Abstract
The objectives of this study were to lay the methodological groundwork for field studies of microRNA analysis in exosomes from small sample volumes of human milk, and assess exosome and microRNA content in infant formulas. When human milk was stored at 4°C for four weeks, the count of exosome-sized vesicles decreased progressively to 49% ± 13% of that in fresh milk. Exosomes were purified from 1 mL of fresh human milk and their microRNA content was assessed by microRNA-sequencing analysis and compared to that in infant formulas. We identified 221 microRNAs in exosomes from three samples of fresh human milk; 84 microRNAs were present in all three samples. MicroRNAs were not detectable in infant formulas and their exosome-sized vesicles, which appeared to be casein micelles. We conclude that large scale studies of microRNAs in human milk exosomes are feasible, and exosomes and microRNAs are not detectable in formulas.
Keywords: Bovine milk, exosomes, extracellular vesicles, human milk, infant formula, micro RNA, storage stability, casein, fat globules, protocols, stability
Introduction
Exosomes are extracellular vesicles (EVs) with an important function in cell-to-cell communication ([1], Supplemental Table 1, Supplemental Digital Content 1). Communication is achieved through the transfer of exosome cargos from donor cells to recipient cells. MicroRNA cargos are of particular importance because they regulate the expression of more than 60% of human genes [4]. Exosomes and microRNAs may be absorbed from milk [5]. The objectives of this study were to lay the methodological groundwork for studies of microRNA in exosomes from small sample volumes of human milk in large cohorts of women, and assess exosome and microRNA content in infant formulas. This study is significant because it enhances the confidence that future large scale studies of microRNAs in human milk exosomes are feasible and can contribute new information to the Human Milk Composition Database curated by the United States Department of Agriculture [6].
Methods
Participants
Five healthy non-Hispanic women, ages 26-36 years, participated in this study (1 African, 3 Asians, 1 Caucasian). The women gave birth to healthy singletons and had been lactating for 2-10 months when milk was collected. Two women had given birth to one or two babies before, three women were primiparous. This study was approved by the Institutional Review Board at the University of Nebraska-Lincoln (protocol 14302); written informed consent was obtained from participants.
Storage Stability Analysis
Midstream breast milk was collected. Full-fat bovine milk was purchased from a local grocery store; the date of purchase was denoted time zero but does not represent fresh milk. Aliquots were kept under different conditions [storage at 4°C, as well as −80°C without or with preservatives, glycerol (Sigma-Aldrich, St. Louis, MO) or dimethylsulfoxide (DMSO, Sigma-Aldrich)] in 1.5-mL polypropylene microcentrifuge tubes. At timed intervals samples were diluted with water and exosome-sized vesicles were analyzed using a NanoSight NS300 tracking device (Malvern, Westborough, MA) to assess effects of storage on vesicle count and size (syringe speed, 50; camera level, 10-11; detection threshold, 3; five 1-minute videos per sample).
Exosome Isolation and Authentication
Exosomes were isolated from 1 mL human and bovine milk and three brands of milk-based infant formulas (Supplemental Table 2, Supplemental Digital Content 2) as described previously with minor modifications [7, 8]. Samples were centrifuged at 16,000 g for 30 minutes at 4°C to remove fat layer, cells and debris. Fat globules and microvesicles were removed by ultracentrifugation at 83,000 g for 60 min at 4°C (Fiberlite F37L-8×100 rotor; ThermoFisher Scientific, Waltham, MA). Exosomes were collected by ultracentrifugation at 130,000 g for 90 min and re-suspended in 200 μL PBS. Proteins were extracted from exosomes using ice-cold radioimmunoprecipitation assay buffer (Sigma-Aldrich) with protease inhibitor cocktail (Sigma-Aldrich). Antibodies against CD-9 (Biovision, Milpitas, CA), CD63 (Santa Cruz, Dallas, TX), HSP70 (Santa Cruz) and Alix (Santa Cruz) were used as positive controls; anti-histone H3 (Santa Cruz) was used as a negative control [3]. Antibodies against alpha-tubulin (EMD Millipore, Burlington, MA) and integrin-β (Abcam, Cambridge, MA) were used to assess contamination with microvesicles. We probed bovine and human caseins in EV preparations from human milk and infant formulas with anti-bovine α-casein (Abcam) and anti-human β-casein (Novus Biologicals, Littleton, CO) [2]. Anti-α-lactalbumin (Abcam) and anti-butyrophilin (Origene, Rockville, MD) were used to probe whey proteins and lipid globules, respectively.
MicroRNA Sequencing
We used protocols endorsed by the Extracellular RNA Consortium [9]. RNA was extracted from human milk exosomes (1 mL milk) using the miRNeasy Micro Kit (Qiagen, Germantown, MD). MicroRNAs were sequenced in the DNA Sequencing Core at the University of Nebraska Medical Center (Omaha, NE). RNA integrity was confirmed using a Fragment Analyzer Automated CE System (Advanced Analytical Technologies, Ankeny, IA). Libraries were prepared using the NEXTflex Small RNA-Seq Kit v3 (Bioo Scientific, Austin, TX) and sequenced using Illumina Hiseq 2500 (~6 million reads/sample). Quality control was performed using FastQC [10]. Adaptor sequences and reads containing ambiguous bases or having an average quality score less than 30 were removed using Cutadapt [11]. Reads were aligned against the human microRNA database, miRBase version 22 using miRDeep2 [12, 13]. Data were deposited in the BioProject database (accession number PRJNA477819).
Quantitative Reverse Transcription PCR (RT-qPCR)
RT-qPCR analysis was performed to assess contamination of spin columns with small RNAs and confirm findings from sequencing analysis. RNeasy MinElute spin columns (Qiagen) might be contaminated with small RNAs, thereby causing artifacts in microRNA analysis [14]. RNeasy MinElute columns were treated with sodium hypochlorite to remove possible contamination [14]. We dissolved a non-natural microRNA (miSpike; IDT DNA, Coralville, IA; 5’-rCrUrCrArGrGrArUrGrGrCrGrGrArGrCrGrGrUrCrU-3’) in 200 μL RNase-free, molecular biology grade water and extracted miSpike using MinElute columns. Eluted miSpike (100 ng) was reverse transcribed using the miScript II RT Kit (Qiagen). In addition, miR-30d-5p, miR-125a-5p and miR-423-5p were analyzed in hypochlorite-treated and untreated columns. RT-qPCR was performed using miScript SYBR Green (Qiagen), the universal reverse primer in the miScript II RT Kit and sequence-specific forward primers (Supplemental Table 3, Supplemental Digital Content 3). Hypochlorite-treated columns were used when confirming sequencing data by RT-qPCR. Melting curve analyses suggested formation of a single product (not shown). Ct values greater than 29 were considered not detectable [15].
Statistical Analyses
Stability data were analyzed using repeated measures ANOVA. Dunnett’s post hoc test was used to compare data from all storage conditions to that of fresh human milk. Vesicle counts in timed samples of stored bovine milk were analyzed by one-way ANOVA and Tukey’s post hoc test for multiple comparisons. R version 3.5.0 (R Foundation for Statistical Computing) was used for analyses. Data are expressed as means ± SD or means ± SEM. Differences were considered significant if P < 0.05.
Results
Storage Stability of Exosome-sized Vesicles
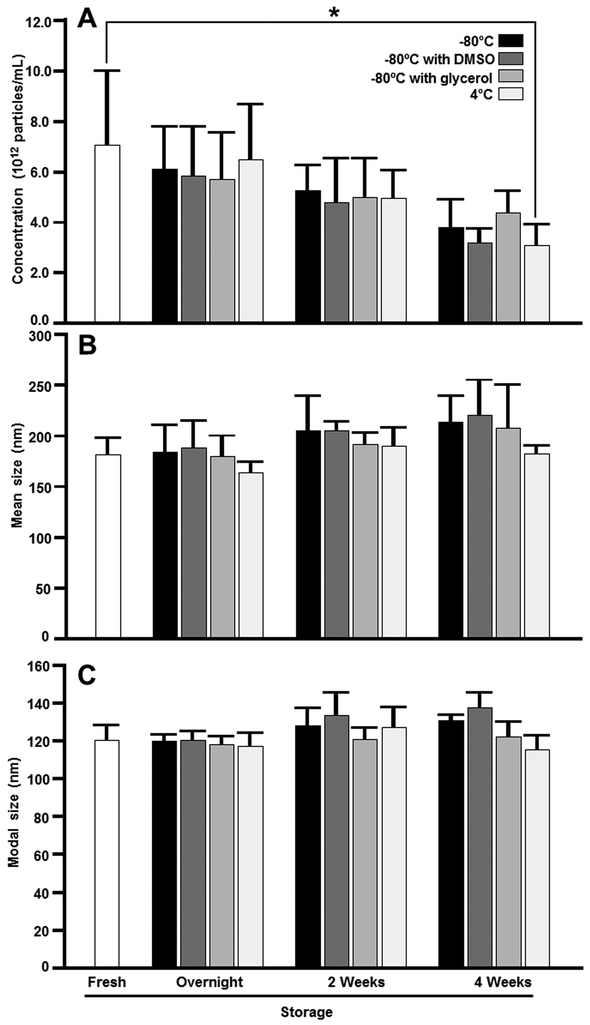
Storage caused a loss of exosome-sized vesicles in human milk. When human milk was stored at 4°C, the vesicle count decreased progressively to 99% ± 17% (overnight), 84% ± 27% (2 weeks), and 49% ± 13% (4 weeks, P = 0.03) of that in fresh milk (Figure 1A). The losses of exosome-sized vesicles at 4 weeks were not statistically significant in frozen samples or in the presence of preservatives: 71% ± 45%, 59% ± 29% and 78% ± 35% of exosome-sized vesicles were recovered if human milk was stored at −80°C, −80°C with DMSO and −80°C with glycerol, respectively. We conducted a power calculation (α = 0.05, β = 0.8) using the mean ± SD of exosome-sized vesicles in fresh milk and the null hypothesis that storage does not affect the count of exosome-sized vesicles in human milk. One hundred forty-eight samples are needed to detect a 25% loss of vesicles. The moderate increases in vesicle size during storage were not statistically significant (Figure 1B, C). Storage did not affect the count and size of exosome-sized vesicles in bovine milk (Supplemental Figure 1; Supplemental Digital Content 4).
Figure 1.
Concentration (A), mean size (B) and modal size (C) of exosome-sized vesicles in human milk stored at −80°C, −80°C with 5% DMSO, −80°C with 10% glycerol, and 4°C compared with fresh human milk. Data are expressed as means ± SEM (n = 3 biological repeats, *P = 0.03).
Exosome Authentication
Vesicles purified from human milk are exosomes, whereas those from formulas are mostly casein micelles. Human milk contained 2.18 ± 1.53 × 1011 vesicles per mL with mean and modal sizes of 149.7 ± 20.7 nm and 126.7 ± 6.7 nm, respectively (n = 3). Formula 1 contained 2.72 ± 0.13 × 1010 vesicles per mL with mean and modal sizes of 140.8 ± 4.5 nm and 128.6 ± 9.6 nm, respectively (n = 3). Formula 2 contained 1.22 ± 0.08 × 1011 vesicles per mL with mean and modal sizes of 199.8 ± 4.5 nm and 170.6 ± 22.6 nm, respectively (n =3). Formula 3 contained 2.14 ± 0.22 × 1010 vesicles per mL with mean and modal sizes of 142.7 ± 7.2 nm and 118.6 ± 6.0 nm, respectively (n = 3). Exosomes purified from human milk tested positive for CD9, CD63, and Alix, and negative for histone H3 and alpha-tubulin (Supplemental Figure 2A, B; Supplemental Digital Content 5). There was little evidence of contamination with EVs larger than expected for exosomes (Supplemental Figure 2C). The vesicles purified from infant formulas tested negative for exosomes markers CD63, HSP70 and Alix (Supplemental Figure 2D and Supplemental Figure 3, Supplemental Digital Content 6), and tested positive for bovine caseins for formulas 2 and 3 (Supplemental Figure 2B). Alpha-lactalbumin was not detected in milk formula 1, was abundant in formula 2 and was present in trace amounts in formula 3. Consistent with this observation, formula 1 is prepared by using hydrolyzed whey protein hydrolysate, whereas formulas 2 and 3 list nonfat milk as major ingredient (Supplemental Table 2, Supplemental Digital Content 2). We detected trace amounts of alpha-tubulin (marker for microvesicles), integrin β (found in most classes of EVs) and butyrophilin (marker for fat globules) in vesicles purified from infant formulas and human and bovine milk (Supplemental Figure 2B). Human β-casein was detected in human milk but not in vesicles purified from human milk.
MicroRNA Expression
Two hundred twenty-one mature microRNAs were identified in the three samples of exosomes isolated from fresh human milk; 84 microRNAs were common among samples (Supplemental Figure 4, Supplemental Digital Content 7). The ten most abundant microRNAs accounted for more than 70% of the total sequencing reads (Table 1). Milk samples stored at 4°C for less than 24 h prior to exosome isolation yielded quantities of small RNAs sufficient for microRNA-sequencing analysis (~ 12 ng/μL), whereas the yield was low if milk was stored at −80°C or for longer than 24 h (< 1 ng/μL).
Table 1.
Normalized counts for top ten microRNAs detected in human milk exosomes (n = 3)
| microRNA | Normalized Counts1 | Cumulative % |
|---|---|---|
| hsa-miR-30d-5p* | 238,233 ± 122,360 | 29.9% |
| hsa-let-7b-5p | 111,414 ± 47,752 | 43.9% |
| hsa-let-7a-5p | 72,111 ± 17,294 | 53.0% |
| hsa-miR-125a-5p* | 24,679 ± 2,485 | 56.1% |
| hsa-miR-21-5p | 23,222 ± 26,043 | 59.0% |
| hsa-miR-423-5p* | 20,667 ± 8,638 | 61.6% |
| hsa-let-7g-5p | 19,890 ± 5,877 | 64.1% |
| hsa-let-7f-5p | 19,022 ± 951 | 66.5% |
| hsa-miR-30a-5p | 18,589 ± 16,448 | 68.8% |
| hsa-miR-146b-5p | 17,246 ± 10,660 | 71.0% |
In expression analysis by miRDeep2, values are normalized by library size and multiplied by a factor of 106 which corresponds to counts per million mapped microRNA reads.
Confirmed by RT-qPCR.
Column Contamination
No contamination with microRNAs was detected in MinElute columns. All Ct values above the detection limit in RT-qPCR. Ct values of miR-30d-5p were 34.4 ± 0.3 and 34.0 ± 0.2 in non-treated and hypochlorite-treated spin columns, respectively. Ct values of miR-423-5p were 32.5 ± 2.4 and 35.0 ± 1.0 in non-treated and hypochlorite-treated spin columns, respectively. miR-125a-5p was not detectable after 40 cycles in both non-treated and hypochlorite-treated spin columns. The external standard, miSpike produced Ct values close to the detection limit in non-treated (28.5 ± 1.1) and hypochlorite-treated (29.9 ± 0.5) spin columns in RT-qPCR (n = 3, P > 0.05).
MicroRNA Analysis in Human Milk, Milk Exosomes and Infant Formulas
RT-qPCR analysis of microRNAs produced expression patterns similar to those observed by sequencing analysis, although the expression of miR-30d-5p and miR-423-5p were near the detection limit of RT-qPCR (Supplemental Figure 5A, Supplemental Digital Content 8). Analysis of exosome-sized vesicles from infant formulas resulted in Ct values above detection limit for miR-30d-5p and miR-423-5p (> 29 cycles) and no signal after 40 cycles for miR-125a-5p. MicroRNAs were detected in whole human milk but not in non-fractionated formulas (Supplemental Figure 5B). Expression was greater in unfractionated milk compared to milk exosomes (Supplemental Figure 5A vs. 5B).
Discussion
This study advances the field of human milk exosomes and their microRNA cargos as follows. First, human milk exosomes and microRNA cargos can be analyzed using small volumes of human milk. Second, storage conditions need to be taken into account before engaging in large-scale studies of exosomes and their cargos in human milk. Third, exosome-sized particles in infant formulas are not exosomes. Fourth, the concentration of microRNAs is below detection limit in infant formulas and in exosome-sized particles isolated from formulas. Fifth, we could not reproduce observations that RNeasy MinElute spin columns are contaminated with microRNAs [14].
To date, only a few studies have explored the phenotypes of milk exosome depletion in infants or suckling animals. Infants fed soy formula experienced a slight delay in cognitive development compared with breastfed infants in a longitudinal study [16]. Assessment of exosome-dependent phenotypes will benefit from the availability of microRNA data collected using protocols that adhered to current standards of sample storage and exosome isolation and authentication. This paper laid the groundwork for future studies of exosomes and their microRNA cargos in large cohorts of women with a high degree of confidence that discoveries will stand the test of time.
Supplementary Material
Supplemental Figure 1, Supplemental Digital Content 4. Concentration (A), mean size (B) and modal size (C) of exosome-sized vesicles in bovine milk stored at −80°C, −80°C with 5% DMSO, −80°C with 10% glycerol, and 4°C. Data are expressed as means ± SEM (n = 3).
Supplemental Figure 2, Supplemental Digital Content 5. Characterization of extracellular vesicles isolated from human milk (HM) and infant formulas (F1, F2 and F3). (A) CD63, CD9 and Alix (exosome markers) and histone H3 (negative control) in human milk exosomes. (B) α-Lactalbumin, α-casein, β-casein, α-tubulin, integrin β and butyrophilin in vesicles purified from infant formulas and human milk exosomes (HME). (C) Size distribution of human milk exosomes. (D) CD63, heat shock protein 70 (HSP70) and Alix in vesicles purified from infant formulas and human and bovine milk exosomes (BME, controls). D, cell debris; HMF, human milk fat; M, protein marker; MVs, microvesicles; SPs, supernatant from HME precipitation.
Supplemental Figure 3, Supplemental Digital Content 6. Exosome marker proteins in human and bovine milk exosomes and infant formulas. Images depict the full gels of the images depicted in Supplemental Figure 2D. CD63, HSP70 and Alix were analyzed by Western blotting in vesicles purified from three milk formulas and human and bovine milk exosomes (controls). BME, bovine milk exosomes; F1, F2 and F3, milk formulas 1, 2 and 3, respectively; HME, human milk exosomes; HSP70, heat shock protein 70; M, protein marker.
Supplemental Figure 4, Supplemental Digital Content 7. Venn diagram of mature microRNAs detected in human milk exosomes by microRNA-sequencing analysis (n = 3).
Supplemental Figure 5, Supplemental Digital Content 8. MicroRNA analysis by quantitative reverse transcriptase PCR. (A) Analysis of microRNAs in human milk and three brands of infant formula. (B) Analysis of microRNAs in human milk exosomes and in exosome-sized vesicles isolated from three brands of infant formula. Data are expressed as mean ± SD (n = 4 milk and milk exosomes; n = 3 formulas and exosome-sized vesicles from formulas). N.D. denotes not detectable.
Supplemental Table 1, Supplemental Digital Content 1. Definition of extracellular vesicles in this paper and their characteristics [1-3].
Supplemental Table 2, Supplemental Digital Content 2. Macronutrient content (per 148 mL reconstitution volume) and ingredients as per packing label.
Supplemental Table 3, Supplemental Digital Content 3. Primers used in real-time PCR analysis.
What is Known
Exosomes and their microRNA cargos from milk are bioavailable.
Dietary depletion of milk exosomes and their microRNA cargos elicits phenotypes in infants and piglets.
Exosome preparations may be contaminated with non-exosome particles of similar size.
What is New
Storage of human milk is a confounder in the analysis of exosomes and their cargos.
MicroRNA-sequencing analysis suggests that 10 microRNAs account for more than 70% of the 221 mature microRNAs detected in exosomes from human milk.
Exosome-sized particles in infant formulas are likely casein micelles, and the microRNA content in formulas is below detection limit.
This paper provides guidelines for the analysis of vesicles and RNA in human milk.
Acknowledgements
The authors acknowledge the use of the Biomedical and Obesity Research Core and the assistance of Dr. Steven Kachman in the Nebraska Center for the Prevention of Obesity Disease through Dietary Molecules (NIH 1P20GM104320) at the University of Nebraska-Lincoln, and the services provided by the DNA Sequencing Core at the University of Nebraska Medical Center (NIH P20GM103427, 1P30GM110768 and P30CA036727). The authors acknowledge the Holland Computing Center at the University of Nebraska-Lincoln for providing computational support.
Source of Funding: JZ receives research support from the National Institute of Food and Agriculture (NIFA), U.S. Department of Agriculture, under award numbers 2015-67017-23181 and NIFA2016-67001-25301, National Institutes of Health (NIH) grant 1P20GM104320, the University of Nebraska Agricultural Research Division (Hatch Act), U.S. Department of Agriculture multistate group W3002, and the University of Nebraska President’s Office. JC receives research support from the University of Nebraska President’s Office. JZ serves as a consultant for PureTech Health, Inc.
Abbreviations:
- bta
Bos taurus
- Ct
cycle threshold
- DMSO
dimethylsulfoxide
- EVs
extracellular vesicles
- hsa
Homo sapiens
- miR
microRNA
- N.D.
not detectable
- RT-qPCR
quantitative reverse transcription PCR
Footnotes
Conflicts of Interest: None declared.
References
- [1].Abels ER, Breakefield XO. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol Neurobiol 2016;36:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zonneveld MI, Brisson AR, van Herwijnen MJ, et al. Recovery of extracellular vesicles from human breast milk is influenced by sample collection and vesicle isolation procedures. J Extracell Vesicles 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lotvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zempleni J, Sukreet S, Zhou F, et al. Milk-Derived Exosomes and Metabolic Regulation. Annu Rev Anim Biosci 2018. (in press). [DOI] [PubMed] [Google Scholar]
- [6].National Institutes of Health, Human milk composition - biological environmental, nutritional, and methodological considerations, Bethesda, MD, 2017. https://www.niddk.nih.gov/news/events-calendar/Pages/workshop-human-milk-composition-biological-environmental-nutritional-methodological-considerations.aspx (accessed 12/8/2017). [Google Scholar]
- [7].Wolf T, Baier SR, Zempleni J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma caco-2 cells and rat small intestinal IEC-6 cells. J Nutr 2015;145:2201–06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kusuma Jati R, Manca S, Friemel T, et al. Human vascular endothelial cells transport foreign exosomes from cow’s milk by endocytosis. Am J Physiol Cell Physiol 2016;310:C800–C07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].National Institutes of Health, Extracellular RNA Consortium, Bethesda, MD, 2016, http://commonfund.nih.gov/Exrna/index (accessed 12/8/2016). [Google Scholar]
- [10].Babraham Bioinformatics, FastQC, 2017, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed 8/24/2017).
- [11].Martin M, Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads, EMBnetjournal, vol. 17, 2011, pp. 10–12. [Google Scholar]
- [12].Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Friedlander MR, Mackowiak SD, Li N, et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 2012;40:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Heintz-Buschart A, Yusuf D, Kaysen A, et al. Isolation of nucleic acids from low biomass samples: detection and removal of sRNA contaminants. BMC biology 2018;16:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang L, Sadri M, Giraud D, et al. RNase H2-dependent polymerase chain reaction and elimination of confounders in sample collection, storage, and analysis strengthen evidence that microRNAs in bovine milk are bioavailable in humans. J Nutr 2018;148:153–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Andres A, Cleves MA, Bellando JB, et al. Developmental status of 1-year-old infants fed breast milk, cow’s milk formula, or soy formula. Pediatrics 2012;129:1134–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1, Supplemental Digital Content 4. Concentration (A), mean size (B) and modal size (C) of exosome-sized vesicles in bovine milk stored at −80°C, −80°C with 5% DMSO, −80°C with 10% glycerol, and 4°C. Data are expressed as means ± SEM (n = 3).
Supplemental Figure 2, Supplemental Digital Content 5. Characterization of extracellular vesicles isolated from human milk (HM) and infant formulas (F1, F2 and F3). (A) CD63, CD9 and Alix (exosome markers) and histone H3 (negative control) in human milk exosomes. (B) α-Lactalbumin, α-casein, β-casein, α-tubulin, integrin β and butyrophilin in vesicles purified from infant formulas and human milk exosomes (HME). (C) Size distribution of human milk exosomes. (D) CD63, heat shock protein 70 (HSP70) and Alix in vesicles purified from infant formulas and human and bovine milk exosomes (BME, controls). D, cell debris; HMF, human milk fat; M, protein marker; MVs, microvesicles; SPs, supernatant from HME precipitation.
Supplemental Figure 3, Supplemental Digital Content 6. Exosome marker proteins in human and bovine milk exosomes and infant formulas. Images depict the full gels of the images depicted in Supplemental Figure 2D. CD63, HSP70 and Alix were analyzed by Western blotting in vesicles purified from three milk formulas and human and bovine milk exosomes (controls). BME, bovine milk exosomes; F1, F2 and F3, milk formulas 1, 2 and 3, respectively; HME, human milk exosomes; HSP70, heat shock protein 70; M, protein marker.
Supplemental Figure 4, Supplemental Digital Content 7. Venn diagram of mature microRNAs detected in human milk exosomes by microRNA-sequencing analysis (n = 3).
Supplemental Figure 5, Supplemental Digital Content 8. MicroRNA analysis by quantitative reverse transcriptase PCR. (A) Analysis of microRNAs in human milk and three brands of infant formula. (B) Analysis of microRNAs in human milk exosomes and in exosome-sized vesicles isolated from three brands of infant formula. Data are expressed as mean ± SD (n = 4 milk and milk exosomes; n = 3 formulas and exosome-sized vesicles from formulas). N.D. denotes not detectable.
Supplemental Table 1, Supplemental Digital Content 1. Definition of extracellular vesicles in this paper and their characteristics [1-3].
Supplemental Table 2, Supplemental Digital Content 2. Macronutrient content (per 148 mL reconstitution volume) and ingredients as per packing label.
Supplemental Table 3, Supplemental Digital Content 3. Primers used in real-time PCR analysis.