Abstract
Purpose of review:
To relate genomic changes to phenotypic adaptation and evolution from environmental bacteria to obligate human pathogens, focusing on the examples within Bordetella species.
Recent findings:
Recent studies showed that animal-pathogenic and human-pathogenic Bordetella species evolved from environmental ancestors in soil. The animal-pathogenic B. bronchiseptica can hijack the life cycle of the soil-living amoeba Dictyostelium discoideum, surviving inside single-celled trophozoites, translocating to the fruiting bodies and disseminating along with amoeba spores. The association with amoeba may have been a ‘training ground’ for bacteria during the evolution to pathogens. Adaptation to an animal-associated life style was characterized by decreasing metabolic versatility and genome size and by acquisition of ‘virulence factors’ mediating the interaction with the new animal hosts. Subsequent emergence of human-specific pathogens such as B. pertussis from zoonoses of broader host range progenitors was accompanied by a dramatic reduction in genome size, marked by the loss of hundreds of genes.
Summary:
The evolution of Bordetella from environmental microbes to animal-adapted and obligate human pathogens was accompanied by significant genome reduction with large scale gene loss during divergence.
Keywords: Bordetella, environmental origin, amoebic predation, bacterial pathogen, host specialization
Introduction:
Genomic plasticity has enabled bacteria to rapidly adapt to ever-changing environments. The imprints of these adaptive changes are often retained in the genome and provide a blueprint on how bacteria adapt to, and evolve, within newly established niches. These imprints can often be identified by comparative study of closely related bacterial genomes, providing insight on the subtle genetic changes that accompany phenotypic adaptations. The bordetellae are ideally suited for these studies, owing to the broad environmental niches they occupy, the diverse life styles they have, and their phylogenetic relatedness.
The genus Bordetella belongs to the Betaproteobacteria and contains several pathogenic species that include the so-called ‘classical’ bordetellae consisting of B. bronchiseptica, B. pertussis and B. parapertussis. B. pertussis is the etiological agent of whooping cough, which is known for the characteristic symptoms of paroxysmal cough, whooping, and post-tussive vomiting, and kills hundreds of thousands of children annually [1]. B. parapertussis is the common name of two different lineages: a human-adapted lineage that causes whooping cough-like disease in children [2], hereafter referred to as Bpphu, and an ovine-adapted lineage that causes pneumonia in sheep, referred to as Bppov [3]. The third species, B. bronchiseptica, has a broader host range, causing respiratory diseases that can be mild and chronic or acute and severe: kennel cough in dogs, bronchopneumonia or atrophic rhinitis in pigs [4]. These three species can be considered subspecies as their shared genes possess over 98% nucleotide identity. Multi Locus Sequence Typing (MLST) and whole genome sequences revealed that B. pertussis and B. parapertussis independently evolved from a B. bronchiseptica-like ancestor [5–7], human-specific pathogens emerging from likely zoonoses of the broader host range progenitor.
In addition to the ‘classical’ bordetellae, the genus contains numerous other, phylogenetically distinct species collectively referred to as ‘non-classical’. The emerging, human-restricted B. holmesii was first isolated in 1983 [8], and has since been isolated from blood of septicemic patients and from nasopharyngeal samples of patients with pertussis-like disease [9–12]. B. avium causes respiratory disease in poultry with the clinical symptoms known as coryza or bordetellosis [13] and also infects other wild and domesticated birds [14]. B. hinzii is generally regarded as a commensal in the respiratory tract of poultry but causes disease during experimental infection of turkey chicks [15] and opportunistic infection in immunocompromised patients [16,17]. The closely related B. pseudohinzii naturally infects laboratory raised mice [18,19] and was also identified in a wild rat in Malaysia [20]. B. trematum and B. ansorpii are rare and generally found associated with infected wounds of immunocompromised patients [21–23], and B. bronchialis, B. sputigena and B. flabilis have recently been isolated from respiratory samples collected from patients with cystic fibrosis [24]. B. petrii shares the most ancient common ancestor and also has been recovered from quite diverse environmental settings, including soils and immunocompromised patients [25,26], providing some perspective on the natural history of the bordetellae, an intriguing “origin story” explored herein.
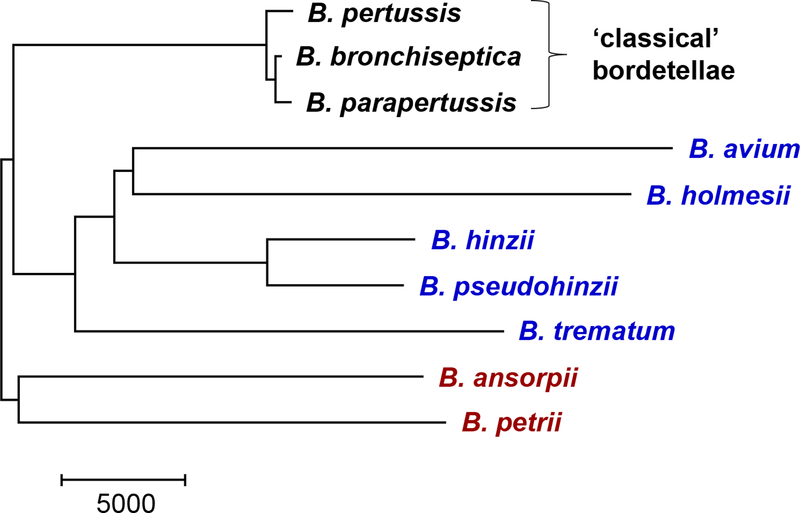
In a whole genome-based phylogenetic tree Bordetella species form three main clades (Fig. 1). One clade contains the three ‘classical’ species B. bronchiseptica, B. parapertussis and B. pertussis, by far the best studied species. A second clade consists of B. trematum, B. hinzii, B. pseudohinzii, B. avium and B. holmesii, and the third clade includes B. petrii and B. ansorpii [7,19]. The genomes of the classical bordetellae are closely related and show a low between-species genetic diversity, which reflects the relatively recent emergence of B. parapertussis and B. pertussis from B. bronchiseptica-like ancestors [7]. Indeed, B. pertussis is a very young species. A global genome-based phylogeny of a worldwide collection of B. pertussis strains revealed two deep branches that coalesce about 2,300 years ago [27]. Subsequently, one of the branches in the phylogenetic tree, which contains over 98 % of all analyzed strains, appears to have started to expand about 500 years ago [27], which coincides with the first descriptions of whooping cough outbreaks in Persia and Europe [27,28].
Fig 1. Heading: Whole genome phylogeny of ten Bordetella species based on a genome-wide sequence alignment.
Legend: The Neighbor-joining tree shows three clades of species that consist of B. bronchiseptica, B. parapertussis and B. pertussis (black), of B. avium, B. holmesii, B. hinzii, B. pseudohinzii and B. trematum (blue), and of B. ansorpii and B. petrii (brown). The tree was rooted according to Linz et al., 2016 [7].
Environmental origin of Bordetella species
For most of the last century Bordetella species were considered to be host-restricted pathogens, with variable host specificity. However, in 2001, B. petrii was isolated from a mixed anaerobic, dechlorinating culture enriched with river sediment [25]. In 2015, three new species named B. muralis, B. tumulicola and B. tumbae were isolated from the plaster wall surface of 1,300-year-old mural paintings inside the stone chamber of the Takamatsuzuka Tomb in Japan [29]. Since then, bordetellae have been identified in several metagenomic analyses of environmental samples: A microbial ecosystem in a bioreactor degrading thiocyanate and cyanide contained a Bordetella-like organism [30], and a metagenomic analysis of a consortium of biphenyl-degrading soil bacteria revealed Bordetella as a key player in the degradation of benzoate [31]. Sequences from numerous Bordetella-like organisms were found among environmental sources by probing 16S ribosomal RNA sequences available in the NCBI database [32]. A phylogenetic comparison of 16S rRNA sequences from animal-associated species and from strains recovered from various sources, including soil and water, provided evidence for an environmental origin of the bordetellae. First, the genetic diversity of the environmental samples was significantly larger than the diversity of the animal- and human-associated samples. And second, sequences from environmental sources were present in all ten phylogenetic clades in the tree, including sequence clades at the root of the tree, whereas sequences from animal samples were only found in four clades near the top of the tree. The branching order in the phylogenetic tree indicated that the genus Bordetella originated from an environmental source [32].
A detailed genome analysis of B. petrii strain DSMZ12804 has identified a core set of 2,049 genes that are shared with the genomes of both B. bronchiseptica (strain RB50) and B. avium (strain 197N) and thus are likely part of the ancestral genome inherited from a common ancestor [33]. Among these are genes encoding the central metabolic pathways that are shared between B. petrii as an example of an environmental species and the animal-pathogenic Bordetella species, including the TCA cycle and synthesis of amino acids, fatty acids and nucleotides. Interestingly, B. petrii has a large set of 1,825 unique genes, many of which encode auxiliary pathways that may be involved in the metabolic breakdown of plant material and other organic components. In this regard, B. petrii is remarkably versatile. It possesses genes to utilize gluconate, other plant products such as pectin, cyanate and a vast variety of aromatic compounds, including phthalate, phenylacetate, benzoate, benzylalcohol, chlorobenzenes, and phenol [33]. B. petrii harbors numerous peripheral pathways to preprocess and channel those and other aromatic compounds into at least eight different central metabolic pathways. Some of these pathways are encoded by genes on genomic islands with an atypical GC content, indicating an important role of horizontal gene transfer (HGT) in the metabolic versatility. In addition, several of the central pathways are represented by multiple gene paralogs, such as three sets of chlorocatechol pathway genes for the degradation of chlorobenzenes [33]. Likewise, the genome of a biphenyl-degrading Bordetella isolate from soil possesses genes of three central pathways for the degradation of benzoate, with most of the genes present as two to six distinct copies, emphasizing the vast metabolic diversity [30]. In contrast, most of the pathways for the degradation of aromatic compounds are not present in the genome of the animal- pathogenic bordetellae. Still, both B. bronchiseptica and B. hinzii are known to associate with animal hosts, yet are able to grow efficiently in soil and thus to survive under environmental conditions [32].
Comparative genomics suggested that B. petrii may be well equipped for competition with other microbes. The B. petrii genome harbors two different type 6 secretion systems (T6SS) [7], which encode a syringe-like apparatus that mediates injection of effectors into both bacterial competitors and eukaryotic host cells and are commonly found in many soil bacteria [34–36]. One T6SS was also found in the genomes of B. bronchiseptica, B. parapertussis and B. ansorpii, suggesting that it may have been present in the Bordetella ancestor. In contrast, the second T6SS was uniquely present in the genome of the environmental B. petrii, implying gene acquisition or repeated loss in all other divergent lineages [7]. Together, these findings suggest, that a high genome plasticity has provided the tools to cope with environmental challenges, allowing acquisition of nutrients and competition against other bacteria.
Adaptation from the environment to ancient hosts
The ability to persist, replicate and disseminate can provide critical advantages in competitive soil/water environments. Numerous bacteria have been reported to form endosymbiotic relationships with amoeba, including Escherichia coli O157, Francisella tularensis, Legionella pneumophila and Mycobacterium spp. [37–41]. Besides providing protection against external dangers and an apparent competitive advantage against other bacteria, symbiosis with amoebae can enhance bacterial dissemination along with the amoebic host, some of which have evolved highly effective mechanisms of travel. Our recent study showed that the animal-pathogenic B. bronchiseptica can hijack the life cycle of the soil-living amoeba Dictyostelium discoideum [42], resisting digestion by single-celled trophozoites and translocating to the fruiting bodies to spread along with its spores. This relationship is stable over multiple amoeba life cycles without affecting B. bronchiseptica’s ability to subsequently infect a mammalian host. By not just residing but growing within the amoeba fruiting body, the bacteria can be disseminated along with the spores to new geographic locations, where they can either maintain a stable association with the amoebic host or infect a new mammalian host, revealing two independent but interconnected life cycles of B. bronchiseptica in protozoan and mammalian hosts [42]. The bacteria and spores can be spread by wind and passing animals, such as ants and flies, and a recent metagenomic study indeed detected B. bronchiseptica and B. hinzii sequencing reads in the microbiome of flies [43].
The similarities between amoeba trophozoites and mammalian phagocytic cells have led some authors to speculate that the interaction between bacterial pathogens and amoebic hosts could have served as a ‘training ground’ for bacteria before they became animal pathogens [40]. Our recent examination of the potential environmental origin of the Bordetella genus [32] and description of complex B. bronchiseptica interaction with D. discoideum [42] also supported the view that bacterial interaction with amoebae may have enabled members of this genus to evolve from environmental microbes to become pathogenic [41].
Species-specific virulence and host specialization
Bordetella species have evolved and successfully adapted to infect and transmit in animal hosts. But what did it take to become animal pathogens? The paucity of genomes from environmental sources makes this a challenge to address. A comparison of the relatively small number of genomes available [7] indicated that the potential ancestor of the pathogenic bordetellae likely possessed several protein secretion systems, including a T2SS, a T3SS, and one or two T6SS, but lacked the best studied Bordetella toxins, including pertussis toxin, adenylate cyclase toxin and dermonecrotic toxin (DNT). The genes encoding those toxins appear to have been acquired by the ancestor of the ‘classical’ bordetellae, with DNT being independently acquired by B. avium at a different chromosomal location. Species-specific putative adhesins such as autotransporters appear to have been acquired both vertically and horizontally. All pathogenic bordetellae possess heme receptors, but those are absent in the genome of the environmental B. petrii and likely represent an adaptation to an animal-associated life style. In general, gain and loss of multiple genes, including many encoding bacterial toxins, protein secretion systems and other virulence factors, accompanied the diversification and speciation in the genus. The loss of so many genes associated with pathogenesis in specialized lineages reveals that there were many more of these in the apparent progenitor than are necessary for the remarkable success of some lineages, including the most prominent human pathogens [7].
B. bronchiseptica, the apparent progenitor of the classical Bordetella species, can infect the respiratory track of a wide range of mammals, including dogs, cats, sheep, rabbit, swine, and mice [4]. Yet, it retains the ability to establish a successful symbiosis with amoeba. Other classical Bordetella spp. have specialized at causing disease and spreading within a single mammalian host. For example, the ovine-specific B. parapertussis (Bppov) successfully colonizes and persists in the respiratory track of sheep but is not observed in other animals and is severely defective in its ability to infect mice [3]. While accumulation of mutations in genes involved in O-antigen production have rendered Bppov highly susceptible to complement mediated killing in mice [44], loss of O-antigen is also well described in human restricted pathogen B. pertussis, suggesting that the lack of O-antigen confers a selective advantage in their respective hosts, although it may also limit them to that particular host. Another example of adaptation to a particular mammalian host is the recently discovered B. pseudohinzii, which was isolated from laboratory raised mice with no obvious clinical symptoms [18,19]. Interestingly, all B. pseudohinzii strains to date have been isolated from mice [18–20,45–48] and a wild rat [20], but the sister species B. hinzii can infect poultry, rabbits and immunocompromised humans [16,17].
Genome reduction during host specialization
Several Bordetella species have evolved and adapted to infect only humans, undergoing large scale gene loss and inactivation in the process [5–7], similar to the genome reduction associated with host-specialization of other pathogens [49]. During speciation, B. pertussis and Bpphu experienced genome reduction of about 22% and 9% respectively, which is reflected in the much smaller genome size of B. pertussis (~ 4.1 Mb) and B. parapertussis (~ 4.8 Mb) compared to B. bronchiseptica (~ 5.3 Mb). B. pertussis genome reduction was mediated by homologous recombination between identical copies of IS481 insertion sequence elements, which effectively deleted large parts of the genome and resulted in loss of over 1,000 genes [5]. Genome reduction was less severe in B. parapertussis, as genome comparisons of Bpphu strain 12822 and Bppov strain Bpp5 against B. bronchiseptica strain RB50 showed large colinear regions between the genomes, with a limited number of chromosomal breakpoints flanked by IS1001 [5,6]. Interestingly, the genome of B. holmesii, the only human-restricted species of the ‘non-classical’ bordetellae, also appears to have undergone substantial reduction in size, mediated by IS407, IS481 and ISL3 [7]. As a result, the genome of B. holmesii (3.7 Mb) is much smaller than that of its closest relative B. hinzii (4.9 Mb), as is the genome of the poultry-specific pathogen B. avium (3.7 Mb). Thus, it appears that host specialization of several Bordetella species is associated with acquisition, expansion and subsequent recombination between IS elements mediating large-scale gene loss. In addition, the extremely low genetic diversity within these species suggests that these speciation events were associated with severe population bottlenecks that drastically reduced the genetic diversity, followed by expansion of a few successful clones [5,7,27].
Genome size and life cycle
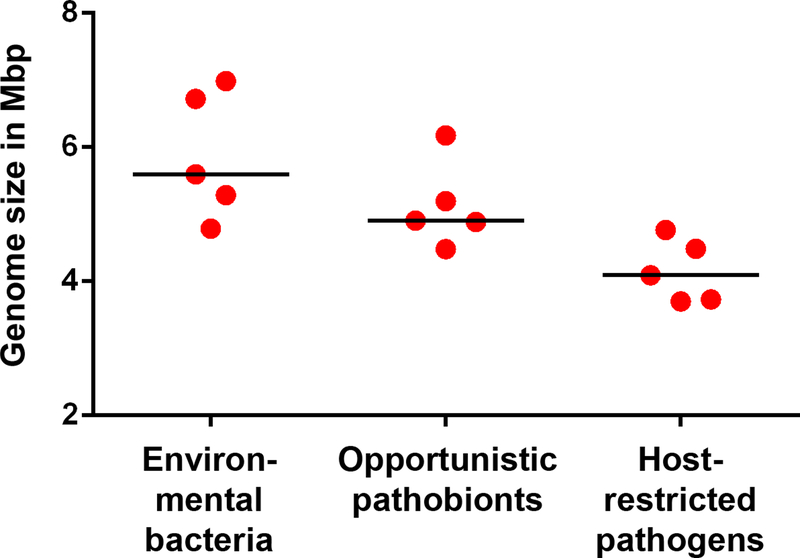
Environmental species need to possess genomes that can facilitate adaption to changing availability of various carbon and nitrogen sources by encoding multiple metabolic pathways. B. petrii’s versatile pathways for the degradation for aromatic compounds [31,33] is likely to provide a valuable fitness advantage in these competitive environmental microbial communities. Diverse metabolic pathways provide a substantial advantage in naturally variable environments but require relatively large genomes. In contrast, the environment and nutrient sources within the airways of hosts are relatively stable and do not require such metabolic versatility. However, opportunistic pathobionts and host-restricted pathogens need other sets of genes that mediate their interactions with the host, including adhesins and virulence factors to manipulate the host’s immune response. Interestingly, the genomes of these animal-adapted Bordetella species are substantially smaller than those of environmental species (Fig. 2). Just like other host-restricted bacteria, host-specialized bordetellae appear to have undergone dramatic genome reduction during speciation that led to the loss of hundreds of genes and resulted in genomes that may be more than 1 Mb smaller than those of their inferred ancestors. These results are consistent with the explanation that a sustained, closed life cycle in a specific host allows for the loss of the many genes involved in success outside of that host.
Fig 2. Heading: Genome size of environmental bacteria, pathobionts and host-restricted pathogens in the genus Bordetella.
Legend: Bordetella sp. SCN 68–11 (accession: MEFS00000000), B. sp. SCN 67–23 (MEDQ00000000), B. sp. BFMG2 (PKCD00000000), B. petrii (NC_010170), and B. sp. N (NZ_CP013111), isolated from environmental sources possess the largest genomes in the genus Bordetella. In contrast, the genomes of the obligate host-restricted pathogens B. holmesii (NZ_CP007494), B. avium (NC_010645.1), B. pertussis (NC_002929), B. parapertussishu (NC_002928) and B. pseudohinzii (NZ_CP016440) featured substantial reduction. Pathobionts B. trematum (NZ_LT546645), B. hinzii (NZ_CP012076), B. ansorpii (NZ_FKIF00000000), and B. bronchiseptica (NC_002927), have been isolated from multiple animal and human sources.
Relatively frequent loss or mutation of genes required in the extra-host environment are likely to result in host-restricted pathogens. But such spontaneous mutants would be rapidly lost from nature when they cannot sustain a stable chain of transmission that lasts over extended evolutionary time. Bordetella species that are observed in a single host must have established a successful medium to long term strategy, which is only possible in a host population that is large and connected enough to sustain transmission for long periods of time. Interestingly, several Bordetella species appear to have emerged as host-restricted in the relatively recent Anthropocene era, when human activities have had a major impact on various animal populations. Thus B. pertussis and Bpphu are highly successful in large human populations. Similarly, B. avium and Bppov are highly successful in poultry and sheep populations, respectively, that human husbandry has allowed to reach sizes and densities not present before the Anthropocene. Together these observations support speculation that the emergence of these host restricted Bordetella species is anthropogenic; human changes to host populations provided an environment in which these species could emerge with host-specific closed life cycles and persist over time.
Ongoing changes in clinical B. pertussis genomes
All Bordetella species are continuing to evolve. In the best studied of these there is regular speculation about the pressures shaping this evolution. Recent B. pertussis isolates that are deficient in expression of components of the current acellular pertussis (aP) vaccines, have led to extensive speculation about ‘vaccine-driven evolution’. Precisely because this is such a compelling and scary idea, it is particularly important to remain impartial and skeptical, to consider other explanations, to look for all types of evidence for and against, and to put genotypic and phenotypic changes in the context of recent evolution of the genus. Increasing numbers of isolates lacking a particular factor, such as Pertactin, correlate with increased aP vaccine use in some populations, but these are observations of correlations, and are quite emphatically NOT evidence of vaccine-driven evolution. The evolutionary history of the genus demonstrates substantial genome reduction in several lineages, especially B. pertussis, resulting in loss of a large proportion of genes during speciation, including several autotransporter genes. In addition, the observed switches in allele frequencies of aP vaccine component genes cannot be directly linked to aP vaccines, because a comparative genome analysis of 343 B. pertussis strains isolated between 1920 and 2010 revealed that the new alleles were already present in the pre-vaccine era (ptxA1) or in the whole cell vaccine era (ptxP3, prn2, fim2–2, fim3–2) and thus did not originate under aP vaccine pressure [27]. The argument that loss of one of five antigens would somehow allow escape from all five antigens in the vaccines is tenuous. In addition, a true vaccine-escape mutant would be expected to sweep across aP vaccinated populations across the globe, resulting in substantially decreased vaccine efficacy and newly increased epidemics in many countries simultaneously, which is not the prevailing pattern. In the absence of compelling evidence of such a sweep, Ocham’s Razor requires that we consider a simpler explanation; B. pertussis continues to undergo rapid and profound genome reduction associated with its relatively recent loss of an environmental reservoir and commitment to a closed life cycle in humans. It is critical to better understand the genotypic and phenotypic variation amongst these species as they diverge, evolve and adapt to different ecological niches. This will, in turn, provide the broader context in which to consider and understand their ongoing evolution.
Conclusion:
Recent isolates from environmental sources provide a new perspective on the natural history of Bordetella, showing that pathogenic bordetellae likely evolved from ancestors in soil. The evolutionary pressure to evade amoebic predators may have selected for adaptations that enable survival of phagocytosis by either protists or mammalian immune cells. As more genomes from environmental sources become available, comparative genomics and metabolomics will reveal the genomic changes that accompany bacterial adaptation to eukaryotic hosts, which paved the way for the emergence of pathogens. Subsequent specialization of some lineages to a closed host-specific lifecycle involved substantial genome reduction, resulting in loss of genes required only in the environmental reservoir.
Key points.
Environmental Bordetella species have significantly larger genetic diversity than animal- and human-associated samples.
Association with amoebic hosts may have been an intermediate step in the evolution from environmental microbes to pathogenic bacteria.
Host specialization was accompanied by significant genome reduction.
Anthropogenic factors enable closed life cycles in populous host species, with pathogenesis mediating efficient transmission between hosts.
Acknowledgements:
None.
Financial support: This study was supported by grants AI104399 and AI142678 of the National Institutes of Health to ETH.
Footnotes
Conflict of interest: None.
References and recommended reading:
- 1.Mattoo S, Cherry JD: Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 2005, 18:326–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heininger U, Stehr K, Schmitt-Grohe S, Lorenz C, Rost R, Christenson PD, Uberall M, Cherry JD: Clinical characteristics of illness caused by Bordetella parapertussis compared with illness caused by Bordetella pertussis. Pediatr Infect Dis J 1994, 13:306–309. [DOI] [PubMed] [Google Scholar]
- 3.Porter JF, Connor K, Donachie W: Isolation and characterization of Bordetella parapertussis-like bacteria from ovine lungs. Microbiology 1994, 140 (Pt 2):255–261. [DOI] [PubMed] [Google Scholar]
- 4.Goodnow RA: Biology of Bordetella bronchiseptica. Microbiological Reviews 1980, 44:722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MTG, Churcher CM, Bentley SD, Mungall KL, et al. : Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nature Genetics 2003, 35:32–40. [DOI] [PubMed] [Google Scholar]
- 6.Park J, Zhang Y, Buboltz AM, Zhang XQ, Schuster SC, Ahuja U, Liu MH, Miller JF, Sebaihia M, Bentley SD, et al. : Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics 2012, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linz B, Ivanov YV, Preston A, Brinkac L, Parkhill J, Kim M, Harris SR, Goodfield LL, Fry NK, Gorringe AR, et al. : Acquisition and loss of virulence-associated factors during genome evolution and speciation in three clades of Bordetella species. BMC Genomics 2016, 17:767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weyant RS, Hollis DG, Weaver RE, Amin MF, Steigerwalt AG, O’Connor SP, Whitney AM, Daneshvar MI, Moss CW, Brenner DJ: Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J Clin Microbiol 1995, 33:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Njamkepo E, Bonacorsi S, Debruyne M, Gibaud SA, Guillot S, Guiso N: Significant finding of Bordetella holmesii DNA in nasopharyngeal samples from French patients with suspected pertussis. J Clin Microbiol 2011, 49:4347–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamiya H, Otsuka N, Ando Y, Odaira F, Yoshino S, Kawano K, Takahashi H, Nishida T, Hidaka Y, Toyoizumi-Ajisaka H, et al. : Transmission of Bordetella holmesii during pertussis outbreak, Japan. Emerg Infect Dis 2012, 18:1166–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodgers L, Martin SW, Cohn A, Budd J, Marcon M, Terranella A, Mandal S, Salamon D, Leber A, Tondella ML, et al. : Epidemiologic and laboratory features of a large outbreak of pertussis-like illnesses associated with cocirculating Bordetella holmesii and Bordetella pertussis--Ohio, 2010–2011. Clin Infect Dis 2013, 56:322–331. [DOI] [PubMed] [Google Scholar]
- 12.Harvill ET, Goodfield LL, Ivanov Y, Smallridge WE, Meyer JA, Cassiday PK, Tondella ML, Brinkac L, Sanka R, Kim M, et al. : Genome Sequences of Nine Bordetella holmesii Strains Isolated in the United States. Genome Announc 2014, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kersters K, Hinz KH, Hertle A, Segers P, Lievens A, Siegmann O, Deley J: Bordetella avium sp. nov., isolated from the respiratory tracts of turkeys and other birds. International Journal of Systematic Bacteriology 1984, 34:56–70. [Google Scholar]
- 14.Raffel TR, Register KB, Marks SA, Temple L: Prevalence of Bordetella avium infection in selected wild and domesticated birds in the eastern USA. Journal of Wildlife Diseases 2002, 38:40–46. [DOI] [PubMed] [Google Scholar]
- 15.Register KB, Kunkle RA: Strain-specific virulence of Bordetella hinzii in poultry. Avian Dis 2009, 53:50–54. [DOI] [PubMed] [Google Scholar]
- 16.Vandamme P, Hommez J, Vancanneyt M, Monsieurs M, Hoste B, Cookson B, Wirsing von Konig CH, Kersters K, Blackall PJ: Bordetella hinzii sp. nov., isolated from poultry and humans. Int J Syst Bacteriol 1995, 45:37–45. [DOI] [PubMed] [Google Scholar]
- 17.Register KB, Ivanov YV, Harvill ET, Brinkac L, Kim M, Losada L: Draft Genome Sequences of Six Bordetella hinzii Isolates Acquired from Avian and Mammalian Hosts. Genome Announc 2015, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov YV, Shariat N, Register KB, Linz B, Rivera I, Hu K, Dudley EG, Harvill ET: A newly discovered Bordetella species carries a transcriptionally active CRISPR-Cas with a small Cas9 endonuclease. BMC Genomics 2015, 16:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov YV, Linz B, Register KB, Newman JD, Taylor DL, Boschert KR, Le Guyon S, Wilson EF, Brinkac LM, Sanka R, et al. : Identification and taxonomic characterization of Bordetella pseudohinzii sp. nov. isolated from laboratory-raised mice. Int J Syst Evol Microbiol 2016, 66:5452–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loong SK, Che-Mat-Seri NA, Abdulrazak O, Douadi B, Ahmad-Nasrah SN, Johari J, Mohd-Zain SN, Abubakar S: Recovery of Bordetella bronchiseptica sequence type 82 and B. pseudohinzii from urban rats in Terengganu, Malaysia. J Vet Med Sci 2018, 80:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]; * first finding of B. pseudohinzii in a wild animal.
- 21.Vandamme P, Heyndrickx M, Vancanneyt M, Hoste B, De Vos P, Falsen E, Kersters K, Hinz KH: Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Ruger and Tan 1983. Int J Syst Bacteriol 1996, 46:849–858. [DOI] [PubMed] [Google Scholar]
- 22.Daxboeck F, Goerzer E, Apfalter P, Nehr M, Krause R: Isolation of Bordetella trematum from a diabetic leg ulcer. Diabet Med 2004, 21:1247–1248. [DOI] [PubMed] [Google Scholar]
- 23.Ko KS, Peck KR, Oh WS, Lee NY, Lee JH, Song JH: New species of Bordetella, Bordetella ansorpii sp. nov., isolated from the purulent exudate of an epidermal cyst. J Clin Microbiol 2005, 43:2516–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandamme PA, Peeters C, Cnockaert M, Inganas E, Falsen E, Moore ER, Nunes OC, Manaia CM, Spilker T, LiPuma JJ: Bordetella bronchialis sp. nov., Bordetella flabilis sp. nov. and Bordetella sputigena sp. nov., isolated from human respiratory specimens, and reclassification of Achromobacter sediminum Zhang et al. 2014 as Verticia sediminum gen. nov., comb. nov. Int J Syst Evol Microbiol 2015, 65:3674–3682. [DOI] [PubMed] [Google Scholar]
- 25.von Wintzingerode F, Schattke A, Siddiqui RA, Rosick U, Gobel UB, Gross R: Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int J Syst Evol Microbiol 2001, 51:1257–1265. [DOI] [PubMed] [Google Scholar]
- 26.Nagata JM, Charville GW, Klotz JM, Wickremasinghe WR, Kann DC, Schwenk HT, Longhurst CA: Bordetella petrii sinusitis in an immunocompromised adolescent. Pediatr Infect Dis J 2015, 34:458. [DOI] [PubMed] [Google Scholar]
- 27.Bart MJ, Harris SR, Advani A, Arakawa Y, Bottero D, Bouchez V, Cassiday PK, Chiang CS, Dalby T, Fry NK, et al. : Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. MBio 2014, 5:e01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aslanabadi A, Ghabili K, Shad K, Khalili M, Sajadi MM: Emergence of whooping cough: notes from three early epidemics in Persia. Lancet Infectious Diseases 2015, 15:1480–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tazato N, Handa Y, Nishijima M, Kigawa R, Sano C, Sugiyama J: Novel environmental species isolated from the plaster wall surface of mural paintings in the Takamatsuzuka tumulus: Bordetella muralis sp. nov., Bordetella tumulicola sp. nov. and Bordetella tumbae sp. nov. International Journal of Systematic and Evolutionary Microbiology 2015, 65:4830–4838. [DOI] [PubMed] [Google Scholar]
- 30.Kantor RS, van Zyl AW, van Hille RP, Thomas BC, Harrison ST, Banfield JF: Bioreactor microbial ecosystems for thiocyanate and cyanide degradation unravelled with genome-resolved metagenomics. Environ Microbiol 2015, 17:4929–4941. [DOI] [PubMed] [Google Scholar]
- 31.Garrido-Sanz D, Manzano J, Martin M, Redondo-Nieto M, Rivilla R: Metagenomic Analysis of a Biphenyl-Degrading Soil Bacterial Consortium Reveals the Metabolic Roles of Specific Populations. Front Microbiol 2018, 9:232. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** an interesting insight into the metabolic diversity of an environmental Bordetella.
- 32.Hamidou Soumana I, Linz B, Harvill ET: Environmental Origin of the Genus Bordetella. Frontiers in Microbiology 2017, 8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** of central importance for our hypothesis that bordetellae evolved from environmental ancestors in soil.
- 33.Gross R, Guzman CA, Sebaihia M, dos Santos VA, Pieper DH, Koebnik R, Lechner M, Bartels D, Buhrmester J, Choudhuri JV, et al. : The missing link: Bordetella petrii is endowed with both the metabolic versatility of environmental bacteria and virulence traits of pathogenic Bordetellae. BMC Genomics 2008, 9:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho BT, Dong TG, Mekalanos JJ: A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 2014, 15:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD: Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 2010, 6:e1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma LS, Hachani A, Lin JS, Filloux A, Lai EM: Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 2014, 16:94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abu Kwaik Y: The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl Environ Microbiol 1996, 62:2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bozue JA, Johnson W: Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect Immun 1996, 64:668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newsome AL, Baker RL, Miller RD, Arnold RR: Interactions between Naegleria fowleri and Legionella pneumophila. Infect Immun 1985, 50:449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y: Amoebae as training grounds for intracellular bacterial pathogens. Applied and Environmental Microbiology 2005, 71:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor-Mulneix DL, Hamidou Soumana I, Linz B, Harvill ET: Evolution of Bordetellae from Environmental Microbes to Human Respiratory Pathogens: Amoebae as a Missing Link. Front Cell Infect Microbiol 2017, 7:510. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** an interesting opinion on how the ability to resist amoeba phagocytic cells may have enabled bordetellae to adapt to mammalian hosts.
- 42.Taylor-Mulneix DL, Bendor L, Linz B, Rivera I, Ryman VE, Dewan KK, Wagner SM, Wilson EF, Hilburger LJ, Cuff LE, et al. : Bordetella bronchiseptica exploits the complex life cycle of Dictyostelium discoideum as an amplifying transmission vector. PLoS Biology 2017, 15:e2000420. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** a ground-breaking study on an enrironmental niche and vector for Bordetella, and on the function for the Bvg-phase.
- 43.Junqueira ACM, Ratan A, Acerbi E, Drautz-Moses DI, Premkrishnan BNV, Costea PI, Linz B, Purbojati RW, Paulo DF, Gaultier NE, et al. : The microbiomes of blowflies and houseflies as bacterial transmission reservoirs. Sci Rep 2017, 7:16324. [DOI] [PMC free article] [PubMed] [Google Scholar]; * an interesting insight into transmission of bacterial pathogens by flies.
- 44.Hester SE, Goodfield LL, Park J, Feaga HA, Ivanov YV, Bendor L, Taylor DL, Harvill ET: Host Specificity of Ovine Bordetella parapertussis and the Role of Complement. PLoS One 2015, 10:e0130964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark SE, Purcell JE, Sammani S, Steffen EK, Crim MJ, Livingston RS, Besch-Williford C, Fortman JD: Bordetella pseudohinzii as a Confounding Organism in Murine Models of Pulmonary Disease. Comp Med 2016, 66:361–366. [PMC free article] [PubMed] [Google Scholar]
- 46.Loong SK, Tan KK, Sulaiman S, Wong PF, AbuBakar S: Draft genome of Bordetella pseudohinzii BH370 isolated from trachea and lung tissues of a laboratory mouse. Genom Data 2017, 12:69–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perniss A, Schmidt N, Gurtner C, Dietert K, Schwengers O, Weigel M, Hempe J, Ewers C, Pfeil U, Gartner U, et al. : Bordetella pseudohinzii targets cilia and impairs tracheal cilia-driven transport in naturally acquired infection in mice. Sci Rep 2018, 8:5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spilker T, Darrah R, LiPuma JJ: Complete Genome Sequences of Bordetella flabilis, Bordetella bronchialis, and “Bordetella pseudohinzii”. Genome Announc 2016, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moran NA: Microbial minimalism: genome reduction in bacterial pathogens. Cell 2002, 108:583–586. [DOI] [PubMed] [Google Scholar]