Abstract
Background
Quantifying reproducibility of native T1 and T2 mapping over a long period (> 1 year) is necessary to assess whether changes in T1 and T2 over repeated sessions in a longitudinal study are associated with variability due to underlying tissue composition or technical confounders.
Objectives
To carry out a single-center phantom study to 1) investigate measurement reproducibility of slice-interleaved T1 (STONE) and T2 mapping over 20 months, 2) quantify sources of variability, and 3) compare reproducibility and measurements against reference spin-echo measurements.
Methods
MR imaging was performed on a 1.5 Tesla Philips Achieva scanner every 2–3 weeks over 20 months using the T1MES phantom. In each session, slice-interleaved T1 and T2 mapping was repeated 3 times for 5 slices, and maps were reconstructed using both 2-parameter and 3-parameter fit models. Reproducibility between sessions, and repeatability between repetitions and slices were evaluated using coefficients of variation (CV). Different sources of variability were quantified using variance decomposition analysis. The slice-interleaved measurement was compared to the spin-echo reference and MOLLI.
Results
Slice-interleaved T1 had excellent reproducibility and repeatability with a CV < 2%. The main sources of T1 variability were temperature in 2-parameter maps, and slice in 3-parameter maps. Superior between-session reproducibility to the spin-echo T1 was shown in 2-parameter maps, and similar reproducibility in 3-parameter maps. Superior reproducibility to MOLLI T1 was also shown. Similar measurements to the spin-echo T1 were observed with linear regression slopes of 0.94–0.99, but slight underestimation. Slice-interleaved T2 showed good reproducibility and repeatability with a CV < 7%. The main source of T2 variability was slice location/orientation. Between-session reproducibility was lower than the spin-echo T2 reference and showed good measurement agreement with linear regression slopes of 0.78–1.06.
Conclusions
Slice-interleaved T1 and T2 mapping sequences yield excellent long-term reproducibility over 20 months.
Introduction
Cardiovascular magnetic resonance (CMR) native T1 and T2 mapping have emerged as promising techniques for myocardial tissue characterization [1]. Studies have reported increased native T1 times in the presence of myocardial fibrosis, inflammation, amyloids, and decreased T1 in the presence of Anderson-Fabry disease, and iron overload [2]. Increased T2 times have also been reported in the presence of edema or inflammation [3–5]. Assessing T1 and T2 measurement reproducibility is a necessary step toward their clinical utility as quantitative imaging biomarkers [6].
Various cardiac mapping techniques have been proposed for T1 [7–11] and T2 mapping [12–16]. The most widely used T1 mapping sequence is the Modified Look-Locker inversion recovery (MOLLI) [7], which is based on sampling the inversion recovery of the longitudinal relaxation signal. Other types of T1 mapping sequences, such as the Saturation recovery single-shot acquisition (SASHA), are based on sampling the saturation recovery curve [9]. A hybrid sequence combining inversion and saturation recovery curves, such as the Saturation pulse prepared heart rate independent inversion recovery (SAPPHIRE), has also been proposed [10]. The most widely used T2 mapping sequences are based on T2-preparation (T2prep) [17–20] with balanced steady-state free precession (bSSFP) imaging [5, 12] or spoiled gradient echo (GRE) [14] acquired with at least 3 different echo times. Other types of T2 mapping sequences are based on turbo spin echo (TSE) [15] or gradient spin echo (GraSE) [16].
In longitudinal studies, understanding technical variability is critical to determining if observed changes over time are biological and therefore clinically significant or only related to measurement variation [21]. Furthermore, higher reproducibility means fewer patients are necessary to achieve statistical significance in clinical trials, ultimately reducing study costs [22]. Several prior studies have investigated the reproducibility of various T1 and T2 mapping sequences, however they are test/retest studies carried out within several weeks [23–26]. Reproducibility studies using MOLLI and shortened MOLLI (ShMOLLI) have demonstrated that both sequences are highly reproducible [24, 26–29]. SASHA and SAPPHIRE were reported to have similar reproducibility as inversion recovery-based sequences [23]. The reproducibility of T2 mapping of multi-echo-spin-echo T2, T2prep-bSSFP, and GraSE T2 mapping sequences were also reported to be excellent [25].
The free-breathing slice-interleaved T1 [30, 31] and T2 [32] mapping techniques have been proposed and used in various clinical scenarios [33–37]. Slice-interleaved T1 (STONE) acquires data for different slices within one inversion recovery curve to allow more accurate measurement with a bSSFP (STONE-bSSFP) [30] or spoiled gradient echo (STONE-GRE) [31]. Slice-interleaved T2 uses slice-selective T2prep with an interleaved slice acquisition scheme which permits increased time efficiency [32]. Slice-interleaved T1 and T2 mapping sequences provide highly reproducible measurements in test/retest studies of healthy subjects [22], however the long-term reproducibility (> 1 year) has not yet been studied. Long-term reproducibility of T1 and T2 measurements using slice-interleaved T1 and T2 mapping needs to be investigated prior to utilization of these sequences in longitudinal studies monitoring disease progression or treatment efficacy.
Various confounders can impact the accuracy and reproducibility of myocardial tissue characterization. Therefore, performance assessment of the myocardial tissue characterization techniques requires rigorous in-vivo or phantom validation. While in-vivo studies are the ideal experimental setting, a phantom study is necessary in cases where in-vivo experiments are not feasible or scenarios in which the reference standard can only be measured in a phantom setting. Phantom studies are also necessary for assessing long-term measurement variability when scanning volunteers for extended periods over multiple sessions is not feasible. T1 or T2 accuracy and temperature sensitivity, for example, can only be measured in the phantom setting. Although a phantom experiment may not address all relevant confounding factors of an in-vivo setting, it provides valuable information that may not be easily attainable from an in-vivo experiment.
The aim of this study was to carry out a single-center phantom study to 1) investigate the measurement reproducibility of slice-interleaved T1 and T2 mapping over 20 months, 2) quantify sources of variability, and 3) compare the performance of each in terms of reproducibility and measurement against reference spin-echo measurements.
Materials and methods
Experiments were performed using T1 Mapping and ECV Standardization Program (T1MES) phantom [38]. This Food and Drug Administration (FDA)-cleared/Conformité Européene (CE)-marked MR phantom enables stable quality measures to study measurement variability over time. T1MES contains 9 vials (NiCl2 doped agarose) covering the physiological ranges of T1 and T2 in the blood and myocardium pre- and post-Gadolinium-based contrast agents (GBCA; for a 1.5 T phantom: T1:255ms to 1489ms, T2:44ms to 243ms, referenced from the T1MES manual measured by slow inversion-recovery/spin-echo methods at 1.5T) (Fig 1A). The T1MES phantom volume is 2L with an inner dimension size of 197 × 122 × 122 mm, and the vials have a minimum diameter of 20 mm [38]. For T1 mapping, all 9 vials were studied given that the phantom is designed to include all relevant T1 ranges of myocardium and blood pre- and post-GBCA. For T2 mapping we only studied vial ‘F’ (Fig 1A) which modulates “Medium” native myocardial T1 and T2 times at 1.5 T. Our T2 mapping sequence is not designed to handle high T2 values over 100 ms found in the blood, and all remaining vials had no variability (44–50 ms).
Fig 1. T1MES phantom used in this study, and the reproducibility study protocol.
a) The T1MES phantom used in this study consists of 9 vials of NiCl2 doped agarose covering T1 and T2 ranges in the blood and myocardium before and after Gadolinium-based contrast agents. b) An imaging session was repeated every 2–3 weeks over 20 months (between-session reproducibility). Within each session, slice-interleaved T1 and T2 mapping sequences were repeated 3 times (between-repetition repeatability) for five slices (between-slice repeatability). SE T1 and T2 measurements and MOLLI were performed for comparison. STONE-bSSFP, slice-interleaved T1 with balanced steady‐state free precession; STONE-GRE, slice-interleaved T1 with spoiled gradient echo; SE, spin-echo.
Reproducibility is defined as the measurement precision between replicate measurements under varying conditions, and repeatability is defined as the measurement precision between replicate measurements under constant conditions [21]. In this study, we use ‘reproducibility’ when referring to measurement precision over multiple sessions, and ‘repeatability’ when referring to scanning in the same session. We defined a ‘session’ as a ‘single CMR imaging with identical image localization’.
The study design schematic is shown in Fig 1B. Reproducibility over several weeks was reported for between-session reproducibility. Images were acquired using STONE-bSSFP T1 [30], STONE-GRE T1 mapping [31], and slice-interleaved T2 mapping [32] sequences. Within each session, imaging was repeated 3 times to allow repeatability assessment within each session and between repetitions. For multi-slice sequences, between-slice repeatability was also studied. Additionally, we acquired spin-echo (SE) T1 and T2 measurements and MOLLI in each imaging session for comparison; MOLLI was repeated 2 times.
CMR imaging
CMR imaging was performed using a 1.5 T scanner (Philips Achieva, Best, The Netherlands) with a 32-element cardiac phased-array receiver coil. The phantom was stored and scanned at room temperature in the scanner room. We assumed temperature and subsequently diffusion was uniform along vials in our study. Scanning was strictly performed according to the T1MES phantom user manual [38]. All acquisitions were performed with a simulated electrocardiogram (ECG) at a RR (interval time between two R-waves) period of 900 ms (heart rate 67 bpm). The positioning process was consistent for all sessions throughout the study. The book used to lift the phantom, large towel, coil, software version of the scanner, and air-flow setting of the scanner room remained constant throughout the study.
T1 mapping
The STONE-bSSFP sequence was acquired with the following parameters: 5 slices, in-plane resolution = 2.1 × 2.1 mm2, slice thickness = 8 mm, slice gap = 4 mm, field-of-view = 280 × 280 mm2, TR/TE/flip angle = 2.8 ms / 1.39 ms / 70°, a sensitivity encoding (SENSE) rate = 2, linear ordering, 10 linear ramp-up pulses and bandwidth = 1894 Hz, acquisition duration = 1 min 38 sec. Eleven inversion images were acquired with inversion times of ∞, 130, 1030, 1930, 2830, 3730, 350, 1250, 2150, 3050, and 3950 ms. The STONE-GRE sequence was acquired with the following parameters: 5 slices, in-plane resolution = 2 × 2 mm2, slice thickness = 8 mm, slice gap = 4 mm, field-of-view = 280 × 280 mm2, TR/TE/flip angle = 4.7 ms / 2.3 ms / 10°, a SENSE rate = 2.5, half-scan factor = 0.75, linear ordering, 10 linear ramp-up pulses and bandwidth = 383 Hz, acquisition duration = 1 min 38 sec. Eleven inversion images were acquired with inversion times of ∞, 109, 1009, 1909, 2809, 3709, 350, 1250, 2150, 3050, and 3950 ms. For both STONE-bSSFP and STONE-GRE sequences, the inversion preparation pulse was an adiabatic hyperbolic secant pulse with an 11 ms pulse duration. The radiofrequency (RF) excitation pulse was a slice-selective Sinc-Gauss pulse with a duration of 0.43 ms. Images were acquired without prospective slice tracking, and the order of slices was 1-4-2-5-3. The MOLLI 5b(3s)3b [39] sequence was acquired with the following parameters: single slices, in-plane resolution = 2 × 2 mm2, slice thickness = 8 mm, field-of-view = 280 × 280 mm2, TR/TE/flip angle = 2.6 ms / 1.30 ms / 35°, a SENSE rate = 2.5, linear ordering, 10 linear ramp-up pulses and bandwidth = 1786 Hz, acquisition duration = 8 sec. Eight inversion images were acquired with inversion times of 79, 979, 1879, 2779, 3679, 350, 1250, and 2150 ms. SE T1 times were obtained using inversion-recovery SE acquisitions with 16 inversion times of 50, 100, 200, 300, 400, 500, 600, 700, 800, 900, 1000, 1250, 1500, 1750, 2000, and 3000 ms with the following imaging parameters: single slice, in-plane resolution = 1.2 × 1.2 mm2, slice thickness = 8 mm, field-of-view = 140 × 140 mm2, TR/TE/flip angle = 10 s / 11 ms / 90° and bandwidth = 510 Hz, acquisition duration = 5 hour 18 min.
T2 mapping
The slice-interleaved T2 mapping sequence was acquired with the following parameters: 5 slices, in-plane resolution = 2 × 2 mm2, slice thickness = 8 mm, slice gap = 4 mm, slice ordering = 1-3-5-2-4, field-of-view = 280 × 280 mm2, TR/TE/flip angle = 2.8 ms / 1.42 ms / 55°, a SENSE rate = 2.5, linear ordering, 10 linear ramp-up pulses and bandwidth = 1786 Hz, acquisition duration = 1 min 26 sec. Ten T2prep images were acquired with T2prep echo times of 0, 25, 35, 45, 55, 65, 75, 85, 95 ms, and ∞ was simulated with a saturation pulse. For T2 mapping with 4 echo times, T2prep images of 0, 25, 55, and ∞ were used for map reconstruction, and the results are reported as T2 4echo. A T2-prep pulse consists of a tip-down slice-selective 90° pulse, followed by four non-selective 180° refocusing pulses that end with a closing tip-up slice-selective 90° pulse [32, 40]. SE T2 times were obtained using a Carr-Purcell-Meiboom-Gill (CPMG) SE sequence with 32 TEs of 10, 20, 30, …, 320 ms. The imaging parameters were as follows: single slice, in-plane resolution = 1.16 × 1.16 mm2, slice thickness = 8 mm, field-of-view = 140 × 140 mm2, TR/TE/flip angle = 10 s / 10 ms / 90°, number of signals averaged = 4, bandwidth = 1029 Hz, acquisition duration = 1 hour 21 min.
Map reconstruction
Slice-interleaved T1 and T2 maps were reconstructed using both 2-parameter (2P) and 3-parameter (3P) fit models and all results were reported for both 2P and 3P maps. T1 and T2 maps were reconstructed offline using MATLAB (MathWorks Inc., Natick, Massachusetts, USA). STONE-bSSFP and STONE-GRE maps were estimated by voxel-wise curve-fitting of the signal with a 2-parameter () and 3-parameter () model of the inversion-recovery signal [30]. MOLLI and SE T1 values were obtained using . For MOLLI, apparent T1 values were corrected using Look-Locker correction based on the fitted parameters [7].
For slice-interleaved T2 mapping, a 2-parameter () and 3-parameter () curve fitting model of the T2 signal capturing the effect of imaging pulses on the magnetization was used [41]. SE T2 values were estimated using . All parameters were estimated using a Levenberg-Marquardt optimizer [42].
Data analysis
A region-of-interest (ROI) was manually contoured once for each vial, and identical ROIs were programmatically applied to all slice-interleaved T1, T2 and MOLLI maps throughout all experiments. A graphical illustration of the ROI is shown in S1 Fig. The mean area of the elliptical ROIs of each vial was 73 mm2. A separate ROI was manually contoured once and identical ROIs were used for all SE T1 and T2 maps throughout all sessions. The mean area of the elliptical SE ROIs of each vial was 90 mm2. A linear translation of ROIs less than 1cm in the imaging plane directions was applied in case of offsets from the isocenter. The measurement was defined for each vial as the mean T1 or T2 in each ROI and was acquired separately for all slices, repetitions, sessions, vials, and sequences. Data analysis was performed using MATLAB (MathWorks Inc., Natick, Massachusetts, USA).
Statistical analysis
To investigate T1 and T2 measurement drift over 20 months, a linear regression was performed for each vial over sessions, and the regression slope and 95% confidence interval (CI) of the slopes were reported. We carried out three analyses to assess the reproducibility and repeatability of the observed slice-interleaved T1 and T2 measurements via coefficients of variation, variance component decompositions, linear regressions, and Bland-Altman plots.
Estimation of coefficient of variation
The coefficient of variation (CV), defined as the ratio of the standard deviation to the mean multiplied by 100, was performed to assess reproducibility between sessions, repeatability between repetitions and within a session, and repeatability between slices and within single repetitions. CV was reported as the mean ± standard deviation and visualized by bar plots. To further study variability in T1 mapping due to different T1 times, a CV scatter plot for each vial, sorted from shortest T1 to longest T1 time, and a Spearman correlation between the CV and T1 time (vials) was reported. For T2 mapping, between-session reproducibility CV was estimated for a single vial and therefore no standard deviation among sessions was reported. CV was considered excellent at 0–5%, and good at 5–10%.
Variance decomposition analysis
We considered the observed T1 and T2 measurements as random variables whose variability originates from experimental factors and measurement errors. We considered temperature, session, repetition, and slice as the experimental factors and studied how much T1 and T2 variability is due to each of these factors. Variance component decomposition analysis [43] yielded an estimation of variance components for each factor. The mean square variance and the variance component to total variance ratio was multiplied by 100, yielding the variability percentage of the respective experimental factor. The analysis was performed for each vial, and we reported the averaged variance and variance ratio of all vials respectively.
Performance analysis against the spin echo
For T1 mapping, a t-test was performed to assess between-session reproducibility differences between reference SE T1 measurements and MOLLI vs. slice-interleaved T1 sequences. Measurement comparison analysis of each sequence to the SE was also performed by using the Pearson correlation between the SE and each sequence. Linear regression was performed and slopes between the sequences and the 95% CI of the slopes were reported. Finally, Bland-Altman analysis was performed to study measurement bias between the two sequences, and the percentage of data points outside of the 95% limits of agreement (mean ± 2 standard deviations) was reported.
For T2 mapping, the relative CV percentage difference between slice-interleaved T2 and SE T2 was reported to assess differences in between-session reproducibility. A measurement comparison analysis to the SE was performed using the Pearson correlation, linear regression, and Bland-Altman analysis. Since only one vial was used for T2 mapping analyses, slice-interleaved T2 was averaged over all slices/ repetitions for each of the 37 sessions and compared to the SE T2 measurement of 37 sessions.
For all analyses, type-I error was set to 0.05. All statistical analyses were performed with SAS software (SAS Institute Inc., Cary, North Carolina, USA).
Results
Thirty-seven imaging sessions were performed from March 7, 2016 to October 31, 2017 The interval between successive sessions was 17±4 days. One session was excluded from the analysis due to incomplete acquisition of the SE T1 sequence. The isocenter cross marker of the phantom bottle enabled consistent positioning of the phantom throughout the study. Linear translations of ROIs were applied in 6 sessions with the offsets from the isocenter of 2.19±1.20 mm. Examples of T1 and T2 weighted images of each sequence are shown in S2 Fig. The temperature of the scanner room over the 20 months duration of experiments was 20.22±1.12°C (range 18–22°C). No measurement drift was observed in vials with low T1 (<1000 ms) over the 20 month study duration; increased T1 measurements were observed in vials with high T1 (>1000 ms) (S3 Fig; S1 Table). No drift in the T2 measurements was observed over the 20 month study duration (S4 Fig; S2 Table).
T1 Mapping
Estimation of coefficient of variation
Excellent reproducibility between sessions, and excellent repeatability between repetitions and slices of slice-interleaved T1 mapping sequences were observed with a CV less than 2% (Fig 2). There was a positive association between the T1 value and the CV, with longer T1 times corresponding to higher variability (Fig 3). The Spearman correlation between the T1 of each vial and the variability of each sequence was as follows: SE T1 = 0.88, MOLLI = 0.37, STONE-bSSFP 2P = 0.48, STONE-bSSFP 3P = 0.48, STONE-GRE 2P = 0.60, and STONE-GRE 3P = 0.60.
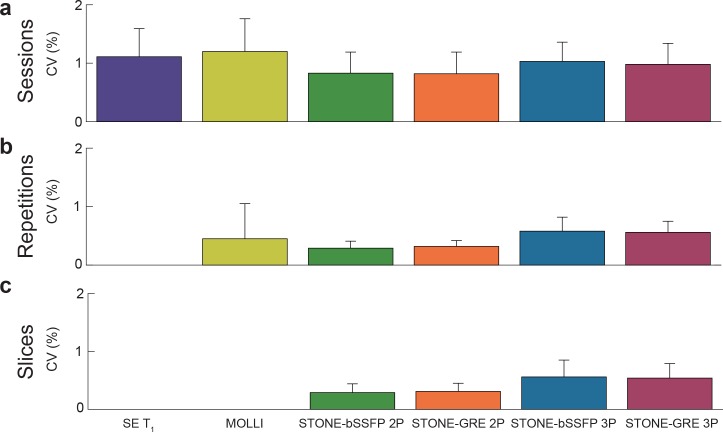
Fig 2. Reproducibility between sessions, and repeatability between repetitions and slices of slice-interleaved T1 mapping sequences were assessed using coefficients of variation (CV).
Slice-interleaved T1 mapping sequences showed excellent between-session reproducibility (CV: SE T1 = 1.1±0.5%, MOLLI = 1.2±0.6%, STONE-bSSFP 2P = 0.8±0.4%, STONE-GRE 2P = 0.8±0.4%, STONE-bSSFP 3P = 1.0±0.3%, STONE-GRE 3P = 1.0±0.4%), between-repetition repeatability (CV: MOLLI = 0.5±0.6%, STONE-bSSFP 2P = 0.3±0.1%, STONE-GRE 2P = 0.3±0.1%, STONE-bSSFP 3P = 0.6±0.2%, STONE-GRE 3P = 0.6±0.2%), and between-slice repeatability (CV: STONE-bSSFP 2P = 0.3±0.2%, STONE-GRE 2P = 0.3±0.1%, STONE-bSSFP 3P = 0.6±0.3%, STONE-GRE 3P = 0.5±0.3%).
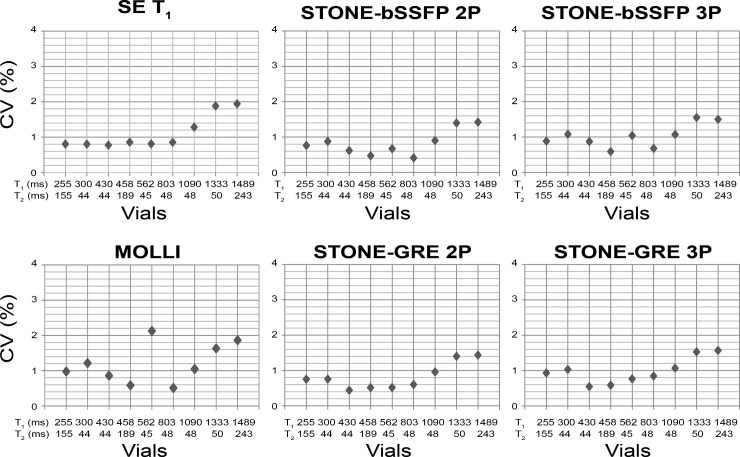
Fig 3. Coefficients of variations (CV) shown as scatter plots for each vial.
Vials are sorted from shortest T1 to longest T1 time (reference T1 from the T1MES manual measured by slow inversion-recovery/spin-echo methods at 1.5T: 255, 300, 430, 458, 562, 803, 1090, 1333, and 1489 ms). Vials with higher T1 time show higher variability.
Variance decomposition analysis
The sources of variability for slice-interleaved T1 mapping sequences are summarized in Table 1. The main source of variability was temperature when reconstructed with a 2-parameter fit model, and slice location/ orientation when reconstructed with a 3-parameter fit model. Repeated measurements within the same session at the same slice location did not contribute to variability (variance decompositions less than 1%).
Table 1. Sources of variability in T1 mapping defined by variance decomposition analysis.
| T1 Mapping, variance [ms2] (variance ratio [%]) | ||||||
|---|---|---|---|---|---|---|
| SE T1 | MOLLI | STONE-bSSFP 2P | STONE-bSSFP 3P | STONE-GRE 2P | STONE-GRE 3P | |
| Temperature | 151.8 (26.5) | 78.9 (30.8) | 91.1 (52.8) | 108.2 (39.4) | 98.3 (51.0) | 116.4 (38.3) |
| Session | 99.0 (73.5) | 64.0 (32.2) | 29.6 (28.5) | 35.5 (19.2) | 27.3 (26.0) | 30.2 (20.4) |
| Repetition | N/A | 52.5 (37.0) | 0.0 (0.0) | 0.0 (0.1) | 0.00 (0.1) | 0.0 (0.3) |
| Slice | N/A | N/A | 9.0 (18.8) | 22.1 (41.4) | 9.9 (22.9) | 26.9 (40.9) |
In slice-interleaved T1 mapping, the main source of variability is temperature when reconstructed with a 2-parameter fit model, and slice when reconstructed with a 3-parameter fit. Variability due to repetition is minimal with variance decompositions less than 1%. SE, spin-echo; STONE-bSSFP, slice-interleaved T1 with balanced steady‐state free precession; STONE-GRE, slice-interleaved T1 with spoiled gradient echo.
Performance analysis against the spin echo
Between-session reproducibility and comparison of slice-interleaved T1 mapping sequences against SE T1 and MOLLI are summarized in Table 2. Slice-interleaved T1 mapping sequences provided superior between-session reproducibility compared to SE T1 when reconstructed with a 2-parameter fit model (p<0.05). There were no statistically significant differences between the slice-interleaved T1 and the reference when reconstructed with a 3-parameter fit model (p>0.05). Slice-interleaved T1 mapping sequences provided superior between-session reproducibility compared to MOLLI (p<0.05).
Table 2. Between-session reproducibility and the comparison of slice-interleaved T1 mapping sequences against SE T1 and MOLLI.
| SE T1 | MOLLI | STONE-bSSFP 2P | STONE-bSSFP 3P | STONE-GRE 2P | STONE GRE 3P | |
|---|---|---|---|---|---|---|
| Between-session Reproducibility (CV, %) | 1.1±0.5 | 1.2±0.6 | 0.8±0.4 | 1.0±0.3 | 1.0±0.4 | 1.0±0.4 |
| p-value (vs. SE T1) | N/A | N/A | 0.005 | 0.377 | 0.001 | 0.117 |
| p-value (vs. MOLLI) | N/A | N/A | 0.011 | 0.031 | 0.010 | 0.024 |
Slice-interleaved T1 mapping sequences provided superior reproducibility compared to SE T1 when reconstructed with a 2-parameter fit model, and no statistically significant difference when reconstructed with a 3-parameter fit model. Slice-interleaved T1 mapping sequences provided superior reproducibility compared to MOLLI.
Slice-interleaved T1 mapping showed good agreement to the SE measurement with Pearson correlation coefficients of 1.00 (p<0.001) for all STONE-bSSFP 2P, STONE-GRE 2P, STONE-bSSFP 3P, and STONE-GRE. MOLLI also showed good agreement to the SE with Pearson correlation coefficients of 1.00 (p<0.001). All sequences showed good correlation to SE measurements with regression slopes as follows: MOLLI = 0.94 (95% CI: 0.936–0.945), STONE-bSSFP 2P = 0.95 (95% CI: 0.949–0.958), STONE-GRE 2P = 0.96 (95% CI: 0.957–0.961), STONE-bSSFP 3P = 0.97 (95% CI: 0.971–0.976), STONE-GRE 3P = 0.99 (95% CI: 0.987–0.992) (Table 3).
Table 3. Linear regression analysis of slice-interleaved T1 mapping sequences against reference SE T1 measurements.
| Regression Slope (Standard Error) | 95% Confidence Interval | |
|---|---|---|
| MOLLI | 0.9407 (0.0023) | 0.9362, 0.9452 |
| STONE-bSSFP 2P | 0.9536 (0.0024) | 0.9489, 0.9583 |
| STONE-bSSFP 3P | 0.9737 (0.0011) | 0.9715, 0.9759 |
| STONE-GRE 2P | 0.9591 (0.0011) | 0.9569, 0.9613 |
| STONE GRE 3P | 0.9894 (0.0011) | 0.9872, 0.9916 |
All sequences show strong agreement with the reference measurements with regression slopes of 0.9–1.0 and tight 95% confidence limits.
Bland-Altman analysis results for all vials are shown in Fig 4, and the result per each vial is shown in S3 Table. STONE-GRE 3P showed very close T1 values to the SE with an underestimation less than 1 ms. T1 bias between SE and other T1 mapping sequences were as follows: MOLLI = -29.6 ms, STONE-bSSFP 2P = -27.9 ms, STONE-bSSFP 3P = -10.2 ms, STONE-GRE 2P = -25.7 ms. The % of data points outside the 95% limits of agreement were as follows: MOLLI = 3.7%, STONE-bSSFP 2P = 3.4%, STONE-bSSFP 3P = 5.6%, STONE-GRE 2P = 5.3%, STONE-GRE 3P = 5.3%.
Fig 4. Bland-Altman analyses of slice-interleaved T1 mapping sequences against SE measurements.
The mean difference (bias) is presented as the red line, and the 95% limits of agreement (mean ± 2 standard deviations) are presented as dashed lines. Each data point represents one study time point which was averaged for all repetitions and slices within each session. The T1 mapping sequences show underestimation compared to the reference measurement. STONE-GRE 3P shows strongest agreement with the reference measurement with an underestimation less than 1 ms.
T2 mapping
Estimation of coefficient of variation
High reproducibility between sessions, and high repeatability between repetitions and slices of slice-interleaved T2 mapping sequences were observed with a CV less than 7% (Fig 5).
Fig 5. Reproducibility between sessions, and repeatability between repetitions and slices of slice-interleaved T2 mapping sequences were estimated using coefficients of variation (CV).
Slice-interleaved T2 mapping had good between-session reproducibility (CV: SE T2 = 2.5%, slice-interleaved T2 2P = 3.5%, slice-interleaved T2 3P = 6.3%, slice-interleaved T2 4-T2preps 2P = 4.7%, slice-interleaved T2 4-T2preps 3P = 6.7%), between-repetition repeatability (CV: slice-interleaved T2 2P = 2.7±0.2%, slice-interleaved T2 3P was 6.0±0.2%, slice-interleaved T2 T2preps 2P = 4.2±0.2%, slice-interleaved T2 T2preps 3P was 6.5±0.2%), and good between-slice repeatability (CV: slice-interleaved T2 2P = 2.9±0.2%, slice-interleaved T2 3P = 6.5±0.3%, slice-interleaved T2 T2preps 2P = 4.5±0.4%, slice-interleaved T2 T2preps 3P was 7.0±0.4%).
Variance decomposition analysis
The sources of variabilities are summarized in Table 4. The main source of variability was the slice location/ orientation, which represents variability due to spatial location and B0, B1 field inhomogeneity. The second source of variability was the temperature. Variability in repeated measurements was minimal with variance decompositions of 0%.
Table 4. Sources of variability in T2 mapping defined by the variance decomposition analysis.
| T2 Mapping, variance [ms2] (variance ratio [%]) | |||||
|---|---|---|---|---|---|
| SE T2 | Slice-interleaved T2 2P | Slice-interleaved T2 3P | T2 4echo 2P | T2 4echo 3P | |
| Temperature | 1.8 (83.5) | 2.2 (35.1) | 1.4 (12.5) | 2.0 (21.4) | 1.3 (10.4) |
| Session | 0.4 (16.5) | 0.6 (9.7) | 0.3 (3.0) | 0.5 (5.0) | 0.3 (2.4) |
| Repetition | N/A | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) |
| Slice | N/A | 3.4 (55.3) | 9.3 (84.5) | 7.0 (73.6) | 10.9 (87.2) |
In slice-interleaved T2 mapping, the main source of variability is slice, representing different spatial locations and different B0 and B1 field inhomogeneity. The variability in repeated measurements is minimal with variance decompositions of 0%.
Performance analysis against the spin echo
Slice-interleaved T2 mapping yielded lower between-session reproducibility than SE T2 (3.5 vs. 2.5% for slice-interleaved T2 2P vs. SE T2; 6.3 vs. 2.5% for slice-interleaved T2 3P vs. SE T2; 4.7 vs. 2.5% for T2 4echo 2P vs. SE T2; 6.7 vs. 2.5% for T2 4echo 3P vs. SE T2).
Slice-interleaved T2 mapping showed good correlation with Pearson correlation coefficients of 0.92 for slice-interleaved T2 2P, 0.91 for slice-interleaved T2 3P, 0.93 for T2 4echo 2P, and 0.91 for T2 4echo 3P (p <0.001 for all). Slice-interleaved T2 mapping showed good correlation to SE T2 with regression slopes as follows: slice-interleaved T2 2P = 1.06 (95% CI: 0.91–1.21), slice-interleaved T2 3P = 0.78 (95% CI: 0.66–0.90), T2 4echo 2P = 1.04 (95% CI: 0.90–1.18), and T2 4echo 3P = 0.78 (95% CI: 0.66–0.90) (Table 5).
Table 5. Linear regression of slice-interleaved T2 mapping sequences against the reference SE T2 measurements.
| Regression Slope (Standard Error) | 95% Confidence Interval | |
|---|---|---|
| Slice-interleaved T2 2P | 1.0594 (0.0767) | 0.9091, 1.2097 |
| Slice-interleaved T2 3P | 0.7830 (0.0606) | 0.6642, 0.9018 |
| T2 4echo 2P | 1.0436 (0.0707) | 0.9050, 1.1822 |
| T2 4echo 3P | 0.7828 (0.0623) | 0.6607, 0.9049 |
Slice-interleaved T2 sequences show good agreement with reference measurements with regression slopes of 0.8–1.1.
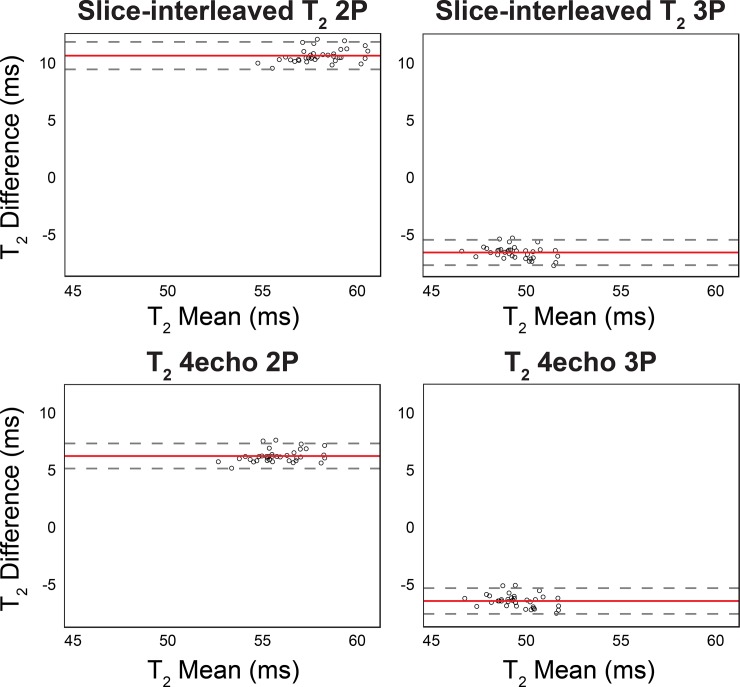
Bland-Altman analysis of slice-interleaved T2 mapping sequences showed different estimation biases depending on the fitting model (Fig 6). The slice-interleaved T2 showed overestimation when reconstructed with a 2-parameter fit model (slice-interleaved T2 2P = 10.6 ms, T2 4echo 2P = 6.3 ms), and an underestimation when reconstructed with a 3-parameter fit model (slice-interleaved T2 3P = -6.4 ms, T2 4echo 3P = -6.3 ms) against the SE T2 (Fig 6). The % of data points outside the 95% limits of agreement were as follows: slice-interleaved T2 2P = 5.6%, slice-interleaved T2 3P = 8.3%, T2 4echo 2P = 5.6%, T2 4echo 3P = 5.6%.
Fig 6. Bland-Altman plots of slice-interleaved T2 mapping sequences against the reference SE T2 measurements.
Bland-Altman analyses of slice-interleaved T2 mapping shows an overestimation when the map is reconstructed with a 2-parameter fit model, and an underestimation when reconstructed with a 3-parameter fit model. Each data point represents one study time point which was averaged for all repetitions and slices within session. The mean difference (bias) is presented as the red line, and the 95% limits of agreement (mean ± 2 standard deviations) are presented as dashed lines.
Discussion
In this study, we demonstrate highly reproducible long-term measurements of slice-interleaved T1 and T2 mapping with a CV less than 2% for T1 and less than 7% for T2. Reproducible measurements are essential to detect subtle changes in T1 and T2 times due to pathological processes. In particular, assessing long-term measurement stability is necessary for confidently differentiating variability due to disease progression or treatment efficacy over an extended period in a longitudinal study. The current phantom study reports rigorous long-term technical performance of slice-interleaved T1 and T2 mapping sequences to better understand baseline variations under controlled conditions.
Regular phantom-based quality control is recommended to ensure stability of a CMR system. Our study reports baseline long term variability which can be used to assess the stability of a CMR system for quality control, and to establish normal and clinical values with expected ranges of variability due to technical confounders. In the long-term time span, factors such as scanner performance can result in systematic differences compared to a shorter time interval. Furthermore, phantom-based quality control allows for T1 or T2 accuracy assessment and temperature sensitivity measurements as monitored for each session in our study.
T1 and T2 measurements with identical imaging parameters can still vary across session, repetition, and slice due to various factors. Our study showed that the main source of variability in T1 mapping was temperature when reconstructed with a 2-parameter fit model, and slice when reconstructed with a 3-parameter fit model. Temperature impacts diffusion coefficients [44], which can in turn impact T1 and T2. In vials with longer T1 times where a concentration of Ni2+ is low, T1 becomes more sensitive to temperature due to the temperature sensitivity of the T1 of water in gel [38]. Imperfect inversion pulses due to field inhomogeneity can be modeled using a 3-parameter fit model [9, 30, 45]. In turn, variability due to slice, representative of B0, B1 inhomogeneity, becomes dominant. For T2 mapping, slice was the main source of variability, which may be associated with differences in B0 and B1 field inhomogeneity. Variability in repeated measurements was negligible.
In this phantom study, we used in-vivo protocols currently used in our laboratory to mimic a clinically-relevant setting. Identical in-plane resolution was used to maintain similar TEs for similar performance. We used extra padding around the phantom to create distance from the RF coils to approximate coil geometry and proximity when imaging the human heart. Slice-interleaved T2 mapping was acquired with 10 T2prep echo times, previously evaluated in-vivo [20, 22, 32].
The variability observed in the current study shows a similar CV magnitude range as that shown in in-vivo reproducibility studies. Recent shorter-term reproducibility studies in T1 mapping yield CV magnitude ranges similar to our results, where the CV of ShMOLLI and MOLLI are reported as 2% for 35 patients undergoing repeated measurements the following day [46]. Slice-interleaved T1 and T2 show between-day CVs of 2.1% and 6.3%, respectively, in 11 healthy subjects on a 2-day test/retest study [22]. Higher variation is expected in the in-vivo study performed in a longer time span over multiple sessions due to patient-related artifacts such as respiratory and cardiac motion.
We performed long-term between-session reproducibility assessment including SE measurements. Even though SE is typically used as the reference, no study has evaluated its measurement variability over an extended period. Between-session reproducibility of SE measurements was excellent, and slice-interleaved T1 mapping sequences showed superior between-session reproducibility compared to SE. In particular, STONE-GRE 3P had excellent agreement to SE with similar reproducibility and an underestimation of only < 1ms. Considering the long scan time of SE sequences (typically 5–6 hours for T1 and 1–2 hours for T2), alternative sequences for the reference measurement are desirable.
We performed Bland-Altman analyses in each individual vial and for all vials to reflect the unique dependence of T1 on the bias. Longer T1 times corresponded with higher T1 error as previously reported [45]. We studied the measurement variability of slice-interleaved T1 and T2 maps reconstructed using both 2-parameter and 3-parameter fit models. For all sequences, higher reproducibility and repeatability was achieved when reconstructed with the 2-parameter fit model; however, the measurement bias was smaller when reconstructed with the 3-parameter fit model. This is in line with previous studies showing higher accuracy but lower precision when fitted with additional parameters [45]. Previous study demonstrated higher precision and reproducibility is achieved by increasing the number of T2prep echo times from 3 to 14, where the effect nearly saturates above 10 echo times in both phantom and in-vivo studies [47]. Our result shows higher reproducibility for T2 mapping with 9 T2prep images compared to 4 T2prep images as previously reported. We observed higher variability in T2 mapping, which may be due to lower SNR of the T2prep sequence due to field inhomogeneities and spoiling gradient.
Our study has several limitations. We studied slice-interleaved T1 and T2 mapping sequences on a single MRI scanner at a field strength of 1.5 T. The T1MES phantom used in this study is not optimally designed for studying T2 mapping; therefore, T2 analysis was carried out in a single vial with similar myocardial T1 and T2 values. A phantom with a different T2 range needs to be developed to study T2 reproducibility. The CPMG SE used as a reference of T2 measurements may be susceptible to stimulated-echo related bias. Our data shows 10.6±1.5% T2 difference compared to the T2 measurements by slow SE acquired with 8 TEs from 10–640 ms [38]. We did not study the impact of SNR, although with a relatively large region of interest in the current study, the impact may be negligible. Respiratory and cardiac motion could degrade T1 and T2 mapping reproducibility and were not simulated in our phantom study. Future long-term reproducibility studies in humans are warranted to enhance our understanding of measurement variability in a more clinically relevant setting.
Conclusions
Slice-interleaved T1 and T2 mapping sequences demonstrate highly reproducible measurement with a coefficient of variation less than 2% for T1, and 7% for T2 ranges of < 100 ms measured beyond one year. Slice-interleaved T1 mapping offers superior reproducibility than both MOLLI and SE T1 when reconstructed with a 2-paremeter fit model, and slice-interleaved T2 mapping shows lower reproducibility than SE T2. All sequences demonstrate strong agreement with reference SE measurements.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Dr. Warren J. Manning, Dr. Peter Gatehouse, and Jennifer Rodriguez for their editorial comments.
Data Availability
All DICOM files are available from the Harvard Dataverse database (doi:10.7910/DVN/ATXC5B).
Funding Statement
This work was supported by the National Institutes of Health [grant number R01HL129157, R01HL129122; https://www.nih.gov/] to RN; and the American Heart Association [grant number 15EIA22710040; https://www.heart.org/] to RN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Messroghli DR, Moon JC, Ferreira VM, Grosse-Wortmann L, He T, Kellman P, et al. Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: A consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging (EACVI). J Cardiovasc Magn Reson. 2017;19(1):75 10.1186/s12968-017-0389-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moon JC, Messroghli DR, Kellman P, Piechnik SK, Robson MD, Ugander M, et al. Myocardial T1 mapping and extracellular volume quantification: a Society for Cardiovascular Magnetic Resonance (SCMR) and CMR Working Group of the European Society of Cardiology consensus statement. J Cardiovasc Magn Reson. 2013;15:92 10.1186/1532-429X-15-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thavendiranathan P, Walls M, Giri S, Verhaert D, Rajagopalan S, Moore S, et al. Improved detection of myocardial involvement in acute inflammatory cardiomyopathies using T2 mapping. Circ Cardiovasc Imaging. 2011:CIRCIMAGING. 111.967836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verhaert D, Thavendiranathan P, Giri S, Mihai G, Rajagopalan S, Simonetti OP, et al. Direct T2 quantification of myocardial edema in acute ischemic injury. JACC Cardiovasc Imaging. 2011;4(3):269–78. 10.1016/j.jcmg.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giri S, Chung Y-C, Merchant A, Mihai G, Rajagopalan S, Raman SV, et al. T2 quantification for improved detection of myocardial edema. J Cardiovasc Magn Reson. 2009;11(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raunig DL, McShane LM, Pennello G, Gatsonis C, Carson PL, Voyvodic JT, et al. Quantitative imaging biomarkers: a review of statistical methods for technical performance assessment. Stat Methods Med Res. 2015;24(1):27–67. 10.1177/0962280214537344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Reson Med. 2004;52(1):141–6. 10.1002/mrm.20110 . [DOI] [PubMed] [Google Scholar]
- 8.Piechnik SK, Ferreira VM, Dall'Armellina E, Cochlin LE, Greiser A, Neubauer S, et al. Shortened Modified Look-Locker Inversion recovery (ShMOLLI) for clinical myocardial T1-mapping at 1.5 and 3 T within a 9 heartbeat breathhold. J Cardiovasc Magn Reson. 2010;12:69 10.1186/1532-429X-12-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow K, Flewitt JA, Green JD, Pagano JJ, Friedrich MG, Thompson RB. Saturation recovery single-shot acquisition (SASHA) for myocardial T(1) mapping. Magn Reson Med. 2014;71(6):2082–95. 10.1002/mrm.24878 . [DOI] [PubMed] [Google Scholar]
- 10.Weingärtner S, Akcakaya M, Basha T, Kissinger KV, Goddu B, Berg S, et al. Combined saturation/inversion recovery sequences for improved evaluation of scar and diffuse fibrosis in patients with arrhythmia or heart rate variability. Magn Reson Med. 2014;71(3):1024–34. 10.1002/mrm.24761 . [DOI] [PubMed] [Google Scholar]
- 11.Slavin GS, Stainsby JA. True T1 mapping with SMART1Map (saturation method using adaptive recovery times for cardiac T1 mapping): a comparison with MOLLI. J Cardiovasc Magn Reson. 2013;15(1):1–3.23324167 [Google Scholar]
- 12.Huang TY, Liu YJ, Stemmer A, Poncelet BP. T2 measurement of the human myocardium using a T2-prepared transient-state trueFISP sequence. Magn Reson Med. 2007;57(5):960–6. 10.1002/mrm.21208 [DOI] [PubMed] [Google Scholar]
- 13.Giri S, Shah S, Xue H, Chung YC, Pennell ML, Guehring J, et al. Myocardial T2 mapping with respiratory navigator and automatic nonrigid motion correction. Magn Reson Med. 2012;68(5):1570–8. 10.1002/mrm.24139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Heeswijk RB, Feliciano H, Bongard C, Bonanno G, Coppo S, Lauriers N, et al. Free-breathing 3 T magnetic resonance T 2-mapping of the heart. JACC Cardiovasc Imaging. 2012;5(12):1231–9. 10.1016/j.jcmg.2012.06.010 [DOI] [PubMed] [Google Scholar]
- 15.Mcnamara MT, Higgins CB, Schechtmann N, Botvinick E, Lipton MJ, Chatterjee K, et al. Detection and characterization of acute myocardial infarction in man with use of gated magnetic resonance. Circulation. 1985;71(4):717–24. 10.1161/01.cir.71.4.717 [DOI] [PubMed] [Google Scholar]
- 16.Sprinkart AM, Luetkens JA, Träber F, Doerner J, Gieseke J, Schnackenburg B, et al. Gradient Spin Echo (GraSE) imaging for fast myocardial T2 mapping. J Cardiovasc Magn Reson. 2015;17(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brittain JH, Hu BS, Wright GA, Meyer CH, Macovski A, Nishimura DG. Coronary angiography with magnetization-prepared T2 contrast. Magn Reson Med. 1995;33(5):689–96. [DOI] [PubMed] [Google Scholar]
- 18.Nezafat R, Ouwerkerk R, Derbyshire AJ, Stuber M, McVeigh ER. Spectrally selective B1-insensitive T2 magnetization preparation sequence. Magn Reson Med. 2009;61(6):1326–35. 10.1002/mrm.21742 [DOI] [PubMed] [Google Scholar]
- 19.Nezafat R, Stuber M, Ouwerkerk R, Gharib AM, Desai MY, Pettigrew RI. B1-insensitive T2 preparation for improved coronary magnetic resonance angiography at 3 T. Magn Reson Med. 2006;55(4):858–64. 10.1002/mrm.20835 [DOI] [PubMed] [Google Scholar]
- 20.Wang C, Jang J, Neisius U, Nezafat M, Fahmy A, Kang J, et al. Black blood myocardial T 2 mapping. Magnetic resonance in medicine. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan DC, Obuchowski NA, Kessler LG, Raunig DL, Gatsonis C, Huang EP, et al. Metrology standards for quantitative imaging biomarkers. Radiology. 2015;277(3):813–25. 10.1148/radiol.2015142202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellm S, Basha TA, Shah RV, Murthy VL, Liew C, Tang M, et al. Reproducibility of myocardial T1 and T2 relaxation time measurement using slice‐interleaved T1 and T2 mapping sequences. J Magn Reson Imaging. 2016;44(5):1159–67. 10.1002/jmri.25255 [DOI] [PubMed] [Google Scholar]
- 23.Roujol S, Weingärtner S, Foppa M, Chow K, Kawaji K, Ngo LH, et al. Accuracy, precision, and reproducibility of four T1 mapping sequences: A head-to-head comparison of MOLLI, ShMOLLI, SASHA, and SAPPHIRE. Radiology. 2014:140296 Epub 2014/04/08. 10.1148/radiol.14140296 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messroghli DR, Plein S, Higgins DM, Walters K, Jones TR, Ridgway JP, et al. Human myocardium: single-breath-hold MR T1 mapping with high spatial resolution—reproducibility study. Radiology. 2006;238(3):1004–12. 10.1148/radiol.2382041903 [DOI] [PubMed] [Google Scholar]
- 25.Baeßler B, Schaarschmidt F, Stehning C, Schnackenburg B, Giolda A, Maintz D, et al. Reproducibility of three different cardiac T2‐mapping sequences at 1.5 T. J Magn Reson Imaging. 2016;44(5):1168–78. 10.1002/jmri.25258 [DOI] [PubMed] [Google Scholar]
- 26.Fontana M, White SK, Banypersad SM, Sado DM, Maestrini V, Flett AS, et al. Comparison of T1 mapping techniques for ECV quantification. Histological validation and reproducibility of ShMOLLI versus multibreath-hold T1 quantification equilibrium contrast CMR. J Cardiovasc Magn Reson. 2012;14(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piechnik SK, Ferreira VM, Lewandowski AJ, Ntusi NA, Banerjee R, Holloway C, et al. Normal variation of magnetic resonance T1 relaxation times in the human population at 1.5 T using ShMOLLI. Journal of Cardiovascular Magnetic Resonance. 2013;15(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham-Brown MP, Rutherford E, Levelt E, March DS, Churchward DR, Stensel DJ, et al. Native T1 mapping: inter-study, inter-observer and inter-center reproducibility in hemodialysis patients. Journal of Cardiovascular Magnetic Resonance. 2017;19(1):21 10.1186/s12968-017-0337-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pica S, Sado DM, Maestrini V, Fontana M, White SK, Treibel T, et al. Reproducibility of native myocardial T1 mapping in the assessment of Fabry disease and its role in early detection of cardiac involvement by cardiovascular magnetic resonance. Journal of Cardiovascular Magnetic Resonance. 2014;16(1):99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weingärtner S, Roujol S, Akçakaya M, Basha TA, Nezafat R. Free-breathing multislice native myocardial T1 mapping using the slice-interleaved T1 (STONE) sequence. Magn Reson Med. 2015;74(1):115–24. 10.1002/mrm.25387 [DOI] [PubMed] [Google Scholar]
- 31.Jang J, Bellm S, Roujol S, Basha TA, Nezafat M, Kato S, et al. Comparison of spoiled gradient echo and steady‐state free‐precession imaging for native myocardial T1 mapping using the slice‐interleaved T1 mapping (STONE) sequence. NMR in Biomedicine. 2016;29(10):1486–96. 10.1002/nbm.3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basha TA, Bellm S, Roujol S, Kato S, Nezafat R. Free‐breathing slice‐interleaved myocardial T2 mapping with slice‐selective T2 magnetization preparation. Magn Reson Med. 2016;76(2):555–65. 10.1002/mrm.25907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamori S, Alakbarli J, Bellm S, Motiwala SR, Addae G, Manning WJ, et al. Native T1 value in the remote myocardium is independently associated with left ventricular dysfunction in patients with prior myocardial infarction. Journal of Magnetic Resonance Imaging. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah RV, Kato S, Roujol S, Murthy V, Bellm S, Kashem A, et al. Native myocardial T1 as a biomarker of cardiac structure in non-ischemic cardiomyopathy. The American journal of cardiology. 2016;117(2):282–8. 10.1016/j.amjcard.2015.10.046 [DOI] [PubMed] [Google Scholar]
- 35.Kato S, Nakamori S, Bellm S, Jang J, Basha T, Maron M, et al. Myocardial Native T 1 Time in Patients With Hypertrophic Cardiomyopathy. The American journal of cardiology. 2016;118(7):1057–62. 10.1016/j.amjcard.2016.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamori S, Bui AH, Jang J, El‐Rewaidy HA, Kato S, Ngo LH, et al. Increased myocardial native T1 relaxation time in patients with nonischemic dilated cardiomyopathy with complex ventricular arrhythmia. Journal of magnetic resonance imaging. 2018;47(3):779–86. 10.1002/jmri.25811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato S, Nakamori S, Roujol S, Delling FN, Akhtari S, Jang J, et al. Relationship between native papillary muscle T 1 time and severity of functional mitral regurgitation in patients with non-ischemic dilated cardiomyopathy. Journal of cardiovascular magnetic resonance. 2017;18(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Captur G, Gatehouse P, Keenan KE, Heslinga FG, Bruehl R, Prothmann M, et al. A medical device-grade T1 and ECV phantom for global T1 mapping quality assurance—the T 1 Mapping and ECV Standardization in cardiovascular magnetic resonance (T1MES) program. J Cardiovasc Magn Reson. 2016;18(1):58 10.1186/s12968-016-0280-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kellman P, Arai AE, Xue H. T1 and extracellular volume mapping in the heart: estimation of error maps and the influence of noise on precision. Journal of Cardiovascular Magnetic Resonance. 2013;15(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brittain JH, Hu BS, Wright GA, Meyer CH, Macovski A, Nishimura DG. Coronary angiography with magnetization‐prepared T2 contrast. Magn Reson Med. 1995;33(5):689–96. [DOI] [PubMed] [Google Scholar]
- 41.Akçakaya M, Basha TA, Weingärtner S, Roujol S, Berg S, Nezafat R. Improved quantitative myocardial T2 mapping: Impact of the fitting model. Magn Reson Med. 2015;74(1):93–105. 10.1002/mrm.25377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lourakis M. levmar: Levenberg-Marquardt nonlinear least squares algorithms in C/C++. http://wwwicsforthgr/~lourakis/levmar. Published 2004, Updated November 29, 2011, Accessed June 1, 2013. [Google Scholar]
- 43.Gaylor D, Lucas H, Anderson R. Calculation of expected mean squares by the abbreviated Doolittle and square root methods. Biometrics. 1970:641–55. [Google Scholar]
- 44.Le Bihan D, Delannoy J, Levin RL. Temperature mapping with MR imaging of molecular diffusion: application to hyperthermia. Radiology. 1989;171(3):853–7. 10.1148/radiology.171.3.2717764 [DOI] [PubMed] [Google Scholar]
- 45.Kellman P, Hansen MS. T1-mapping in the heart: accuracy and precision. J Cardiovasc Magn Reson. 2014;16:2 10.1186/1532-429X-16-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sado DM, Maestrini V, Piechnik SK, Banypersad SM, White SK, Flett AS, et al. Noncontrast myocardial T1 mapping using cardiovascular magnetic resonance for iron overload. J Magn Reson Imaging. 2015;41(6):1505–11. 10.1002/jmri.24727 [DOI] [PubMed] [Google Scholar]
- 47.Basha T, Akcakaya M, Roujol S, Nezafat R. Precision and reproducibility of T 2 quantifications in myocardial T 2 mapping: impact of the number of echoes and reconstruction model. Journal of Cardiovascular Magnetic Resonance. 2015;17(1):W9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All DICOM files are available from the Harvard Dataverse database (doi:10.7910/DVN/ATXC5B).