Abstract
The present study investigated the methylation of CpG sites in the cyclooxygenase (COX)-2 promoter via nuclear factor (NF)-κB transcriptional regulation and elucidated its effect on the COX-2 transcriptional expression in a ketamine-induced ulcerative cystitis (KIC) animal model. The results of the present study revealed that ketamine treatment induced NF-κB p65 translocation to nuclei and activated COX-2 expression and prostaglandin (PGE)2 production in bladder tissue, whereas COX-2 inhibitor suppressed the inflammatory effect. Moreover, DNA hypomethylation of the COX-2 promoter region located from -1,522 to -829 bp might contribute to transcriptional regulation of COX-2 expression and induce a pro-inflammatory response in KIC. Ketamine treatment increased the binding of NF-κB and permissive histone H3 lysine-4 (H3K4)m3, but caused a decrease in the repressive histone H3K27m3 and H3K36m3 on the COX-2 promoter ranging from -1,522 to -1,331 bp as determined by a chromatin immunoprecipitation assay. Moreover, in the ketamine group, the level of Ten-Eleven-Translocation methylcytosine dioxygenase for demethylation as determined by reverse transcription-quantitative PCR assay was increased in comparison with the control group, but that was not the case for the level of DNA methyltransferases for methylation. The present findings revealed that there was a hypomethylation pattern of the COX-2 promoter in association with the level of COX-2 transcription in KIC.
Keywords: bladder, ketamine, ulcerative cystitis, cyclooxygenase-2 methylation, nuclear factor-κB, methyltransferase
Introduction
The symptoms and signs of ketamine-induced ulcerative cystitis (KIC) include increased voiding frequency, non-voiding contraction, hematuria and dysuria. Previous experimental observations revealed that KIC occurred in bladder mucosa and decreased bladder capacity, enhanced detrusor hyperactivity and thereafter induced interstitial fibrosis (1). The symptoms of KIC are similar to interstitial cystitis (2,3). Histological features of ketamine-associated damage in a rat bladder were characterized by an ulcerated urothelium, erythrocyte accumulation (hemorrhages), mononuclear cell infiltration and an increased interstitial fibrosis between detrusor smooth muscle bundles (1,4-6). Clinical characteristics of ketamine abusers exhibit urinary frequency, urgency and at times urinary incontinence. Clinical studies in bladder tissue of KIC patients have shown increases in the infiltration of eosinophil and mast cells as well as serum immunoglobulin (Ig)E level, which displayed an association with hypersensitivity and/or allergic reactions (7,8). In spite of the above progress, the pathophysiological mechanism of the bladder voiding dysfunction in KIC patients is still unclear.
The mechanism of epigenetic regulation involves the CpG site methylation of promoter regions and the modification of DNA and histones by altering chromatin structure (9-11). DNA meth-ylation represses transcription by interfering with transcription factor binding and indirectly by recruiting methyl-CpG-binding proteins and reducing chromatin remodeling activities (12). Moreover, the CpG site methylation is mediated by DNA methyltransferases (DNMTs), which catalyze the addition of a methyl group to cytosine (13,14). The DNMT family enzymes include DNMT1, DNMT3a and DNMT3b. DNMT1 preserves the methyltransferase by binding to hemi-methylated CpG sites and methylates the cytosine on the newly synthesized strand after DNA replication, whereas DNMT3a/DNMT3b are required for the de novo genomic methylation of DNA (15). In contrast, the Ten-Eleven-Translocation (TET) dioxygenase family, including TET1, TET2 and TET3, mediates active DNA demethylation and hydroxylate-methylated DNA by converting 5-methylcyto-sine to 5-hydroxymethylcytosine to regulate DNA methylation status. The role of DNMT and TET proteins in regulating the epigenetic mechanisms includes DNA methylation at CpG sites and histone methylation, particularly histone H3 lysine-4 (H3K4) and H3K27 (16). Histone-lysine methylation is associated with either gene activation or repression depending on the histone residue modification. For example, methylation of H3K4 is associated with transcriptional activation, whereas H3K9, H3K27 and H3K36 are related to transcriptional repression (17-19).
DNA methylation plays a critical role in normal development, while aberrant hypermethylation of 5′ CpG sites are implicated in the transcriptional silencing of the cyclooxygenase (COX)-2 gene in the pathogenesis of the inflammatory diseases (20-22) and neoplastic disorders (23-25). Two types of DNA methylation changes are observed in cancer: Hypomethylation, linked to chromosomal instability and activity (26), and hyper-methylation that can lead to transcriptional silencing (27). In pathological diseases, overexpression of COX-2 and abnormal production of COX-induced prostaglandin E2 (PGE2) have been reported (28). Certain periodontal bacteria can induce epigenetic alterations in the DNA methylation of the COX-2 gene promoter and affect the transcriptional regulation of COX-2 in chronic periodontitis (20). Helicobacter pylori infection causes aberrant DNA methylation of the COX-2 gene promoter in the gastric mucosa of patients. Treatment with the DNA-demethylating drug 5-aza-deoxycytidine (a DNA methyl-transferase inhibitor) was found to increase COX-2 expression and prostaglandin synthesis (29,30).
The COX-2 gene promoter has several potential response elements for transcription factors, such as nuclear factor (NF)-κB, nuclear factor of activated T cells/NF-interleukin (IL)-6 (NFAT/NF-IL6) and activator protein-1 (AP-1) (6,31). Inflammatory stimuli cause NF-κB dimers (p65 and p50 subunits) to dissociate from cytoplasmic inhibitors, followed by NF-κB p65 translocation and binding to specific gene promoter sequences (32,33). An animal study with chemically-induced hemorrhagic cystitis revealed that COX-2 upregulation played an important role in bladder inflammation (34). Moreover, the authors' previous results demonstrated that ketamine and norketamine accelerated NF-κB p65 translocation and induced the upregulation of COX-2 expression in bladder urothelium (6). Promoter-deletion analysis of the rat COX-2 promoter region ranging from −918 to −250 bp suggested that NF-κB was a crucial transcription factor for COX-2 gene activation (6). However, promoter deletion analysis did not provide any conclusion with respect to which specific binding sites for NF-κB were involved in the COX-2 modification response to ketamine and metabolites norketamine.
In the present study, specific binding sequences (sites) of the COX-2 promoter responding to NF-κB were identified by focusing on the promoter ranging from -1,522 to -71 bp. The authors hypothesized that ketamine-induced chronic inflammation is associated with an altered DNA methylation level within the COX-2 promoter and with change in transcriptional modification. To test this hypothesis, the potential alteration in DNA methylation status of the COX-2 promoter via NF-κB activation and its effect on the transcriptional COX-2 expression in KIC were investigated. Moreover, to improve an understanding of the epigenetic regulation of the COX-2 gene via NF-κB activation, it was important to determine specific NF-κB binding sequences within the COX-2 promoter and to investigate methylation associated enzymes responsible for the promoter COX-2 activity.
Materials and methods
Animals and ketamine administration
A total of 45 adult female Sprague-Dawley (S-D) rats weighing 250 g were purchased from the Animal Center of BioLASCO. All S-D rats were housed in a standard room with constant temperature (23±2°C), a relative humidity of 70%, and a 12 h light-dark cycle (light on at 07:00 a.m., light off at 07:00 p.m.; 100 lux at cage level) conditions with ad libitum access to food and water. S-D rats were divided into three experimental groups: i) The control (saline) group, ii) the ketamine-treated group and iii) the ketamine combined with COX-2 inhibitor (ketamine+COX-2 inhibitor) group, which received 0.9% saline, ketamine (30 mg/kg/day, Pfizer, Inc.) intraperitoneal (IP) injection, and ketamine combined with COX-2 inhibitor (Parecoxib sodium, Dynastat®, Pfizer, Inc.; 10 mg/kg/day) IP injection for 3 months respectively (4-6). At the end of the experimental period, bladder tissues and whole blood were collected from all animals. After induction of anesthesia with 4% isoflurane, rats were subjected to cardiac puncture or euthanasia and total blood collection. Blood samples were left to clot for 2 h on the ice, then centrifuged at 1,500 × g at 4°C for 15 min to separate out the serum samples which were preserved at −80°C in the refrigerator for further detection. S-D rats were perfused with 09% saline solution through the left ventricle and the bladders were removed and washed in ice-cold PBS and carefully dissected in a horizontal plane into two portions: One was fixed in 4% paraformaldehyde solution at 4°C for 24 h for histological analysis; another was placed in liquid nitrogen for mRNA, protein and chromatin immunoprecipitation (ChIP) analysis. This study was approved by the Animal Care and Treatment Committee of Kaohsiung Medical University. All experiments were conducted according to the guidelines for laboratory animal care.
Western blot analysis
Nuclear and cytoplasmic extracts of rat bladder tissues were prepared according to the protocols modified from the method described by NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific, Inc.) (6). The protein concentration was determined using Bicinchoninic Acid Protein Assay kits (Pierce Biotechnology; Thermo Fisher Scientific, Inc.). A total of 50 µg of protein from the bladders were loaded on a 10% SDS polyacrylamide electrophoresis gel and transferred to PVDF membranes (Immobilon-P; EMD Millipore). Afterward, the PVDF membrane was blocked with 5% non-fat-milk in PBS with Tween-20 (PBST) for 2 h at room temperature. Primary antibodies COX-2 (cat. no. 160126; Cayman Chemical Company; rabbit IgG, 1:1,000; MW, 72 kDa) and NF-κB p65 (cat. no. NBP1-48427; Novus Biologicals LLC; rabbit IgG; 1:1,000; MW, 65 kDa) were used for determining protein expression at 4°C overnight, and were incubated with the horseradish peroxidase-conjugated goat anti-mouse (cat. no. 115-035-003; Jackson ImmunoResearch Laboratories; 1:7,000-1:100,000) and goat anti-rabbit (cat. no. 111-035-003; Jackson ImmunoResearch Laboratories; 1:7,000) secondary antibody for 1 h at room temperature. Lamin A/C (cat. no. 2032S; Cell Signaling Technology, Inc.; mouse IgG; 1:5,000; MW, 70 kDa) served as the internal control for nuclei extract and β-actin (cat. no. MAB1501; Upstate Biotechnology, Inc.; mouse IgG; 1:5,000; MW, 41-43 kDa) served as the internal control for total protein and cytoplasmic extract. The band intensity was quantified by densitometry using image analysis software (ImageJ, version 1.49; National Institutes of Health). In each experiment, negative control without the primary antibody was analyzed.
ELISA
The level of PGE2 production was used to serve as determining COX activity. Determination of PGE2 levels in serum was performed by Prostaglandin E2 ELISA kit following the manufacturer's protocol (Abcam; cat. no. ab133021). A mouse IgG antibody was precoated onto 96-well plates. Serum samples were added to the wells, along with an alkaline phosphatase (AP) conjugated-PGE2 antibody. After reagent incubation, substrate was added and catalyzed by AP to produce a yellow color. The intensity of the yellow coloration was inversely proportional to the amount of PGE2.
Immunofluorescence
For in vivo bladder section, immunostaining was performed according to published methods (6). The sections were then double-stained with the primary antibody to NF-κB p65 (cat. no. 6956S; Cell Signaling Technology, Inc.; mouse IgG2b; 1:50-100) and COX-2 (cat. no. 4212-1; Epitomics; rabbit IgG; 1:50) at 4°C overnight, then incubated with goat anti-mouse IgG Alexa Fluor 568 (cat. no. A-11004; Invitrogen; Thermo Fisher Scientific, Inc.; 1:800) and goat anti-rabbit IgG Alexa Fluor 488 secondary antibody (cat. no. A-27034; Invitrogen; Thermo Fisher Scientific, Inc.; 1:800) conjugated to fluorescein isothiocyanate (FITC) for NF-κB and rhodamine for COX-2 for 1 h at room temperature. The nuclei of the cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; 1:5,000) for 10 min at room temperature. Immunofluorescence images were observed using a Leica DMI6000 inverted microscope (Leica Microsystems, Inc.).
Analysis and design of rat COX-2 promoter
The DNA sequence of COX-2 promoter was analyzed with MethPrimer software (version 2.0) (35). The promoter regions of the rat COX-2 gene (accession number NM_017232) ranging from −1,522 to −71 contains 1,452 bp (nt) and 44 CpG sites were identified by nucleotide sequence analysis for potential methylation (www.genome.ucsc.edu). In this region, a COX-2 promoter construct was designed for five subregions, including Region I (−71/−299), II (−286/−551), III (−548/−830), IV (−829/−1,195) and V (−1,181/−1,522), for bisulfite methylation-specific PCR (MSP), cloning, and genomic sequencing. Additionally, five potential motifs similar to the consensus binding sites for NF-κB in the region were identified by TFBIND software (version 8.3; http://tfbind.hgc.jp/; Table I).
Table I.
Primer sequences used in methylation-specific polymerase chain reaction, bisulfite DNA sequencing, ChIP assay and RT-qPCR.
| A, Primers for methylation-specific transcription PCR
| ||||
|---|---|---|---|---|
| Name | Primer sequences | Sequence range | Product | Methylated NF-κB binding sequence |
| Rat-COX2 region I Bisulfite-sequence |
F: 5′-TTGTTTTTATGGGTATTATGTAATTGG-3′ R: 5′-ACAAAACACAAAACTAAATTCCTTC-3′ |
−299 to −71 | 229 bp | 5′-ggggaaagtcga-3′ (−235 to −224) |
| Rat-COX2 region II Bisulfite-sequence |
F: 5′-AAGGGGATTTTTTTAGTTAGGATTT-3′ R: 5′-ACCCATAAAAACAAACTTTACTCAC-3′ |
−551 to −286 | 266 bp | 5′-ggggattttt-3′ (-549 to -540) |
| Rat-COX2 region III Bisulfite-sequence |
F: 5′-TAGGGAGGAAAATATTTTAAAGTAATG-3′ R: 5′-CCTTACCTCTCCCCACTAAAAC-3′ |
−830 to −548 | 283 bp | 5′-ggaaaatattt-3′ (−824 to −814) |
| Rat-COX2 region IV Bisulfite-sequence |
F: 5′-AGTTTTTTATTTTTTTGTTTTATT-3′ R: 5′-TACTATTACATAACTTTTATCATTTTAATC-3′ |
−1,195 to −829 | 367 bp | - |
| Rat-COX2 region V Bisulfite-sequence |
F: 5′-AGTATGTATATGAAGTAAATAGTTAAAAA-3′ R: 5′-AAAAATAAAAAAACTAAAACATTCAATTAA-3 |
−1,522 to −1,181 | 342 bp | 5′-gtcgattttt-3′ (−1,413 to −1,404) 5′-cggtagttttc-3′ (−1,442 to −1,432) |
| COX2-Methylation | F: 5′-AAGGGGATTTTTTTAGTTAGGATTTC-3′ R: 5′-TCCAAACGCCCTATAATTCG-3′ |
−551 to −393 | 159 bp | |
| COX2-Unmethylation | F: 5′-AGGGGATTTTTTTAGTTAGGATTTT-3′ R: 5′-TTCCAAACACCCTATAATTCACT-3′ |
−550 to −392 | 159 bp | |
|
B, ChIP primers for NF-κB binding sites of Cox-2 promoter sequence | ||||
| Name | Primer sequences | Sequence range | Product | Predicted NF-κB binding sequence |
|
| ||||
| Rat-COX2 -Primer I | F: 5′-TGCCCCTATGGGTATTATGC-3′ R: 5′-CTGAAGCTCTCCGCTCAGTT-3′ |
−298 to −117 | 182 bp | 5′-ggggaaagccga-3′ (−235 to −224) |
| Rat-COX2 -Primer II | F: 5′-GACAGCAGCCCTCTCATTTC-3′ R: 5′-CGGAGGAGCAAGAGAATGTC-3′ |
−660 to −484 | 177 bp | 5′-ggggattccc-3′ (−549 to −540) |
| Rat-COX2 -Primer III | F: 5′-TGTAAACGTAAACGTGGACAAAA-3′ R: 5′-CCTTTCCCAGAGACAGATGC-3′ |
−948 to −751 | 198 bp | 5′-ggaaaatacct-3′ (−824 to −814) |
| Rat-COX2 -Primer IV | F: 5′-AGCATGCACATGAAGCAAAC-3′ R: 5′-GCCCTGCTCAAAAGAAAACA-3′ |
−1,522 to −1,331 | 192 bp | 5′-gtcgattccc-3′ (−1,413 to −1,404) 5′-cggtagtttcc-3′ (−1,442 to −1,432) |
|
C, RT-qPCR primers for DNMT and TET methylcytosine dioxygenase | ||||
| Gene | Primer sequences | Accession no.a | Tm (°C) | Product size (bp) |
|
| ||||
| Rat-DNMT1 | F: 5′-GGAGGTGTCCTAACTTGGC-3′ R: 5′-GGGTGACGGCAACTCTGGTA-3′ |
NM_053354.3 | 60 | 80 |
| Rat-DNMT3a | F: 5′-GTGCTTACCAATACGATGACGA-3′ R: 5′-ATCCACACACTCCACACAAAAG-3′ |
NM_001003958.1 | 60 | 122 |
| Rat-DNMT3b | F: 5′-GATGATGGAGATGGCTCTGATA-3′ R: 5′-GGCTGGAGATACTGTTGCTGTT-3′ |
NM_001003959.1 | 60 | 121 |
| Rat-TET1 | F: 5′-GAAACCCTGAATTGGCAAAA-3′ R: 5′-GGGTGAGCTTTCTGATCGAC-3′ |
XM_017601794.1 | 60 | 157 |
| Rat-TET2 | F: 5′-TCGGAGGAGAAGAGTCAGGA-3′ R: 5′-TAGGGCTTGCATTTTCCATC-3′ |
XM_006224264.3 | 60 | 167 |
| Rat-TET3 | F: 5′-ATGGCATGAAACCACCCAAC-3′ R: 5′-ACTTGATCTTCCCCTCCAGC-3′ |
XM_008763094.2 | 60 | 81 |
Available at https://www.ncbi.nlm.nih.gov/nuccore. F, forward; R, reverse; Tm, temperature; DNMT, DNA methyltransferase; TET, Ten-Eleven-Translocation; COX, cyclooxygenase; ChIP, chromatin immunoprecipitation; RT-q, reverse transcription-quantitative; NF, nuclear factor.
MSP, cloning and genomic sequencing
The DNA methylation pattern in the COX-2 promoter was analyzed by MSP. Genomic DNAs from rat bladder tissues were isolated using the PureLink Genomic DNA Mini kit (Invitrogen; Thermo Fisher Scientific, Inc.). Total extracted DNA was used for bisulfite-mediated conversion of unmethylated cytosines to uracils using an EpiTect Bisulfite kit (Qiagen, Inc.). Bisulfite-treated genomic DNA was amplified with primers (Table I) specific to the CpG sites within COX-2 promoter sequence. After bisulfite treatment, unmethylated cytosine residues were changed into uracil residues, whereas methylated cytosine remained unmodified. The differentiation between methylated and unmethylated sequences could be amplified using specific primers that targeted the uracil or the cytosine nucleotide. COX-2 promoter temperature profiles for amplification were 5 min at 95°C, 30 cycles of 45 sec at 94°C, 45 sec at 63°C and 45 sec at 72°C. All reactions were repeated three times to ensure reproducibility of results. Colonies showing positive PCR fragment insertion were selected and the purified fragments were cloned into a PCR 2.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. PCR products from 4 clones for each individual sample were sequenced. The completeness of the conversion of unmethylated cytosines to uracils was confirmed by evaluation of non-CpG cytosine conversion. The DNA methylation patterns were analyzed by sequencing.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from bladder tissues with the use of the RNeasy Mini kit (Qiagen, Inc.). The cDNA was then synthesized from 1 µg of total RNA using the Omniscript kit (Qiagen, Inc.) by random decamer primers (Applied Biosystems/Ambion; Thermo Fisher Scientific, Inc.). The reaction was incubated at 37°C for 60 min, inactivated by heating at 95°C for 5 min, and stored at -20°C. An SYBR-Green I kit (Takara Biotechnology, Co., Ltd.) was used and all the primers of rat DNA methyltransferase (DNMT1, 3a, and 3b) and TET enzymes (TET1, TET2, and TET3) were listed in Table I. RT-qPCR was performed in a 7500 Sequence Detection System apparatus (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the following thermocycling conditions: Initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The relative expression levels of each targeted gene were normalized by subtracting the corresponding β-actin threshold cycle (Cq) values using the ΔΔCq comparative method (36). Six samples for each group were used and run in triplicate.
ChIP
A total of 70 mg bladder tissues were homogenized into small pieces and treated with 1% formaldehyde for 10 min at 37°C, followed by sonication of DNAs. ChIP was performed with primary antibodies for anti-Histone H3K4 (tri methyl K4; cat. no. AB8580; Abcam; rabbit polyclonal IgG; 1:100), H3K9 (cat. no. AB8898; Abcam; rabbit polyclonal IgG; 1:100), H3K27 (cat. no. AB6002; Abcam; mouse monoclonal IgG; 1:100), H3K36 (cat. no. AB9050; Abcam; rabbit polyclonal IgG; 1:100) and H3K79 (cat. no. AB2621; Abcam; rabbit poly-clonal IgG; 1:100), NF-κB p65 (cat. no. 6956S; Cell Signaling Technology, Inc.; mouse monoclonal IgG2b; 1:100), and IgG (cat. no. 6180-01; negative control; SouthernBiotech; rabbit IgG; 1:500) overnight. Immune complexes were collected using a protein G agarose, Fast flow (50% slurry; EMD Millipore) and the DNA was reverse cross-linked, extracted, and quantified on a Taqman SDS 7900HT. All the primers were listed in Table I. PCR conditions were as follows: Initial denaturation at 95°C for 5 min followed by 30 cycles of denaturation at 94°C for 45 sec, annealing at 63°C for 45 sec and extension at 72°C for 45 sec; final extension at 72°C; and holding at 4°C. PCRs run on the ABI 7700 Taqman thermocycler. All Taqman reagents were purchased from Applied Biosystems; Thermo Fisher Scientific, Inc. The relative intensities of the amplified products were normalized according to the input DNAs.
Statistical analysis
Each experiment was carried out at least three times independently. Data were expressed as the mean ± standard deviation and subjected to one-way analysis of variance to find the significant difference in various parameters between the control and the treated groups. The post hoc Tukey honest significant difference tests were used to make comparisons between the control and each of the treated groups and to calculate P-values for comparison, P<0.05 was considered to indicate a statistically significant difference. The statistical analyses were implemented using the IBM SPSS Statistics package (version 21.0; IBM, Corps.).
Results
Increases in COX-2 expression and NF-κB p65 translocation after ketamine treatment
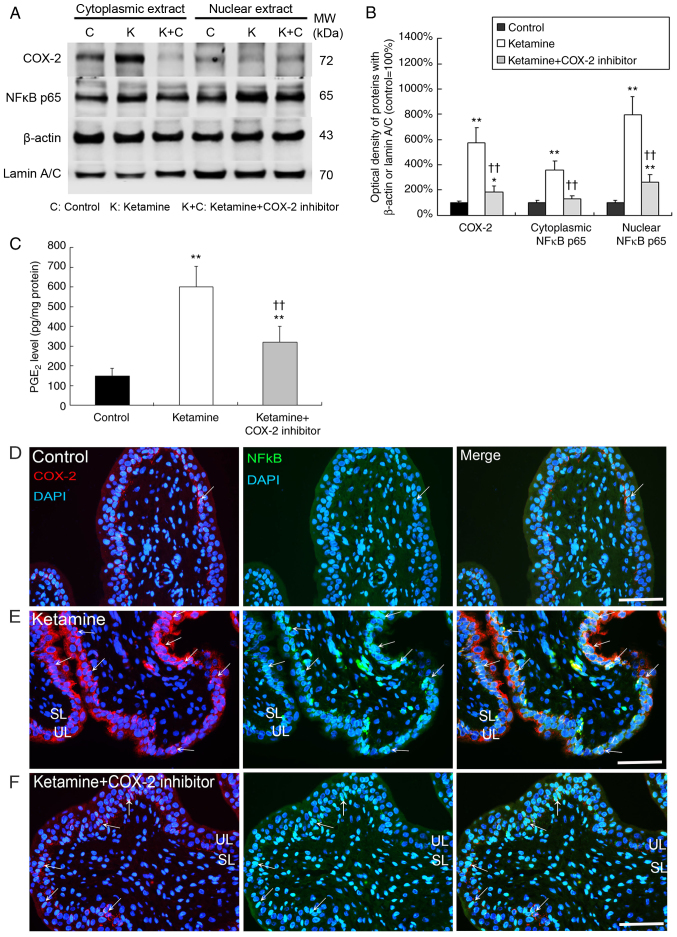
In Fig. 1A and B, western blot analysis showed that the expression of COX-2, cytoplasmic NF-κB p65 and nuclear NF-κB p65 was significantly increased by 5.8-, 3.7- and 8.0-fold in the bladder tissue of the ketamine groups respectively, as compared with the control group (P<0.01). In the ketamine+COX-2 inhibitor group, the expression of COX-2 and nuclear NF-κB p65 was significantly increased by 2.0- and 2.9-fold respectively, compared with the control group (P<0.05). These data indicated that the ketamine+COX-2 inhibitor group resulted in more significant decline in NF-κB p65 expression in nucleus and cytoplasm compared with the ketamine treatment group.
Figure 1.
Ketamine treatment induced the translocation of NF-κB p65 to activate COX-2 expression and PGE2 production. The COX-2 and NF-κB p65 expression of bladder tissue was measured by (A) western blot analysis and (B) quantified against β-actin or Lamin A/C. Results were normalized as the control group=100%. (C) An ELISA was applied to assess the level of PGE2 production for COX-2 enzyme activity in the bladder. Values are presented as the mean ± standard deviation for n=6. *P<0.05 and **P<0.01 vs. the control group. ††P<0.01 vs. the ketamine group. Double immunostaining of COX-2 (red, left panels) and NF-κB p65 (green, middle panels) for bladder is shown in the (D) control group, the (E) ketamine group and the (F) ketamine+COX-2 inhibitor group. Nuclear DNA was labeled with DAPI (blue). The merged image from left and middle panels (yellow, right panels) is shown. Most COX-2 staining co-labeled with NF-κB p65 (arrows) in the UL and SL was identified in the ketamine group. Scale bar=100 µm. SL, suburothelial layer; UL, urothelial layer; COX, cyclooxygenase; NF, nuclear factor; PGE, prostaglandin.
In Fig. 1C, PGE2 production in the control group was 150 pg/mg, which was significantly increased by 3.9- and 2.1-fold in the ketamine and the ketamine+COX-2 inhibitor groups respectively (P<0.01). These findings demonstrated that ketamine treatment induced NF-κB p65 translocation and activated COX-2 expression and PGE2 production in bladder tissue. Whereas the COX-2 inhibitor suppressed the level of NF-κB p65 translocation and COX-2 expression as well as PGE2 production.
Double-staining for the distribution of COX-2 (red) and NF-κB p65 (green) proteins was performed. In the control group, weak staining for the co-labeling of NF-κB p65 and COX-2 was found (Fig. 1D). However, in the ketamine group, most NF-κB p65/DAPI cells co-stained with COX-2 expression were enhanced and restricted to the thinner and disrupted urothelium (Fig. 1E; arrows). In the ketamine+COX-2 inhibitor group, double-staining of NF-κB p65 and COX-2 was decreased, as compared to the ketamine group (Fig. 1F; arrows). These observations revealed that COX-2 expression coincided with NF-κB p65 after ketamine treatment, implying that COX-2 and NF-κB p65 were synthesized during the inflammatory process of KIC, whereas COX-2 inhibitor reduced NF-κB p65 translocation and COX-2 expression.
Methylation sequencing analysis for the upstream sequences of COX-2 promoter region
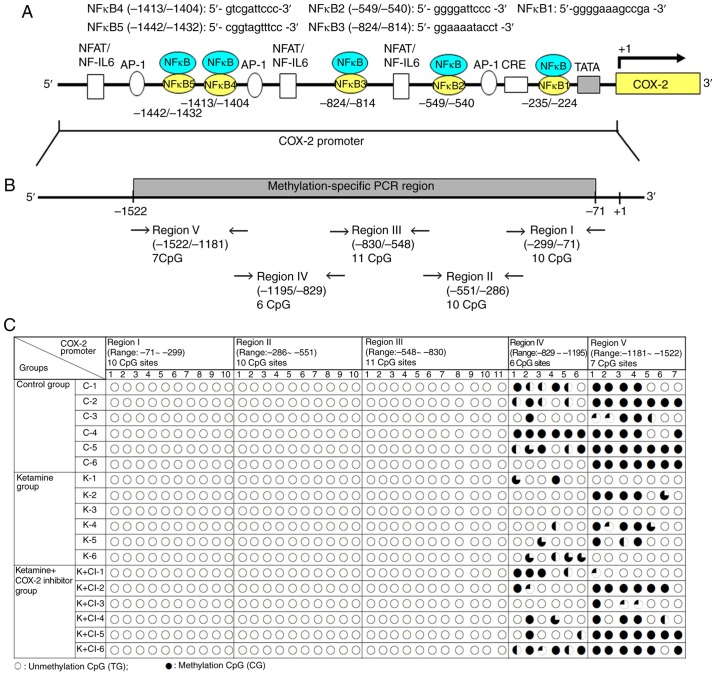
The present study also explored whether NF-κB translocation after ketamine treatment was associated with COX-2 hypomethylation and transcriptional modification in KIC. To determine which NF-κB binding sites within COX-2 promoter were involved in COX-2 activation and DNA methylation in KIC, the methylation level of CpG sites within the COX-2 promoter region ranging from -1,522 to -71 bp in relation to the transcriptional starting site was analyzed by bisulfite MSP and genomic sequencing analysis. A schematic diagram for COX-2 promoter spanning the region with respect to the ATG start site (+1) is shown in Fig. 2A. Locations of NF-κB binding sequences within the region for methylation analysis were denoted in Fig. 2A and Table I. According to rat COX-2 gene (accession number NM_017232) sequence analysis, bisulfite-COX-2 sequence in the promoter had a size of 1,452 bp and contained 44 CpG sites, as identified by nucleotide sequence analysis for potential methylation (Fig. 2B and Table I). The bisulfite genomic sequence located from -1,522 to -71 bp was denoted as regions I-V and designed to amplify with the primers that were specific to the CpG site within COX-2 promoter. The bisulfite sequencing chromatogram of the methylated cytosine of these CpG sites for bladder tissue is presented in Fig. 2C.
Figure 2.
Methylation status of the CpG site in the COX-2 promoter region in ketamine-induced ulcerative cystitis. (A) Schematic diagram of COX-2 promoter spanning the region from -1,522 to -71 bp with respect to the ATG start site (+1) (translation initiation site) and targeted sites for methylation-specific PCR region and bisulfite genomic sequencing analysis are shown. Additionally, five putative NF-κB DNA binding sites in the region were identified by TFBIND software (http://tfbind.hgc.jp/). The broad arrow indicated the direction of transcription. A total of five putative NF-κB binding sequences for methylation analysis are denoted. (B) The bisulfite-COX-2 sequence in the promoter had a size of 1,452 bp and contained 44 CpG sites as identified by nucleotide sequence analysis for potential methylation (http://www.genome.ucsc.edu). There were 10 CpG sites in the region I fragment of COX-2 promoter ranging from −299 to −71 bp, 10 CpG sites in the region II ranging from −551 to −286 bp, 11 CpG sites in the region III ranging from -830 to -548 bp, 6 CpG sites in the region IV ranging from -1,195 to -829 bp and 7 CpG sites in the region V ranging from -1,522 to -1,181 bp for methylation sequencing analysis. A total of five sets of primers were designed to amplify the bisulfite genomic sequence. (C) Bisulfite sequencing chromatogram of CpG sites in bladder tissue. Each circle denoted as one CpG site and represented the methylation status of the CpGs. The clone was sequenced from PCR products generated from amplification of bisulfite-treated DNA. The percentage of methylation at a single CpG site was calculated from the sequencing results of four independent clones. ○, unmethylated cytosines; •, methylated cytosines. NF, nuclear factor; IL, interleukin; NFAT/NF-IL6, nuclear factor of activated T cells/NF-IL-6; AP-1, activator protein 1; CRE, cAMP-response element; SP-1, specificity protein 1; TATA, TATA box motif; COX, cyclooxygenase.
The present study's data showed DNA unmethylation (white circles) in regions I-III of COX-2 promoter fragments in three experimental groups. However, in regions IV and V, the CpG site methylation in the control group, and hypomethylation in the ketamine and the ketamine+COX-2 group was found. Although the cloned sequences exhibited considerable heterogeneity in methylation, it was also demonstrated that bisulfite MSP and genomic sequencing data displayed the hypomethylation at the CpG sites in the ketamine and the ketamine+COX-2 inhibitor groups.
Methylation analysis of rat COX-2 promoter ranging from −830 to −71 bp by methylation-specific PCR and bisulfite genomic sequencing
Schematic diagrams of the promoter fragment region I ranging from −299 to −71 bp and region II ranging from −551 to −286 bp in the COX-2 promoter are presented in Figs. S1A and S2A. Both regions I and II contained 10 methylatable CpG sites marked with red numbers. A total of two potential NF-κB binding sequences spanning from −235 to −224 and −549 to −540 bp were marked with a square box (Figs. S1B and S2B). Sequence traces were obtained from the PCR products from bisulfite-treated DNA using primers (blue color in bisulfate sequence) and detailed normal and bisulfite genomic sequences of regions I and II were presented (Figs. S1C and D, S2C and D). Consistent with the data, it was found that the CpG sites displayed unmethylation at the CpG sites in the control, the ketamine and the ketamine+COX-2 groups.
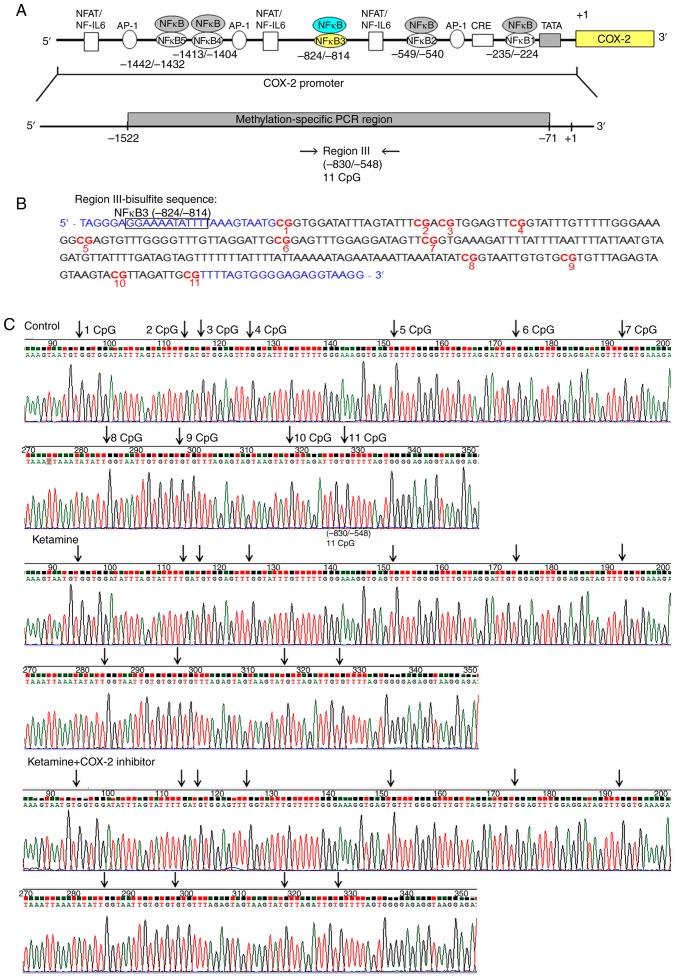
A schematic diagram of the fragment region III spanning from −830 to −548 bp containing 11 methylated CpG sites and the potential NF-κB binding sequence spanning from −824 to −814 bp was marked with square box. Bisulfite methylated NFκB3 (mNFκB3) sequence was located at −824 to −814 bp, 5′-GGA AAA TAT TT-3′. Sequence traces were obtained from the PCR products from bisulfite-treated DNA (Fig. 3A and B), and the CpG sites were highlighted as black arrows (Fig. 3C). Detailed normal and bisulfite genomic sequences of region III in COX-2 promoter were presented (Fig. S3). Based on the above findings, the control, the ketamine and the ketamine+COX-2 inhibitor groups all displayed unmethylation at the CpG sites in regions I-III ranging from −830 to −71 bp.
Figure 3.
Methylation status of the CpG site in the COX-2 promoter region III in ketamine-induced ulcerative cystitis. (A) A schematic depiction of COX-2 promoter region III and targeted sites for methylation-specific PCR region is presented. Region III sequence spanned from -830 to -548 relative to the transcription start site (+1) and contained 11 CpG dinucleotides. (B) Bisulfite sequencing data for region III of rat COX-2 promoter were analyzed. This promoter fragment contained 11 CpG sites that were marked with a red color and number. The potential site for NF-κB binding was marked with black square box and bisulfite methylated NFκB3 sequence was located at −824 to −814 bp. Bisulfite genomic sequencing traces were obtained from the PCR products from bisulfite-treated DNA using primers indicated in blue font, including forward and reverse. (C) The trace represented an approximation of the 'average' methylation status at each CpG residue. CpG sites are highlighted as black arrows. Sample number (n)=6. COX, cyclooxygenase; NF, nuclear factor.
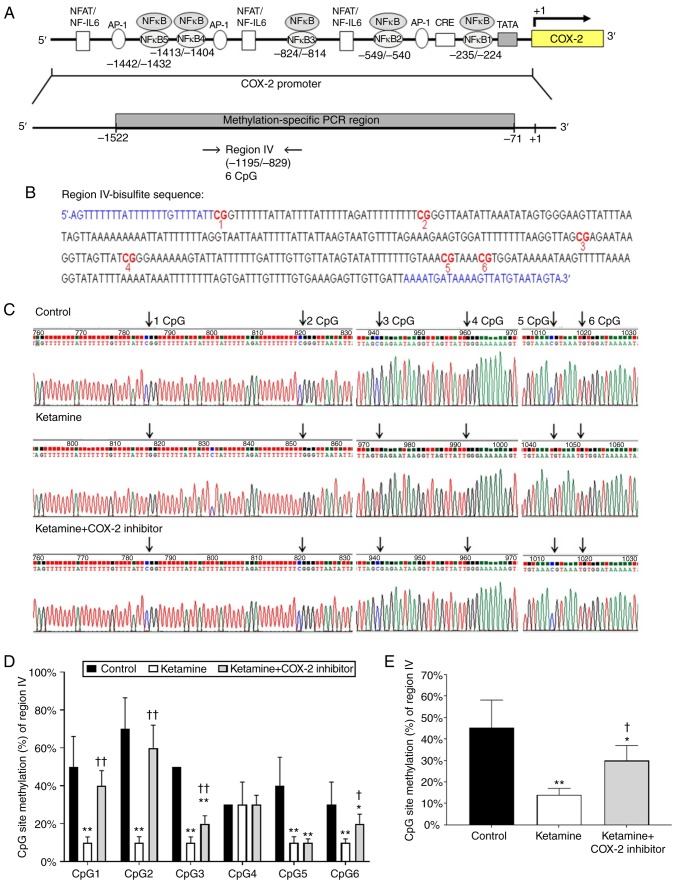
Ketamine treatment caused hypomethylation of the COX-2 promoter ranging from −1,195 to −829 bp
Diagrammatic presentation of the region IV ranging from −1,195 to −829 bp is shown in Fig. 4A. The promoter fragment region IV contained 6 methylatable CpG sites marked with red numbers (Fig. 4B). There was no potential sequence for NF-κB binding site in region IV fragment. Sequence traces were obtained from the PCR products of bisulfite-treated DNA using primers marked with blue color in bisulfite sequence (Fig. 4B) and the CpG sites were highlighted as black arrows in Fig. 4C. Detailed normal and bisulfite genomic sequences of region IV were shown in Fig. S4. The association between the percentage of methylation at CpG sites within the fragment region and the COX-2 transcriptional level was evaluated. The percentage of methylation level for each individual CpG site from bladder tissue was presented in Fig. 4D. The percentage of methylation at a single CpG site was calculated from the sequencing results of four independent clones. The percentage of CpG sites in the control group was 50.0±16.1% in CpG1, 70.0±16.5% in CpG2, 50.0±0% in CpG3, 30.0±0% in CpG4, 40.0±15.1% in CpG5 and 30.0±12.0% in CpG6. In comparison, the percentage of CpG sites in the ketamine group was significantly decreased: 10.0±3.0% in CpG1, 10.0±3.1% in CpG2, 10.0±2.0% in CpG3, 30.0±12.0% in CpG4, 10.0±3.1% in CpG5, and 10.0±2.0% in CpG6. However, such ketamine-induced percentage decrease was largely restored by COX-2 inhibitor as revealed by the percentage of CpG sites in the ketamine+COX-2 inhibitor group: 40.0±8.0% in CpG1, 60.0±12.0% in CpG2, 20.0±4.2% in CpG3, 30.0±5.0% in CpG4, 10.0±2.0% in CpG5 and 20.0±5.0% in CpG6. Moreover, the methylated percentage of total six CpG sites in COX-2 promoter located from −1,195 to −829 bp was 45.2±13.0% in the control group, 14.0±3.0% in the ketamine group and 30.0±7.0% in the ketamine+COX-2 inhibitor group. Similarly, ketamine caused significant reduction (P<0.01) in the methylated percentage, while such reduction was largely restored by COX-2 inhibitor (Fig. 4E). These observations demonstrated that ketamine reduced the methylated level of region IV and such the reduction was largely restored by the COX-2 inhibitor.
Figure 4.
Detailed methylation analysis of region IV in the COX-2 promoter. (A) A diagrammatic representation of COX-2 promoter region IV and targeted sites for methylation-specific PCR is shown. Region IV sequence spanned from −1,195 to −829 and contained 6 CpG sites. (B) Bisulfite genomic sequencing data for region IV in the COX-2 promoter were present. This promoter fragment in region IV contained 6 CpG sites that were marked with a red color and number. Sequence traces obtained from the PCR products of bisulfite-treated DNA using primers (blue color in bisulfite sequence) were determined by bisulfite sequencing analysis. (C) Methylation patterns of the bisulfite-COX-2 region IV in three groups. Arrows indicate the positions of CpG sites. (D) The methylation level of each individual CpG site in region IV within the COX-2 promoter. (E) The methylated percentage of total six CpG sites in region IV was analyzed. The methylated level of region IV from the control group showed a 3.22-fold increase compared with that in the ketamine group and a 1.51-fold increase compared with that in the ketamine+COX-2 group Values were the mean ± standard deviation for n=6. *P<0.05 and **P<0.01 vs. the control group. †P<0.05 and ††P<0.01 vs. the ketamine group. COX, cyclooxygenase; NF, nuclear factor.
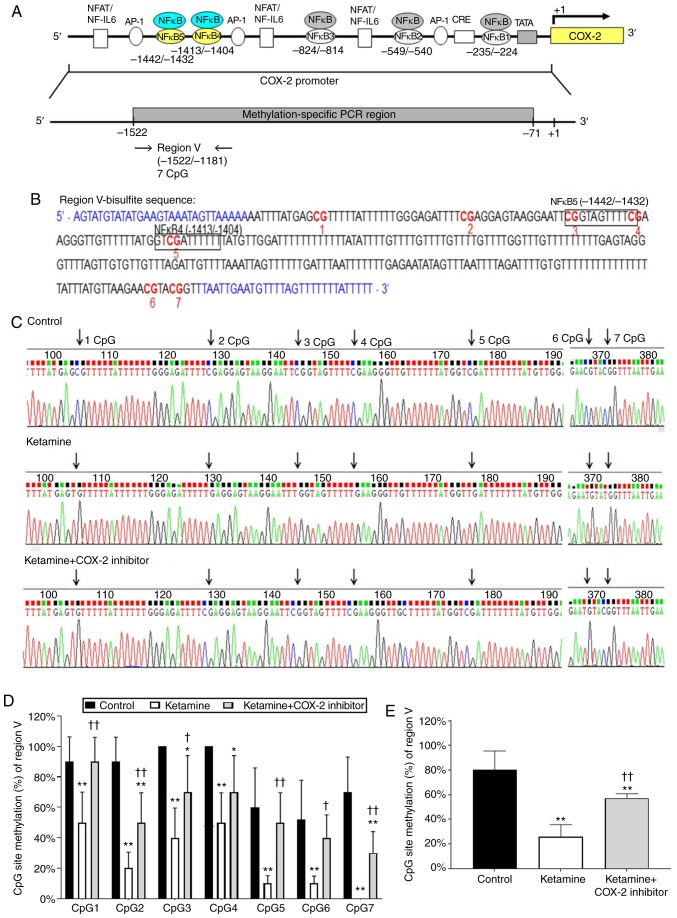
Genomic sequencing ranging from −1,522 to −1,181 bp influences the COX-2 transcriptional level
Diagrammatic representation of the region V ranging from −1,522 to −1,181 bp is shown in Fig. 5A. The fragment region V containing 7 CpG sites was marked with a red number (Fig. 5B). There are two potential binding sequences for NF-κB transcriptional factor spanning from -1,442 to −1,432 and −1,413 to −1,404 bp marked with a square box (Fig. 5B). The CpG sites by bisulfite genomic sequencing data are highlighted as arrows (Fig. 5C). Detailed normal and bisulfite genomic sequences for region V are shown in Fig. S5. The percentage of methylation level for each individual CpG site is presented in Fig. 5D. The percentage of CpG sites in the control group was 90.0±16.3% in CpG1, 90.0±16.1% in CpG2, 100.0±0% in CpG3, 100.0±0% in CpG4, 60.0±25.9% in CpG5, 52.0±25.8% in CpG6, and 70.0±23.0% in CpG7. In comparison, the percentage of CpG sites in the ketamine group was significantly decreased (P<0.01): 50.0±20.0% in CpG1, 20.0±10.5% in CpG2, 40.0±19.5% in CpG3, 50.0±19.6% in CpG4, 10.0±5.0% in CpG5, 10.0±4.8% in CpG6 and unmethylation in CpG7. However, such ketamine-induced percentage decrease was largely restored by COX-2 inhibitor as revealed by the percentage of CpG sites: 90.0±16.0% in CpG1, 50.0±19.5% in CpG2, 70.0±24.0% in CpG3, 70.0±24.0% in CpG4, 50.0±9.6% in CpG5, 40.0±15.0% in CpG6 and 30.0±14.0% in CpG7 in the ketamine+COX-2 inhibitor group. Additionally, the methylated percentage of total seven CpG sites in the COX-2 promoter region located from −1,522 to −1,181 bp was 80.0±15.4% in the control group, 25.7±10.0% in the ketamine group and 57.1±3.7% in the ketamine+COX-2 inhibitor group. Similarly, ketamine caused a significant reduction in the methylated percentage, while such reduction was largely restored by COX-2 inhibitor (P<0.01; Fig. 5E). The above findings implied that the hypo-methylation of CpG sites in the region ranging from −1,522 to −829 bp might increase the sequence-specific binding affinity of NF-κB and enhance the COX-2 gene activation.
Figure 5.
Methylation status of the CpG site in COX-2 promoter region ranging from −1,181 to −1,522 in ketamine-induced ulcerative cystitis. (A) A schematic depiction of region V and sequence ranging from -1,181 to −1,522 relative to the transcription start site (+1) is presented. (B) Bisulfite genomic sequencing analysis for region V in the COX-2 promoter. Genomic sequencing of bisulfite-COX-2 region V-associated 7 CpG sites was obtained from the PCR products from bisulfite-treated DNA using primers (blue color). The site-specific methylation of the CpG sites (dinucleotides) at −1,442, −1,432 and −1,411 bp located the sequence of NF-κB binding sites in the COX- promoter regions ranging from -1,442 to -1,404 bp. The potential site for NF-κB binding was marked with black square box and the genomic NFκB4 was located at -1,413 to -1,404 bp, 5′-GTC GAT TCC C-3′ (Bisulfite sequence of mNFκB4: 5′-GTC GAT TTT T-3′) and NFκB5 located at -1,442 to -1,432 bp, 5′-CGG TAG TTT CC-3′ (Bisulfite sequence of mNFκB5: 5′-CGG TAG TTT TC-3′). (C) Methylation patterns of the bisulfite-COX-2 region V in three groups. CpG sites were highlighted as arrows. (D) The methylation level of region V in the COX-2 promoter. The methylated percentage of each individual CpG site from bladder tissue was presented. (E) The percentage of total seven CpG sites methylation in region IV. The methylation level of region V from the control group showed a 3.11-fold increase compared with in the ketamine group and a 1.40-fold increase compared with in the ketamine+COX-2 group. Values were the mean ± standard deviation for n=6. *P<0.05 and **P<0.01 vs. the control group. †P<0.05 and ††P<0.01 vs. the ketamine group. COX, cyclooxygenase; NF-κB4, nuclear factor-κB binding site.
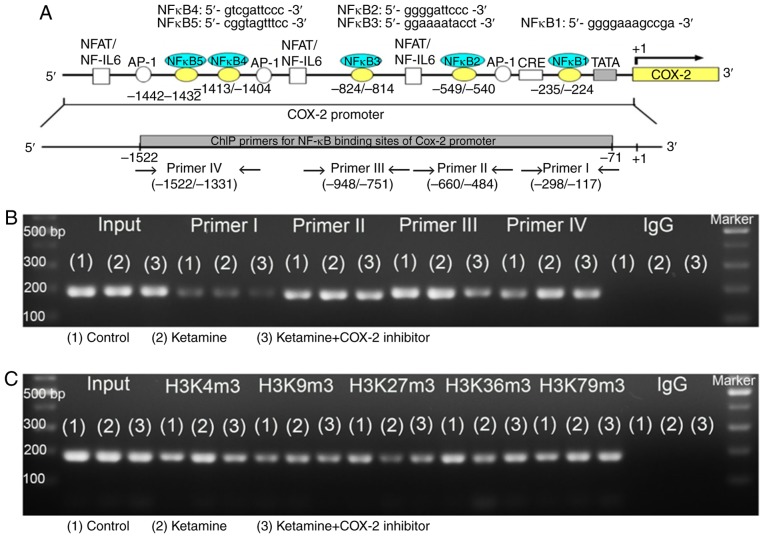
Analysis of NF-κB binding sequences and DNA methylation in association with enzymes responsible for COX-2 promoter activity
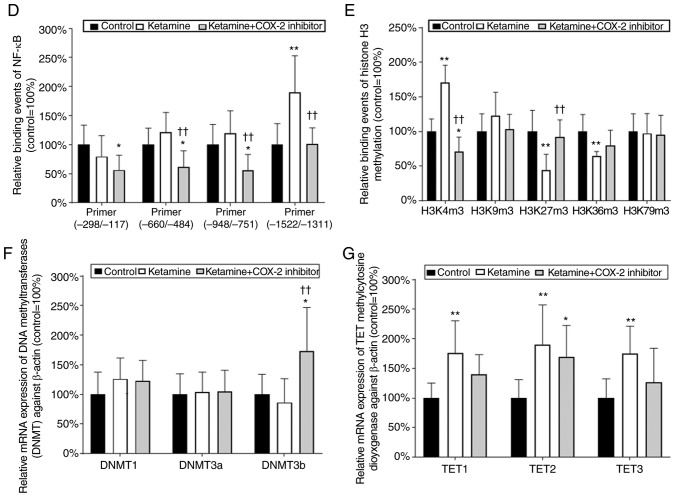
To determine which NF-κB binding sequence within the COX-2 promoter was involved in the COX-2 expression in KIC, the potential NF-κB binding sequence was analyzed by ChIP assay. Schematic diagram for COX-2 promoter region ranging from −1,522 to −71 bp is shown (Fig. 6A). These data suggest that ketamine treatment was able to increase the binding of NF-κB on the COX-2 promoter region ranging from -1,522 to -1,331 bp as compared with the control group. However, the ketamine+COX-2 inhibitor group reduced the level of NF-κB binding in COX-2 promoter in comparison with the ketamine group (Fig. 6B and D).
Figure 6.
Analyses of NF-κB binding sequences and DNA methylation-associated enzymes responsible for the promoter activity of COX-2 gene. (A) Schematic diagram of the COX-2 promoter and targeted sites ranging from -1,522 to -71 bp for ChIP analysis was presented. A total of five potential NF-κB binding sequences in the COX-2 promoter are denoted in the diagram. DNA fragments from ChIP assay were amplified using primer sets to amplify the sequence of the ChIP product. A total of four sets of primers were denoted as primer I-IV, which included five NF-κB binding sequences. ChIP analysis of the potential NF-κB binding sequence in the COX-2 promoter is shown. Formaldehyde cross-linked protein chromatin complexes were immunoprecipitated and genomic DNA was analyzed by (B) PCR and (D) qPCR using primers that recognized the COX-2 promoter region. Input for each reaction was used for internal control of samples loading. IgG was employed as negative control. ChIP analysis of histone H3 methylation on the COX-2 promoter was analyzed by (C) PCR and (E) qPCR. ChIP assay was also applied to anti-tri methyl histone H3 (H3K4m3, H3K9m3, H3K27m3, H3K36m3 and H3K79m3) specific for histone H3 tri-methylated at Lysine (K4, K9, K27, K36, and K79). These antibodies were used to provide direct evidence that histone H3 modulated COX-2 transcriptional regulation by modulating methylation of the COX-2 promoter region ranging from -1,522 to -1,331 bp. Results were normalized as the control group=100%. The mRNA levels of DNMTs (DNMT1, 3a and 3b) for evaluation of the DNA methylation (F) and TETs (TET1-3) for evaluation of the DNA demethylation (G) by reverse transcription-qPCR. The level of TETs expression in the ketamine group was represented as the number of fold increases compared with the control group. Data were expressed as the mean ± standard deviation for n=8. *P<0.05 and **P<0.01 vs. the control group. ††P<0.01 vs. the ketamine group. ChIP, chromatin immunoprecipitation; TET, Ten-Eleven-Translocation; DNMT, DNA methyltransferase; COX, cyclooxygenase; q, quantitative.
A ChIP assay using anti-tri methyl histone H3 (H3K4m3, H3K9m3, H3K27m3, H3K36m3 and H3K79m3) antibodies was used to provide evidence to demonstrate that histone H3 methylation on the COX-2 promoter region ranging from −1,522 to −1,331 bp affected the COX-2 mRNA expression. A significant amount of methylated histone H3 associated with the COX-2 promoter was detected (Fig. 6C and E). In the ketamine group, the level of histone H3K4m3 was increased compared with the control group, but the level of histone H3K27m3 and H3K36m3 was decreased. However, in the ketamine+COX-2 inhibitor group, the level of H3K4m3 was reduced in comparison with in the control group. However, there was no difference in the expression level of H3K79m3 among these three groups.
To determine which enzyme was involved in COX-2 the methylation and transcription, RT-qPCR was performed to analyze the mRNA expression levels of rat DNMT1, 3a and 3b and TET dioxygenase (TET1, TET2, and TET3) for the evaluation of the methylation/demethylation (Fig. 6F-G) isolated from bladders. In the ketamine group, the level of TETs was increased compared with in the control group, but not for DNMTs. However, in the ketamine+COX-2 inhibitor group, the level of DNMT3b and TET2 was increased compared with that in the control group. These observations demonstrated that ketamine induced NF-κB transcriptional regulation of COX-2 expression by enhancing TET dioxygenase-mediated demethylation and increasing the histone H3K4m3 binding affinity of COX-2 promoter.
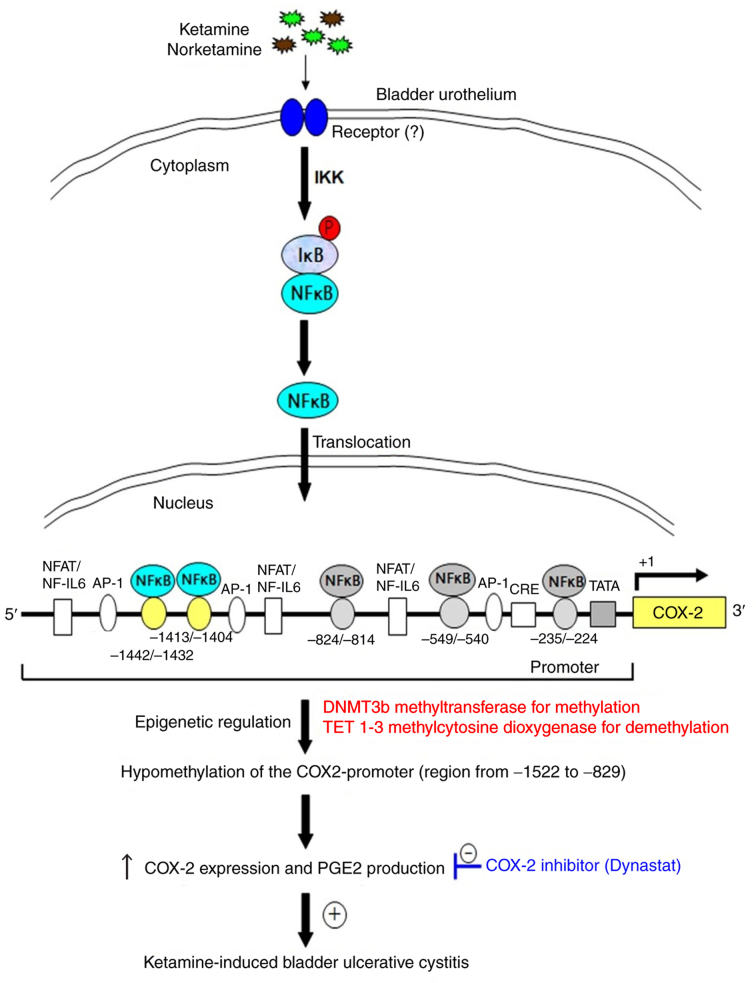
Proposed model for epigenetic regulation of COX-2 expression by DNA methylation via NF-κB activation in KIC
The present results revealed that ketamine treatment caused significant hypomethylation of the COX-2 promoter, whereas ketamine+COX-2 inhibitor improved the inflammatory effect (Figs. 1, 4 and 5). The hypomethylation of CpG sites in the COX-2 promoter region ranging from −1,522 to −829 bp increased the sequence-specific binding affinity of NF-κB transcriptional factor and enhanced the COX-2 gene activation. The COX-2 promoter hypomethylation was modulated by increasing TET enzymes for demethylation. Based on the above findings, a potential model for illustrating epigen-etic regulation of COX-2 expression by DNA methylation mediated via NF-κB activation in KIC was proposed in Fig. 7. Moreover, using TFBIND software and MethPrimer software analysis showed that two potential NF-κB binding sites of human COX-2 promoter had potential methylation sites (Fig. S6A), which implied that the transcriptional silencing of COX-2 by the epigenetic mechanisms might have potential applications for the identification of molecular biomarkers and clinical therapy for KIC.
Figure 7.
A model illustrating ketamine treatment promoting the hypomethylation of COX-2 gene promoter via NF-κB in ketamine-induced ulcerative cystitis of rat bladder. Inflammatory stimuli of ketamine induced nuclear translocation of NF-κB and increased nuclear NF-κB level. The activated nuclear NF-κB bound to specific sequences of COX-2 promoter and thereafter enhanced hypomethylation of COX-2 gene promoter. The hypomethylation of COX-2 promoter increased COX-2 expression and the level of PGE2 production. PGE, prostaglandin; IKK, IκB kinase; P, phosphorylation; NFAT/NF-IL6, nuclear factor of activated T cells/NF-IL-6; AP-1, activator protein 1; CRE, cAMP-response element; SP-1, specificity protein 1; TATA, TATA box motif.
Discussion
The results of the present study demonstrated that ketamine treatment induced the translocation of NF-κB p65 to activate COX-2 expression and promote PGE2 production in bladder tissue in KIC, whereas the COX-2 inhibitor suppressed the inflammatory effect mediated by KIC. Furthermore, whether epigenetic regulation of COX-2 promoter by DNA hypomethylation could alter NF-κB translocation and transcriptional signaling of COX-2 expression was also determined in KIC. Bisulfite MSP and genomic sequencing data showed that all the three experimental groups exhibited un-methylation at CpG sites within the COX-2 promoter region ranging from -830 to -71 bp. However, ketamine treatment caused COX-2 promoter hypomethylation ranging from -1,522 to -829 bp, which contributed to the COX-2 transcriptional regulation, whereas ketamine combined with COX-2 inhibitor improved such alterations. Ketamine treatment increased the binding of NF-κB and permissive histone H3K4m3, but decreased the repressive histone H3K27m3 and H3K36m3 binding on the COX-2 promoter region ranging from -1,522 to -1,331 bp as determined by ChIP assay. These observations suggested that there was a hypomethylation pattern of the COX-2 promoter via NF-κB p65 translocation in association with the level of COX-2 transcription after ketamine treatment.
Following ketamine injection, translocated NF-κB p65 bound to the specific sequence of COX-2 promoter region ranging from −1,522 to −829 bp, which transcriptionally induced COX-2 expression to activate PGE2 production. Synthesized PGE2 might act on its specific receptor subtype (EP2) and further feedback activate NF-κB (37-39). The treatment of COX-2 inhibitor reduced COX-2 transcriptional expression and decreased PGE2 production, leading to an inhibited inflammatory reaction (40). Additionally, previous studies indicated that NF-κB could control COX-2 transcription (6,41), however cyclopentenone prostaglandins could inhibit NF-κB activation via IκB kinase (IKK) inhibition (42), suggesting cyclopentenone feedback could inhibit continued nuclear accumulation of NF-κB (38). Following ketamine injection combined with the COX-2 inhibitor, the COX-2 inhibitor was converted to an anti-inflammatory compound (valdecoxib). The pharmacological effect of valdecoxib was to inhibit COX-2-mediated prostaglandin synthesis. In the present study, it was proposed that the pharmacological effect of COX-2 inhibitor might suppress PGE2 synthesis and block the nuclear accumulation of NF-κB, leading to inhibition of the positive feedback loop of the COX-2-PGE2-EP2-NFκB pathway. The prostaglandins might inhibit NF-κB activity through IKK inhibition, restrict NF-κB to the cytoplasm and this negative feedback loop might result in reducing NF-κB-dependent COX-2 gene expression in KIC. COX-2 overexpression affected the activity of TET (43), which implied that COX-2 inhibitor might affect the methylation status within COX-2 promoter.
Previous studies reported that COX-2 expression might be related to the methylation status of 5′-CpG site of COX-2 gene in cancer cell lines (25,44), which exhibited hypermethylation of CpG sites in the promoter or exon 1 region and methylation-inhibiting agents restored the expression of COX-2 (28). Toyota et al (23) suggested that aberrant methylation of the 5′ CpG island to reduce COX-2 expression was presented in colorectal cancer and a detailed methylated mapping of 24 CpG sites revealed the methylated region closest to the transcription start site. However, the results of the present study indicated that ketamine treatment induced the translocation of NF-κB p65 to activate COX-2 expression and PGE2 production in bladder tissue. Meanwhile, bisulfite MSP and genomic sequencing data showed that DNA hypomethylation at CpG sites within COX-2 promoter region located from −1,522 to −829 bp might contribute to the COX-2 transcriptional regulation in the ketamine group. Ketamine treatment was able to increase the binding of NF-κB and transcriptionally permissive histone H3K4m3, but decrease the repressive histone H3K27m3 and H3K36m3 on the COX-2 promoter ranging from −1,522 to −1,331 bp as determined by ChIP assay. Thus, such epigenetic regulation of COX-2 activity for PGE2 production is interesting and deserves further investigations.
The hypermethylation of CpG sites in the COX-2 promoter for the suppression of gene expression was shown to reduce the binding affinity of sequence-specific transcription factors (30,45). Kelavkar et al (11) reported that the increase in the methylation status of specific CpG sites of COX-2 promoter might affect NF-κB binding affinity. In the present study, there were 7 CpG sites and two potential sequences for NF-κB transcriptional factor binding within the COX-2 promoter located from -1,522 to -1,181 bp. Two potential genomic sequences for NFκB4 binding site in the COX-2 promoter were located at -1,413 to -1,404 bp, 5′-GTC GAT TCC C-3′ and NFκB5 located at −1,442 to −1,432 bp, 5′-CGG TAG TTT CC-3′. The site-specific methylation of 5′-CpG dinucleotides at sites -1,442, 1,432, -1,411 bp (3, 4 and 5 CpG site of region V) was located at the COX-2 promoter sequence ranging from −1,442 to 1,404 bp. The observed increase in the methylation at these specific CpG sites might impair NF-κB binding affinity to affect COX-2 expression in the control group. The potential NF-κB binding sequence involved in the COX-2 expression in KIC by ChIP analysis was also analyzed. The results of the present study suggested that the NF-κB binding sequence ranging from -1,522 to -1,331 bp modulated the COX-2 mRNA expression as observed. ChIP assay used anti-trimethyl histone H3 antibodies to provide direct evidence that histone H3 methylation on the COX-2 promoter affected the COX-2 mRNA expression in KIC.
The double helical strands forming the DNA backbone contained a major groove and a minor groove and the major groove interacted with more functional groups than the minor groove, which could interact with sequence-specific DNA-binding proteins. Moreover, transcription factors were composed of DNA-binding domains and had a specific affinity for single- or double-stranded DNA (46). Cytosine methylation could either influence the interactions between promoter DNA and transcription factors or exhibit no effect, depending on the context (14). Despite being only a minor chemical change, addition of a methyl group to cytosine could affect nucleotide readout via hydrophobic contacts in the major groove and shape readout via electrostatic contacts in the minor groove (47,48). 5-methylcytosines changed the nucleosome stability and affected the chromatin structure and the affinity of transcription factor bound to genomic DNA (14,49,50). The results of the present study demonstrated that ketamine treatment caused significant COX-2 hypomethylation ranging from −1,522 to −829 bp, which contributed to the COX-2 transcriptional regulation and increased NF-κB binding to the promoter region located from −1,442 to −1,404 bp. It was also found that the NF-κB binding sequence ranging from -1,522 to -1,331 bp modulated the COX-2 mRNA expression as observed by ChIP assay. These data indicated that the COX-2 promoter region located from −1,522 to −829 bp resided in the major groove of DNA and played a pivotal role for regulation COX-2 gene expression in KIC. Furthermore, the authors' previous study demonstrated the involvement of the regulation of COX-2 via the NF-κB pathway in the inflammatory signaling of KIC in rat urinary bladder (6). Misoprostol, the FDA approved clinical medication, is beneficial to various immunological diseases, e.g., interstitial cystitis (51). It attenuates the transcriptional activity of NF-κB and recruitment of p65 (52). The combination of those studies with the present study's findings suggested that the inhibitor of NF-κB transcription factor might provide a potential application for therapeutic strategy for KIC.
The COX-2 promoter region, NF-κB binding sites and the potential methylation of CpG sites were compared between rat and human species. In rats, the promoter of COX-2 gene (accession number NM_017232) ranging from -1,522 to -71 including 1,452 bp and 44 CpG sites were identified by nucleotide sequence analysis for potential methylation. There were 7 CpG sites and 2 potential sequences for NF-κB transcriptional factor binding within the COX-2 promoter located from -1,522 to −1,181 bp. A total of two potential genomic sequences for NF-κB binding sites in the COX-2 promoter were located at -1,413 to −1,404 bp, 5′-GTC GAT TCC C-3′ and at -1,442 to -1,432 bp, 5′-CGG TAG TTT CC-3′. The site-specific methylation of 5′-CpG dinucleotides at sites -1,442, -1,432 and -1,411 bp was located the COX-2 promoter sequence ranging from -1,442 to -1,404 bp (Table I). Moreover, in humans the promoter regions of the COX-2 gene (accession number NM_000963) ranging from -2,000 to -1 including 2,000 bp and 59 CpG sites were identified by nucleotide sequence analysis for potential methylation. Seven potential motifs similar to the consensus binding sites for NF-κB in the region were identified by TFBIND software. Two potential genomic sequences for NF-κB binding sites in the COX-2 promoter were located at -320 to -310 bp, 5′-CTG GGT TTC CG-3′ and at -640 to -631 bp, 5′-GTG ACT T CC T-3′. The site-specific methylation of 5′-CpG dinucleotides at sites -640, -310 bp was located in the COX-2 promoter. The present study showed that NF-κB binding sequence ranging from -1,442 to 1,404 modulated the COX-2 mRNA expression as observed in rats. These findings suggested that the transcriptional silencing of COX-2 by epigenetic mechanisms might be involved in the pathogenesis of KIC. However, further investigations of the epigenetic mechanisms in the pathogenesis of KIC in human are needed.
DNA hypomethylation of COX-2 promoter region might contribute to COX-2 transcriptional regulation and induce a pro-inflammatory response in KIC. However, COX-2 inhibitor treatment increased the methylation of COX-2 promoter and reduced COX-2 transcriptional expression. These results implied that the transcriptional silencing of COX-2 by the epigenetic mechanisms might be involved in the pathogenesis of KIC, which could have potential application for the molecular biomarker and clinical therapy of KIC.
Supplementary Materials
Acknowledgments
The authors would like to thank Professor Chang-Hwei Chen of the University at Albany, State University of New York for his valuable comments on this manuscript.
Funding
The present study was supported by the Ministry of Science and Technology NSC (grant no. 105-2314-B-037-043-MY3); Ministry of Health and Welfare (grant no. MOHW107-TDU-B-212-123006); Department of Medical Research, Kaohsiung Medical University Hospital (grant nos. KMUH105-5R44 and KMUH106-6R60); and from the Chi-Mei Medical Center and the Kaohsiung Medical University Research (grant no. 107CM-KMU-12).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
SMC, JHL, CYL, KLL, YCL, HPH, CCT, WJW, HJY and YSJ designed the study; SMC, JHL, CYL, KLL, YCL, HPH, CCT, WJW, and YSJ conducted review and editing; SMC, JHL and YSJ had full access to all the data in the study and took responsibility for the integrity of the data and the accuracy of the data analysis. SMC, JHL and YSJ wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Animal Care and Treatment Committee of Kaohsiung Medical University. All experiments were conducted according to the guidelines for laboratory animal care.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lee YL, Lin KL, Chuang SM, Lee YC, Lu MC, Wu BN, Wu WJ, Yuan SF, Ho WT, Juan YS. Elucidating mechanisms of bladder repair after hyaluronan instillation in ketamine-induced ulcerative cystitis in animal model. Am J Pathol. 2017;187:1945–1959. doi: 10.1016/j.ajpath.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Shahani R, Streutker C, Dickson B, Stewart RJ. Ketamine-associated ulcerative cystitis: A new clinical entity. Urology. 2007;69:810–812. doi: 10.1016/j.urology.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 3.Middela S, Pearce I. Ketamine-induced vesicopathy: A literature review. Int J Clin Pract. 2011;65:27–30. doi: 10.1111/j.1742-1241.2010.02502.x. [DOI] [PubMed] [Google Scholar]
- 4.Chuang SM, Liu KM, Li YL, Jang MY, Lee HH, Wu WJ, Chang WC, Levin RM, Juan YS. Dual involvements of cyclooxygenase and nitric oxide synthase expressions in ketamine-induced ulcerative cystitis in rat bladder. Neurourol Urodyn. 2013;32:1137–1143. doi: 10.1002/nau.22367. [DOI] [PubMed] [Google Scholar]
- 5.Liu KM, Chuang SM, Long CY, Lee YL, Wang CC, Lu MC, Lin RJ, Lu JH, Jang MY, Wu WJ, et al. Ketamine-induced ulcerative cystitis and bladder apoptosis involve oxidative stress mediated by mitochondria and the endoplasmic reticulum. Am J Physiol Renal Physiol. 2015;309:F318–331. doi: 10.1152/ajprenal.00607.2014. [DOI] [PubMed] [Google Scholar]
- 6.Juan YS, Lee YL, Long CY, Wong JH, Jang MY, Lu JH, Wu WJ, Huang YS, Chang WC, Chuang SM. Translocation of NF-κB and expression of cyclooxygenase-2 are enhanced by ketamine-induced ulcerative cystitis in rat bladder. Am J Pathol. 2015;185:2269–2285. doi: 10.1016/j.ajpath.2015.04.020. [DOI] [PubMed] [Google Scholar]
- 7.Jhang JF, Hsu YH, Jiang YH, Kuo HC. Elevated serum IgE may be associated with development of ketamine cystitis. J Urol. 2014;192:1249–1256. doi: 10.1016/j.juro.2014.05.084. [DOI] [PubMed] [Google Scholar]
- 8.Lee CL, Jiang YH, Kuo HC. Increased apoptosis and suburothelial inflammation in patients with ketamine-related cystitis: A comparison with non-ulcerative interstitial cystitis and controls. BJU Int. 2013;112:1156–1162. doi: 10.1111/bju.12256. [DOI] [PubMed] [Google Scholar]
- 9.Jones PL, Wolffe AP. Relationships between chromatin organization and DNA methylation in determining gene expression. Semin Cancer Biol. 1999;9:339–347. doi: 10.1006/scbi.1999.0134. [DOI] [PubMed] [Google Scholar]
- 10.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 11.Kelavkar UP, Harya NS, Hutzley J, Bacich DJ, Monzon FA, Chandran U, Dhir R, O'Keefe DS. DNA methylation paradigm shift: 15-lipoxygenase-1 upregulation in prostatic intraepithelial neoplasia and prostate cancer by atypical promoter hypermethylation. Prostaglandins Other Lipid Mediat. 2007;82:185–197. doi: 10.1016/j.prostaglandins.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 12.Curradi M, Izzo A, Badaracco G, Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol. 2002;22:3157–3173. doi: 10.1128/MCB.22.9.3157-3173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urnov FD, Wolffe AP. Chromatin remodeling and transcriptional activation: The cast (in order of appearance) Oncogene. 2001;20:2991–3006. doi: 10.1038/sj.onc.1204323. [DOI] [PubMed] [Google Scholar]
- 14.Dantas Machado AC, Zhou T, Rao S, Goel P, Rastogi C, Lazarovici A, Bussemaker HJ, Rohs R. Evolving insights on how cytosine methylation affects protein-DNA binding. Brief Funct Genomics. 2015;14:61–73. doi: 10.1093/bfgp/elu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin B, Robertson KD. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol. 2013;754:3–29. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deneberg S, Guardiola P, Lennartsson A, Qu Y, Gaidzik V, Blanchet O, Karimi M, Bengtzén S, Nahi H, Uggla B, et al. Prognostic DNA methylation patterns in cytogenetically normal acute myeloid leukemia are predefined by stem cell chromatin marks. Blood. 2011;118:5573–5582. doi: 10.1182/blood-2011-01-332353. [DOI] [PubMed] [Google Scholar]
- 17.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Völkel P, Angrand PO. The control of histone lysine methylation in epigenetic regulation. Biochimie. 2007;89:1–20. doi: 10.1016/j.biochi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Sims RJ, III, Reinberg D. Histone H3 Lys 4 methylation: Caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Barros SP, Niculescu MD, Moretti AJ, Preisser JS, Offenbacher S. Alteration of PTGS2 promoter methylation in chronic periodontitis. J Dent Res. 2010;89:133–137. doi: 10.1177/0022034509356512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo WT, Jin L, Cheung MN, Wang M, Chow LW. Epigenetic change in E-cadherin and COX-2 to predict chronic periodontitis. J Transl Med. 2010;8:110. doi: 10.1186/1479-5876-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedre X, Mastronardi F, Bruck W, Lopez-Rodas G, Kuhlmann T, Casaccia P. Changed histone acetylation patterns in normal-appearing white matter and early multiple sclerosis lesions. J Neurosci. 2011;31:3435–3445. doi: 10.1523/JNEUROSCI.4507-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyota M, Shen L, Ohe-Toyota M, Hamilton SR, Sinicrope FA, Issa JP. Aberrant methylation of the Cyclooxygenase 2 CpG island in colorectal tumors. Cancer Res. 2000;60:4044–4048. [PubMed] [Google Scholar]
- 24.Kikuchi T, Itoh F, Toyota M, Suzuki H, Yamamoto H, Fujita M, Hosokawa M, Imai K. Aberrant methylation and histone deacetylation of cyclooxygenase 2 in gastric cancer. Int J Cancer. 2002;97:272–277. doi: 10.1002/ijc.1612. [DOI] [PubMed] [Google Scholar]
- 25.Ma X, Yang Q, Wilson KT, Kundu N, Meltzer SJ, Fulton AM. Promoter methylation regulates cyclooxygenase expression in breast cancer. Breast Cancer Res. 2004;6:R316–R321. doi: 10.1186/bcr793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 27.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 28.Chell S, Kaidi A, Williams AC, Paraskeva C. Mediators of PGE2 synthesis and signalling downstream of COX-2 represent potential targets for the prevention/treatment of colorectal cancer. Biochim Biophys Acta. 2006;1766:104–119. doi: 10.1016/j.bbcan.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y, Hu Z. Genome-wide demethylation by 5-aza-2′-deoxycytidine alters the cell fate of stem/progenitor cells. Stem Cell Rev. 2015;11:87–95. doi: 10.1007/s12015-014-9542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murata H, Tsuji S, Tsujii M, Sakaguchi Y, Fu HY, Kawano S, Hori M. Promoter hypermethylation silences cyclooxygenase-2 (Cox-2) and regulates growth of human hepatocellular carcinoma cells. Lab Invest. 2004;84:1050–1059. doi: 10.1038/labinvest.3700118. [DOI] [PubMed] [Google Scholar]
- 31.Liou JT, Chen ZY, Ho LJ, Yang SP, Chang DM, Liang CC, Lai JH. Differential effects of triptolide and tetrandrine on activation of COX-2, NF-kappaB, and AP-1 and virus production in dengue virus-infected human lung cells. Eur J Pharmacol. 2008;589:288–298. doi: 10.1016/j.ejphar.2008.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poligone B, Baldwin AS. Positive and negative regulation of NF-kappaB by COX-2: Roles of different prostaglandins. J Biol Chem. 2001;276:38658–38664. doi: 10.1074/jbc.M106599200. [DOI] [PubMed] [Google Scholar]
- 33.Glinghammar B, Rafter J. Colonic luminal contents induce cyclooxygenase 2 transcription in human colon carcinoma cells. Gastroenterology. 2001;120:401–410. doi: 10.1053/gast.2001.21188. [DOI] [PubMed] [Google Scholar]
- 34.Hu VY, Malley S, Dattilio A, Folsom JB, Zvara P, Vizzard MA. COX-2 and prostanoid expression in micturition pathways after cyclophosphamide-induced cystitis in the rat. Am J Physiol Regul Integr Comp Physiol. 2003;284:R574–R585. doi: 10.1152/ajpregu.00465.2002. [DOI] [PubMed] [Google Scholar]
- 35.Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Aoki T, Nishimura M, Matsuoka T, Yamamoto K, Furuyashiki T, Kataoka H, Kitaoka S, Ishibashi R, Ishibazawa A, Miyamoto S, et al. PGE(2) -EP(2) signalling in endothelium is activated by haemodynamic stress and induces cerebral aneurysm through an amplifying loop via NF-κB. Br J Pharmacol. 2011;163:1237–1249. doi: 10.1111/j.1476-5381.2011.01358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul AG, Chandran B, Sharma-Walia N. Cyclooxygenase-2-prostaglandin E2-eicosanoid receptor inflammatory axis: A key player in Kaposi's sarcoma-associated herpes virus associated malignancies. Transl Res. 2013;162:77–92. doi: 10.1016/j.trsl.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rundhaug JE, Simper MS, Surh I, Fischer SM. The role of the EP receptors for prostaglandin E2 in skin and skin cancer. Cancer Metastasis Rev. 2011;30:465–480. doi: 10.1007/s10555-011-9317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller N. COX-2 inhibitors as antidepressants and antipsychotics: Clinical evidence. Curr Opin Investig Drugs. 2010;11:31–42. [PubMed] [Google Scholar]
- 41.Ackerman WE, IV, Summerfield TL, Vandre DD, Robinson JM, Kniss DA. Nuclear factor-kappa B regulates inducible pros-taglandin E synthase expression in human amnion mesenchymal cells. Biol Reprod. 2008;78:68–76. doi: 10.1095/biolreprod.107.061663. [DOI] [PubMed] [Google Scholar]
- 42.Rossi A, Kapahi P, Natoli G, Takahashi T, Chen Y, Karin M, Santoro MG. Anti-inflammatory cyclopentenone prostaglandins are direct inhibitors of IkappaB kinase. Nature. 2000;403:103–108. doi: 10.1038/47520. [DOI] [PubMed] [Google Scholar]
- 43.Chen H, Cai W, Chu ESH, Tang J, Wong CC, Wong SH, Sun W, Liang Q, Fang J, Sun Z, Yu J. Hepatic cyclooxygenase-2 overexpression induced spontaneous hepatocellular carcinoma formation in mice. Oncogene. 2017;36:4415–4426. doi: 10.1038/onc.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akhtar M, Cheng Y, Magno RM, Ashktorab H, Smoot DT, Meltzer SJ, Wilson KT. Promoter methylation regulates Helicobacter pylori-stimulated cyclooxygenase-2 expression in gastric epithelial cells. Cancer Res. 2001;61:2399–2403. [PubMed] [Google Scholar]
- 45.Clark SJ, Harrison J, Molloy PL. Sp1 binding is inhibited by (m)Cp(m)CpG methylation. Gene. 1997;195:67–71. doi: 10.1016/S0378-1119(97)00164-9. [DOI] [PubMed] [Google Scholar]
- 46.Pabo CO, Sauer RT. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 47.Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harb Perspect Biol. 2014;6:a019133. doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chuang TJ, Chen FC. DNA methylation is associated with an increased level of conservation at nondegenerate nucleotides in mammals. Mol Biol Evol. 2014;31:387–396. doi: 10.1093/molbev/mst208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/S0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 50.Bestor TH. Gene silencing. Methylation meets acetylation. Nature. 1998;393:311–312. doi: 10.1038/30613. [DOI] [PubMed] [Google Scholar]
- 51.Davies NM, Longstreth J, Jamali F. Misoprostol therapeutics revisited. Pharmacotherapy. 2001;21:60–73. doi: 10.1592/phco.21.1.60.34442. [DOI] [PubMed] [Google Scholar]
- 52.Gobejishvili L, Ghare S, Khan R, Cambon A, Barker DF, Barve S, McClain C, Hill D. Misoprostol modulates cytokine expression through a cAMP pathway: Potential therapeutic implication for liver disease. Clin Immunol. 2015;161:291–299. doi: 10.1016/j.clim.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.