Abstract
Background: Endometrial carcinoma (EC) still threatens the health of women. Thus, to explore how long intergenic non-protein coding RNA 01220 regulates the development of EC.
Methods: Whole genome expression profile data of EC and paracancerous tissues in TCGA database were downloaded. LINC01220 expression in EC and paracancerous tissues of patients in our hospital were detected by qRT-PCR. Furthermore, the relationship between LINC01220 expression and clinicopathological features of EC patients was analyzed. After transfection with sh-LINC01220 and pcDNA-MAPK11 (mitogen-activated protein kinase) in EC cells, proliferative, colony formation abilities and apoptosis were determined by cell counting kit-8 (CCK-8), colony formation assay and flow cytometry, respectively. Western blot was conducted to determine the regulatory role of LINC01220 on MAPK11.
Results: TCGA data showed that LINC01220 expression is markedly higher in EC tissues than that of paracancerous tissues, which was consistent without detection in EC patients of our hospital. LINC01220 expression was positively correlated to pathological grade and International Federation of Gynecology and Obstetrics (FIGO) stage of EC patients. After knockdown of LINC01220 in EC cells, proliferative and colony formation abilities decreased, whereas apoptotic rate increased. Cor function analysis revealed the positive correlation between LINC01220 and MAPK11 in EC. MAPK11 expression was regulated by LINC01220 in EC cells. Overexpression of MAPK11 can reverse the tumor suppressing effect of LINC01220 on EC.
Conclusions: LINC01220 promotes EC development by stimulating proliferation and inhibiting apoptosis of EC cells through up-regulating MAPK11.
Keywords: Apoptosis, endometrial carcinoma, LINC01220, MAPK11, Proliferation
Background
Endometrial carcinoma (EC) belongs to malignant tumor occurring in the endometrium, and most of EC originates in the endometrial glands. EC is common in the female reproductive tract, accounting for 20–30% of female germ cell tumors [1]. In developed countries, EC ranks fourth in female malignant tumors, second only to breast cancer, colorectal cancer and lung cancer. It ranks seventh in female tumors in developing countries [2]. Current treatments of EC mainly include hormone therapy, hysterectomy and combination therapy of chemotherapy and radiotherapy. These measures are effective for early-stage EC. However, advanced, poorly differentiated or special types of EC are difficult to be treated and may ultimately produce poor results [3,4]. Most EC patients experience tumor recurrence or metastasis [5]. Therefore, exploring the molecular mechanisms, identifying new diagnostic and prognostic markers, and exploring new therapeutic strategies for EC are critical to improve the clinical outcomes.
Long noncoding RNA (lncRNA) is a kind of RNA molecule in eukaryotes with a transcript of over 200 nt in length, and it could not encode protein. LncRNA could regulate gene expressions at epigenetic, transcriptional and post-transcriptional levels [6–8]. LncRNAs are divided into five types according to the positions, including sense lncRNA (sense lncRNA), antisense lncRNA, bidirectional lncRNA (bidirectional lncRNA), intronic lncRNA (intronic lncRNA) and larger intergenic noncoding RNA (lincRNA). The ratio of lncRNAs in all RNAs is far more higher than those of mRNAs and miRNAs, which accounts for over 90%, whereas the ratio of mRNAs is only approximately 2% [7,9]. Accumulating studies have shown that lncRNA exerts an important role in tumor development, which undoubtedly brings new hopes in tumor treatment [10–12]. Clarification of the specific mechanism of lncRNAs in EC development contributes to improve EC treatment, which is of great significance in enhancing survival rate of EC patients.
Mitogen-activated protein kinases (MAPKs) are a group of protein kinases containing a threonine/tyrosine residue with a molecular weight of 40–46 kDa [13]. MAPKs are regulated by stimuli, such as extracellular growth factors and differentiation factors, and exert a key role in cell proliferation and differentiation. In the absence of stimulation, intracellular MAPK is dephosphorylated. MAPK is activated only when its threonine and tyrosine residues are phosphorylated. Meanwhile, MAPK is a common pathway or junction for the transmission of intracellular information about the transcription and expressions of genes to the nucleus. MAPK consists of four subunits, namely MAPK14, MAPK11, MAPK12 and MAPK13, of which MAPK14 and MAPK11 are the most important types. Studies have shown that MAPKs are greatly involved in the occurrence and development of various tumors. MAPK11 is highly expressed in breast cancer cells, and is capable of enhancing osteoclast formation and bone resorption [14]. Inhibition of MAPK14 is effective in reducing metastasis of breast cancer cells [15,16]. In glioblastoma, apoptosis can be inhibited by the MAPK and p53 pathways [17]. MAPK can be used as a potential target for the treatment of hematological malignancies [18]. However, the mechanism of MAPK in EC is rarely explored.
Methods
Sample collection
Thirty EC patients treated in The Second Affiliated Hospital of Nanjing Medical University from the years 2014 to 2018 were enrolled and they did not receive any preoperative treatments. All harvested EC tissues were pathologically diagnosed. Paracancerous tissues were harvested 2 cm away from the distal end of the EC tissue and confirmed without any infiltration of tumor tissues. Samples were preserved in liquid nitrogen for extracting RNA and proteins. Sample collection was obtained from the written informed consent of patients and approved by the Ethics Committee of Nanjing Medical University.
Data acquisition
LncRNA expression data of EC were downloaded from the TCGA database (https://tcga-data.nih.gov/tcga/) using the Bioconductor/TCGA biolinks function package. Differentially expressed lncRNAs were analyzed using the edger package.
Gene set enrichment analysis
Analysis was performed using GSEA 2.2.1 software. Dataset of c2.cp.kegg.v5.1.symbols.gmt was obtained from the MsigDB database in Gene set enrichment analysis (GSEA) website and the enrichment analysis was conducted using the default weighted enrichment statistics method. The random combination number was set as 1000 times.
Cell culture and transfection
Three EC cell lines (KLE, Ishikawa and RL95-2) were obtained from ATCC. Cells were cultured in DMEM containing 10% FBS (fetal bovine serum), 100 U/ml penicillin and 100 μg/ml streptomycin (HyClone, South Logan, UT, U.S.A.). Cells were incubated in a 5% CO2 incubator at 37°C.
For cell transfection, 1.5 ml of serum-free DMEM and 500 μl mixture containing transfection reagent and Lipofectamine™ 2000 were added in each well. After 4–6 h, complete medium was replaced. Plasmids of sh-LINC01220, shRNA-NC, pcDNA-MAPK11 and pcDNA-NC were constructed by GenePharma (Shanghai, China). Plasmid sequence was: sh-LINC01220: 5′-GAGGCCUAUAAUGCAAAGATT-3′.
qRT-PCR
Total RNA in 30 mg EC tissues or 2 × 105 EC cells was extracted using TRIzol method for reverse transcription according to the instructions of PrimeScript RT reagent Kit (Takara, Tokyo, Japan). RNA concentration was detected using spectrometer. qRT-PCR was then performed based on the instructions of SYBR Premix ExTaq™ (Takara, Tokyo, Japan). The relative gene expression was calculated using 2−ΔCt method. Primers used in the study were as follows:
GAPDH (forward): 5′-CGGAGTCAACGGATTTGGTCGTAT-3′;
GAPDH (reverse): 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′;
LINC01220 (forward): 5′-CCTTCCATGCTGAGCTGCT-3′;
LINC01220 (reverse): 5′-TGTTGGTGGGATCCAGGAAA-3′;
MAPK11 (forward): 5′-GCTGTCTCGCCCTTTCCAATC-3′;
MAPK11 (reverse): 5′-CGTGCTTCAGGTGCTTGAGTAG-3′.
Western blot
EC cells were lysed to harvest total cellular protein, followed by determination of total protein concentration. An equal amount of protein sample was loaded on to a 10% SDS/PAGE and then transferred to a polyvinylidene fluoride (PVDF) membrane. Membranes were blocked with skim milk, and incubated with primary antibody (Cell Signaling Technology, Danvers, MA, U.S.A.) overnight at 4°C. On other day, membranes were incubated with horseradish peroxidase (HRP)–conjugated secondary antibody for 2–3 h at room temperature. Finally, an image of the protein band was captured by the Tanon detection system using ECL reagent (Thermo, Waltham, MA, U.S.A.).
Cell counting kit-8
Transfected EC cells for 24 h were seeded into 96-well plates with 2 × 105 per well. Ten microliters of cell counting kit-8 solution (CCK-8, Dojindo, Kumamoto, Japan) was added in each well after cell culture for 2 days. The absorbance at 450 nm of each sample was measured by a microplate reader (Bio-Rad, Hercules, CA, U.S.A.).
Apoptotic determination
EC cells were seeded in the six-well plates and transfected for 72 h. Cells were incubated with 5 ml of pre-cooled 70% ethanol overnight at 4°C. On other day, 3 μl of RNase-A was added until the dose of 50 μg/ml for digestion in warm water bath for 30 min. Propidium iodide was then added until the dose of 100 μg/ml and incubated in ice without light for 30 min. Apoptotic rate was determined using flow cytometry.
Colony formation assay
EC cells in the logarithmic growth phase were seeded in the culture dish with 1000 cells per dish, respectively. After cell culture for 2–3 weeks until visible colony formation, cells were washed with PBS and fixed with 4% paraformaldehyde. Colonies were stained with Crystal Violet for 10–30 min and captured using the microscope.
Statistical analyses
SPSS 22.0 statistical software was used for data analysis. Data were expressed as mean ± standard deviation (x ® ± s). Differences between the two groups were compared with the t test. Cor function was used for comparison of gene correlations. A gene set with a false discovery rate (FDR) < 0.25 was utilized as a significantly enriched gene set in the GSEA. P<0.05 considered the difference was statistically significant.
Results
High expression of LINC01220 in EC
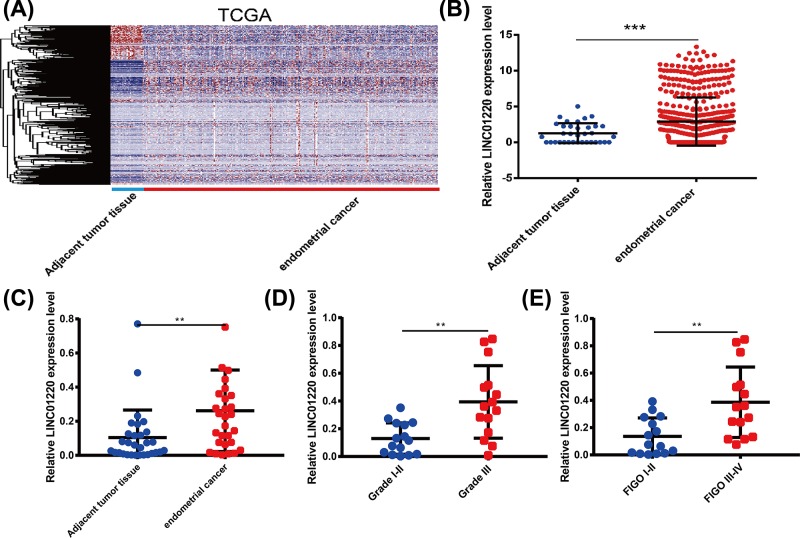
To explore the role of lncRNAs in EC, we first downloaded the whole genome expression profile data of EC and paracancerous tissues in TCGA database (Figure 1A). Among them, LINC01220 expression was markedly higher in EC tissues than that of paracancerous tissues, and the differential expression ratio was the highest (Figure 1B). Next, qRT-PCR was performed to detect the expression of LINC01220 in EC and paracancerous tissues of patients in our hospital. The results also showed the higher expression of LINC01220 in EC tissues than that of paracancerous tissues in 30 EC patients enrolled in our hospital (Figure 1C). Furthermore, the relationship between LINC01220 expression and clinicopathological features of EC patients was analyzed. As the data indicated, LINC01220 expression was associated with International Federation of Gynecology and Obstetrics (FIGO) of EC patients. EC patients with Grade III had higher expression of LINC01220 than those with Grades I–II (Figure 1D). Besides, EC patients with FIGO III–IV showed higher expression of LINC01220 compared with those with FIGO I–II (Figure 1E). These data suggested that LINC01220 may be closely related to the development of EC.
Figure 1. High expression of LINC01220 in EC.
(A) Heat map of differentially expressed lncRNAs in EC of TCGA database. (B) TCGA data showed that LINC01220 expression is markedly higher in EC tissues than that of paracancerous tissues. (C) LINC01220 expression was higher in EC tissues than that of paracancerous tissues of 30 EC patients enrolled at our hospital. (D) EC patients with Grade III had higher expression of LINC01220 than those with Grade I–II. (E) EC patients with FIGO III–IV showed higher expression of LINC01220 compared with those with FIGO I–II. **P<0.01, ***P<0.001.
Knocking down of LINC01220 inhibited proliferation and induced apoptosis of EC cells
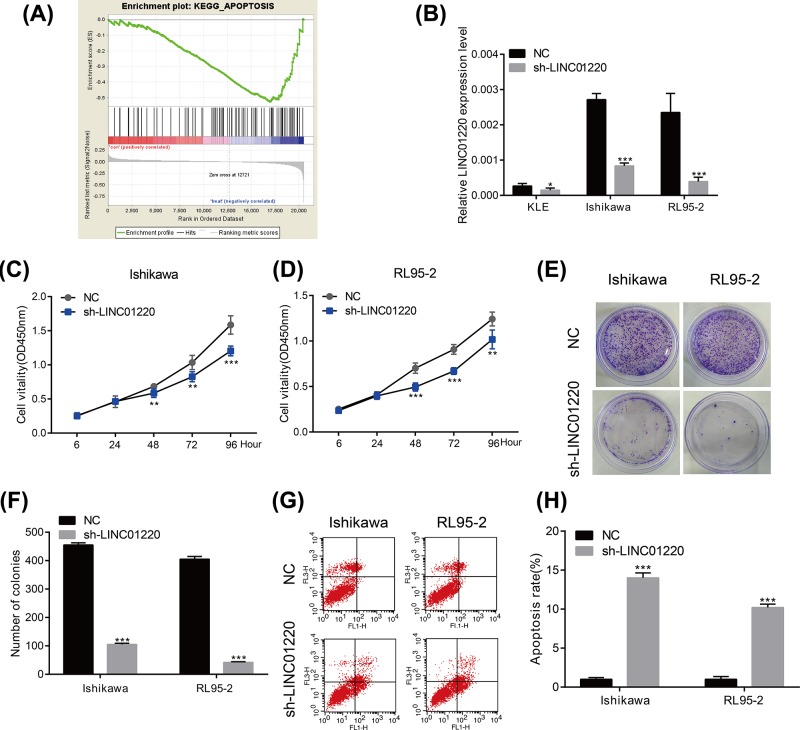
To further explore the biological function of LINC01220, GSEA was conducted. The results showed that the main function of LINC01220 is enriched in apoptosis (Figure 2A). Subsequently, sh-LINC01220 was constructed and verified for its transfection efficacy in three EC cell lines. Among them, LINC01220 expression markedly decreased in Ishikawa and RL95-2 cells, which were selected for the following experiments (Figure 2B). CCK-8 results revealed that OD value of EC cells transfected with sh-LINC01220 gradually decreased after cell culture for 6, 24, 48, 72 and 96 h, respectively (Figure 2C,D). Similarly, colony formation abilities of Ishikawa and RL95-2 cells reduced after LINC01220 knockdown (Figure 2E,F). Moreover, apoptotic rate of EC cells remarkably increased after LINC01220 knockdown (Figure 2G,H). These results indicated that knockdown of LINC01220 can inhibit the proliferation and clonality of EC cells and promote apoptosis in vitro.
Figure 2. LINC01220 promoted proliferation and inhibited apoptosis of EC cells.
(A) GSEA results showed that the main function of LINC01220 is enriched in apoptosis. (B) Transfection efficacy of sh-LINC01220 in KLE, Ishikawa and RL95-2 cells. (C) OD value of Ishikawa cells transfected with sh-LINC01220 gradually decreased after cell culture for 6, 24, 48, 72 and 96 h, respectively. (D) OD value of RL95-2 cells transfected with sh-LINC01220 gradually decreased after cell culture for 6, 24, 48, 72 and 96 h, respectively. (E,F) Colony formation abilities of Ishikawa and RL95-2 cells reduced after LINC01220 knockdown. (G,H) Apoptotic rate of Ishikawa and RL95-2 cells remarkably increased after LINC01220 knockdown. *P<0.05, **P<0.01, ***P<0.001.
MAPK11 promoted EC development and is regulated by LINC01220
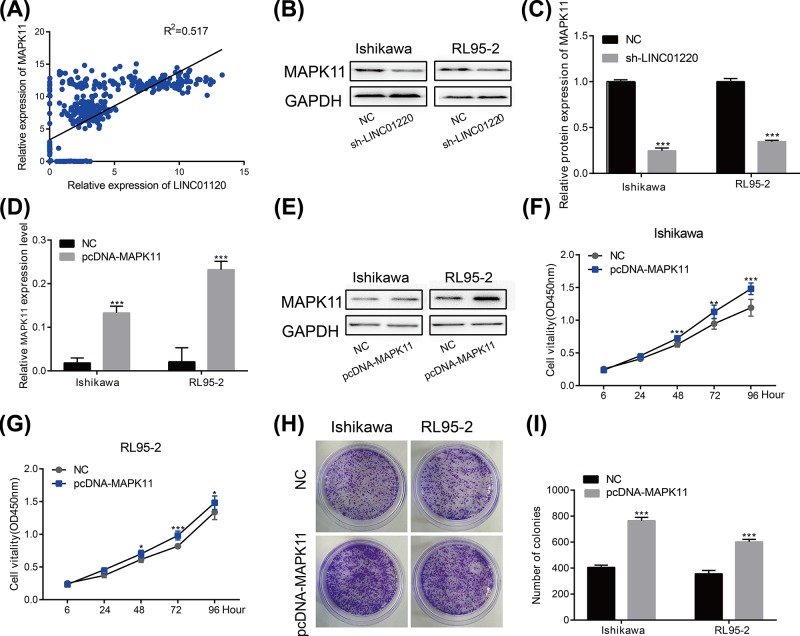
We evaluated the correlation between LINC01220 expression and the genome-wide gene in EC expression profiles by the Cor package. The results showed that LINC01220 expression is positively correlated with MAPK11 (Figure 3A). A large number of studies have reported that MAPK11 serves as an oncogene in tumors [19]. In this study, LINC01220 knockdown down-regulated protein level of MAPK11 in Ishikawa and RL95-2 cells (Figure 3B,C). Then the pcDNA-MAPK11 was transfected to Ishikawa and RL95-2 cell lines and the results showed that the mRNA and protein levels of MAPK11 were significantly up-regulated (Figure 3D,E). Furthermore, MAPK11 overexpression enhanced proliferative rate of EC cells (Figure 3F,G), as well as their colony formation ability (Figure 3H,I). The above results indicated that MAPK11 promotes the progression of EC and is regulated by LINC01220.
Figure 3. MAPK11 promoted EC development.
(A) Cor function results showed that LINC01220 expression is positively correlated with MAPK11. (B,C) LINC01220 knockdown down-regulated protein level of MAPK11 in Ishikawa and RL95-2 cells. (D,E) LINC01220 knockdown down-regulated mRNA level of MAPK11 in Ishikawa and RL95-2 cells. (F,G) MAPK11 overexpression enhanced proliferative rate of Ishikawa and RL95-2 cells. (H,I) MAPK11 overexpression enhanced colony formation ability of Ishikawa and RL95-2 cells. *P<0.05, **P<0.01, ***P<0.001.
MAPK11 overexpression reversed the knockdown effect of LINC01220 on EC cells
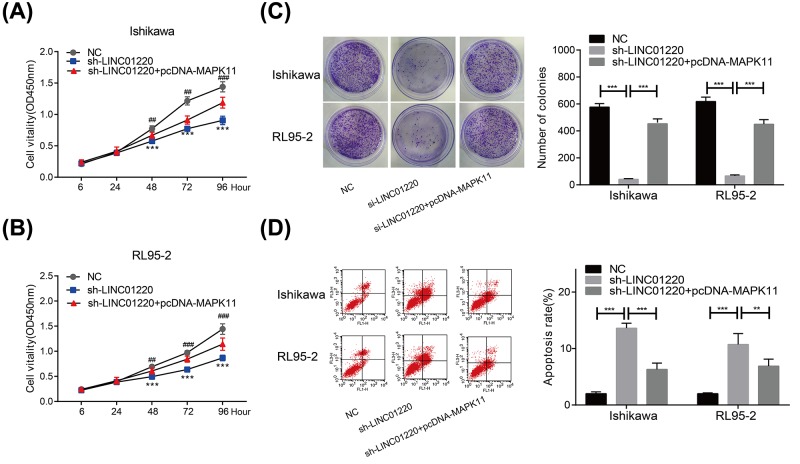
To further examine the roles of MAPK11 and LINC01220 in EC, we knocked down LINC01220 and overexpressed MAPK11 in EC cells at the same time. LINC01220 knockdown reduced proliferative ability of Ishikawa cells, which was enhanced after MAPK11 overexpression (Figure 4A). Similar results were obtained in RL95-2 cells as well (Figure 4B). Subsequently, colony formation ability was determined in co-transfected EC cells. LINC01220 knockdown markedly decreased colony formation ability of EC cells, whereas co-transfected cells showed an enhanced ability (Figure 4C). Rescue experiments were also conducted to determine cell apoptosis in co-transfected EC cells. Previous experiments have already found the stimulated cell apoptosis after LINC01220 knockdown. However, apoptotic rate of EC cells markedly decreased after co-transfection with sh-LINC01220 and pcDNA-MAPK11 (Figure 4D).
Figure 4. MAPK11 overexpression reversed the effect of LINC01220 on EC cells.
(A,B) LINC01220 knockdown reduced proliferative ability of Ishikawa and RL95-2 cells, which was enhanced after co-transfection with sh-LINC01220 and pcDNA-MAPK11. (C) LINC01220 knockdown reduced colony formation ability of Ishikawa and RL95-2 cells, which was enhanced after co-transfection with sh-LINC01220 and pcDNA-MAPK11. (D) LINC01220 knockdown induced apoptosis of Ishikawa and RL95-2 cells, which was inhibited after co-transfection with sh-LINC01220 and pcDNA-MAPK11. *P<0.05, **P<0.01, ***P<0.001, #P<0.01 (vs. sh-LINC01220), #P<0.001 (vs. sh-LINC01220).
The above results suggested that MAPK11 can reverse the tumor suppressing effect of LINC01220 on EC.
Discussion
EC is a malignant tumor of the female reproductive system. The incidence of EC has increased year by year, which has seriously threatened women’s physical and mental health. It is found that approximately 20% of EC patients have a clear family history. Besides, long-term sustained estrogen stimulation, obesity, infertility, polycystic ovary syndrome and other factors are related to the occurrence of EC. Similar to other malignant tumors, EC results from a slow and complex process involving oncogene activation and tumor-suppressor gene inhibition. Therefore, it is imperative to explore the pathogenesis of EC, so as to develop new targeted therapy.
LncRNA was originally thought to be a ‘transcriptional noise’. As it has been discovered to be dysregulated in a variety of tumor tissues, and involved in apoptosis, invasion and metastasis of tumor tissues, lncRNAs are expected to become new targets for tumor diagnosis and treatment [20]. According to the different roles of lncRNAs in tumor development, they are divided into oncogenes and tumor-suppressor genes. LncRNAs as oncogenes are usually up-regulated in tumor tissues that promote tumorigenesis. For example, lncRNA MALAT1 is highly expressed in prostate cancer, which promotes the development of prostate cancer by activating EZH2 [21]. LncRNA H19 is highly expressed in colorectal cancer cells, and can be utilized as miRNA sponges to promote the transformation of epithelial cells into stromal cells, as well as promote the proliferation and carcinogenic activities of colorectal cancer cells [22]. LncRNA-UCA1 is highly expressed in non-small cell lung cancer cells and promotes its development by targeting miR-193a-3p [23]. The second type of lncRNAs are tumor-suppressor genes, which are usually down-regulated in tumor tissues and inhibit tumorigenesis and development. For example, lncRNA NBAT-1 is lowly expressed in ovarian cancer tissues, and inhibits proliferation, migration and invasion of ovarian cancer cells [23]. LncRNA MEG3 is lowly expressed in breast cancer cells, which is capable of inhibiting proliferation, migration and transformation of epithelial cells into stromal cells by targeting miR-412 [24].
Relative studies have shown that lncRNA ABHD11-AS1 is highly expressed in EC tissues, and promotes proliferation, invasion, migration and progression into G1/S phase by up-regulating cyclin D1, CDK1, CKD2, CDK4, vEGF and other factors of EC cells [25]. LncRNA TDRG1, MEG3 and MIR22HG have been found to exert important roles in EC [26–28]. LINC01220 is located on chromosome 14q24.3 and has not been reported in tumor diseases yet. In the present paper, TCGA data analyses found that LINC01220 is highly expressed in EC tissues than paracancerous tissues, which was consistent with the detection in EC patients of our hospital. Besides, LINC01220 expression was positively correlated with pathological grade and FIGO staging of EC patients. In vitro experiments indicated that LINC01220 knockdown decreased proliferative and colony formation abilities, but induced apoptosis of EC cells. These results strongly suggested that LINC01220 is expected to be a new target for the diagnosis of EC. MAPKs are a family of conserved serine/threonine protein kinases that are involved in basic cellular development processes, such as proliferation, differentiation, stress response, apoptosis and survival [29]. Abnormal activation of the MAPK pathway has been found in a variety of human tumors, indicating its vital role in tumorigenesis. MAPK activation promotes cancer progression and resistance to chemotherapeutic drugs [30,31]. On the contrary, inhibition of MAPK pathway inhibits tumor metastasis and tumor-related angiogenesis [32,33]. However, studies of MAPK pathway in EC have rarely been reported. Cor function analysis conducted in the present study showed that LINC01220 is positively correlated with MAPK11 in expression profiles of EC. Ater LINC01220 knockdown in EC cells, both protein and mRNA levels of MAPK11 markedly decreased. Overexpression of MAPK11 in EC cells elevated proliferative and colony formation abilities. More importantly, MAPK11 can reverse the effect of LINC01220 on endometrial cancer cells. These results suggested that LINC01220 can promote the progression of EC by regulating MAPK11.
However, the present study still had certain deficiencies. First of all, this research was mainly carried out by in vitro cell model. In vivo functional researches could further improve the credibility of our results. Second, the way that LINC01220 regulated MAPK11 in EC cells has not been fully explained. In the future experiments, cell localization and potential binding protein of LINC01220 are needed to be explored.
In summary, LINC01220 promotes EC development by stimulating proliferation and inhibiting apoptosis of EC cells through up-regulating MAPK11.
Acknowledgments
We would like to express our sincere thanks to all those patients who donated to this work.
Abbreviations
- ABHD11-AS1
ABHD11 antisense RNA 1
- ATCC
American type culture collection
- CCK-8
cell counting kit-8
- CDK
cyclin dependent kinase
- EC
endometrial carcinoma
- EZH2
enhancer of zeste 2 polycomb repressive complex 2 subunit
- FIGO
International Federation of Gynecology and Obstetrics
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GSEA
gene set enrichment analysis
- LINC01220
long intergenic non-protein coding RNA 1220
- lncRNA
long noncoding RNA
- MAPK
mitogen-activated protein kinase
- MEG3
maternally expressed 3
- MIR22HG
microRNA-22 host gene
- NBAT1
neuroblastoma associated transcript 1
- OD
optical density
- qRT-PCR
quantitative real time polymerase chain reaction
- TCGA
the cancer genome atlas
- TDRG1
testis development related 1
- vEGF
vascular endothelial growth factor
Author Contribution
X.Y. designed the experiments. Y.L. and C.K. conducted the experiments. C.W. and B.X. provided research materials and methods. Y.W. and S.L. analyzed data. Y.L. wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Key Projects of Science and Technology Development Fund of Nanjing Medical University [grant number 2015NJMUZD083].
References
- 1.Liao Y., Lu W., Che Q., Yang T., Qiu H., Zhang H.. et al. (2014) Sharp1 suppresses angiogenesis of endometrial cancer by decreasing hypoxia-inducible factor-1α level. PLoS ONE 9, e99907. 10.1371/journal.pone.0099907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schreuders E.H., Ruco A., Rabeneck L., Schoen R.E., Sung J.J.Y., Young G.P.. et al. (2015) Colorectal cancer screening: a global overview of existing programmes. Gut 64, 1637–1649 10.1136/gutjnl-2014-309086 [DOI] [PubMed] [Google Scholar]
- 3.Humber C., Tierney J., Symonds P., Collingwood M., Kirwan J., Williams C.. et al. (2005) Chemotherapy for advanced, recurrent or metastatic endometrial carcinoma. Cochrane Database Syst. Rev. 5, D3915. [DOI] [PubMed] [Google Scholar]
- 4.Hill E.K. and Dizon D.S. (2012) Medical therapy of endometrial cancer: current status and promising novel treatments. Drugs 72, 705. 10.2165/11631840-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 5.Oza A.M., Elit L., Tsao M.S., Kamel-Reid S., Biagi J., Provencher D.M.. et al. (2011) Phase II study of temsirolimus in women with recurrent or metastatic endometrial cancer: a trial of the ncic clinical trials group. J. Clin. Oncol. 29, 3278–3285 10.1200/JCO.2010.34.1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo X., Gao L., Wang Y., Chiu D.K., Wang T. and Deng Y. (2015) Advances in long noncoding rnas: identification, structure prediction and function annotation. Brief Funct. Genomics 15, 38–46 10.1093/bfgp/elv022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Xu G., Chen W., Pan Q., Huang K., Pan J.. et al. (2015) Detection of long-chain non-encoding rna differential expression in non-small cell lung cancer by microarray analysis and preliminary verification. Mol. Med. Rep. 11, 1925–1932 10.3892/mmr.2014.2944 [DOI] [PubMed] [Google Scholar]
- 8.Ma C., Shi X., Zhu Q., Li Q., Liu Y., Yao Y.. et al. (2016) The growth arrest-specific transcript 5 (gas5): a pivotal tumor suppressor long noncoding rna in human cancers. Tumour Biol. 37, 1437–1444 10.1007/s13277-015-4521-9 [DOI] [PubMed] [Google Scholar]
- 9.Li Z., Zhao X., Zhou Y., Liu Y., Zhou Q., Ye H.. et al. (2015) The long non-coding rna hottip promotes progression and gemcitabine resistance by regulating hoxa13 in pancreatic cancer. J. Transl. Med. 13, 84. 10.1186/s12967-015-0442-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J., Mo J., Luo M., Yu Q., Zhou S., Li T.. et al. (2015) C-myc-activated long non-coding rna h19 downregulates mir-107 and promotes cell cycle progression of non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 8, 12400. [PMC free article] [PubMed] [Google Scholar]
- 11.Guo W., Liu S., Cheng Y., Lu L., Shi J., Xu G.. et al. (2016) Icam-1-related non-coding rna in cancer stem cells maintains icam-1 expression in hepatocellular carcinoma. Clin. Cancer Res. 22, 2041. 10.1158/1078-0432.CCR-14-3106 [DOI] [PubMed] [Google Scholar]
- 12.Liu G., Xiang T., Wu Q.F. and Wang W.X. (2016) Long noncoding rna h19-derived mir-675 enhances proliferation and invasion via runx1 in gastric cancer cells. Oncol. Res. 23, 99–107 10.3727/096504015X14496932933575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beardmore V.A., Hinton H.J., Eftychi C., Apostolaki M., Armaka M., Darragh J.. et al. (2005) Generation and characterization of p38β (mapk11) gene-targeted mice. Mol. Cell. Biol. 25, 10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Z., He J., Liu Z., Xu J., Yi S.F., Liu H.. et al. (2014) Mapk11 in breast cancer cells enhances osteoclastogenesis and bone resorption. Biochimie 106, 24–32 10.1016/j.biochi.2014.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Q.M., Wang S., Zhang H., Lu Y.Y., Wang X.F., Motoo Y.. et al. (2009) The combination of baicalin and baicalein enhances apoptosis via the ERK/p38 MAPK pathway in human breast cancer cells. Acta Pharmacol. Sin. 30, 1648–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suarezcuervo C., Merrell M.A., Watson L., Harris K.W., Rosenthal E.L., Väänänen H.K.. et al. (2004) Breast cancer cells with inhibition of p38alpha have decreased mmp-9 activity and exhibit decreased bone metastasis in mice. Clin. Exp. Metastasis 21, 525–533 10.1007/s10585-004-3503-x [DOI] [PubMed] [Google Scholar]
- 17.Wu N., Lin X., Zhao X., Zheng L., Xiao L., Liu J.. et al. (2013) Mir-125b acts as an oncogene in glioblastoma cells and inhibits cell apoptosis through p53 and p38mapk-independent pathways. Br. J. Cancer 109, 2853–2863 10.1038/bjc.2013.672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaundar S.S. and Bendall L.J. (2010) The potential and limitations of p38mapk as a drug target for the treatment of hematological malignancies. Curr. Drug Targets 11, 823–833 [DOI] [PubMed] [Google Scholar]
- 19.Browne A.J., Göbel A., Thiele S., Hofbauer L.C., Rauner M. and Rachner T.D. (2016) P38 mapk regulates the wnt inhibitor dickkopf-1 in osteotropic prostate cancer cells. Cell Death Dis. 7, e2119. 10.1038/cddis.2016.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang G., Lu X. and Yuan L. (2014) LncRNA: a link between RNA and cancer. Biochim. Biophys. Acta 1839, 1097–1109 10.1016/j.bbagrm.2014.08.012 [DOI] [PubMed] [Google Scholar]
- 21.Wang D., Ding L., Wang L., Yu Z., Sun Z., Karnes R.J.. et al. (2015) Lncrnamalat1enhances oncogenic activities of ezh2 in castration-resistant prostate cancer. Oncotarget 6, 41045–41055 10.18632/oncotarget.5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang W.C., Fu W.M., Wong C.W., Wang Y., Wang W.M., Hu G.X.. et al. (2015) The lncrna h19 promotes epithelial to mesenchymal transition by functioning as mirna sponges in colorectal cancer. Oncotarget 6, 22513–22525 10.18632/oncotarget.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nie W., Ge H.J., Yang X.Q., Sun X., Huang H., Tao X.. et al. (2016) LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting mir-193a-3p. Cancer Lett. 371, 99–106 10.1016/j.canlet.2015.11.024 [DOI] [PubMed] [Google Scholar]
- 24.Zhang W., Shi S., Jiang J., Li X., Lu H. and Ren F. (2017) LncRNA meg3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed. Pharmacother. 91, 312–319 10.1016/j.biopha.2017.04.085 [DOI] [PubMed] [Google Scholar]
- 25.Liu Y., Wang L.L., Chen S., Zong Z.H., Guan X. and Zhao Y. (2018) Lncrna abhd11-as1 promotes the development of endometrial carcinoma by targeting cyclin d1. J. Cell. Mol. Med. 22, 3955–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S., Wang L.L., Sun K.X., Liu Y., Guan X., Zong Z.H.. et al. (2018) LncRNA tdrg1 enhances tumorigenicity in endometrial carcinoma by binding and targeting vegf-a protein. Biochim. Biophys. Acta 1864, 3013–3021 [DOI] [PubMed] [Google Scholar]
- 27.Guo Q., Qian Z., Yan D., Li L. and Huang L. (2016) LncRNA-meg3 inhibits cell proliferation of endometrial carcinoma by repressing notch signaling. Biomed. Pharmacother. 82, 589–594 10.1016/j.biopha.2016.02.049 [DOI] [PubMed] [Google Scholar]
- 28.Cui Z., An X., Li J., Liu Q. and Liu W. (2018) LncRNA miR22hg negatively regulates miR-141-3p to enhance dapk1 expression and inhibits endometrial carcinoma cells proliferation. Biomed. Pharmacother. 104, 223–228 10.1016/j.biopha.2018.05.046 [DOI] [PubMed] [Google Scholar]
- 29.Dhillon A.S., Hagan S., Rath O. and Kolch W. (2007) Map kinase signalling pathways in cancer. Oncogene 26, 3279–3290 10.1038/sj.onc.1210421 [DOI] [PubMed] [Google Scholar]
- 30.Lu M., Chen W.H., Wang C.Y., Mao C.Q. and Wang J. (2017) Reciprocal regulation of mir-1254 and c-myc in oral squamous cell carcinoma suppresses emt-mediated metastasis and tumor-initiating properties through mapk signaling. Biochem. Biophys. Res. Commun. 484, 801–807 [DOI] [PubMed] [Google Scholar]
- 31.Roberts P.J. and Der C.J. (2007) Targeting the raf-mek-erk mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 26, 3291–3310 10.1038/sj.onc.1210422 [DOI] [PubMed] [Google Scholar]
- 32.Liu Y., Zhi D., Li M., Liu D., Wang X., Wu Z.. et al. (2016) Shengmai formula suppressed over-activated ras/mapk pathway in c. Elegans by opening mitochondrial permeability transition pore via regulating cyclophilin d. Sci. Rep. 6, 38934. 10.1038/srep38934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maolin H., Chen P.N., Chu S.C., Dongyih K., Wuhsien K., Chen J.Y.. et al. (2010) Peonidin 3-glucoside inhibits lung cancer metastasis by downregulation of proteinases activities and mapk pathway. Nutr. Cancer 62, 505–516 [DOI] [PubMed] [Google Scholar]