon upregulation of adhesion molecules, activation of NF-κB, a transcriptional factor known to regulate ICAM1 expression, was then assessed. Indeed, SIRT1 depletion by sirtinol or silencing of ABCD1 or HSD17B4 in HBMECs led to increased IκBα phosphorylation and subsequent NF-κB accumulation in the nucleus under basal conditions (P<0.05; Fig. 4A and B) and following TNF-α treatment (P<0.01; Fig. 5A and B), suggesting activation of the NF-κB signaling pathway. When SIRT1 activity was enhanced by resveratrol treatment, a significant reduction of IκBα phosphorylation and nuclear NF-κB levels was observed in the control as well as in the ABCD1 or HSD17B4 gene silencing groups under basal conditions (P<0.01; Fig. 4C and D) and following TNF-α treatment (P<0.01; Fig. 5C and D), indicating that restoration of the function of SIRT1 is able to reverse NF-κB activation and significantly inhibit the expression of adhesion molecules, including ICAM1. A recent study reported that SIRT1 deacetylated KLF4 to activate Claudin 5 transcription in ovarian cancer cells (25). The present results indicated that ABCD1 and HSD17B4 silencing significantly reduced nuclear KLF4 levels under basal (P<0.05) and TNF-α treatment conditions (P<0.001), and the same effect was observed following SIRT1 inhibition using sirtinol (Figs. 4A and B; 5A and C). Increased SIRT1 activation by resveratrol significantly increased nuclear KLF4 levels in the control group and also restored the levels of KLF4 that were reduced by ABCD1 and HSD17B4 silencing under basal (P<0.05; Fig. 4C and D) and TNF-α treatment conditions (P<0.01; Fig. 5C and D). The direct interaction between SIRT1 and KLF4 and subsequent regulation of Claudin 5 expression was assessed by IP and silencing of KLF4. Reduction of SIRT1 binding to KLF4 protein and increase in KLF4 protein acetylation following ABCD1 and HSD17B4 silencing were confirmed (P<0.01; Fig. 6A-D). Finally, KLF4 silencing significantly inhibited Claudin 5 transcription in HBMECs (P<0.05; Fig. 6E and F).
Figure 4.
Regulation of SIRT1 on NF-κB and KLF4 following ABCD1 or HSD17B4 silencing. (A) Representative western blot images as well as (B) intensity quantification of p-IκBα, nuclear NF-κB and nuclear KLF4 in ABCD1 or HSD17B4 silenced HBMECs with or without sirtinol (20 µM) treatment at basal condition. (C) Representative western blot images as well as (D) intensity quantification of p-IκBα, nuclear NF-κB and nuclear KLF4 in ABCD1 or HSD17B4 silenced HBMECs with or without resveratrol (20 µM) treatment at basal condition. *P<0.05 and **P<0.01 vs. NT control group. #P<0.05. NT, non-targeting; ABCD, ATP binding cassette subfamily D member 1; HBMECs, human brain microvascular endothelial cells; TNF, tumor necrosis factor; NF, nuclear factor; si, small interfering; KLF4, Krüppel-like factor 4; NF, nuclear factor; p-IκBα, phosphorylated-inhibitor of NF-κB; NT, non-targeting.
Figure 5.
Regulation of SIRT1 on NF-κB and KLF4 following ABCD1 or HSD17B4 silencing at TNF-α treated condition. (A) Representative western blot images as well as (B) intensity quantification of p-IκBα, nuclear NF-κB and nuclear KLF4 in ABCD1 or HSD17B4 silenced HBMECs with or without sirtinol (20 µM) treatment at TNF-α (10 ng/ml) treated condition. (C) Representative western blot images as well as (D) intensity quantification of p-IκBα, nuclear NF-κB and nuclear KLF4 in ABCD1 or HSD17B4 silenced HBMECs with or without resveratrol (20 µM) treatment at TNF-α (10 ng/ml) treated condition. **P<0.01 and ***P<0.001 vs. the NT control group. #P<0.05 and ##P<0.05. NT, non-targeting; ABCD, ATP binding cassette subfamily D member 1; HBMECs, human brain microvascular endothelial cells; TNF, tumor necrosis factor; NF, nuclear factor; si, small interfering; KLF4, Krüppel-like factor 4; p-IκBα, phosphorylated-inhibitor of NF-κB; NT, non-targeting.
Figure 6.
Regulation of SIRT1 on KLF4 and subsequent Claudin 5 expression. (A) Representative western blot images as well as (B) intensity quantification showing reduced binding of SIRT1 to KLF4 in ABCD1 or HSD17B4 silenced HBMECs. (C) Representative western blot images as well as (D) intensity quantification showing increased KLF4 acetylation in ABCD1 or HSD17B4 silenced HBMECs. (E) Representative western blot images as well as (F) intensity quantification showing KLF4 silencing reduced Claudin 5 expression at both mRNA and protein level. *P<0.05 and **P<0.01 vs. NT control group. NT, non-targeting; ABCD, ATP binding cassette subfamily D member 1; HBMECs, human brain microvascular endothelial cells; TNF, tumor necrosis factor; NF, nuclear factor; KLF4, Krüppel-like factor 4; IgG, immunoglobulin G.
Discussion
The present study demonstrated that dysfunction of ABCD1 and HSD17B4, two peroxisomal disorders manifesting with neurodegeneration, caused brain microvascular endothelium dysfunction in the in vitro cell model of the BBB. This dysfunction was correlated with a reduction of SIRT1 levels in HBMECs. Inhibition of SIRT1 activity by sirtinol mimicked the molecular and functional changes caused by ABCD1 and HSD17B4 silencing, while improvement of SIRT1 function by resveratrol ameliorated HBMEC dysfunction.
The BBB constitutes specialized endothelial cells, pericytes and astrocytes, and tightly regulates the communication between the immune and nervous systems (26). Leukocyte infiltration is considered a critical step in the pathogenesis of numerous CNS diseases (27-30), but under normal conditions, brain leukocyte traffic into the brain is limited due to the tight endothelial barrier (30-32).
A leukocyte may cross the BBB and enter the brain parenchyma through several steps, consisting of the initial rolling and binding of leukocytes on the endothelium by selectins, and subsequent adhesion of leukocytes and their transendothelial egress by ICAM1 and VCAM1 (33,34). Elevated levels of soluble E-selectin, VCAM1 and ICAM1 have been reported in the sera and cerebrospinal fluid of patients multiple sclerosis (MS), and are correlated with disease activity (35). Tight junctions are essential to the transendothelial egress of leukocytes. It is known that abnormalities in ZO1 and occludin occur in the brain of subjects with MS, causing interruption of junctional integrity (36). Dysfunctional tight junctions may allow for a greater influx of blood-borne cells and cytokines, thus amplifying inflammation and further parenchymal damage. In the present study, it was demonstrated that ABCD1 silencing [as previously reported (12)] and HSD17B4 silencing increase ICAM1 and VCAM1 expression in HBMECs, while reducing Claudin 5 expression, which facilitates monocyte adhesion and subsequent transmigration, a potential mechanism contributing to neuroinflammation in cerebral ALD and MFP deficiency.
It is well known that impaired bioenergetics and mitochondrial metabolism, together with oxidative stress, are factors underlying a variety of neurodegenerative diseases, including Parkinson's, Huntington's and Alzheimer's disease, or amyotrophic lateral sclerosis (37-40). CALD and MFP deficiency are peroxisomal disorders that lead to a deficit of VLCFA metabolism. Disrupted energy homeostasis with diminished ATP, NADH and GSH levels, accompanied by dysregulation of oxidative phosphorylation, was discovered in patients with X-ALD as well as ABCD1−/− mouse tissue samples (41-43). Similarly, increased oxidative stress and disrupted energy homeostasis was also reported in MFP deficiency (44). As a NAD+-dependent protein deacetylase and the key metabolic sensor, SIRT1 tightly regulates numerous cellular activities involved in systemic metabolic homeostasis. In the present study, increased NADP/NADPH levels and oxidative stress were detected in HBMECs after ABCD1 or HSD17B4 silencing. The disturbance of metabolic homeostasis may be the potential trigger of SIRT1 inhibition upon ABCD1 or HSD17B4 deficiency.
The crucial role of SIRT1 in maintaining normal vascular endothelial function has been well established in a variety of vascular disorders (19-21). Jia et al (45) reported the inhibitory effect of SIRT1 on ICAM1 expression in human umbilical vein endothelial cells by suppressing its promoter activity. Transcriptional factor NF-κB activation and SIRT1 are closely regulated by each other and have an essential role in the maintenance of endothelial function (11,12). It has been well established that ICAM1 has a binding site for NF-κB in its promoter region and its expression is largely controlled by NF-κB activation (45-47). The present results first suggest that depletion of SIRT1 by ABCD1 or HSD17B4 silencing or sirtinol causes activation of NF-κB signaling in HBMECs and reciprocally, increases in SIRT1 expression caused by resveratrol treatment prevent NF-κB activation, and subsequently ameliorates endothelial dysfunction. Another protective effect of SIRT1 on barrier function was via regulation of tight-junction proteins in endothelial cells (48,49). The present study indicated that upregulation of SIRT1 increases Claudin 5 expression. In terms of the mechanism, Zhang et al (25) reported that, in ovarian cancer cells, activation of Claudin 5 transcription was mediated by SIRT1-induced deacetylation of KLF4 and subsequent transport of KLF4 to the nucleus. In the present study, it was proved that ABCD1 and HSD17B4 silencing down-regulates Claudin 5 transcription in HBMECs via reducing SIRT1 and its binding to KLF4. Furthermore, resveratrol effectively ameliorates brain endothelial dysfunction, while pleiotropic effects other than NF-κB and KLF4 signaling may also occur. Regarding those possible effects, resveratrol is well known as an anti-oxidant, which is able to inhibit mitochondria-derived oxidative damage (50,51) and regulate inflammatory and immune responses in endothelial cells (52). Further studies are necessary to demonstrate its beneficial effects in more complex model systems of peroxisomal dysfunction, including in vitro multicellular BBB models and in vivo mouse models.
In conclusion, the present study demonstrates, for the first time, that brain endothelial peroxisomal dysfunction secondary to X-ALD and MFP deficiency causes dysregulation of SIRT1, which alters the transcriptional regulation of adhesion molecules and tight junction proteins via the NF-κB and KLF4 signaling pathways. SIRT1 depletion in brain endothelial cells, which is most likely caused by disruption of β-oxidation and oxidative stress, may be one of the underlying mechanisms for monocyte and microglia activation during neuroinflammation in peroxisomal disorders. Restoration of endothelial function by normalizing SIRT1 levels with substances including resveratrol or by targeting its downstream signaling pathways may give rise to novel therapeutic strategies for these devastating neurodegenerative disorders.
Acknowledgments
We sincerely thank the help provided by the Department of Neurobiology, Xuzhou Medical University and The Affiliated Hospital of Xuzhou Medical University.
Abbreviations
- X-ALD
X-linked adrenoleukodystrophy
- MFP2
multifunctional protein 2
- HSD17B4
hydroxysteroid 17-β dehydrogenase 4
- ABCD1
ATP binding cassette subfamily D Member 1
- SIRT1
sirtuin1
- ATP
adenosine triphosphate
- VLCFA
very long chain fatty acid
- BBB
blood brain barrier
- BMECs
brain microvascular endothelial cells
- HBMECS
human brain microvascular endothelial cells
- NAD
nicotinamide adenosine dinucleotide
- NADP
Nicotinamide adenine dinucleotide phosphate
- TNF-α
tumor necrosis factor-α
- ICAM
intercellular adhesion molecule 1
- VCAM1
vascular cell adhesion molecule 1
- NF-κB
nuclear factor κ-light-chain-enhancer of activated B cells
- KLF4
Krüppel-like factor 4
- MDA
malondialdehyde
- SOD
superoxide dismutase
- RIPA
radioimmunoprecipitation assay
- SD
standard deviation
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81501038).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YG and YW were responsible for conception and design of the study, YZ, YW and YG were responsible for data acquisition and analysis. YG and YZ were responsible for drafting manuscript and figures. GC was responsible for providing general guidance. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Trompier D, Vejux A, Zarrouk A, Gondcaille C, Geillon F, Nury T, Savary S, Lizard G. Brain peroxisomes. Biochimie. 2014;98:102–110. doi: 10.1016/j.biochi.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 3.Mosser J, Douar AM, Sarde CO, Kioschis P, Feil R, Moser H, Poustka AM, Mandel JL, Aubourg P. Putative X-linked adrenoleukodystrophy gene shares unexpected homology with ABC transporters. Nature. 1993;361:726–730. doi: 10.1038/361726a0. [DOI] [PubMed] [Google Scholar]
- 4.Eichler F, Van Haren K. Immune response in leukodystrophies. Pediatr Neurol. 2007;37:235–244. doi: 10.1016/j.pediatrneurol.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 5.van der Voorn JP, Pouwels PJ, Powers JM, Kamphorst W, Martin JJ, Troost D, Spreeuwenberg MD. Correlating quantitative MR imaging with histopathology in X-linked adrenoleukodystrophy. AJNR Am J Neuroradiol. 2011;32:481–489. doi: 10.3174/ajnr.A2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichler FS, Ren JQ, Cossoy M, Rietsch AM, Nagpal S, Moser AB, Frosch MP, Ransohoff RM. Is microglial apoptosis an early pathogenic change in cerebral X-linked adrenoleukodystrophy? Ann Neurol. 2008;63:729–742. doi: 10.1002/ana.21391. [DOI] [PubMed] [Google Scholar]
- 7.Musolino PL, Rapalino O, Caruso P, Caviness VS, Eichler FS. Hypoperfusion predicts lesion progression in cerebral X-linked adrenoleukodystrophy. Brain. 2012;135:2676–2683. doi: 10.1093/brain/aws206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger J, Dorninger F, Forss-Petter S, Kunze M. Peroxisomes in brain development and function. Biochim Biophys Acta. 2016;1863:934–955. doi: 10.1016/j.bbamcr.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verheijden S, Beckers L, De Munter S, Van Veldhoven PP, Baes M. Central nervous system pathology in MFP2 deficiency: Insights from general and conditional knockout mouse models. Biochimie. 2014;98:119–126. doi: 10.1016/j.biochi.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Ferdinandusse S, Denis S, Mooyer PA, Dekker C, Duran M, Soorani-Lunsing RJ, Boltshauser E, Macaya A, Gärtner J, Majoie CB, et al. Clinical and biochemical spectrum of D-bifunctional protein deficiency. Ann Neurol. 2006;59:92–104. doi: 10.1002/ana.20702. [DOI] [PubMed] [Google Scholar]
- 11.Zhang W, Huang Q, Zeng Z, Wu J, Zhang Y, Chen Z. Sirt1 inhibits oxidative stress in vascular endothelial cells. Oxid Med Cell Longev. 2017;2017:7543973. doi: 10.1155/2017/7543973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H, Zhang W, Pan H, Feldser HG, Lainez E, Miller C, Leung S, Zhong Z, Zhao H, Sweitzer S, et al. SIRT1 activators suppress inflammatory responses through promotion of p65 deacetylation and inhibition of NF-κB activity. PLoS One. 2012;7:e46364. doi: 10.1371/journal.pone.0046364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubin LL, Staddon JM. The cell biology of the blood-brain barrier. Annu Rev Neurosci. 1999;22:11–28. doi: 10.1146/annurev.neuro.22.1.11. [DOI] [PubMed] [Google Scholar]
- 14.Ueno M. Molecular anatomy of the brain endothelial barrier: An overview of the distributional features. Curr Med Chem. 2007;14:1199–1206. doi: 10.2174/092986707780597943. [DOI] [PubMed] [Google Scholar]
- 15.Pardridge WM. Blood-brain barrier delivery. Drug Discov Today. 2007;12:54–61. doi: 10.1016/j.drudis.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: Understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 17.Morató L, Ruiz M, Boada J, Calingasan NY, Galino J, Guilera C, Jové M, Naudí A, Ferrer I, Pamplona R, et al. Activation of sirtuin 1 as therapy for the peroxisomal disease adrenoleukodystrophy. Cell Death Differ. 2015;22:1742–1753. doi: 10.1038/cdd.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamatovic SM, Martinez-Revollar G, Hu A, Choi J, Keep RF, Andjelkovic AV. Decline in Sirtuin-1 expression and activity plays a critical role in blood-brain barrier permeability in aging. Neurobiol Dis. 2019;126:105–116. doi: 10.1016/j.nbd.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orimo M, Minamino T, Miyauchi H, Tateno K, Okada S, Moriya J, Komuro I. Protective role of SIRT1 in diabetic vascular dysfunction. Arterioscler Thromb Vasc Biol. 2009;29:889–894. doi: 10.1161/ATVBAHA.109.185694. [DOI] [PubMed] [Google Scholar]
- 20.Zhang QJ, Wang Z, Chen HZ, Zhou S, Zheng W, Liu G, Wei YS, Cai H, Liu DP, Liang CC. Endothelium-specific overexpression of class III deacetylase SIRT1 decreases atherosclerosis in apolipoprotein E-deficient mice. Cardiovasc Res. 2008;80:191–199. doi: 10.1093/cvr/cvn224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vassallo PF, Simoncini S, Ligi I, Chateau AL, Bachelier R, Robert S, Morere J, Fernandez S, Guillet B, Marcelli M, et al. Accelerated senescence of cord blood endothelial progenitor cells in premature neonates is driven by SIRT1 decreased expression. Blood. 2014;123:2116–2126. doi: 10.1182/blood-2013-02-484956. [DOI] [PubMed] [Google Scholar]
- 22.Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donmez G, Outeiro TF. SIRT1 and SIRT2: Emerging targets in neurodegeneration. EMBO Mol Med. 2013;5:344–352. doi: 10.1002/emmm.201302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Chen J, Sun L, Xu Y. SIRT1 deacetylates KLF4 to activate Claudin-5 transcription in ovarian cancer cells. J Cell Biochem. 2018;119:2418–2426. doi: 10.1002/jcb.26404. [DOI] [PubMed] [Google Scholar]
- 26.Obermeier B, Daneman R, Ransohoff RM. Development, maintenance and disruption of the blood-brain barrier. Nat Med. 2013;19:1584–1596. doi: 10.1038/nm.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wekerle H, Schwab M, Linington C, Meyermann R. Antigen presentation in the peripheral nervous system: Schwann cells present endogenous myelin autoantigens to lymphocytes. Eur J Immunol. 1986;16:1551–1557. doi: 10.1002/eji.1830161214. [DOI] [PubMed] [Google Scholar]
- 28.Cross AH, Dolich S, Raine CS. Antigen processing of myelin basic protein is required prior to recognition by T cells inducing EAE. Cell Immunol. 1990;129:22–31. doi: 10.1016/0008-8749(90)90183-R. [DOI] [PubMed] [Google Scholar]
- 29.Raine CS, Cannella B, Duijvestijn AM, Cross AH. Homing to central nervous system vasculature by antigen-specific lymphocytes. II Lymphocyte/endothelial cell adhesion during the initial stages of autoimmune demyelination Lab Invest. 1990;63:476–489. [PubMed] [Google Scholar]
- 30.Zlokovic BV. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron. 2008;57:178–201. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Wilson EH, Weninger W, Hunter CA. Trafficking of immune cells in the central nervous system. J Clin Invest. 2010;120:1368–1379. doi: 10.1172/JCI41911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Man S, Ubogu EE, Williams KA, Tucky B, Callahan MK, Ransohoff RM. Human brain microvascular endothelial cells and umbilical vein endothelial cells differentially facilitate leukocyte recruitment and utilize chemokines for T cell migration. Clin Dev Immunol. 2008;2008:384982. doi: 10.1155/2008/384982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller DW. Immunobiology of the blood-brain barrier. J Neurovirol. 1999;5:570–578. doi: 10.3109/13550289909021286. [DOI] [PubMed] [Google Scholar]
- 34.Minagar A, Alexander JS. Blood-brain barrier disruption in multiple sclerosis. Mult Scler. 2003;9:540–549. doi: 10.1191/1352458503ms965oa. [DOI] [PubMed] [Google Scholar]
- 35.Droogan AG, McMillan SA, Douglas JP, Hawkins SA. Serum and cerebrospinal fluid levels of soluble adhesion molecules in multiple sclerosis: Predominant intrathecal release of vascular cell adhesion molecule-1. J Neuroimmunol. 1996;64:185–191. doi: 10.1016/0165-5728(95)00174-3. [DOI] [PubMed] [Google Scholar]
- 36.Plumb J, McQuaid S, Mirakhur M, Kirk J. Abnormal endothelial tight junctions in active lesions and normal-appearing white matter in multiple sclerosis. Brain Pathol. 2002;12:154–169. doi: 10.1111/j.1750-3639.2002.tb00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calabrese V, Cornelius C, Dinkova-Kostova AT, Calabrese EJ, Mattson MP. Cellular stress responses, the hormesis paradigm, and vitagenes: Novel targets for therapeutic intervention in neurodegenerative disorders. Antioxid Redox Signal. 2010;13:1763–1811. doi: 10.1089/ars.2009.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Domenico F, Perluigi M, Butterfield DA, Cornelius C, Calabrese V. Oxidative damage in rat brain during aging: Interplay between energy and metabolic key target proteins. Neurochem Res. 2010;35:2184–2192. doi: 10.1007/s11064-010-0295-z. [DOI] [PubMed] [Google Scholar]
- 39.Leuner K, Hauptmann S, Abdel-Kader R, Scherping I, Keil U, Strosznajder JB, Eckert A, Müller WE. Mitochondrial dysfunction: The first domino in brain aging and Alzheimer's disease? Antioxid Redox Signal. 2007;9:1659–1675. doi: 10.1089/ars.2007.1763. [DOI] [PubMed] [Google Scholar]
- 40.Martinez A, Portero-Otin M, Pamplona R, Ferrer I. Protein targets of oxidative damage in human neurodegenerative diseases with abnormal protein aggregates. Brain Pathol. 2010;20:281–297. doi: 10.1111/j.1750-3639.2009.00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galino J, Ruiz M, Fourcade S, Schlüter A, López-Erauskin J, Guilera C, Jove M, Naudi A, García-Arumí E, Andreu AL, et al. Oxidative damage compromises energy metabolism in the axonal degeneration mouse model of X-adrenoleukodystrophy. Antioxid Redox Signal. 2011;15:2095–2107. doi: 10.1089/ars.2010.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlüter A, Espinosa L, Fourcade S, Galino J, López E, Ilieva E, Morató L, Asheuer M, Cook T, McLaren A, et al. Functional genomic analysis unravels a metabolic-inflammatory interplay in adrenoleukodystrophy. Hum Mol Genet. 2012;21:1062–1077. doi: 10.1093/hmg/ddr536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fourcade S, López-Erauskin J, Galino J, Duval C, Naudi A, Jove M, Kemp S, Villarroya F, Ferrer I, Pamplona R, et al. Early oxidative damage underlying neurodegeneration in X-adrenoleukodystrophy. Hum Mol Genet. 2008;17:1762–1773. doi: 10.1093/hmg/ddn085. [DOI] [PubMed] [Google Scholar]
- 44.Ferdinandusse S, Finckh B, de Hingh YC, Stroomer LE, Denis S, Kohlschütter A, Wanders RJ. Evidence for increased oxidative stress in peroxisomal D-bifunctional protein deficiency. Mol Genet Metab. 2003;79:281–287. doi: 10.1016/S1096-7192(03)00108-2. [DOI] [PubMed] [Google Scholar]
- 45.Jia Y, Gao P, Chen H, Wan Y, Zhang R, Zhang Z, Yang R, Wang X, Xu J, Liu D. SIRT1 suppresses PMA and ionomycin-induced ICAM-1 expression in endothelial cells. Sci China Life Sci. 2013;56:19–25. doi: 10.1007/s11427-012-4407-7. [DOI] [PubMed] [Google Scholar]
- 46.Melotti P, Nicolis E, Tamanini A, Rolfini R, Pavirani A, Cabrini G. Activation of NF-kB mediates ICAM-1 induction in respiratory cells exposed to an adenovirus-derived vector. Gene Ther. 2001;8:1436–1442. doi: 10.1038/sj.gt.3301533. [DOI] [PubMed] [Google Scholar]
- 47.Ledebur HC, Parks TP. Transcriptional regulation of the inter-cellular adhesion molecule-1 gene by inflammatory cytokines in human endothelial cells. Essential roles of a variant NF-kappa B site and p65 homodimers. J Biol Chem. 1995;270:933–943. doi: 10.1074/jbc.270.2.933. [DOI] [PubMed] [Google Scholar]
- 48.Ma Y, Xu C, Wang W, Sun L, Yang S, Lu D, Liu Y, Yang H. Role of SIRT1 in the protection of intestinal epithelial barrier under hypoxia and its mechanism. Zhonghua Wei Chang Wai Ke Za Zhi. 2014;17:602–606. In Chinese. [PubMed] [Google Scholar]
- 49.Ma J, Wang P, Liu Y, Zhao L, Li Z, Xue Y. Krüppel-like factor 4 regulates blood-tumor barrier permeability via ZO-1, occludin and claudin-5. J Cell Physiol. 2014;229:916–926. doi: 10.1002/jcp.24523. [DOI] [PubMed] [Google Scholar]
- 50.Liu CW, Sung HC, Lin SR, Wu CW, Lee CW, Lee IT, Yang YF, Yu IS, Lin SW, Chiang MH, et al. Resveratrol attenuates ICAM-1 expression and monocyte adhesiveness to TNF-α-treated endothelial cells: Evidence for an anti-inflammatory cascade mediated by the miR-221/222/AMPK/p38/NF-κB pathway. Sci Rep. 2017;7:44689. doi: 10.1038/srep44689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaisar MA, Prasad S, Cucullo L. Protecting the BBB endothelium against cigarette smoke-induced oxidative stress using popular antioxidants: Are they really beneficial? Brain Res. 2015;1627:90–100. doi: 10.1016/j.brainres.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwager J, Richard N, Widmer F, Raederstorff D. Resveratrol distinctively modulates the inflammatory profiles of immune and endothelial cells. BMC Complement Altern Med. 2017;17:309. doi: 10.1186/s12906-017-1823-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.