Abstract
The diverse bacterial populations that comprise the commensal microbiome of the human intestine play a central role in health and disease. A method that sustains complex microbial communities in direct contact with living human intestinal cells and their overlying mucus layer in vitro would thus enable investigations of host–microbiome interactions. Here, we show the extended co-culture of living human intestinal epithelium with stable communities of aerobic and anaerobic human gut microbiota, enabled by a microfluidic intestine-on-a-chip that permits the control and real-time assessment of physiologically relevant oxygen gradients. When compared to aerobic co-culture conditions, the establishment of a transluminal hypoxia gradient in the chip increased intestinal barrier function and sustained a physiologically relevant level of microbial diversity, consisting of over 200 unique operational taxonomic units from 11 different genera, and of an abundance of obligate anaerobic bacteria with ratios of Firmicutes and Bacteroidetes similar to those observed in human faeces. The intestine-on-a-chip may serve as a discovery tool for the development of microbiome-related therapeutics, probiotics and nutraceuticals.
One of the major recent paradigm shifts in medicine relates to the recognition of the central role that the microbiome, composed of host-specific communities of commensal microbes, plays in human health and disease1. While human microbiota colonize mucosal surfaces of various tissues, the gastrointestinal tract supports the greatest mass and diversity of microorganisms2. Aerobic and anaerobic commensal gut microbes are essential for the maintenance of normal nutrient absorption, drug metabolism, and immune responses, as well as for protection against infectious pathogens3. Conversely, changes or imbalances in the microbial community within the intestine can contribute to the development of a broad range of pathological disorders within and beyond the gastrointestinal system, including inflammatory bowel disease, colorectal cancer, radiation enteropathy, diabetes, hepatic steatosis, obesity, and rheumatoid arthritis4,5. Thus, the establishment and preservation of balanced host–intestinal microbiome interactions are key requirements for maintaining gut homeostasis and human health.
Analysis of gut-microbiome crosstalk has almost exclusively relied on genomic or metagenomic analysis of samples collected in vivo because no method exists to establish stable complex communities of gut commensal microbes in direct contact with intestinal epithelium and their overlying mucus layer in vitro6,7. While animal models have been used to analyse host-microbiome interactions and their contributions to pathophysiology8-10, there are no in vitro systems available to verify these interactions in human cells cultured with complex human microbiome. Thus, there is a great need for experimental models that can sustain complex populations of human aerobic and anaerobic microbiota in contact with living human tissues to analyse dynamic and physiologically relevant human host-microbiome interactions.
Existing in vitro models, such as Transwell inserts, have been used to study human host-microbe interactions; however, these studies can only be carried out over a period of hours before bacterial overgrowth leads to cell injury and death11-13. Organoid cultures, have shown great promise for studying host-microbiome interactions, but they also cannot be co-cultured with living microbes for more than ~1 day; they also do not provide a vascular interface nor can they sustain luminal oxygen levels below 0.5%, which is required for co-culture of certain obligate anaerobes14,15. Specialized bioreactor models, such as the mucosal-simulator of the human intestinal microbial ecosystem (M-SHIME), have been developed to sustain growth of luminal and mucosal gut microbes in vitro, but they do not include living human intestinal epithelium16. A human-microbiota interaction (HMI) module was developed that permits analysis of aerobic and anaerobic microbes, including complex living microbiome derived from a SHIME reactor, when co-cultured with human Caco2 intestinal epithelial cells under an oxygen gradient; however, the microbes need to be separated from the human cells by a nanoporous membrane with an artificial mucus layer, and even under these conditions, the co-cultures were only maintained for 48 hours17. We previously described a two-channel microfluidic Organ Chip device lined by human Caco2 intestinal epithelial cells cultured under dynamic fluid flow and peristalsis-like mechanical deformations, which enabled establishment of stable co-cultures of a mucus-producing human villus intestinal epithelium with up to 8 different strains of human commensal gut microbes for weeks in vitro under aerobic conditions17-19. But the human gut microbiome contains hundreds of different types of bacteria, many of which are obligate anaerobes that will not grow in this environment. Thus, no existing in vitro model enables analysis of direct interactions among complex communities of anaerobic and aerobic gut bacteria, human intestinal epithelium, and its overlying mucus layer when cultured for multiple days in vitro, which is crucial for analysing gut health and disease20-22.
In this study, we therefore set out to develop an experimental intestine-on-a-chip (Intestine Chip) system that can support dynamic interactions between living, mucus-producing, human intestinal epithelium and a directly apposed complex community of living human aerobic and anaerobic commensal gut microbes with a population diversity similar to that observed in living human intestine. To meet this challenge, we first modified the human Caco2 Intestine Chip by integrating microscale oxygen sensors into the devices for in situ oxygen measurements, and placing the chips within an engineered anaerobic chamber to establish a physiologically relevant oxygen gradient across a human intestinal epithelium as well as a microvascular endothelium that are cultured in parallel channels separated by a porous matrix-coated membrane within the device. To ensure a stable source of complex human intestinal gut-microbiota, we used complex microbiota originally derived from healthy human stool specimens, which have been maintained stably in gnotobiotic mice for multiple years, and closely resemble the relative abundance of major bacterial phyla patterns in their respective inocula23,24. We also applied the same method to co-culture fresh gut microbiome isolated from human infant stool samples in a primary human Intestine Chip lined by cells isolated from normal human ileum. Here we describe how establishing a hypoxia gradient across the engineered tissue-tissue (endothelium-epithelium) interface of the Intestine Chip allows for stable co-culture of highly complex communities of anaerobic and aerobic human commensal gut bacteria in the same channel as mucus-producing human villus intestinal epithelium while simultaneously monitoring oxygen levels and intestinal barrier function for at least 5 days in vitro.
Results
Establishing an oxygen gradient across the lumen of the Intestine Chip.
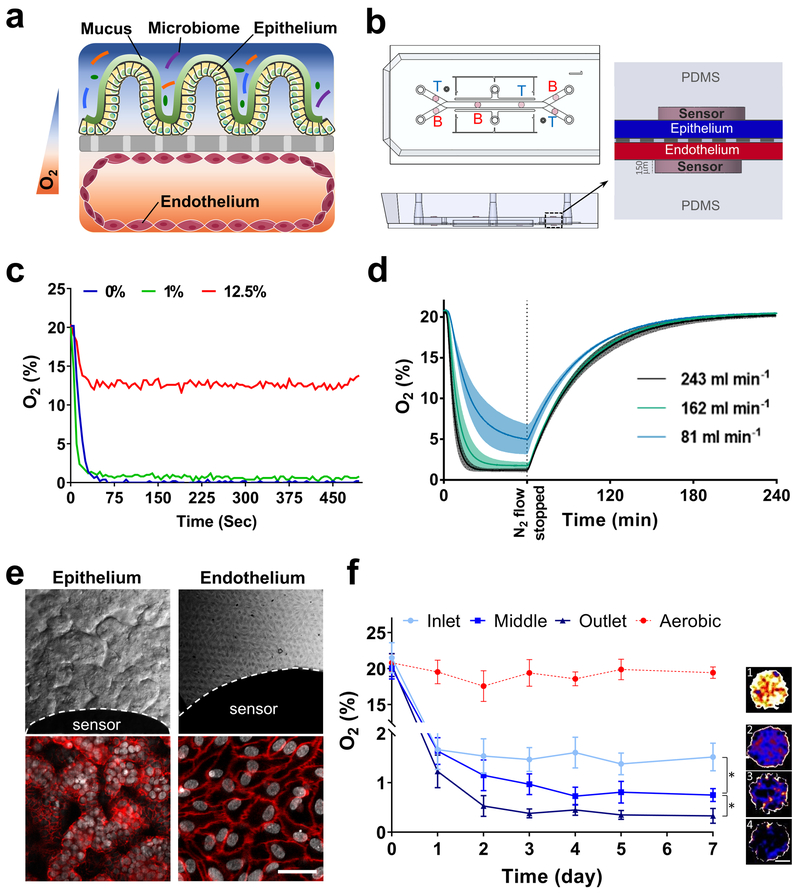
To recapitulate a physiologically relevant intestinal oxygen gradient profile inside Intestine Chips (Fig. 1a), we fabricated an oxygen-sensing, dual channel, human Organ Chip composed of optically clear and flexible poly(dimethyl siloxane) (PDMS) polymer (Fig. 1b; Supplementary Fig. S1a), as well as an anaerobic chamber (Supplementary Fig. S2). For real-time, non-invasive, monitoring of oxygen tension, six sensor spots containing oxygen-quenched fluorescent particles were embedded in the top and bottom portions of the chip beneath the central microchannels (Fig. 1b; Supplementary Fig. S1b). Changes in the fluorescent intensities of these sensors in response to oxygen tension were captured by a VisiSens camera (Supplementary Fig. S1b) and translated into oxygen concentrations by comparison with a standard Oxy-4 probe system (Supplementary Fig. S1c). As both the chips and sensors are composed of highly gas-permeable PDMS, the sensors respond rapidly (< 30 sec) to changes in oxygen concentrations (Fig. 1c).
Fig. 1 ∣. Oxygen sensitive human Intestine Chip microfluidic culture device.
a, Schematic showing the position of the human intestinal epithelium overlaid with its own mucus layer and complex gut microbiota on top, with vascular endothelium on bottom side of the same ECM-coated porous membrane, within a 2-channel microfluidic Organ Chip device in presence of oxygen gradients. Orange and blue colors indicated high and low levels of oxygen concentration, respectively. b, Schematic representation of the Intestine Chip with 6 oxygen quenched fluorescent particles embed in inlet, middle and outlet of top and bottom channels. (T, top channel; B, bottom channel). c, Sensitivity analysis of oxygen spots located in the Intestine Chip in response to defined, standard oxygen concentrations. d, Anaerobic chamber validation at various N2 inflow pressures; N2 was introduced into the chamber at 81 mL min−1, 162 mL min−1, or 243 mL min−1 for 1 h before gas flow was stopped and the chamber was allowed to recover (n=3, shaded regions indicate standard deviation; data are presented as mean ± s.d.). e, Microscopic views showing the villus morphology of the human Caco-2 intestinal epithelium (top left; scale bar, 100 μm) and vascular endothelium (top right; scale bar, 100 μm) cultured for 6 days in the Intestine Chip under anaerobic conditions, when viewed from above by DIC and phase contrast imaging, respectively, or by immunofluorescence staining for the tight junction protein, ZO-1 (red, bottom left; scale bar, 100 μm) and endothelial cell junction-associated protein, VE-cadherin (red, bottom right; scale bar, 20 μm). Gray indicates DAPI-stained nuclei; white dashed lines indicate the border of the oxygen sensor spot). f, Oxygen concentration profiles within aerobically- and anaerobically-cultured Intestine Chips. Representative pseudocolor insets indicate average oxygen concentration in aerobic chip (1), and inlet (2), middle (3) and outlet (4) of the anaerobically-cultured epithelium channel, at day 7 of culture. Scale bar, 200 μm. (n=3 individual chips; data are presented as mean ± s.d.; significance was calculated by one-way analysis of variance; *P = 0.046).
To simultaneously provide adequate oxygen for maintaining human cells and an anaerobic microenvironment suitable for culturing complex human microbiota while establishing a functional host-microbiome interface, we flushed the custom anaerobic chamber (Supplementary Fig. S2a,b) continually with humidified 5% CO2 in nitrogen gas. This setup enables us to maintain low oxygen levels within the lumen of the upper chamber (Fig. 1d), while the epithelium is sustained via diffusion of oxygen through the permeable PDMS membrane from the well-oxygenated medium flowing through the lower endothelium-lined vascular channel from external oxygenated medium reservoirs (Supplementary Fig. S2a,b). Using this method, physiologically relevant anaerobic conditions (<0.5%) can be generated within less than 30 min at 243 ml min−1 of nitrogen flow into the anaerobic chamber (Fig. 1d). The chamber also can sustain low oxygen levels (<5.0%) for about 15 min after it is disconnected from the nitrogen source (Fig. 1d). This allows the chamber to be temporarily moved from the incubator for imaging or into a bacterial glove box (e.g. to replenish culture medium or add microbiota) without significantly disturbing the low oxygen environment.
When human Caco-2 intestinal epithelial cells are cultured for 5 to 7 days under aerobic conditions and dynamic flow, they undergo villus differentiation and express multiple features of the ileum portion of the human small intestine, including secretion of a mucus layer overlying the apical surface of the epithelium and establishment of barrier function25,20,26. Endothelial cells also can be co-cultured on the bottom of the central porous membrane in the lower channel of the same device, where they form a hollow vascular lumen lined by cells joined by VE cadherin-containing cell-cell junctions under aerobic conditions26. The co-culture of endothelium has been shown to enhance barrier function and mucus production (e.g., expression of MUC2 and MUC5AC), as well as influence villi development and cytokine production by intestinal Caco2 epithelium under these conditions26,27.
When we cultured Intestine Chips lined by Caco2 intestinal epithelial cells and human intestinal microvascular endothelial cells (HIMECs) under a hypoxia gradient using our chamber, differential interference contrast (DIC) and immunofluorescence microscopic analysis confirmed that the cells again formed a villus intestinal epithelium containing polarized cells joined by ZO-1-containing tight junctions (Fig. 1e, top; Supplementary Fig. S3a-d) that was underlaid by a confluent HIMEC monolayer with cells linked by VE-cadherin-containing tight junctions, even in the presence of this hypoxia gradient (Fig. 1e, bottom). Both cell types also remained viable under these conditions, as measured by quantifying release of the intracellular enzyme lactate dehydrogenase (LDH), which remained relatively unchanged compared to the aerobic control during one week of anaerobic culture (Supplementary Fig. S4a). Quantification of the apparent permeability (Papp) of the intestinal epithelial barrier similarly revealed no changes in the paracellular barrier function, and these human Intestine Chips displayed Papp values of about 1 × 10−7 cm s−1 after 7 days (Supplementary Fig. S4b), which are similar to those previously reported26. Importantly, we also confirmed that both the human intestinal epithelium and endothelium experienced oxygen gradients by analysing the expression of hypoxia-inducible factor 1α (HIF-1α). HIF-1α is a key mediator of oxygen hemostasis and intestinal epithelial cell adaptation to oxygen deprivation,28 which is stabilized in a graded fashion in response to decreasing oxygen concentrations29. Expression of nuclear HIF-1α levels were significantly higher (~3-fold; p < 0.01) in the lumen of the anaerobically-cultured epithelium than in the adjacent oxygenated endothelium-lined channel (Supplementary Fig. S5a,b), which is where our sensors indicated maintenance of a hypoxic environment for up to 7 days in culture (Fig. 1f and Supplementary Fig. S4c). Similar nuclear HIF1α expression has been previously shown in both mouse and human cells cultured under hypoxic (1% O2) condition30.
Co-culture of human intestinal epithelium with an obligate anaerobe on-chip.
We next explored whether the hypoxic environment can support co-culture of the intestinal epithelium with the obligate anaerobe, Bacteroides fragilis (B. fragilis; strain NCTC 9343), which is a human commensal symbiotic bacterium that cannot grow under aerobic (> 0.5% oxygen) conditions31,32. During the co-culture procedure which began after the epithelium had been cultured and differentiated on-chip for 7 days under anaerobic condition (Supplementary Fig. S6a), B. fragilis bacteria (2.5 × 105 CFU; fluorescently labelled with HCC-amino-D-alanine (HADA)33; Supplementary Fig. S6b) were introduced into the lumen of upper channel where they distributed across the surface of the intestinal epithelium (Supplementary Fig. S6c). They were subsequently cultured under either aerobic or anaerobic conditions, while being flushed daily to remove both luminal and tissue-associated microbes, and CFU counts were carried out by plating.
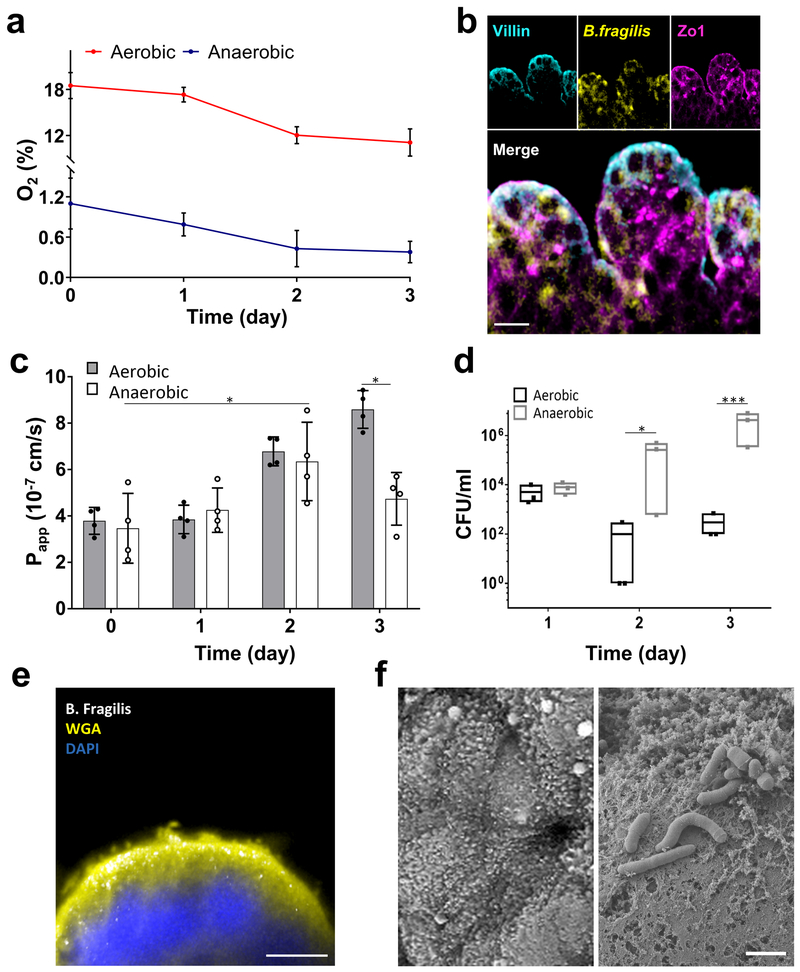
Continuous monitoring of oxygen concentrations from inoculation to day 3 of co-culture revealed that our anaerobic chip setup maintained a low oxygen environment that decreased from ~ 1% oxygen levels to 0.3% in the presence of B. fragilis (Fig. 2a). Yet, the intestinal epithelium maintained its ZO-1-containing tight junctions and apical brush border polarity when co-cultured in direct contact with B. fragilis under these highly anaerobic conditions (Fig. 2b). Interestingly, the presence of this obligate anaerobe enhanced, rather than decreased, barrier function (reduced Papp by 1.8-fold compared to aerobic conditions; *p = 0.033) after 3 days in anaerobic culture (Fig. 2c) and this barrier was maintained for up to at least 8 days in culture (Supplementary Fig. S6d). As expected, the B. fragilis bacteria continued to grow in the anaerobic chips over 3 days (p < 0.001), whereas they started to die off by day 2 and appeared at significantly lower levels at 3 days under aerobic culture conditions (Fig. 2d). These data confirm that our chips that experience a hypoxia gradient support the growth of an obligate anaerobic bacterial species in the same channel as living human intestinal epithelial cells, whereas these bacteria would have otherwise died in a conventional aerobic culture system.
Fig. 2 ∣. Co-culture of human intestinal epithelium and obligate anaerobe, Bacteroides fragilis, on-chip.
a, Oxygen concentration profiles in aerobic and anaerobic Intestine Chips co-cultured with Bacteroides fragilis (n=4 individual chips; data are presented as mean ± s.d.). b, Representative vertical cross-sectional, confocal micrographic views through the intestinal epithelium-microbiome interface within the Intestine Chip cultured under anaerobic conditions, when immunostained for villin (cyan), ZO-1 (magenta) and nuclei with DAPI (blue). (scale bar, 50 μm; B. fragilis was HADA (yellow) labelled; representative images from 4 intestine chips are shown). c, Changes in apparent paracellular permeability (Papp) measured by quantitating cascade blue transport across the tissue-tissue interface within the Intestine Chip microdevices co-cultured with Bacteroides fragilis under aerobic and anaerobic conditions (n=4 individual chips; data are presented as mean ± s.d.; significance was calculated by one-way analysis of variance; *P = 0.042 and 0.033 for anaerobic day 2 vs. day 0 and aerobic vs. anaerobic day 3, respectively). d, CFU counts/mL of Bacteroides fragilis co-cultured in the Intestine Chip under aerobic and anaerobic conditions (n=3 individual chips; data are presented as mean ± s.d.; significance was calculated by one-way analysis of variance; *P = 0.031 for day 2 and ***P<0.001 for day 3). e, Cross-sectional fluorescence microscopic view of the Caco2 epithelium (nuclei stained in blue with DAPI), overlying mucus layer stained with Alexa Fluor 488-conjugated WGA (yellow), and B. fragilis bacteria (GalCCP labelled, white) when co-cultured in the Intestine Chip. Scale bar, 10 μm. f, SEM views of the apical surface of the Caco2 epithelium in the Intestine Chip comparing the morphology on day 4 of culture before it accumulates a mucus layer and when the surface microvilli are visible (top) versus when Bacteroides fragilis have been added on day 12 after the mucus layer has accumulated, which can be seen as a dense mat that separates the bacteria from the epithelial cell surface (bottom). Scale bar, 2 μm.
A mucus layer separates the commensal microbes from the epithelium.
One of the characteristic features of host-microbiome interactions in the living intestine is that they are mediated through an intervening mucus layer that is secreted by the epithelium along its apical surface. Live staining using Wheat Germ Agglutinin (WGA), which has been previously used for mucus visualization in vitro34 and in vivo35, confirmed that B. fragilis resides on top of the mucus layer (Fig. 2e), which is secreted by the intestinal Caco2 epithelium, as previously demonstrated34,36. This was independently confirmed by scanning electron microscopic (SEM), which clearly revealed a continuous and dense mucus blanket that completely covered the surface of the differentiated villus epithelium separating it from overlying bacteria after 12 days of culture (Fig. 2f), much as is and observed in vivo37,38. This was in contrast to SEM analysis of Caco2 Intestine Chips that were only cultured for 4 days before full differentiation occurred and mucus had accumulated where the microvilli-lined surface of the apical epithelium remained clearly detectable (Fig. 2f). Based on these images, the thickness of mucus layer was estimated at ~10 μm, which is similar to that reported with 30-day old mouse ileum37.
Sustaining a complex human intestinal microbiome in vitro.
To optimize growth of a complex microbiome in our culture system, we searched for a source of complex human gut microbes that remains stable over time. Therefore, we inoculated the anaerobic Intestine Chips with a sample of complex gut microbiota originally isolated from human faeces, which has been stably maintained in gnotobiotic mice in isolators for over 30 generations23,24 and that maintains a composition closely resembling the original human stool inoculum at the genera and species levels23. To identify a medium composition that would promote the growth of a complex set of commensal bacteria, we first inoculated the healthy human microbiome (Hmb) stock into 13 different types of culture medium in standard culture tubes, placed the cultures in an anaerobic chamber at 37°C, and then carried out 16S rRNA sequencing after 3 days of culture (Supplementary Fig. S7a). Samples of these 13 types of medium were also added to cultured human Caco2 intestinal epithelial cells to test for toxicity (Supplementary Fig. S7b). The medium that promoted the most diverse set of viable microbes without injuring the epithelium contained DMEM, 20% FBS, 1% glutamine, 1 mg.ml−1 pectin, 1 mg.ml−1 mucin, 5 μg.ml−1 Hemin and 0.5 μg.ml−1 Vitamin K1. The microbiota stock was introduced into this medium (0.1 mg.ml−1) and perfused through the upper Caco2 epithelium-lined channel of the Intestine Chip while oxygenated endothelial culture medium was flowed through the lower channel. The epithelial channel of the chips was flushed daily with a short (2 min) fluid pulse at higher flow rate (50 μl.min−1) to remove adherent and luminal bacteria, and 16S rRNA sequencing was carried out using samples from the effluent to assess bacterial diversity in each condition over 3 days of culture (n = 4 for each condition).
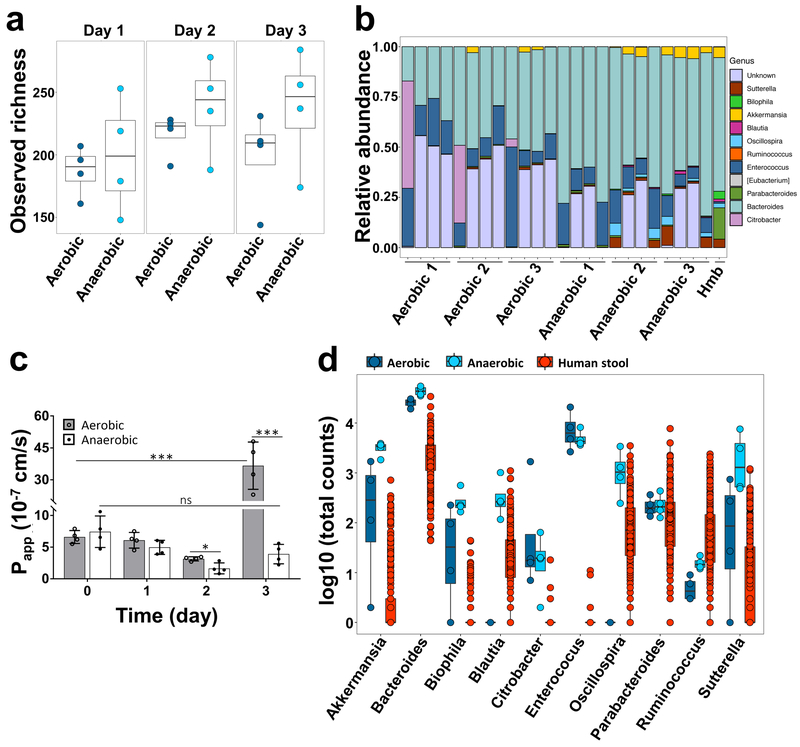
After data processing, we identified a total of 938 operational taxonomic units (OTUs) among all samples, which corresponded to approximately 200 unique OTUs shared between samples of each chip after filtering and removing singletons, which is similar in scale to the number of OTUs previously observed in human intestinal aspirates (280 OTUs)39, although, as expected, with a different phylum distribution. Analysis of the alpha diversity between the two conditions showed that the species diversity in anaerobic chips were statistically different (PERMANOVA, ***p < 0.001) from aerobic chips (Fig. 3a). Although the observed diversity and Shannon Index are lower than what is observed in human stool samples (Supplementary Fig. S8a,b), we observed an increase in richness compared to our starting inoculum over the course of the 3 days of experiment (Supplementary Fig. S9a,b). We identified 11 well characterized genera including Eubacterium, Oscillospira, Blautia, Sutterella, Biophila, Akkermansia, Ruminococcus, Bacteroides, Parabacteroides, Enterococcus and Citrobacter (Fig. 3b), with an additional 8 OTUs of unknown genera from Firmicutes (5 OTUs) and Proteobacteria (3 OTUs) phyla, that were present in our chips (phylum level analysis is shown in Supplementary Fig. S10). Interestingly, in comparison to aerobic conditions, co-culturing diverse microbiota under anaerobic conditions for 3 days in direct contact with the human intestinal epithelium did not compromise intestinal barrier integrity, and, instead, it led to an increase in barrier function by almost 2-fold (i.e., decrease in Papp from 3.1 × 10−7 to 1.6 × 10−7 cm s−1 in aerobic versus anaerobic chips, respectively; *p = 0.048) (Fig. 3c). In contrast, epithelial barrier function decreased (***p < 0.001) after day 3 of co-culture under aerobic conditions when co-cultured with the same complex gut microbiome (Fig. 3c). Furthermore, to explore the durability of these cultures, we then carried out an additional experiment in which we extended aerobic and anaerobic chips for 5 days with and without the same Hmb stock. Sterile chips maintained barrier function over the five days of culture under both anaerobic and aerobic conditions (Supplementary Fig. S11a,b). Again, we observed that while barrier function decreased in aerobic chips with Hmb at day 3 (Supplementary Fig. S11a), no compromise of barrier function was seen in anaerobic chips with Hmb even over the 5 days of culture (Supplementary Fig. S11b) and these cultures could be extended longer if experimental needs require. Moreover, in the presence of complex microbiota, the intestinal epithelium remained viable, as measured by quantifying release of the intracellular enzyme LDH, which remained unchanged compared to the sterile control chips cultures (Supplementary Fig. S11c).
Fig. 3 ∣. Analysis of the diversity and relative abundance of microbiota co-cultured in Intestine Chips under aerobic and aerobic conditions.
a, Observed alpha diversity (richness) in our complex gut microbiome samples when cultured for 1 to 3 days in direct contact with human Caco2 intestinal epithelium (each data point represents one Intestine Chip). b, Relative abundance of genera measured across all samples highlighting changes in the abundance of the different genera observed over time. Data points represent each of the 3 replicates cultured under aerobic or anaerobic conditions at 0, 1, 2 or 3 days of culture (left to right, respectively); Hmb indicates genera abundance in the complex microbiome stock at time 0. c, Changes in apparent paracellular permeability (Papp) measured by quantifying cascade blue transport across the tissue-tissue interface within the Intestine Chip after co-culture with complex gut microbiome under aerobic and anaerobic conditions (n=4 individual chips; data are presented as mean ± s.d.; significance was calculated by one-way analysis of variance; *P = 0.048, ***P<0.001, ns = not significant). d, Differences in microbial abundance between Intestine Chip samples (dark blue: aerobic; light blue: anaerobic) and human microbiome stool sample from the Human Microbiome Project (red). Data are shown as log10 of the total number of reads; each data point corresponds to a single sample (error bars represent the s.d.; data are presented as box plots with individual data points overlaid, where lower or upper edges of the box represent 25th or 75th percentiles and the middle bar is the median).
To further assess the physiological mimicry obtained using the anaerobic Intestine Chip lined by Caco2 epithelium, we compared the genera identified in this study with publicly available data from studies of human stool generated by the Human Microbiome Project40 (Fig. 3d). We did not expect the composition of the microbiome grown on chip to precisely recapitulate that of stool because the microbiome of the human intestine is known to show regional differences41,42. For example previous reports have shown different ratios of the Firmicutes, Bacteroidetes and Actinobacteria phyla in stool samples as compared to small intestine aspirates41,43. Nevertheless, our results show that the anaerobic culture system provides an environment for culture of complex gut microbiota in direct contact with living human intestinal epithelium that sustains a diverse bacterial community, which falls into the range of abundances reported in the Human Microbiome Project. Furthermore, the relative abundances of the phyla that dominate the human gut, Bacteroidetes (Bacteroidetes and Parabacteroides genera) and Firmicutes (Blautia, Enterococcus, Ruminococcus, and Oscillospira genera), were higher in the anaerobic chips than in the aerobic chips with some genera (Blautia and Oscillospira) missing in the aerobic chips altogether (Fig. 3d). Oxygen sensor readouts in aerobic and anaerobic chips cultured with a viable microbiome or under sterile (microbe-free) conditions confirmed that the oxygen concentration was maintained below 1% throughout 5-day co-culture period in the anaerobic co-cultures (Supplementary Fig. S11d). Moreover, these results showed a decrease in oxygen concentration in aerobic chips cultured with microbiome over time (Supplementary Fig. S11d), which is similar to what we observed in co-cultures with B. fragilis. This was likely due to the increased vertical growth of villi we observed in these chips relative to anaerobic chips, as well as to concomitant oxygen utilization by the bacteria, which increase in numbers by day 1 in both aerobic and anaerobic chips. Although the oxygen concentration in the aerobic chip never reached the low levels obtained in anaerobic chips, this decrease in oxygen could explain the presence of some obligate anaerobes, such as Akkermansia, that we observed in the aerobic chips; however, clearly the constant anaerobic conditions are more optimal for maintenance of co-cultures of anaerobic bacteria with viable human cells. Interestingly, the genus Akkermansia, which has been recently implicated as an enhancer of gut barrier function44-46, showed a considerably higher number of total counts in the anaerobic culture system compared to human stool (Supplementary Fig. S12, S13). Additionally, the genus Enterococcus was found to be present at higher levels in both chip culture systems compared to the stool samples, suggesting that some gut microbial species may grow better under conditions that more closely mimic regions of the living intestine than in stool. Taken together, these data confirm that this anaerobic human Intestine Chip system enables living human intestinal epithelium to be co-cultured in the same channel as a complex human gut microbiome containing a range of bacterial genera that come much closer to what is observed in healthy human donors than has ever been possible before.
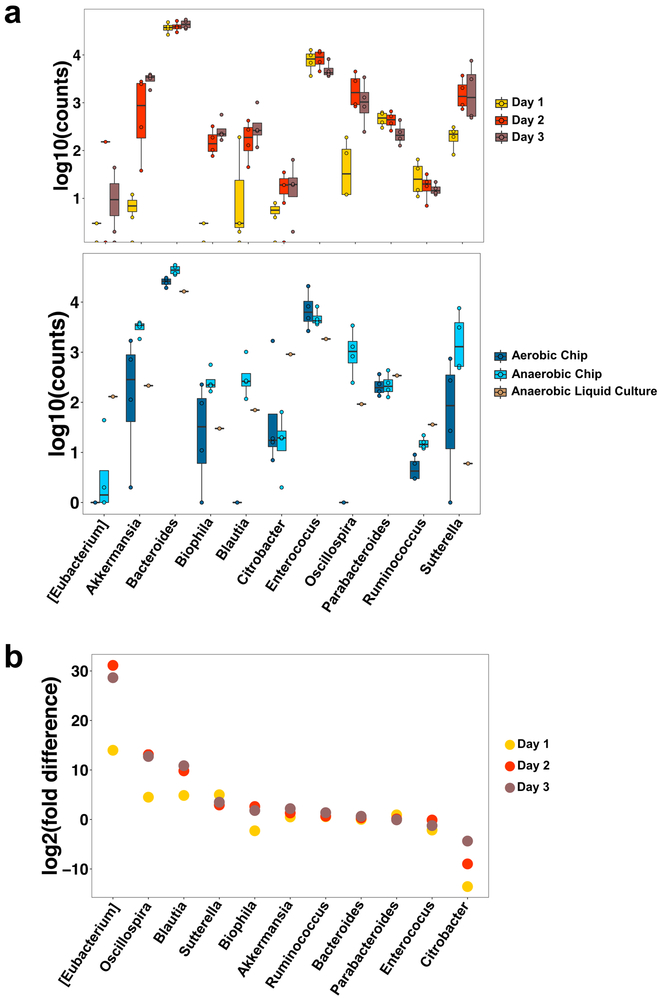
To determine the stability of the microbial communities in the anaerobic Intestine Chip system, we analysed their change in abundance over 3 days of co-culture with human Caco2 intestinal epithelium and underlying endothelium. Our results show that genera composed of obligate anaerobes, such as Akkermansia, Blautia, Bilophila, and Suterella, increased in abundance over time, presumably due to maintenance of low oxygen concentrations (Fig. 4a, top). Bacteroides, the highest abundance genus in the anaerobic Intestine Chips, remained relatively stable over time. Additionally, in a subsequent experiment, we cultured anaerobic Intestine Chips with the same Hmb microbiome stock for 5 days and plated serial dilutions of the flush on anaerobic Brucella culture plates in the presence or absence of vancomycin. Our results show that the bacterial load increased at day 1 compared to the seeding inoculum, and then remained constant throughout the experiment (Supplementary Fig. S11e). Vancomycin kills Gram positive bacteria, and thus the difference in counts between Brucella plates in the presence versus absence of vancomycin suggests both Gram positive bacteria and Gram negative bacteria (i.e., that survived in the vancomycin plates) remained viable over 5 days of co-culture with the intestinal epithelium on-chip. Thus, although serial dilutions of a mixed population of bacteria do not provide accurate total counts, these data show that both Gram positive and Gram negative bacteria remained viable and proliferated (increased in number) over time in the human Intestine Chips.
Fig. 4 ∣. Anaerobic conditions in the Intestine Chip enhance the growth of multiple genera compared to the aerobic chip and conventional liquid culture.
a, Differential abundance bacterial genera in the Caco2 Intestine Chip measured under anaerobic conditions over 3 days of culture (top), or at day 3 in the anaerobic chip compared to the aerobic chip or anaerobic liquid culture (bottom). Data are presented as log10 of the total read counts for each genus; each data point represents one chip.The total read counts for all genera at the bottom are normalized to their counts in liquid culture (data are presented as box plots with individual data points overlaid, where lower or upper edges of the box represent 25th or 75th percentiles and the middle bar is the median). b, Differential abundance in bacterial genera measured over 1 to 3 days of co-culture in the anaerobic versus aerobic Intestine Chip (data are represented as log2 fold change; each data point corresponds to the differential abundance for a given genus at a given day, comparing anaerobic to aerobic cultures; n=4 chips for each group on each day; error bars represent the s.d.).
Importantly, when we compared the growth of microbiota cultured for 3 days in the anaerobic Intestine Chip with that produced by culturing the same microbiome samples in conventional liquid medium culture in an anaerobic chamber, we observed significantly different growth responses for multiple genera (Fig. 4a, bottom). Notably, the genus Akkermansia shows preferential growth in the anaerobic Intestine Chip, presumably due to the presence of high levels of mucus that is produced by the Caco2 intestinal epithelium on-chip46 (Fig. 2e,f), which complements the mucins already present in the medium. Bacteria in the Blautia, Bilophila, Oscillospira, and Suterella genera also showed enhanced growth in the anaerobic chips containing living human intestinal epithelium compared to anaerobic liquid culture, whereas the Gram-negative obligate aerobe, Citrobacter, was less abundant on-chip. Thus, the presence of human intestinal epithelium that secretes a natural mucus layer above its apical surface25 (Fig. 2e,f) is crucial for culturing and sustaining the complex features of human microbiome in vitro.
To complement these analyses, we calculated the differential abundance of the different genera over time in the anaerobic versus aerobic Intestine Chips. Our results show that the obligate anaerobes, Eubacterium, Oscillospira, Blautia, and Suterella were significantly more abundant in the anaerobic chips compared to aerobic chips over our time course (FDR q < 0.05), whereas the obligate aerobe, Citrobacter, consistently showed a lower abundance in the anaerobic chip (Fig. 4b). Whether taking into account the abundance of the various genera in anaerobic liquid culture (Supplementary Fig. S12) or in the Hmb microbiome stock (Supplementary Fig. S13), quantification of the total read counts confirmed that the total numbers of obligate anaerobes, including Sutterella, Blautia, Oscillospira, Bilophila, and Akkermansia, were significantly higher in the anaerobic chips. Taken together, these results confirm that the hypoxia gradient system combined with the presence of a living human intestinal epithelium provides a unique and preferential environment for sustained culture of anaerobic as well as aerobic gut bacteria from diverse genera.
Culture of fresh gut microbiome with primary intestinal epithelium on-chip.
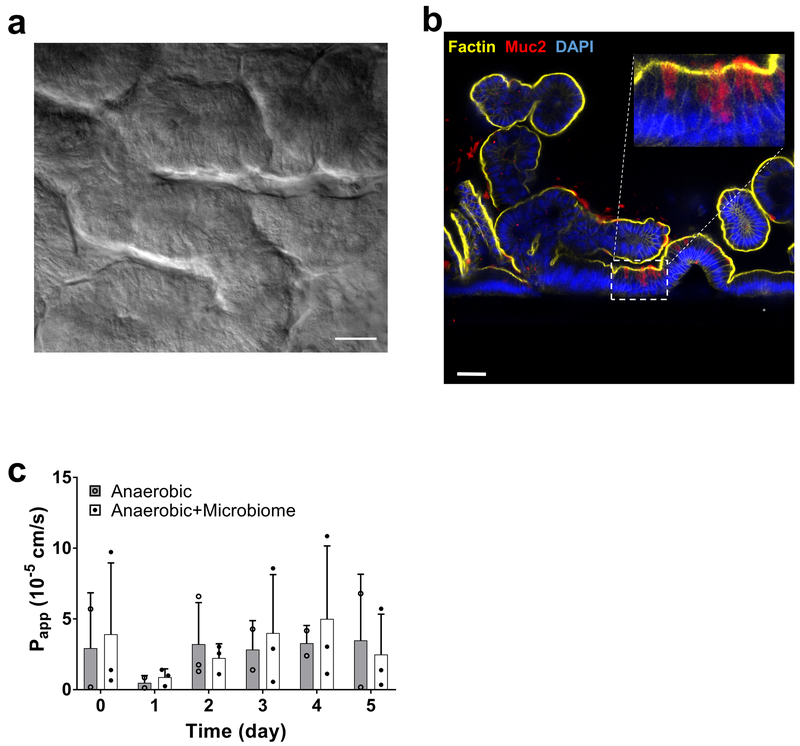
Finally, we explored whether this experimental approach can be used to co-culture complex gut microbiome obtained from fresh human stool specimens in direct contact with primary human intestinal epithelium (i.e., rather than using the established Caco2 intestinal cell line). To do this, we engineered human Intestine Chips lined with intestinal epithelial cells isolated from organoids derived from normal regions of surgical biopsies of human ileum, which exhibit multi-lineage differentiation, villi formation, and mucus production when grown on-chip27. We then inoculated the epithelial channels of 4 different chips with complex microbiome isolated from fresh human stool samples collected from four different infants (one with a corrected gestational age of 30 week and three with an age of 36 week). DIC (Fig. 5a) and confocal fluorescence microscopic (Fig. 5b and Supplementary Fig. S14a) imaging of the primary human Ileum Chips confirmed the presence of a villus intestinal epithelium lined by a continuous polarized epithelium with F-actin- and villin-containing brush borders along its apical membrane, MUC2-producing cells, and basal nuclei. Importantly, when we measured production of secreted mucus using alcian blue staining (Supplementary Fig. S14b), we observed blue stained mucus over the apical surface of the epithelium, and we detected up to 600 ug.ml−1 of mucin in the chip outflow (Supplementary Fig. S14c). As expected, the bacterial richness was reduced in the infant stool stock (586 OTUs) compared to adult human-derived stool (938 OTUs) at the same dilution per gram of materials, and these differences in richness were accurately recapitulated on-chip. We again found that the primary human intestinal epithelium could be co-cultured in direct contact with this complex gut microbiome without compromising epithelial barrier function, and this co-culture was stably maintained for up to at least 5 days on-chip (Fig. 5c), much as we had observed with the Caco2 epithelium. Importantly, the microbiome cultured in these primary Intestine Chips also maintained a high bacterial richness, ranging from 118 to 135 OTUs (Fig. 5d) corresponding to 6 phyla (Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Proteobacteria and Tenericutes) and 32 unique genera. Thus, the hypoxic Intestine Chip method can be used to sustain a complex community of human microbes in direct contact with normal, patient-derived, human intestinal epithelial cells for many days in culture, which could be valuable for personalized medicine in the future.
Fig. 5 ∣. Anaerobic co-culture of gut microbiome obtained from fresh human patient-derived stool with primary human ileal epithelium in the Intestine Chip.
a, Microscopic views showing the villus morphology of the primary ileal epithelium cultured for 5 days in the Intestine Chip under anaerobic conditions when viewed from above by DIC (scale bar, 50μm) or b, shown in cross-section by confocal immunofluorescence imaging for MUC2 (red), F-actin (yellow) and DAPI (blue) (scale bar, 50μm; inset shows area highlighted in white dashed line at higher magnification; representative images from 4 intestine chips are shown). c, Changes in apparent paracellular permeability (Papp) measured by quantifying cascade blue transport across the tissue interface within the primary Intestine Chip during co-culture with or without complex human gut microbiome under anaerobic conditions (bacteria contained with patient-derived stool samples were added on day 0; n=3 individual chips; data are presented as mean ± s.d.). For richness and Shannon diversity of bacteria in effluent samples of primary Intestine Chips refer to Supplementary Table S2.
Discussion
Given the importance of commensal gut microbiome for human health and the lack of any in vitro model that can faithfully mimic the complex epithelial-microbe interactions that occur across the host-microbiome interface, we leveraged human Organ Chip technology to develop a device that enables human intestinal epithelium to be co-cultured with the highly diverse community of commensal microbes that comprises the human gut microbiome under aerobic and anaerobic conditions. Our results show that the anaerobic human Intestine Chip offers a robust modular system for recapitulating the human intestinal-microbiome interface in vitro. Using this device, it is now possible to stably co-culture a complex living microbiome in direct contact with living mammalian cells and their naturally produced overlying mucus layer for 5 days or more in vitro. This is significantly longer than past studies using the HMI model that only sustained co-cultures of intestinal epithelium with complex microbiome for 48 hours, and in which the bacteria had to be physically restricted from contacting the epithelium by a semi-permeable membrane to ensure epithelial viability17,47. Importantly, providing a physiologically-relevant low oxygen microenvironment on-chip also sustained a higher level of microbial diversity (~200 unique OTUs) and increased abundance of obligate anaerobic microbiota compared to aerobically-cultured chips, in addition to maintaining a diverse community of commensal microbes that closely resembled that of the human gut microbiome in vivo. For example, when the complex gut microbiome was cultured in the anaerobic Intestine Chip, it maintained abundant amounts of obligate anaerobic bacteria, with ratios of Firmicutes and Bacteroidetes similar to those observed in human faeces44. Additionally, we found that co-culturing intestinal epithelium with either single commensal gut microbes (e.g., B. fragilis) or over 200 different bacterial OTUs under physiologically relevant anaerobic conditions actually enhanced epithelial barrier function compared to aerobic chips, which again is consistent with in vivo findings39. This is in direct contrast to past studies in which co-culture of pathogenic bacteria (e.g., enteroinvasive E. Coli) was shown to rapidly compromise barrier function in the same Caco2 cells-lined Intestine Chips21. This ability to discriminate between healthy versus injury responses of human intestinal epithelium in the presence of commensal versus pathogenic bacteria, and study these direct intercellular interactions under controlled conditions in vitro, is a significant advantage of this model versus approaches that separate the microbes from the epithelium by nanoporous membranes.
Oxygen tension is one of the main regulators of intestinal function and pathogenesis of GI diseases49,50. By integrating non-toxic oxygen sensors into our devices, we were able to control and measure oxygen levels throughout the microfluidic Intestine Chips without interference with microscopic imaging, device fabrication or cell culture. Use of these sensors, rather than incorporating multiple external oxygen-detecting probes, enables this approach to be more easily scaled so that many miniaturized Organ Chips can be created. The anaerobic chamber we engineered also generates radial oxygen gradients across the endothelium-epithelium-microbiome interface that allows oxygenation of the human tissues while providing an anaerobic environment for growth of the obligate anaerobes. Anaerobic incubators or glove boxes can be used to maintain hypoxic conditions for bacterial cultures, but they commonly provide a single uniform low oxygen concentration, rather than physiologically-relevant oxygen gradients directed across tissue-tissue interfaces. In contrast, our anaerobic chamber is portable, highly customizable, compatible with imaging, and most importantly, capable of engineering oxygen gradients across the endothelial-epithelial interface of any Organ Chip on demand.
Oxygen concentrations in the lumen of the human intestine are known to affect the spatial distribution and metabolism of gut flora51, and most intestinal bacteria are obligate anaerobes, many of which fail to grow at oxygen concentrations greater than ~0.5%52. Any culture system that is designed to recapitulate the host gut-microbiome interface must therefore be able to achieve and sustain oxygen concentrations at these low levels. A microfluidic-based anaerobic culture system has been described previously that maintains oxygen levels as low as 0.8% in the presence of a facultative anaerobe47, but this level is still too high to support obligate anaerobes. Moreover, this past model used both a synthetic mucus layer and a nanoporous membrane to physically separate bacteria from the intestinal epithelium47. Using our custom anaerobic chamber, we were able to attain an oxygen concentration of less than 0.3% in the epithelial channel where we cultured the commensal microbes, which is much closer to that found in the gut lumen in vivo53. Additionally, we validated the relevance of these hypoxic culture conditions by showing that they support the growth of an obligate anaerobe B. fragilis that cannot grow in the presence of greater than ~0.5% dissolved oxygen32,54. Furthermore, the finding that co-culture of the human intestinal epithelium with B. fragilis under anaerobic conditions also increased (rather than decreased) intestinal barrier function on-chip is consistent with the finding that oral delivery of B. fragilis corrects intestinal permeability defects in a mouse autism model55.
More importantly, we found that the hypoxic human Intestine Chip supports co-culture of complex human microbiota composed of over 200 unique OTUs and at least 11 different genera of bacteria for multiple days in co-culture. Bacterial members of the Bacteroidetes and Firmicutes phyla, and to a lesser degree Verrucomicrobia and Proteobacteria, which comprise the human intestinal microbiome in vivo56, also colonized our Caco2 Intestine Chips at similar ratios. Anaerobic chips had increased levels of anaerobic Clostridia, Bacteroides, and Akkermansia, whereas Proteobacteria, which accumulate mainly at more oxygenated regions of the proximal gastrointestinal tract,53,57 dominated the aerobic chips. One limitation of our approach is the need to dilute the complex microbiome inoculum to avoid rapid unrestrained bacterial overgrowth. This may result in exclusion of some rare bacteria; however, this could be ameliorated by using larger Intestine Chips, optimizing the lumen perfusion rate, applying cyclic (peristalsis-like) mechanical deformations that can suppress growth of some commensals21, or altering medium conditions to limit bacterial overgrowth in the future. Nevertheless, these data show that the Intestine Chip with the hypoxia gradient and anaerobic conditions in the lumen of the epithelial channel promoted more bacterial diversity than the aerobic system. Moreover, the anaerobic human Intestine Chip supported a wide range of bacterial genera similar to those found in human stool, which is much more complex than any microbiome community that has been previously cultured in direct contact with mammalian cells, and we could sustain these co-cultures for at least 5 days in vitro.
Others have previously maintained complex microbiota in test tube cultures58, however, our results indicate that the presence of a more in vivo-like intestinal tissue microenvironment significantly influences the composition of the microbial community. For example, the mucus degrading, obligate anaerobe genus Akkermansia was found in higher abundance in the anaerobic Intestine Chips which contain human intestinal epithelial cells that secrete mucus than in liquid cultures maintained under similar anaerobic conditions that were artificially supplemented with mucin. In contrast to liquid cultures, the anaerobic Intestine Chip also allows inferences to be made regarding the effects of commensal microbes on the host epithelium and vice versa. It is interesting that the enhanced growth of Akkermansia in the anaerobic Intestine Chip was accompanied by increased intestinal barrier function since the high abundance of this organism has been previously suggested to enhance gut barrier function in vivo44-46.
HIF-1α is believed to control barrier integrity by regulating multiple barrier-protective genes, and its dysregulation may be involved in GI disorders59,60. Interestingly, although we observed elevated levels of nuclear HIF-1α in anaerobic Intestine Chips, we did not detect any changes in barrier function unless we also co-cultured complex microbiota. Thus, this experimental system allows us to parse out which physiological effects are due to changes in oxygen levels alone and which are due to the presence of commensal microbes under low oxygen conditions. Defining the causative relationship between the abundance of each individual genus of bacteria and distinct functions of the co-cultured human intestinal epithelium is beyond the scope of this study. However, this system could be harnessed to address these types of questions, as well as how different commensal microbes contribute to the pathophysiology of various gastrointestinal diseases61 in the future. This model also could be used to analyse fluctuations in microbiome abundance, function, and location within the chip (e.g., under conditions of antibiotic exposure, hormonal stimulation, etc.) and their effects on intestinal epithelial physiology. Moreover, the clinical relevance of these studies could be further enhanced by developing patient-specific Intestine Chips with both host cells and gut microbiome isolated from the same patients, as well as by obtaining microbiome isolates from different regions of the intestine (e.g., duodenum, jejunum, ileum, colon) and culturing them on chips lined by cells from each of these regions.
The purpose of this study was to develop a method for co-culturing a complex living human gut microbiome, including obligate anaerobes which require strict anaerobic conditions (i.e., < 0.5–1% O2) to survive, in direct contact with human intestinal epithelial cells and their overlying mucus layer for extended times in vitro. While we did not intend to model any specific region of gastrointestinal system using our chips, it should be noted it is possible to create Organ Chips lined by cells from different regions of the intestine (e.g., duodenum, jejunum, ileum, colon) and to use oxygen tensions appropriate for each region (e.g., from 5% to 0.5% moving from duodenum to colon), and potentially introduce the microbiome aspirates from each of these regions. We have previously shown that primary Intestine Chip better recapitulates the morphology, multicellular composition, and gene expression patterns of the intestinal segment from which it was derived than other in vitro intestinal culture systems, such as the Caco2 chip and 3D intestinal organoids27. Furthermore, by integrating primary epithelial cells from intestinal biopsies as we did here, or patient-derived induced pluripotent stem (iPS) cells62, in combination with microbiomes obtained from the same patients, it should be possible to develop patient-, disease-, and location-specific, host-microbiome co-culture models, and thus, pursue a personalized-medicine approach in the future. That said, the Caco2 Intestine Chips also recapitulate many features of human intestinal physiology and pathophysiology, and these cells can be obtained commercially (rather than requiring a patient biopsy), which would enable their widespread use by academic and industrial laboratories, as well as regulatory agencies (e.g., FDA). In addition, the modular nature of the Organ Chip technology allows for the incorporation of additional cell types. In this study, we incorporated intestinal endothelium in our Intestine Chips because it enhances intestinal barrier function, villi development and mucus production26,27,63, but other cell types, such as immune cells and pathogens that play crucial roles in host gut-microbiome interactions64,65could be incorporated as well. Our oxygen sensing chips also have the potential to be combined with on-chip TEER technology66 for real-time monitoring of intestinal barrier function in the presence of different cell types and individual strains of bacteria. Thus, this methodology could be used in the future to unravel complex functional links between intestinal epithelial cells, immune cells, and gut microbes to understand mechanisms of human disease, discover new therapeutics, and advance personalized medicine. It may be possible to modify this Organ Chip to model and study host-microbiome interactions in other organs (e.g., vagina, skin, lung, etc.) as well.
Methods
Fabrication of the oxygen-sensing Organ Chip.
Oxygen sensor spots were prepared by mixing oxygen sensitive and optical isolating particles (PreSens GmbH, Germany) at weight ratio of 1:1 in methanol (sigma, 50 mg ml−1) for 2 h under constant stirring. PDMS prepolymer (Sylgard 184, Dow Corning) was then added to the mixture at 1 g ml−1 and solvent was subsequently removed by applying −70 kPa vacuum at 55 °C for 2 h. PDMS prepolymer was then mixed with curing agent (Sylgard 184, Dow Corning) at weight ratio of 10:1 for 4 min under vacuum, spin-coated (150 μm thick) onto a 5 cm silanized silicon wafer at 800 rpm for 2 min and cured at 60 °C for at least 30 min. The wafer was removed and the 150 µm thick film was punched into 1 mm diameter sensor discs using a biopsy punch. The sensor discs were dip coated in an uncured PDMS (PDMS prepolymer:curing agent 10:1) and embedded into the PDMS channels of a 2-channel Organ Chip by placing them in moulds at the inlet, middle and outlet of both upper (epithelium) and lower (endothelium) channels, and cured in place at 60 °C for 30 min. Organ Chip fabrication was then followed as described previously67. Using this two-step moulding process, these sensors were placed directly on the surface of both the vascular and epithelial channels of the Organ Chips at their inlet, middle and outlet regions. The chip fabrication and sensor integration steps involving plasma treatment did not interfere with sensor function or the functionality of the microfluidic chips, and the thickness of the sensors did not affect the oxygen readouts when maintained between 150 to 300 μm in height.
Anaerobic chamber fabrication and validation.
To fabricate the anaerobic chamber, acrylic parts were cut using a laser cutter (Epilog) and assembled together with an acrylic solvent (SciGrip Acrylic Cement). Gaskets were lasercut from adhesive-backed silicone rubber sheets (20 Shore A hardness, McMaster-Carr) and magnetic clasps were attached using adhesive backed magnets. The anaerobic chamber was tested using a calibrated Oxy-4 optical probe system (PreSens GmbH, Germany) to verify the hypoxic conditions. To do so, the chamber was purged with 5% CO2 in N2 bubbled through deionized water at 81 ml min−1, 162 ml min−1, or 243 ml min−1 for 1 h at which point N2 flow was stopped and the chamber allowed (3 h) to recover to atmospheric oxygen.
Oxygen sensing in the Intestine Chip.
To visualize and quantify the concentration of oxygen throughout the chip, oxygen measurements were performed through non-invasive fluorescence read-out using VisiSens-system (PreSens GmbH, Germany). Using a CCD-camera and the VisiSens software (V1.1.2.10), oxygen amount was detected at sensor spots, and displayed using a computer code in pseudo colours. The software has been designed to calculate oxygen levels on the sensor spots via calibration of fluorescence reading with defined oxygen levels at 0 and 100% air saturation (i.e. 20.9% O2 of all dissolved gas by volume). In all experiments, oxygen levels were quantified after comparing the readings with the calibration values. Air-saturated water and oxygen-free solution (Oakton, WD-00653–00) were used to calibrate the sensor spots. Since the field-of-view of the VisiSens camera is inherently small, we designed a linear positioning system that can position the camera directly beneath the Intestine Chips in the hypoxia chamber. This allows indexed motions of the camera to any sensor spot along the chip or between the chips and thus, facilitates reproducibly imaging multiple chips in one run. The sensors do not obscure regular imaging of the chips as they only cover a small portion of the culture area (~3 mm2), allowing for regular monitoring of cultures throughout the experiment. We also designed a black opaque box that covers entire chip culture chamber and VisiSens camera to block extraneous light. To analyse the accuracy of sensor spots inside the chips Oxygen concentrations were measured in gas phase using calibrated gas tanks with known oxygen concentration, i.e. 0, 1, and 12.5% O2. The VisiSens imaging system was validated using an Oxy-4 optical probe system (PreSens) with optical fibers (POF-L2.5, PreSens, Germany).
Oxygen sensor analysis.
Images of oxygen sensors were processed in MATLAB (Mathworks) and binarized using Otsu’s method 68. Morphological erosion and dilation were preformed to eliminate any spurious artefacts created during binarization. Simulated annealing was applied to find the correct assignment of sensors in each image regardless of the chip alignment. The sum of the distance of each of the sensor’s centroids in the current image between the nearest sensor’s centroid in the original image was minimized. After aligning the images, the sensors in the current image were registered consistently with the sensors in the former image, and colorimetric analyses were computed. The average intensities were calculated for each of the red, blue, and green channels, in each sensor. The uncalibrated signal from each sensor was taken to be the average green intensity divided by the average red intensity. The uncalibrated signal was then fit to a calibration curve. We used a modified Michaelis-Menten two-point calibration as the most generalizable model, Coxy= kmin+ (kmax-kmin) x [xg:r / (krate+xg:r)]; kmax=a x katm, where xg:r denotes the ratio of average green intensity to average red intensity, Coxy is the fraction of atmospheric oxygen, kmin is the sensor signal at anaerobic conditions, kmax is the sensor signal when saturated with oxygen, and the concentration of oxygen is given as Coxy. krate explains the effect that the observed signal, xg:r, has on the concentration of oxygen. The atmospheric oxygen concentration does not fully saturate the sensor with oxygen. To overcome this, actual maximum possible signal from a sensor, kmax, is estimated by multiplying the uncalibrated signal at atmospheric concentration, katm, by a scale factor α. The Michaelis-Menton curve was approximately linear between xg:r=katm and xg:r=kmax, scaling by a linear coefficient did not hamper the equation’s ability to generalize between sensors. The curve was fit using images acquired at known oxygen concentrations. The known concentrations were measured by Oxy-4 optical probe system (PreSens GmbH, Germany). The oxygen concentrations were also validated by flowing oxygen at known concentrations over the probe and sensor. Both krate and α were fit using the data. The model produced a suitable fit for the data (R2=0.990 training, R2=0.997 and 0.998 for testing). The fitted model generalized well for trials repeated in different chips and on different days.
Cell culture procedures.
Prior to cell seeding, microfluidic sensor chips were activated using oxygen plasma (Diener ATTO) and functionalized with (3-Aminopropyl) trimethoxysilane (Sigma, 281778), as reported previously26. Chips were then washed with ethanol, oven-dried at 80°C and coated with 30 μg ml−1 Collagen (Gibco, A10483–01) and 100 μg ml−1 Matrigel (BD Biosciences, 356237) in the serum-free Dulbecco’s Modified Eagle Medium (DMEM; Gibco, 10564011) for 1 h at 37 °C. To fabricate the Caco2 Intestine Chip, human intestinal microvascular endothelial cells (HIMECs; ScienCell) were first seeded (1.5×105 cells cm−2) in the bottom channel of the chips, on opposite side of the porous membrane. Chips were then placed in 37°C incubator for 1.5 h. For HIMECs culture, endothelial growth medium (EGM2-MV) containing human epidermal growth factor, hydrocortisone, vascular endothelial growth factor, human fibroblastic growth factor-B, R3-Insulin-like Growth Factor-1, Ascorbic Acid and 5% fetal bovine serum (Lonza Cat. no. CC-3202) was used. Human intestinal epithelial cells (Caco2 BBE human colorectal carcinoma cell, Harvard Digestive Disease Center) were then seeded into the top microchannel of the chip (1.5×105 cells cm−2) and incubated for 1.5 h. Epithelial cells were fed with DMEM (Gibco, 10564011) containing Pen/Strep and 20% Fetal Bovine Serum (FBS; Gibco, 10082–147). After washing with 200 μl of medium, chips were cultured statically overnight to allow cells to form monolayers on both sides of the membrane. A day after seeding, top and bottom channels were perfused (60 μl h−1) with epithelial medium and reduced-FBS endothelial medium, respectively. Chips were kept in this condition until villus-like intestinal epithelium spontaneously appeared. For anaerobic culture, the same procedure was followed except that after 1 day of perfusion in aerobic conditions, chips were placed in an anaerobic chamber and continuously perfused with 5% CO2 in N2 flowed at 243 mL min-1.
To create primary Intestine Chips, we used cells isolated from human intestinal organoids that were derived from resected tissue of a 15 year-old patient diagnosed with ulcerative colitis and collected from grossly unaffected (macroscopically normal) areas of the terminal ileum during endoscopy procedure, as described previously27. Informed consent and developmentally-appropriate assent were obtained at Boston Children’s Hospital from the donors’ guardian and the donor, respectively. All methods were carried out in accordance with the Institutional Review Board of Boston Children’s Hospital (Protocol number IRB-P00000529) approval. Informed consent and developmentally appropriate assent were obtained at Boston Children’s Hospital from the donors’ guardian and the donor, respectively. All methods were carried out in accordance with the Institutional Review Board of Boston Children’s Hospital (Protocol# IRB-P00000529) approval.
Organ Chips with the same 2-channel design, but obtained from Emulate Inc. (Boston, MA), were used to create the primary human Intestine Chip. The chips were chemically activated using ER1 and ER2 solutions (Emulate Inc.) before introducing type I collagen (200 µg.ml−1) and Matrigel (1% in PBS) into the channels, and incubating in a humidified 37 °C incubator for 2 h to coat the porous membrane with ECM before washing with PBS. Primary human intestinal epithelial cells that were isolated from the ileal organoids using collagenase and mechanical dissociation as described previously27 were then resuspended in expansion medium (EM) consisting of Advanced DMEM/F12 supplemented with L-WRN conditioned medium (50% v/v, ATCC), glutamax, HEPES, murine epidermal growth factor (50 ng.ml−1), N2 supplement, B27 supplement, human [Leu15]-gastrin I (10 nM), n-acetyl cysteine (1 mM), nicotinamide (10 mM), SB202190 (10 µM) and A83–01 (500 nM). A small volume (30µl) of the EM solution containing the primary intestinal cells (6 × 106 cells.ml−1) was used to infuse and fill the apical chamber of each chip resulting in ∼180,000 cells.chip−1; these chips were then incubated overnight under static conditions at 37°C to promote cell adhesion. The following day EM without cells was perfused (60 µl.h−1) through the top and bottom channels and cyclic suction was applied to hollow side chambers using a vacuum pump controlled by an electronic vacuum regulator (ITV009, SMC Corp.) and an Arduino microcontroller to exert peristalsis-like stretching motions (10% cell strain, 0.15Hz frequency) on the porous membrane and attached epithelium. Chips were maintained under these conditions until villus like structures were detected visually under phase contrast imaging (~14 days). Before adding microbiota, peristalsis-like cyclic stretching was stopped and the apical medium was then replaced with antibiotic-free DM containing microbial supplements (1 mg.ml−1 pectin, 1 mg.ml−1 mucin, 5 μg.ml−1 Hemin and 0.5 μg.ml−1 Vitamin K1) and the basal medium was replaced with antibiotic free EM.
Bacterial and Microbiota culture.
B. fragilis (9343 strain) was grown overnight at 37 °C under anaerobic conditions (80% N2, 10% H2, 10% CO2) in rich media containing yeast extract (5 g L−1), proteose peptone (20 g L−1), NaCl (5 g L−1), hemin (5 mg L−1), vitamin K1 (0.5 mg L−1), K2HPO4 (5 g L−1) and HADA (HCC-amino-D-alanine, λem ~ 450 nm; 0.8 mM) or GalCCP (N- cyclopropenyl galactosaminyl carbamate; 250 μm). Hemin, vitamin K1, K2HPO4, and HADA33 were added through a 0.22 μm filter after autoclaving the other ingredients. For HADA labelling, B. fragilis was pelleted at 5000 g, washed once in DMEM, and re-suspended in Caco2 media (DMEM 20% FBS, 1% glutamine, 1 mg ml−1 pectin, 1 mg ml−1 mucin, 5 μg ml−1 Hemin, 0.5 μg ml−1 Vitamin K1) at 1 × 107 CFU ml-1. For GalCCP labelling, B. fragilis was pelleted at 5000 g washed twice with PBS, once with 1% BSA in PBS, then incubated with 5 μM Tetrazine-Cy3 for one hour at 37 °C. Bacteria were pelleted and washed twice with 1% BSA in PBS, one with PBS33, and re-suspended in Caco2 media. For microbiota co-culture, colon and cecum content from five mice colonized with healthy human microbiota (Hmb) 23 were collected and re-suspended in sterile PBS inside an anaerobic chamber (100 mg of content ml−1). The slurry was then filtered (40 μm) and aliquoted and stored at −80 °C as the human microbiome stock, which was diluted 1:100 in epithelial medium when added to Intestine Chips. Animal experiments were performed under IACUC approval (ID# IS00000187–3; Microbial Effect on Mammalian Host). For microbiota co-culture with patient-derived specimens, fecal samples were collected from infants born at Brigham and Women’s Hospital in Boston, MA and cared for in a single-centre Newborn Intensive Care Unit (NICU). For infant fecal samples, informed consent from guardians was obtained at Brigham and Women’s Hospital. All methods were carried out in accordance with a protocol approved by the Institutional Review Board of Brigham and Women’s Hospital (Protocol# 2016-P-001020). Fecal samples were collected from preterm infants born prior to 32 weeks of gestation from birth until discharge. Briefly, diapers with fecal samples were collected daily by the bedside nurse, placed in a specimen bag, and stored at 4 °C for no more than 24 hours. Fecal material was extracted from diapers using sterile procedures and immediately frozen at −80 °C. Selected samples were suspended in Brain Heart Infusion media (100 mg.ml−1) to create a stock solution.
Co-culture with gut microbes and complex microbiome in the Intestine Chips.
One day before adding bacteria, media reservoirs were washed with PBS and antibiotic-free media was then added to Intestine Chips in a tissue culture hood (aerobic conditions) or in an anaerobic chamber (anaerobic conditions). The next day, 25 μl of B. fragilis (1 × 107 CFU ml−1) or diluted microbiota stock was added to the apical side of differentiated Intestine Chips in a tissue culture hood (aerobic conditions) or in an anaerobic chamber (anaerobic conditions). Chips were left static for 30 min and then perfused at 1 μl min-1. Every 24 h, a 2 min flush at 50 μl min−1 was performed and the flush outflow was collected and serial dilutions were plated on Brucella plates incubated at 37°C in an anaerobic chamber (B. fragilis cultures), Brucella plates +/− 6 μg ml−1 vancomycin (Hmb cultures) or sent for 16S rRNA sequencing (Diversigen).
Morphological analyses.
For each experiment, regions of 3 different Intestine Chips were analyzed morphologically at each interval. The villus structures formed by the intestinal epithelium were evaluated using DIC microscopy (Zeiss Axio Observer Z1 2, AXIO2) or immunoflorescence microscopy with a laser scanning confocal microscope (Leica SP5 X MP DMI-6000 and Zeiss TIRF/LSM 710). High-resolution horizontal or vertical cross-sectional images were obtained using deconvolution (Huygens) followed by a 2D projection process. IMARIS (MARIS 7.6 F1 workstation; Bitplane Scientific Software) and ImageJ were used for analysing the obtained images.
For SEM analysis, Intestine Chips were designed in a way that top channel was not irreversibly bonded to the membrane, which permitted the device to be dismantled manually without disturbing the cultured cells. Chips were first treated with Carnoy’s solution for mucus fixation, and then fixed with paraformaldehyde (PFA, 4%; Electron Microscopy Sciences, 157–4) and glutaraldehyde (2.5%; Sigma, G7776) and incubated in osmium tetroxide (0.5%; Electron Microscopy Sciences, 19152) before serial dehydration in ethanol. Samples were then dried using a critical point dryer and imaged with a field emission SEM (Hitachi S-4700).
Immunofluorescence microscopy.
Epithelial and endothelial cells were washed with PBS, fixed with 4% paraformaldehyde (PFA; Electron Microscopy Sciences, 157–4) and subsequently washed with additional PBS. Permeabilization of cells was accomplished with 0.25% Triton X-100 (Sigma, T8787), followed by incubation in blocking buffer containing 1% BSA (Sigma, A4503) and 10% donkey serum (Sigma, D9663) at room temperature. Primary antibodies against ZO1 (Life Technologies, 33–9100, dilution 1:200), VE-cadherin/CD144 (BD Biosciences, 555661, dilution 1:200), Villin (Life Technologies, PA5–29078, dilution 1:100), Muc2 (Santa Cruz Biotechnology, sc-15334, dilution 1:100) or HIF-1α (Abcam, ab16066, dilution 1:100) were added and incubated overnight at 4°C, followed by multiple PBS washes. Cells were then incubated with secondary fluorescent antibodies (Life Technologies) at room temperature and washed with PBS; nuclei were co-stained with DAPI (Invitrogen, D1306). Microscopy was performed with a laser scanning confocal microscope (Leica SP5 X MP DMI-6000 or Zeiss TIRF/LSM 710).
Mucus Detection.
For the mucus visualization, Wheat Germ Agglutinin (WGA) Alexa Fluor 488 conjugate (Thermo Fisher Scientific) was used for the live cell imaging as described previously34. Briefly, WGA solution (25 μg.ml−1 in culture medium) was flowed through the epithelium channel for 30 min. Top channel was washed subsequently with PBS in the dark and counter-stained with DAPI to visualize nuclei. To stain acidic mucopolysaccharides within the intestinal mucus, Intestine Chips were stained with 0.1% (w/v) alcian blue solution (pH 2.5; 8GX, Sigma) in 3% acetic acid (Sigma) by flowing the solution into the microchannels at 50 μL.h−1 for 12 h, and then washing with PBS.
Paracellular permeability measurements.
50 μg ml−1 of cascade blue (5.9 kDa; ThermoFisher, C687) were introduced to the epithelium channel (60 mL hr−1) and fluorescence intensity (390 nm/420 nm) of top and bottom channel effluents were measured using a multi-mode plate reader (BioTek NEO). Apical-to-basolateral flux of the paracellular marker was calculated based on the following equation: Papp=(dQ/dt)/A.dC. Papp (cm s−1) denotes the apparent permeability coefficient, dQ/dt (g s−1) is molecular flux, A (cm2) is the total area of diffusion and dC (mg mL−1) is the average gradient. In some studies, Lucifer Yellow was used instead of Cascade Blue using a previously published method27.
Cellular toxicity.
CytoTox 96 Non-Radioactive Cytotoxicity Assay (LDH; Promega, G1780) was used according to the manufacturer’s instructions to measure epithelium and endothelium viability over time under aerobic and anaerobic culture conditions. In brief, effluents were collected from top and bottom channels, mixed with LDH substrate reagent and incubated for 30 min. The enzymatic reaction was terminated using stop solution (containing acetic acid) and the absorbance at 492 nm was recorded using a multi-mode plate reader (BioTek NEO). The LDH activity was assessed using quadruplicate of each group, calculated after subtracting the background absorbance values and reported as a fold change of the total LDH values of control group.
16S rRNA sequencing analysis.
Following sequencing, read counts were obtained by processing the FASTQ files using QIIME 1.0 under standard protocols and resulting joined reads were aligned to the Greengenes database. A total of 938 OTUs were identified. As one of the steps in our analyses of the 16S sequencing data, we removed OTUs that did not meet certain criteria in terms of representation across all the samples. The data were loaded into R and the phyloseq package69 was used for further processing. Data from a sequencing control (medium only) were used to correct the count data in all of our samples. For any given OTU identified, the read counts for that OTU found in the control sample (i.e., a medium sample without microbes that was run through the sequencer, allowing for the control of sequencing errors) was effectively subtracted. In the cases where the number of reads for a given OTU in a sample of interest was negative (i.e., the number of reads in the control was higher than that of the sample), we listed the number of reads of that OTU as 0. After performing diversity analyses, all singletons were removed from the data set and the OTUs were summarized to the genus level, resulting in a total 42 unique OTUs, corresponding to 9 well characterized genera (34 OTUs) and 8 OTUs of unknown genus (Proteobacteria and Firmicutes phyla). Differential abundance of these genera between the two culture conditions (i.e. aerobic and anaerobic) was done using the DESeq2 package70. OTUs showing a differential abundance with an FDR corrected p-value q < 0.05 were considered significant. The PERMANOVA test was run using in R using the adonis function in the vegan package between aerobic and anaerobic conditions, as well as between the two oxygen conditions across the different days. Comparisons to human stool microbiome was done against the Human Microbiome Project HMP1 data set (data set contains 44,740 OTUs across 4,743 samples from 18 different body sites.)
Statistical analysis.
All experiments were carried out at n=3–6 (see figure captions), and results and error bars indicate mean ± standard deviation (SD). Data analysis was performed with a one-way analysis of variance (ANOVA) with Tukey HSD post hoc tests using Prism (GraphPad). Statistical analysis between two conditions was performed by an unpaired student’s t-test. P values less than 0.05 were considered significant.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The main data supporting the findings of this study are available within the Article and its Supplementary Information. The raw data generated in this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
This research was supported by the U.S. FDA grant (HHSF223201310079C), DARPA THoR grant (W911NF-16-C-0050), Bill & Melinda Gates Foundation, Wyss Institute for Biologically Inspired Engineering at Harvard University, and Fundação para a Ciência e a Tecnologia (FCT) Portugal (project PD/BD/105774/2014 to the Institute for Bioengineering and Biosciences). We thank D. E. Achatz (PreSens Precision Sensing GmbH, Germany) for graciously providing oxygen sensing particles and her expert technical advice, and T. Ferrante for his assistance with imaging.
Footnotes
Competing interests
D.E.I. holds equity in Emulate, Inc., consults to the company, and chairs its scientific advisory board.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cho I & Blaser MJ The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donaldson GP, Lee SM & Mazmanian SK Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pickard JM, Zeng MY, Caruso R & Núñez G Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 279, 70–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sommer F & Bäckhed F The gut microbiota — masters of host development and physiology. Nat. Rev. Microbiol. 11, 227 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Walter J & Ley R The human gut microbiome: ecology and recent evolutionary changes. Annu. Rev. Microbiol. 65, 411–429 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Sommer MO Advancing gut microbiome research using cultivation. Curr. Opin. Microbiol. 27, 127–132 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Eain MMG et al. Engineering Solutions for Representative Models of the Gastrointestinal Human-Microbe Interface. Engineering 3, 60–65 (2017). [Google Scholar]
- 8.Fritz JV, Desai MS, Shah P, Schneider JG & Wilmes P From meta-omics to causality: experimental models for human microbiome research. Microbiome 1, 14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arrieta M-C, Walter J & Finlay BB Human Microbiota-Associated Mice: A Model with Challenges. Cell Host Microbe 19, 575–578 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Nguyen TLA, Vieira-Silva S, Liston A & Raes J How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 8, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadabad MS et al. A simple coculture system shows mutualism between anaerobic faecalibacteria and epithelial Caco-2 cells. Sci. Rep. 5, 17906 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutta D & Clevers H Organoid culture systems to study host–pathogen interactions. Curr. Opin. Immunol. 48, 15–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fatehullah A, Tan SH & Barker N Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 18, 246–254 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Williamson IA et al. A High-Throughput Organoid Microinjection Platform to Study Gastrointestinal Microbiota and Luminal Physiology. Cell. Mol. Gastroenterol. Hepatol. 6, 301–319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bein A et al. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cell. Mol. Gastroenterol. Hepatol. 5, 659–668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.den Abbeele PV et al. Incorporating a mucosal environment in a dynamic gut model results in a more representative colonization by lactobacilli. Microb. Biotechnol. 5, 106–115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marzorati M et al. The HMI™ module: a new tool to study the Host-Microbiota Interaction in the human gastrointestinal tract in vitro. BMC Microbiol. 14, 133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van de Wiele T, Van den Abbeele P, Ossieur W, Possemiers S & Marzorati M The Simulator of the Human Intestinal Microbial Ecosystem (SHIME®) in The Impact of Food Bioactives on Health: in vitro and ex vivo models (eds. Verhoeckx K et al.) (Springer, 2015). [Google Scholar]
- 19.Van den Abbeele P et al. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl. Environ. Microbiol. 76, 5237–5246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HJ, Huh D, Hamilton G & Ingber DE Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab. Chip 12, 2165–2174 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Kim HJ, Li H, Collins JJ & Ingber DE Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. 113, E7–E15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park G-S et al. Emulating Host-Microbiome Ecosystem of Human Gastrointestinal Tract in Vitro. Stem Cell Rev. 13, 321–334 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Chung H et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149, 1578–1593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surana NK & Kasper DL Moving beyond microbiome-wide associations to causal microbe identification. Nature 552, 244–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HJ, Huh D, Hamilton G & Ingber DE Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab. Chip 12, 2165–2174 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Jalili-Firoozinezhad S et al. Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip. Cell Death Dis. 9, 223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasendra M et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 8, 2871 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng L, Kelly CJ & Colgan SP Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A Review in the Theme: Cellular Responses to Hypoxia. Am. J. Physiol. Cell Physiol. 309, C350–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang BH, Semenza GL, Bauer C & Marti HH Hypoxia-inducible factor 1 levels vary exponentially over a physiologically relevant range of O2 tension. Am. J. Physiol. 271, C1172–1180 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Chilov D et al. Induction and nuclear translocation of hypoxia-inducible factor-1 (HIF-1): heterodimerization with ARNT is not necessary for nuclear accumulation of HIF-1alpha. J. Cell Sci. 112, 1203–1212 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Surana NK & Kasper DL The yin yang of bacterial polysaccharides: lessons learned from B. fragilis PSA. Immunol. Rev. 245, 13–26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patrick S, Reid JH & Larkin MJ The growth and survival of capsulate and non-capsulate Bacteroides fragilis in vivo and in vitro. J. Med. Microbiol. 17, 237–246 (1984). [DOI] [PubMed] [Google Scholar]
- 33.Hudak JE, Alvarez D, Skelly A, von Andrian UH & Kasper DL Illuminating vital surface molecules of symbionts in health and disease. Nat. Microbiol. 2, 17099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin W & Kim HJ Intestinal barrier dysfunction orchestrates the onset of inflammatory host–microbiome cross-talk in a human gut inflammation-on-a-chip. Proc. Natl. Acad. Sci. 115, E10539–E10547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo JC-H et al. Detection of colorectal dysplasia using fluorescently labelled lectins. Sci. Rep. 6, 24231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim HJ & Ingber DE Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. Quant. Biosci. Nano Macro 5, 1130–1140 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Rozee KR, Cooper D, Lam K & Costerton JW Microbial flora of the mouse ileum mucous layer and epithelial surface. Appl. Environ. Microbiol. 43, 1451–1463 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lock JY, Carlson TL, Wang C-M, Chen A & Carrier RL Acute Exposure to Commonly Ingested Emulsifiers Alters Intestinal Mucus Structure and Transport Properties. Sci. Rep. 8, 10008 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villmones HC et al. Species Level Description of the Human Ileal Bacterial Microbiota. Sci. Rep. 8, 4736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The Human Microbiome Project Consortium et al. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stearns JC et al. Bacterial biogeography of the human digestive tract. Sci. Rep. 1, 170 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guarner F & Malagelada J-R Gut flora in health and disease. The Lancet 361, 512–519 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Wang M, Ahrné S, Jeppsson B & Molin G Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol. Ecol. 54, 219–231 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Fujio-Vejar S et al. The Gut Microbiota of Healthy Chilean Subjects Reveals a High Abundance of the Phylum Verrucomicrobia. Front. Microbiol. 8, 1221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneeberger M et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep 5, 16643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Everard A et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. 110, 9066–9071 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah P et al. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat. Commun. 7, 11535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedicord VA et al. Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance. Sci. Immunol. 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheridan WG, Lowndes RH & Young HL Intraoperative tissue oximetry in the human gastrointestinal tract. Am. J. Surg. 159, 314–319 (1990). [DOI] [PubMed] [Google Scholar]
- 50.He G et al. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. 96, 4586–4591 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohland CL & Jobin C Microbial Activities and Intestinal Homeostasis: A Delicate Balance Between Health and Disease. Cell. Mol. Gastroenterol. Hepatol. 1, 28–40 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flint HJ, Scott KP, Louis P & Duncan SH The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 9, 577 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Albenberg L et al. Correlation Between Intraluminal Oxygen Gradient and Radial Partitioning of Intestinal Microbiota in Humans and Mice. Gastroenterology 147, 1055–1063.e8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baughn AD & Malamy MH The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427, 441–444 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Hsiao EY et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clemente JC, Ursell LK, Parfrey LW & Knight R The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 148, 1258–1270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shin N-R, Whon TW & Bae J-W Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Goodman AL et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. 108, 6252–6257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karhausen J et al. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J. Clin. Invest. 114, 1098–1106 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manresa MC & Taylor CT Hypoxia Inducible Factor (HIF) Hydroxylases as Regulators of Intestinal Epithelial Barrier Function. Cell. Mol. Gastroenterol. Hepatol. 3, 303–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cirstea M, Radisavljevic N & Finlay BB Good Bug, Bad Bug: Breaking through Microbial Stereotypes. Cell Host Microbe 23, 10–13 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Workman MJ et al. Enhanced Utilization of Induced Pluripotent Stem Cell–Derived Human Intestinal Organoids Using Microengineered Chips. Cell. Mol. Gastroenterol. Hepatol. 5, 669–677.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim HJ, Li H, Collins JJ & Ingber DE Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. 113, E7–E15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hooper LV, Littman DR & Macpherson AJ Interactions Between the Microbiota and the Immune System. Science 336, 1268–1273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rimoldi M et al. Intestinal immune homeostasis is regulated by the crosstalk between epithelial cells and dendritic cells. Nat. Immunol. 6, 507 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Henry OYF et al. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab. Chip 17, 2264–2271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huh D et al. Microfabrication of human organs-on-chips. Nat. Protoc 8, 2135–2157 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Otsu N A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 9, 62–66 (1979). [Google Scholar]
- 69.McMurdie PJ & Holmes S phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLOS ONE 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the findings of this study are available within the Article and its Supplementary Information. The raw data generated in this study are available from the corresponding author upon reasonable request.