Carbapenem-resistant Enterobacteriaceae (CRE) are resistant to most antibiotics, making CRE infections extremely difficult to treat with available agents. Klebsiella pneumoniae carbapenemases (KPC-2 and KPC-3) are predominant carbapenemases in CRE in the United States. Nacubactam is a bridged diazabicyclooctane (DBO) β-lactamase inhibitor that inactivates class A and C β-lactamases and exhibits intrinsic antibiotic and β-lactam “enhancer” activity against Enterobacteriaceae.
KEYWORDS: β-lactam, β-lactamase, K234R, KPC, diazabicyclooctane (DBO), nacubactam
ABSTRACT
Carbapenem-resistant Enterobacteriaceae (CRE) are resistant to most antibiotics, making CRE infections extremely difficult to treat with available agents. Klebsiella pneumoniae carbapenemases (KPC-2 and KPC-3) are predominant carbapenemases in CRE in the United States. Nacubactam is a bridged diazabicyclooctane (DBO) β-lactamase inhibitor that inactivates class A and C β-lactamases and exhibits intrinsic antibiotic and β-lactam “enhancer” activity against Enterobacteriaceae. In this study, we examined a collection of meropenem-resistant K. pneumoniae isolates carrying blaKPC-2 or blaKPC-3; meropenem-nacubactam restored susceptibility. Upon testing isogenic Escherichia coli strains producing KPC-2 variants with single-residue substitutions at important Ambler class A positions (K73, S130, R164, E166, N170, D179, K234, E276, etc.), the K234R variant increased the meropenem-nacubactam MIC compared to that for the strain producing KPC-2, without increasing the meropenem MIC. Correspondingly, nacubactam inhibited KPC-2 (apparent Ki [Ki app] = 31 ± 3 μM) more efficiently than the K234R variant (Ki app = 270 ± 27 μM) and displayed a faster acylation rate (k2/K), which was 5,815 ± 582 M−1 s−1 for KPC-2 versus 247 ± 25 M−1 s−1 for the K234R variant. Unlike avibactam, timed mass spectrometry revealed an intact sulfate on nacubactam and a novel peak (+337 Da) with the K234R variant. Molecular modeling of the K234R variant showed significant catalytic residue (i.e., S70, K73, and S130) rearrangements that likely interfere with nacubactam binding and acylation. Nacubactam’s aminoethoxy tail formed unproductive interactions with the K234R variant’s active site. Molecular modeling and docking observations were consistent with the results of biochemical analyses. Overall, the meropenem-nacubactam combination is effective against carbapenem-resistant K. pneumoniae. Moreover, our data suggest that β-lactamase inhibition by nacubactam proceeds through an alternative mechanism compared to that for avibactam.
INTRODUCTION
Carbapenem-resistant Enterobacteriaceae (CRE) are ranked by the Centers for Disease Control and Prevention (CDC) and the World Health Organization (WHO) as one out of three of the most “urgent” and “critical-priority” microbiological threats, respectively, to humankind (1, 2). The most current β-lactam–β-lactamase inhibitor combinations, namely, ceftazidime-avibactam and meropenem-vaborbactam, are being used to treat highly resistant CRE infections, but even these therapies do not evade resistance (3–11). Klebsiella pneumoniae carbapenemase (KPC) is a predominant underlying resistance determinant in CRE infections, which have an alarmingly high mortality rate (12–16). Despite a steadily increasing number and diversity of β-lactamases (17), KPC-2 and KPC-3 are the carbapenemases within this family that are most often identified in clinical isolates and associated with high levels of resistance (18, 19). An urgent need exists to discover new β-lactam–β-lactamase inhibitor combinations that will overcome KPC-mediated resistance in Enterobacteriaceae.
Nacubactam, formerly RG6080 and OP0595, is a member of the growing class of bridged diazabicyclooctane (DBO) β-lactamase inhibitors that differs from avibactam with the addition of an aminoethoxy group to the carbamoyl side chain present on avibactam (Fig. 1). This addition is likely responsible for the significant intrinsic antibiotic activity of nacubactam alone. Analogously to ETX2514 and the WCK 5153 and zidebactam DBOs (20, 21), nacubactam inhibits Escherichia coli penicillin binding protein 2 (PBP2) (22). Moreover, nacubactam was reported to act synergistically as a β-lactam enhancer, like mecillinam and other DOBs (WCK 5153 and zidebactam), when combined with β-lactams. This enhancer effect is the result of these combinations possessing the ability to target multiple PBPs (22).
FIG 1.
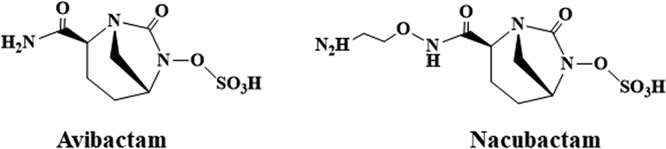
Structure of the diazabicyclooctanes (DBOs) avibactam and nacubactam.
In studies published to date, nacubactam alone was effective against Gram-negative bacteria, including Escherichia coli, Klebsiella spp., Enterobacter spp., and Citrobacter spp. (22–24). When nacubactam is combined with β-lactams, efficacy expands to most Enterobacteriaceae isolates producing extended-spectrum β-lactamases (ESBLs), AmpCs, KPCs, metallo-β-lactamases (MBLs), and OXA-48, as well those ESBL- and AmpC-producing Enterobacteriaceae that lack porins and Pseudomonas aeruginosa strains with derepressed AmpC or PER or VEB ESBLs (22–24). In addition, nacubactam effectively inhibits TEM-1, TEM-10, CTX-M-14, CTX-M-15, CTX-M-44, KPC-2, P99 (Enterobacter cloacae AmpC), PDC-1 (P. aeruginosa AmpC), and CMY-2 β-lactamases (22). The crystal structures of CTX-M-44, PDC-1, and TLA-3 with nacubactam have also been reported (22, 25). A flexible terminal amine of nacubactam that does not bind to the β-lactamase was observed in these studies.
In vivo, nacubactam combined with cefepime was effective in a murine pneumonia model infected with K. pneumoniae and murine thigh infection models using CTX-M-15-containing E. coli, KPC-2-positive K. pneumoniae, or AmpC-depressed P. aeruginosa (26–28). A meropenem-nacubactam combination was also effective in Enterobacteriaceae-infected murine models of complicated urinary tract infections (cUTI) (29). Herein, we set out to determine the efficiency of the meropenem-nacubactam combination against a panel of Klebsiella pneumoniae clinical isolates and to conduct in-depth biochemical and mechanistic studies with nacubactam, namely, to determine the structure-activity relationships (SAR) and efficacy of the DBO against the clinically significant class A carbapenemase KPC.
RESULTS AND DISCUSSION
Susceptibility testing of Klebsiella pneumoniae strains.
To determine the antibiotic susceptibility profiles of carbapenem-resistant clinical strains, 44 K. pneumoniae isolates harboring blaKPC-2 or blaKPC-3 β-lactamases were tested for susceptibility against a panel of antibiotics, including meropenem-nacubactam with the inhibitor concentration at a 1:1 ratio to the β-lactam (Table 1; Fig. 2). Most of the isolates tested were resistant to meropenem, ertapenem, aztreonam, cefepime, piperacillin-tazobactam, and levofloxacin; however, the majority of the strains tested were susceptible to aztreonam-avibactam, ceftazidime-avibactam, colistin, tigecycline, and fosfomycin. Susceptibility results for amikacin were variable across the panel of isolates. Additionally, all of the strains tested susceptible to meropenem-nacubactam (Clinical and Laboratory Standards Institute [CLSI] breakpoint of resistance for meropenem alone, ≥4 mg/liter [30]; European Committee on Antimicrobial Susceptibility Testing [EUCAST] breakpoint of resistance for meropenem alone, >8 mg/liter [31]). One strain, RB1324, that carries blaKPC-3 possessed an aberrantly high nacubactam MIC (>256 mg/liter) (Table 1), likely due to a mutation in PBP2. Despite the elevated nacubactam MIC for this outlier strain, the addition of meropenem still led to an MIC of 0.5 mg/liter. These data suggest that the meropenem-nacubactam combination is an effective combination to overcome highly resistant K. pneumoniae strains compared to other β-lactam antibiotics. To predict how well the meropenem-nacubactam combination will perform against KPC variants that have naturally evolved in the clinic or variants that have been engineered to alter structurally relevant residues that reveal the mechanistic behavior of the β-lactamase inhibition, we next investigated a well-established panel of E. coli isogenic strains, each expressing wild-type KPC-2 or 1 of 53 engineered KPC variants (Table 1).
TABLE 1.
Broth microdilution MICs for K. pneumoniae strains
| Klebsiella pneumoniae strain | β-Lactamase | MIC (mg/liter) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nacubactam | Meropenem-nacubactam (1:1) | Meropenem | Ertapenem | Aztreonam | Aztreonam-avibactama | Ceftazidime-avibactama | Cefepime | Piperacillin-tazobactama | Colistin | Amikacin | Tigecycline | Levofloxacin | Fosfomycin | ||
| RB1188 | KPC-2 | 1 | 1 | 128 | >32 | >32 | 0.12 | 0.5 | >32 | >128 | 0.25 | 32 | 0.5 | >8 | 8 |
| RB1189 | KPC-2 | 1 | 1 | 128 | >32 | >32 | 0.12 | 0.5 | >32 | >128 | 0.25 | 32 | 2 | >8 | 8 |
| RB1190 | KPC-2 | 2 | 0.25 | 32 | 32 | >32 | 0.12 | 0.5 | 32 | >128 | 0.5 | 4 | 1 | >8 | 16 |
| RB1242 | KPC-3 | 1 | 0.25 | 4 | 8 | >32 | 0.12 | 0.5 | 16 | >128 | 0.5 | 0.5 | 1 | >8 | 8 |
| RB1326 | KPC-2 | 1 | 0.25 | 8 | 16 | >32 | 0.12 | 0.5 | 8 | >128 | 0.5 | 0.5 | 1 | >8 | 32 |
| RB1193 | KPC-2 | 2 | 0.25 | 16 | 32 | >32 | 0.12 | 0.5 | 32 | >128 | 0.5 | 64 | 1 | >8 | 4 |
| RB1103 | KPC-2 | 2 | 0.5 | 32 | >32 | >32 | 0.12 | 0.25 | >32 | >128 | 1 | 32 | 0.5 | >8 | 64 |
| RB1243 | KPC-3 | 1 | 0.12 | 8 | 16 | >32 | 0.06 | 0.25 | 16 | >128 | 1 | 0.5 | 0.25 | >8 | 4 |
| RB1104 | KPC-3 | 1 | 0.5 | 16 | 32 | >32 | 0.12 | 1 | >32 | >128 | 1 | 32 | 0.5 | >8 | 8 |
| RB1195 | KPC-2 | 1 | 0.25 | 16 | 16 | >32 | 0.12 | 0.5 | >32 | >128 | 0.5 | 4 | 1 | >8 | 16 |
| RB1197 | KPC-2 | 1 | 1 | 128 | >32 | >32 | 0.12 | 0.5 | >32 | >128 | 0.25 | 32 | 0.5 | >8 | 32 |
| RB1198 | KPC-2 | 2 | 0.25 | 8 | 32 | >32 | 0.12 | 0.5 | 32 | >128 | 1 | 16 | 1 | >8 | 8 |
| RB1324 | KPC-3 | >256 | 0.5 | 8 | 16 | >32 | 0.25 | 2 | 16 | >128 | 0.5 | 2 | 1 | >8 | 32 |
| RB1328 | KPC-2 | 4 | 0.03 | 0.03 | 0.06 | >32 | 0.06 | 0.25 | >32 | 128 | 0.5 | 16 | 0.5 | >8 | 8 |
| RB1340 | KPC-2 | 2 | 0.25 | 8 | 32 | >32 | 0.06 | 0.25 | 32 | >128 | 0.5 | 1 | 0.25 | 0.06 | 8 |
| RB1245 | KPC-3 | 1 | 0.25 | 8 | 16 | >32 | 0.12 | 0.5 | 16 | >128 | 0.25 | 16 | 0.5 | >8 | 16 |
| RB1112 | KPC-3 | 0.5 | 0.06 | 8 | 8 | >32 | 0.03 | 0.06 | 32 | >128 | >8 | 16 | 4 | >8 | 32 |
| RB1200 | KPC-2 | 1 | 1 | 64 | >32 | >32 | 0.06 | 0.015 | >32 | >128 | 1 | 16 | 8 | >8 | 16 |
| RB1201 | KPC-2 | 1 | 0.25 | 32 | 32 | >32 | 0.12 | 0.12 | >32 | >128 | 0.5 | 32 | 1 | >8 | 64 |
| RB1252 | KPC-3 | 2 | 0.25 | 8 | 8 | >32 | 0.25 | 0.5 | 8 | >128 | 1 | 16 | 4 | >8 | 16 |
| RB1105 | KPC-2 | 2 | 2 | >256 | >32 | >32 | 0.5 | 2 | >32 | >128 | 2 | 16 | 1 | >8 | 16 |
| RB1253 | KPC-3 | 1 | 0.5 | 32 | 32 | >32 | 0.12 | 0.5 | 16 | >128 | 0.25 | 8 | 0.5 | >8 | 8 |
| RB1202 | KPC-3 | 1 | 0.5 | 16 | 32 | >32 | 0.12 | 1 | >32 | >128 | 0.25 | 32 | 0.5 | >8 | 8 |
| RB1255 | KPC-3 | 0.5 | 0.12 | 8 | 8 | >32 | ≤0.008 | 0.06 | 16 | >128 | 0.5 | 1 | 4 | >8 | 8 |
| RB1256 | KPC-3 | 0.5 | 0.12 | 8 | 8 | >32 | ≤0.008 | 0.06 | 16 | >128 | 0.5 | 2 | 4 | >8 | 16 |
| RB1106 | KPC-3 | 1 | 0.25 | 8 | 8 | >32 | 0.12 | 0.25 | >32 | >128 | 0.25 | 32 | 0.5 | >8 | 8 |
| RB1325 | KPC-2 | 2 | 0.25 | 8 | 16 | >32 | 0.06 | 0.5 | >32 | >128 | 0.5 | 1 | 0.25 | 1 | 8 |
| RB1107 | KPC-2 | 1 | 1 | 128 | >32 | >32 | 0.12 | 0.5 | >32 | >128 | 2 | 64 | 1 | >8 | 32 |
| RB1133 | KPC-3 | 1 | 0.25 | 8 | 16 | >32 | 0.12 | 0.5 | 8 | >128 | 0.5 | 32 | 1 | >8 | 32 |
| RB1259 | KPC-3 | 1 | 0.06 | 0.5 | 2 | >32 | 0.015 | 0.06 | 16 | >128 | 0.5 | 16 | 1 | >8 | 8 |
| RB1010 | KPC-3 | 4 | 0.12 | 8 | 16 | >32 | 0.12 | 0.25 | 16 | >128 | 0.25 | 16 | 1 | >8 | 16 |
| RB1108 | KPC-2 | 4 | 2 | >256 | >32 | >32 | 0.25 | 1 | >32 | >128 | 0.25 | 32 | 1 | >8 | 16 |
| RB1113 | KPC-3 | 1 | 0.25 | 8 | 16 | >32 | 0.12 | 0.12 | 32 | >128 | 0.5 | 4 | 1 | >8 | 8 |
| RB1114 | KPC-3 | 1 | 0.5 | 64 | 32 | >32 | ≤0.008 | ≤0.015 | >32 | >128 | 0.5 | 2 | 2 | >8 | 64 |
| RB1263 | KPC-3 | 0.5 | 0.12 | 1 | 2 | >32 | ≤0.008 | ≤0.015 | 8 | >128 | 0.5 | 8 | 0.5 | >8 | >256 |
| RB1012 | KPC-3 | 1 | 0.5 | 64 | >32 | >32 | 0.12 | 0.25 | >32 | >128 | 0.5 | 16 | 0.5 | >8 | 64 |
| RB1013 | KPC-3 | 1 | 0.25 | 8 | 32 | >32 | ≤0.008 | ≤0.015 | 32 | >128 | 0.25 | 16 | 8 | >8 | 8 |
| RB1115 | KPC-3 | 1 | 0.12 | 2 | 4 | >32 | 0.12 | 0.25 | 16 | >128 | 0.5 | 16 | 1 | >8 | 16 |
| RB1344 | KPC-2 | 1 | 1 | 128 | >32 | >32 | 0.12 | 0.25 | >32 | >128 | 0.25 | 8 | 0.5 | >8 | 32 |
| RB1109 | KPC-2 | 2 | 1 | 64 | >32 | >32 | 0.12 | 0.5 | >32 | >128 | >8 | 16 | 2 | >8 | 16 |
| RB1329 | KPC-2 | 2 | 0.5 | 16 | 32 | >32 | 0.12 | 0.5 | >32 | >128 | 0.25 | 4 | 1 | >8 | 16 |
| RB1208 | KPC-2 | 1 | 1 | 128 | >32 | >32 | 0.12 | 0.25 | >32 | >128 | 0.25 | 4 | 0.25 | >8 | 8 |
| RB1346 | KPC-3 | 1 | 0.25 | 16 | 32 | >32 | 0.12 | 0.5 | >32 | >128 | 0.5 | 32 | 0.5 | >8 | 8 |
| RB1271 | KPC-2 | 2 | 1 | 128 | >32 | >32 | 0.03 | 0.12 | >32 | >128 | 0.25 | 16 | 0.5 | >8 | 32 |
Avibactam and tazobactam were each used at 4 mg/liter.
FIG 2.
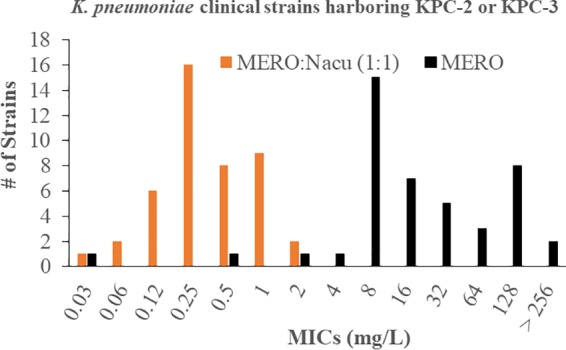
MICs for 44 K. pneumoniae clinical isolates containing the KPC-2 or KPC-3 β-lactamase tested against meropenem (MERO) and meropenem combined with nacubactam (Nacu) at a 1:1 ratio.
Susceptibility of E. coli strains expressing class A KPC β-lactamase variants with single amino acid substitutions.
We hypothesized that the structure and catalytic determinants (i.e., amino acids K73, P104, W105, S130, R164, E166, N170, D179, R220, K234, T235, T237, V240, and E276) that mediate the inactivation of class A β-lactamases by β-lactamase inhibitors, such as oxapenems, sulfones, and DBOs, would also play a role in the inactivation mechanism of nacubactam. Several of these residues comprise evolutionarily conserved motifs in class A β-lactamases. K73 is part of the SXXK motif (Ambler positions 70 to 73) and actively participates in β-lactam acylation. S130 resides in the SDN loop (Ambler positions 130 to 132) and is a critical residue involved in proton shuttling during acylation, deacylation, and recyclization of DBOs. The KTG motif includes K234 and T235, which aid in the binding of ligands; in addition, K234 is postulated to participate in proton shuttling with S130. Residues R164 to D179 constitute the Ω loop; R164 and D179 provide structural integrity via a salt bridge. Within the Ω loop, E166 and N170 position the deacylation water molecule for hydrolysis. Moreover, single amino acid substitutions in the Ω-loop residues notoriously contribute to ceftazidime-avibactam resistance (32, 33). The amide backbone of T237 and S70 forms the oxyanion hole for β-lactam and β-lactamase inhibitor binding. R220 and T237 interact with the carboxylate or sulfonate side chain on β-lactams and β-lactamase inhibitors, while E276 forms long-range second shell interactions with R220. P104, W105, and V240 reside at the opening of the active site and thus impact β-lactam and β-lactamase inhibitor entry and binding.
To address the supposition that these residues mediate inactivation of nacubactam, a panel of isogenic E. coli strains producing KPC variants with amino acid substitutions at the selected positions was subjected to susceptibly testing. As expected for a β-lactamase inhibitor with intrinsic PBP2 activity, all E. coli strains expressing these single β-lactamases were susceptible to meropenem-nacubactam on the basis of the breakpoint for resistance to meropenem alone (CLSI breakpoint, ≥4 mg/liter; EUCAST breakpoint, >8 mg/liter) (Table 2). Aztreonam, which inactivates PBP3, combined with avibactam was also effective against these strains (Table 2).
TABLE 2.
Broth microdilution MICs of β-lactam antibiotics for E. coli DH10B strains with KPC-2 substitutions
| Vector and KPC-2 substitution(s) | MIC (mg/liter) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nacubactam | Meropenem-nacubactam (1:1) | Meropenem | Ertapenem | Aztreonam | Aztreonam-avibactama | Ceftazidime-avibactama | Cefepime | Piperacillin-tazobactama | |
| pBC SK vector | |||||||||
| KPC-2 (wild type) | 1 | 0.12 | 1 | 1 | > 32 | 0.12 | 0.25 | 2 | 128 |
| H274Y (KPC-3) | 2 | 0.06 | 0.5 | 0.5 | 32 | 0.06 | 0.25 | 2 | 64 |
| P104R, V240G (KPC-4) | 2 | 0.12 | 0.25 | 0.5 | >32 | 0.12 | 2 | 4 | 64 |
| P104R (KPC-5) | 2 | 0.06 | 0.12 | 0.12 | >32 | 0.12 | 0.25 | 1 | 32 |
| V240G (KPC-6) | 2 | 0.06 | 0.25 | 0.12 | >32 | 0.12 | 0.5 | 2 | 64 |
| M49I, H274Y (KPC-7) | 2 | 0.06 | 0.25 | 0.25 | 32 | 0.06 | 0.25 | 2 | 64 |
| V240G, H274Y (KPC-8) | 2 | 0.12 | 0.5 | 0.5 | >32 | 0.12 | 1 | 4 | 128 |
| K73A | 2 | 0.015 | 0.015 | 0.008 | 0.25 | 0.12 | 0.5 | 0.06 | 2 |
| K73R | 2 | 0.03 | 0.015 | 0.008 | 0.12 | 0.12 | 0.25 | 0.15 | 4 |
| P104A | 2 | 0.06 | 0.5 | 0.5 | >32 | 0.06 | 0.25 | 2 | 64 |
| P104K | 2 | 0.25 | 1 | 1 | >32 | 0.12 | 0.5 | 4 | 128 |
| W105A | 1 | 0.06 | 0.06 | 0.12 | 32 | 0.06 | 0.12 | 0.25 | 32 |
| S130G | 1 | 0.03 | 0.015 | 0.008 | 0.25 | 0.06 | 0.12 | 0.06 | >128 |
| S130T | 2 | 0.06 | 0.03 | 0.06 | 0.12 | 0.06 | 0.25 | 0.12 | 4 |
| E166A | 2 | 0.03 | 0.06 | 0.015 | 0.25 | 0.06 | 0.25 | 0.12 | 2 |
| E166Y | 2 | 0.03 | 0.03 | 0.06 | 2 | 0.06 | 0.5 | 1 | 1 |
| N170A | 4 | 0.06 | 0.03 | 0.06 | 1 | 0.12 | 2 | 2 | 64 |
| N170P | 2 | 0.06 | 0.03 | 0.25 | 4 | 0.12 | 4 | 4 | 4 |
| R220K | 1 | 0.06 | 0.06 | 0.12 | 2 | 0.06 | 0.12 | 0.12 | 2 |
| R220M | 1 | 0.03 | 0.03 | 0.15 | 0.25 | 0.06 | 0.12 | 0.12 | 32 |
| T235A | 2 | 0.03 | 0.03 | 0.015 | 1 | 0.06 | 0.12 | 0.06 | 32 |
| T235S | 2 | 0.03 | 0.12 | 0.016 | 0.5 | 0.06 | 0.12 | 0.06 | 4 |
| T237A | 1 | 0.03 | 0.06 | 0.25 | 16 | 0.06 | 0.12 | 0.06 | 32 |
| T237S | 1 | 0.06 | 0.5 | 1 | >32 | 0.06 | 0.25 | 1 | 64 |
| V240G | 2 | 0.25 | 4 | 8 | >32 | 0.25 | 2 | 16 | >128 |
| V240K | 2 | 0.25 | 2 | 4 | >32 | 0.12 | 0.5 | 4 | >128 |
| E276A | 1 | 0.12 | 0.5 | 1 | 32 | 0.06 | 0.25 | 1 | 64 |
| E276N | 2 | 0.06 | 0.25 | 0.5 | 16 | 0.12 | 0.12 | 0.5 | 64 |
| pBR322-catI vector | |||||||||
| Wild type | 2 | 0.25 | 8 | 32 | >32 | 0.25 | 1 | 16 | >128 |
| R164A | 2 | 0.25 | 0.5 | 2 | >32 | 0.12 | 4 | 8 | >128 |
| R164H | 2 | 0.25 | 2 | 4 | >32 | 0.12 | 1 | >32 | >128 |
| R164P | 2 | 0.06 | 0.03 | 0.12 | 0.25 | 0.12 | 8 | 0.5 | 2 |
| R164S | 2 | 0.12 | 1 | 2 | >32 | 0.06 | 2 | 8 | >128 |
| D179A | 2 | 0.06 | 0.03 | 0.12 | 1 | 0.12 | 8 | 2 | 2 |
| D179C | 1 | 0.06 | 0.06 | 0.5 | 8 | 0.12 | 8 | 1 | 2 |
| D179E | 1 | 0.03 | 0.015 | 0.03 | 1 | 0.12 | 2 | 0.12 | 2 |
| D179F | 2 | 0.06 | 0.06 | 0.12 | 4 | 0.12 | 8 | 4 | 2 |
| D179G | 4 | 0.12 | 0.12 | 0.25 | 8 | 0.12 | 8 | 8 | 8 |
| D179H | 2 | 0.06 | 0.03 | 0.12 | 0.25 | 0.12 | 8 | 1 | 1 |
| D179I | 1 | 0.015 | 0.015 | 0.15 | 0.25 | 0.12 | 8 | 0.5 | 2 |
| D179K | 2 | 0.03 | 0.03 | 0.03 | 0.12 | 0.12 | 4 | 0.25 | 4 |
| D179L | 2 | 0.06 | 0.03 | 0.03 | 2 | 0.12 | 8 | 2 | 2 |
| D179M | 2 | 0.06 | 0.06 | 0.12 | 1 | 0.12 | 8 | 4 | 2 |
| D179N | 2 | 0.25 | 2 | 4 | >32 | 0.12 | 2 | 8 | >128 |
| D179P | 1 | 0.03 | 0.03 | 0.06 | 1 | 0.12 | 8 | 1 | 1 |
| D179Q | 2 | 0.06 | 0.03 | 0.12 | 0.25 | 0.12 | 4 | 0.25 | 1 |
| D179R | 1 | 0.06 | 0.03 | 0.06 | 0.12 | 0.12 | 4 | 0.12 | 1 |
| D179S | 1 | 0.03 | 0.015 | 0.03 | 1 | 0.06 | 1 | 0.5 | 2 |
| D179T | 1 | 0.06 | 0.03 | 0.12 | 1 | 0.12 | 8 | 1 | 2 |
| D179V | 1 | 0.06 | 0.03 | 0.12 | 1 | 0.12 | 8 | 2 | 1 |
| D179W | 2 | 0.06 | 0.06 | 0.03 | 4 | 0.12 | 8 | 4 | 2 |
| D179Y | 2 | 0.06 | 0.06 | 0.25 | 1 | 0.12 | 8 | 2 | 2 |
| R220A | 2 | 0.12 | 0.12 | 0.25 | 1 | 0.06 | 0.25 | 0.5 | >128 |
| K234A | 1 | 0.015 | 0.015 | 0.008 | 0.12 | 0.06 | 0.12 | 0.03 | 1 |
| K234R | 2 | 0.5 | 2 | 4 | 32 | 0.12 | 0.25 | 8 | >128 |
Avibactam and tazobactam were each used at 4 mg/liter.
The meropenem-nacubactam (1:1 ratio) combination led to a selective and modest attenuation in the meropenem MICs of E. coli strains expressing the KPC variants (Table 2). The most pronounced effect of the addition of nacubactam was observed for the strain expressing the pBR322-KPC-2 wild type, which underwent a 32-fold decrease in MIC compared to the MIC of meropenem alone. Strains expressing the P104K, V240G, V240K, and K234R variants possessed modest increases in the MIC of meropenem-nacubactam relative to that of the strain expressing KPC-2 in the respective vector; however, only the E. coli strain producing the K234R variant lacked a corresponding increase in the meropenem MIC compared to that for the strain producing KPC-2. Importantly, the lysine at position 234 is part of the conserved K234T235G236 motif found in all class A β-lactamases (34), amino acid substitutions at K234 contribute to inhibitor resistance in strains with SHV (35), and K234 was shown to form hydrogen bond interactions with another DBO, avibactam, in strains with CTX-M-15 (36, 37). For KPC-2, the K234R variant was found to play a vital role in carbapenem resistance and resistance to inhibition by avibactam (37). Therefore, the K234R variant was selected for biochemical characterization.
Kinetic characterization of KPC-2 and the K234R variant with nacubactam versus avibactam.
Nacubactam and avibactam were tested against the purified KPC-2 and K234R variant β-lactamases to assess the levels of inhibition using nitrocefin as the reporter substrate. For nitrocefin, the K234R variant demonstrated a higher Km than KPC-2 (29 ± 3 μM for the K234R variant versus 13 ± 1 μM for KPC-2) and a slower kcat than KPC-2 (133 ± 13 s−1 for the K234R variant versus 182 ± 18 s−1 for KPC-2) (Table 3). Similarly, K234R had a higher Km for meropenem (6.3 ± 0.6 μM for the K234R variant versus 4.9 ± 0.5 μM for KPC-2) and a 5-fold lower kcat (1 ± 0.1 s−1 for the K234R variant versus 5 ± 0.5 s−1 for KPC-2).
TABLE 3.
Kinetic parameters for KPC-2 and the K234R variant
| Drug and carbapenemase | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) |
|---|---|---|---|
| Nitrocefin | |||
| KPC-2 | 13 ± 1 | 182 ± 18 | 14 ± 1 |
| K234R | 29 ± 3 | 133 ± 13 | 4.6 ± 0.5 |
| Meropenem | |||
| KPC-2 | 4.9 ± 0.5 | 5 ± 0.5 | 1 ± 0.1 |
| K234R | 6.3 ± 0.6 | 1 ± 0.1 | 0.2 ± 0.02 |
Concentration-dependent inhibition of β-lactamase hydrolytic activity was observed for nacubactam and avibactam. Nacubactam inhibited KPC-2 (apparent Ki [Ki app] = 31 ± 3 μM) more efficiently than it inhibited the K234R variant (Ki app = 270 ± 27 μM) (Table 4). Correspondingly, nacubactam possessed a higher acylation rate (k2/K) and a measurable deacylation rate (koff) for KPC-2 (k2/K = 5,815 ± 582 M−1 s−1, koff = 0.0002 s−1) compared to those for the K234R variant (k2/K = 247 ± 25 M−1 s−1; koff could not be determined with spectrophotometric assays, as an initial velocity of ∼0 μM/s could not be achieved). Nacubactam and avibactam shared a turnover of 1 for both KPC-2 and the K234R variant (Table 4). Nacubactam was less efficient than avibactam at inhibiting KPC-2 (Ki app, 31 ± 3 μM for nacubactam versus 1.0 ± 0.1 μM for avibactam) and K234R (Ki app, 270 ± 27 μM for nacubactam versus 157 ± 15 μM for avibactam) (Table 4). These data suggest that the cellular potency of nacubactam in E. coli and K. pneumoniae may rely more heavily on its significant PBP2 inhibition and enhancer activity (22) (Table 1). Overall, KPC-2 does not turn over either nacubactam or avibactam. Avibactam is more efficient than nacubactam at acylating (higher k2/K) and inhibiting (lower Ki app) KPC-2, but these differences may not result in overt phenotypic outcomes in the cell.
TABLE 4.
DBO inhibition kinetics for KPC-2 and the K234R variant
Mass spectrometry analysis of KPC-2 and the K234R variant with nacubactam versus avibactam.
The integrity and behavior of reaction intermediates between the β-lactamases and DBOs were sought by conducting mass spectrometry (MS) on KPC-2 and the K234R variant after incubation with avibactam and nacubactam for 5 min, 1 h, and 24 h (Fig. 3A, B, and C). Both KPC-2 and the K234R variant caused the desulfation of avibactam (+169 Da); however, the K234R variant seemed to desulfate avibactam more efficiently (Fig. 3C). The sulfate group of the DBO is predicted to anchor the open-ring form of the DBO, thus blocking deacylation; however, the desulfation of avibactam by KPC-2 was previously observed (32, 38). One proposed mechanism of desulfation occurs upon addition of a proton transferred through a water from a base, resulting in a hydroxylamine avibactam intermediate (38). Alternatively, the sulfate could undergo direct β-elimination, possibly initiated by the nearby S130 (38).
FIG 3.
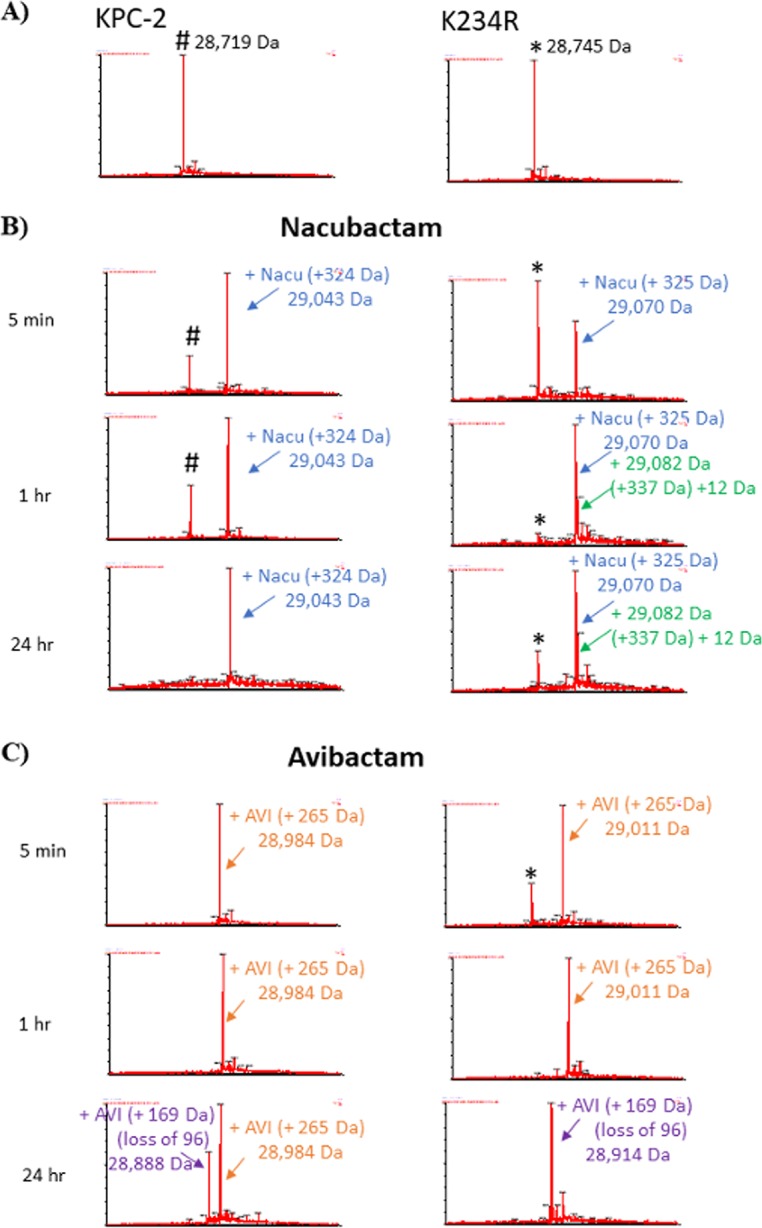
Timed mass spectra of KPC-2 and the K234R variant (A) with nacubactam (B) or avibactam (C). KPC-2 (left) and the K234R variant (right) were incubated at a 1:1 molar ratio of enzyme-DBO for the indicated times. #KPC-2 (28,719 Da) and *K234R (28,745 Da) are the apo-β-lactamases. Acyl complexes with nacubactam are indicated with blue and green arrows. Acyl complexes with avibactam are indicated with orange and purple arrows.
Unlike with avibactam, the desulfation of nacubactam was not observed for KPC-2 or the K234R variant (Fig. 3B). At 24 h, KPC-2 was fully acylated by avibactam and nacubactam. In contrast, the apoenzyme of the K234R variant was detected upon incubation with nacubactam even at the 24-h time point (Fig. 3B). This observation supports the kinetic analysis, as acylation by nacubactam was more efficient with KPC-2 than with the K234R variant. Furthermore, an extra peak (+337 Da, which is a +12-Da addition to the 325 Da of nacubactam) was observed in the K234R variant-nacubactam mass spectra (Fig. 3B). These data suggest that the K234R variant interacts with nacubactam in an alternative way compared to KPC-2. In summary, whereas most of avibactam is desulfated by KPC-2 after 24 h of incubation, nacubactam maintains its sulfate moiety upon incubation with either enzyme.
CD analysis of KPC-2 and the K234R variant with nacubactam versus avibactam.
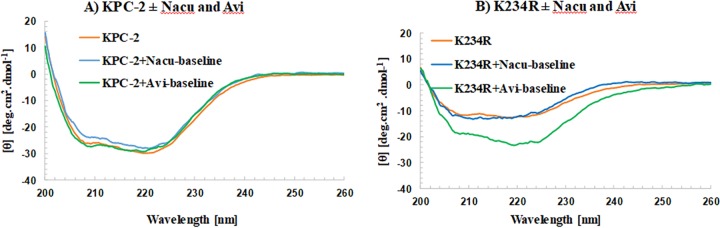
To assess the secondary structure of KPC-2 and the K234R variant with and without nacubactam and avibactam, circular dichroism (CD) experiments were performed. Nacubactam and avibactam did not significantly change the secondary structure or the signal intensity of KPC-2 (Fig. 4A). Avibactam seemed to introduce a minor change into the 220-nm region of KPC-2, and nacubactam seemed to introduce minor changes into the 208-nm and 222-nm regions as well. The minor changes observed in the 208-nm region of KPC-2–nacubactam could be due to the higher signal of the compound in the 205- to 215-nm region as well or due to minor changes in the secondary structure of KPC-2 upon binding (Fig. 4A). However, avibactam introduced significant changes into the K234R variant’s secondary structure (Fig. 4B). The destabilizing effect of the K234R amino acid substitution was revealed by the decreased ellipticity (CD signal) of this variant relative to that of KPC-2. The binding of avibactam seems to partially restore some of the ellipticity (Fig. 4B).
FIG 4.
Circular dichroism (CD) of KPC-2 (10 μM) (A) and the K234R variant (10 μM) (B) with nacubactam (Nacu; 100 μM) and avibactam (Avi; 100 μM). DBO binding to KPC-2 does not induce significant secondary structure changes (A). The binding of avibactam seems to partially restore the stability of the K234R variant, resulting in large secondary structure changes. Nacubactam induces only minor secondary structure changes in the K234R variant (B). θ, ellipticity.
Thermal stability of KPC-2 and K234R with nacubactam versus avibactam.
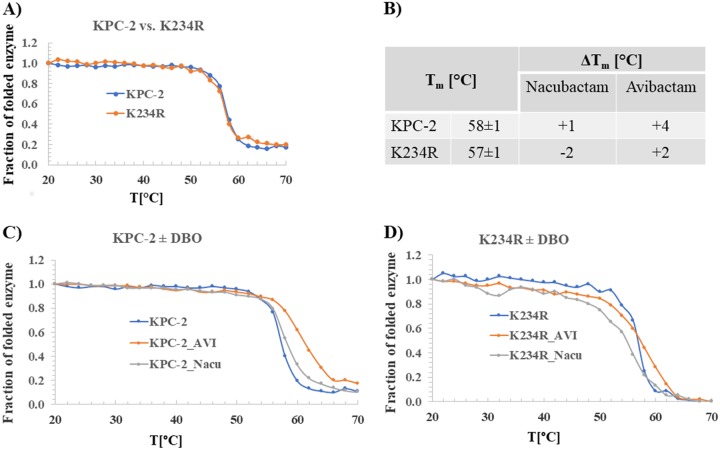
Thermal stability was assessed to determine whether the differences in β-lactamase activity correlate to detectable changes in protein stability. KPC-2 was slightly more stable than the K234R variant (melting temperature [Tm] = 58 ± 1°C versus 57 ± 1°C, respectively) (Fig. 5A and B). The addition of a DBO increased the thermal stability of KPC-2 (by 4°C for avibactam and 1°C for nacubactam). The K234R variant was stabilized by avibactam (increase of 2°C) but less so than KPC-2. Further, the K234R variant exhibited a decrease (−1°C) in stability with nacubactam. The K234R variant seemed to have a less stable secondary structure than KPC-2 during thermal denaturation in the presence of DBOs (Fig. 5C and D).
FIG 5.
Thermal denaturation of KPC-2 (10 μM) and the K234R variant (10 μM) measured alone (A, B) and with nacubactam (100 μM) or avibactam (100 μM) (B, C, D). The K234R variant is slightly less thermodynamically stable than wild-type KPC-2 (A, B). DBOs increase the thermal stability of KPC-2 (B, C), but only avibactam increases the thermal stability of K234R, by 2°C (B, D).
Molecular modeling of KPC-2 and K234R with and without nacubactam.
Molecular modeling using the apo-KPC-2 and the apo-K234R variants displayed dramatic differences as a result of the K234R substitution. The catalytic site of the apoenzyme of the K234R variant was rearranged, which is consistent with the observations that DBO binding and acylation are affected by this amino acid substitution (Fig. 6A and B). The hydrogen bonding interactions of K234 with S130 observed in KPC-2 were replaced by hydrogen bonding interactions of R234 with N214, V127, and T216 (Fig. 6A and B). The larger, positively charged side chain of Arg at position 234 is disruptive to the catalytic-site electrostatic environment of the variant. In the K234R variant model, the lysine was no longer available to serve as an anchor for S130, and as a result, the K73 and S130 residues were shifted up to 1 Å in the K234R variant relative to the same residues in KPC-2. Due to this structural perturbation, K73 lost the hydrogen bond with S70 and established a new hydrogen bond with S130 (Fig. 6A and B). The position and movement of S130 are critical for the integrity of the active-site structure and proton transfer during acylation (39). However, the shift in S130 and R234 allows the K234R variant to accommodate a secondary water, which would enhance the efficiency of avibactam desulfation compared to that in KPC-2.
FIG 6.
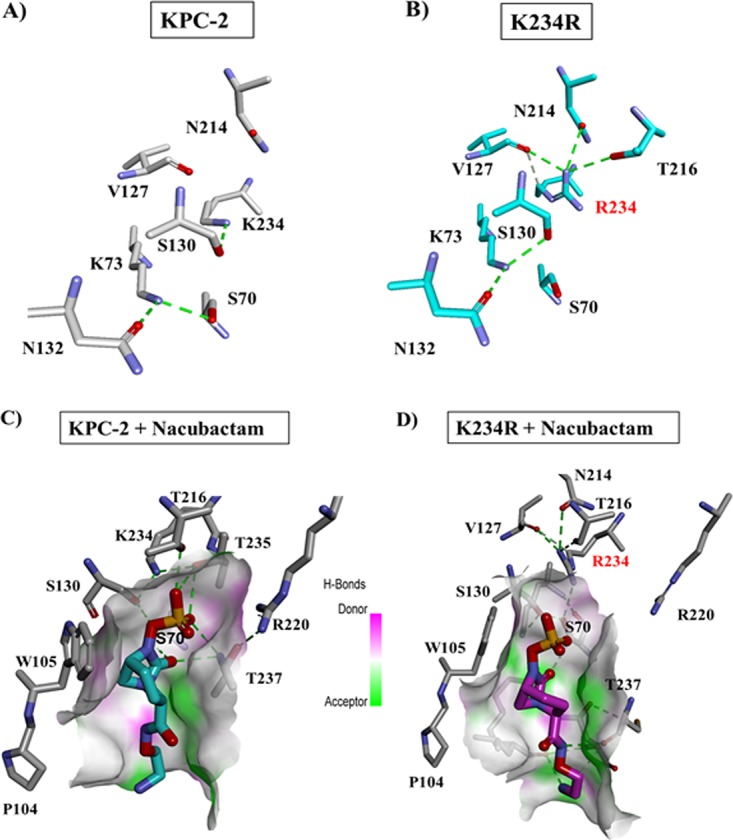
(A and B) Molecular modeling of KPC-2 (A) versus the K234R variant (B). S130 interacts with K234 but not R234. The K73-S70 interaction is also disrupted in the K234R variant, establishing a new hydrogen bond between S130 and K73. (C and D) Molecular docking of nacubactam in KPC-2 (C) and the K234R variant (D). R234 has a different conformation than K234 and forms an alternative network of bonds. The aminoethoxy tail of nacubactam forms additional interactions in K234R.
Docking of nacubactam with KPC-2 suggested a favorable position of nacubactam, with the carbonyl being directed into the oxyanion hole and the sulfate group being within hydrogen-bonding distance of T237, T235, T216, S130, and K234 (Fig. 6C). Nacubactam adopted a series of conformations in KPC-2 suggesting that the aminoethoxy tail conferred flexibility to the DBO structure. Conversely, the docking of nacubactam into the active site of the K234R variant resulted in less favorable conformations. With a shorter carbamoyl side chain, avibactam was more likely than nacubactam to interact with W105 (in the position 102 to 110 loop) and E166 (Fig. 6C and D) (40). The conformations of nacubactam as a result of the aminoethoxy tail resulted in the gross movement of S130 and, subsequently, a decrease in the efficiency of acylation in the K234R variant.
Conclusions.
Nacubactam is effective at restoring the efficacy of meropenem against K. pneumoniae strains producing class A KPC carbapenemases. Interestingly, the K234R substitution was found to result in a disruptive change in the catalytic pocket of KPC-2, leading to a higher Ki app and a lower k2/K for DBOs. Moreover, the inhibition kinetics of nacubactam are less favorable than those of avibactam for KPC-2 and the K234R variant, and nacubactam likely exhibits an altered mechanism of action, as evidenced by mass spectrometry with the K234R variant. Distinct from the characteristics of avibactam, nacubactam demonstrates significant PBP2 inhibitory activity and β-lactam enhancer properties that result in antibiotic synergism. Therefore, nacubactam-meropenem may offer some therapeutic advantage compared to other combinations against highly resistant isolates, such as those producing evolving KPC-2 variants that are resistant to ceftazidime-avibactam. Differences in the clinical outcomes between infections treated with these two potent combinations remain to be determined.
MATERIALS AND METHODS
Critical reagents and strains.
Nitrocefin (catalog number BR0063G) was purchased from Oxoid. For broth microdilution MICs, Sensititre plates were custom ordered (frozen) from Thermo Fisher Scientific (Oakwood Village, OH). Amikacin, cefepime, ertapenem, fosfomycin, levofloxacin, and piperacillin were obtained from Sigma. F. Hoffmann-La Roche Ltd. provided avibactam and nacubactam. Meropenem and tigecycline were procured from Pfizer. Aztreonam, ceftazidime, and colistin were obtained from USP. Tazobactam was obtained from Toku-E. The 44 K. pneumoniae clinical isolates were from the Consortium on Resistance against Carbapenems in Klebsiella and other Enterobacteriaceae (CRACKLE), a multicenter consortium that tracks carbapenem-resistant Enterobacteriaceae (15, 41, 42). Escherichia coli containing blaKPC-2 in the pBR322-catI vector was a gift from Fred Tenover, previously of the Centers for Disease Control and Prevention (43). The pBC SK and pBR322-catI vectors yield reasonable expression levels of KPC (32).
Expression and purification of KPC-2 and the K234R variant.
The KPC-2 and the KPC-2 K234R variant β-lactamases were purified from E. coli Origami 2(DE3) (Novagen) cells carrying the pET24a(+)blaKPC-2 or pET24a(+)blaKPC-2 K234R plasmids, as previously described for KPC-2 (33). Single colonies were used to inoculate 5-ml cultures for overnight growth, and ∼1.5 ml was used to start a 50-ml overnight culture. Ten to 12 ml of an overnight culture was added to each flask of 500 ml superoptimal broth (SOB), grown at 37°C to an optical density at a wavelength (λ) of 600 nm (OD600) of approximately 0.6 to 0.8, and induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for a minimum of 3 h to express the β-lactamase-encoding gene. The cells were pelleted and frozen at −20°C for ≥12 h prior to lysis in 50 mM Tris HCl buffer, pH 7.4, containing 40 mg/ml lysozyme, 0.1 mM magnesium sulfate, 250 U Benzonase nuclease, and 1 mM EDTA. The supernatant was further purified by preparative isoelectric focusing, eluted from the Sephadex resin with 50 mM Tris-HCl (pH 8.8), sterile filtered, and purified once again by fast protein liquid chromatography (FPLC) using a HiTrap Q anion-exchange chromatography column (catalog number 17-1154-01; GE Healthcare Life Sciences). The final sample of protein was concentrated using centrifugal filter units with a molecular weight cutoff of 10,000 daltons (Millipore). A final concentration of 25% glycerol was added to the protein before being stored frozen at −20°C. The purity of the proteins was assessed by quadrupole time of flight (Q-TOF) mass spectrometry. Protein concentrations were determined by measuring the absorbance at a λ of 280 nm and using the protein's extinction coefficient (Δε 39,545 M−1 cm−1 for KPC-2 and the K234R variant), obtained using the ProtParam tool at the ExPASy Bioinformatics Resource Portal.
Susceptibility testing.
MICs were determined by broth microdilution in 96-well plates customized by Thermo Fisher Scientific and analyzed according to CLSI and EUCAST guidelines (30, 31). Nacubactam was tested alone and in a 1:1 ratio to meropenem.
Steady-state kinetic analysis using pure protein.
Steady-state kinetic parameters were determined by using an Agilent 8453 diode array spectrophotometer at room temperature as previously described (37, 44). Each assay was performed in 10 mM phosphate-buffered saline (PBS) at pH 7.4 at room temperature (RT; ∼25°C) in a quartz cuvette with a 1-cm path length. All assays with the K234R variant were conducted in 200 μg/ml bovine serum albumin (BSA) for stabilization of the enzyme as previously described. The kinetic parameters kcat and Km were obtained for KPC-2 (7 nM) and the K234R variant (7 nM) using the chromogenic substrate nitrocefin (Δε482 = 17,400 M−1 cm−1) with a nonlinear least-squares fit of the data (Henri-Michaelis-Menten equation) using Origin (v7.5VR) software (Origin Lab, Northampton, MA).
For Ki app, the KPC-2 (10 nM) hydrolysis of nitrocefin (100 μM) was monitored over time in the presence of increasing concentrations of nacubactam (1 to 100 μM). Similarly, the K234R variant (10 nM) was reacted with nitrocefin (150 μM) in the presence of 200 μg/ml BSA and increasing concentrations of nacubactam (250 to 1,500 μM) and avibactam (122 to 610 μM).
The rate of acylation (k2/K) was measured for KPC-2 (10 nM) with 50 μM nitrocefin and increasing concentrations of nacubactam (6.25 to 100 μM). The acylation rate for the K234R variant (10 nM) was determined using 150 μM nitrocefin and increasing concentrations of nacubactam (100 to 1,000 μM) and avibactam (50 to 400 μM).
The koff value measurements were conducted by preincubating KPC-2 (1 μM) with nacubactam (255 μM) for 5 min, diluting the preincubation mixture 1:100 in PBS, and measuring nitrocefin (50 μM) hydrolysis for 1 h using 10 μl of the diluted mixture (0.1 nM final KPC-2 concentration). koff could not be determined for nacubactam and avibactam with the K234R variant, as an initial rate of zero could not be established after preincubation with either DBO at 2 mM.
The turnover number (kcat/kinact) was determined by preincubating KPC-2 (1 μM) with nacubactam (1 μM) and avibactam (1 μM) for 15 min and adding 10 μl of the preincubation mixture to the cuvette containing 1 ml of PBS in 50 μM nitrocefin (10 nM KPC-2 and 10 nM DBO final concentrations). Similar experiments were conducted for the K234R variant preincubated for 15 min using 150 μM nitrocefin and 200 μg/ml BSA, added to both the preincubation and final reaction mixtures.
Electrospray ionization mass spectrometry (ESI-MS).
Five micrograms of purified β-lactamase (KPC-2 or the K234R variant) was incubated with the DBO at a 1:1 molar ratio of enzyme-inhibitor in 10 mM PBS at pH 7.4 for a total reaction volume of 20 μl for 5 min, 1 h, and 24 h. The reactions were quenched with 10 μl acetonitrile, and the reaction mixtures were added to 1 ml 0.1% formic acid in water. Samples were analyzed using a Q-TOF Waters Synapt-G2-Si mass spectrometer and Waters Acquity UPLC BEH C18 column (2.1 by 50 mm; particle size, 1.7 μm). MassLynx (v4.1) software was used to deconvolute the protein peaks. The tune settings for each data run were as follows: capillary voltage at 3.5 kV, sampling cone at 35, source offset at 35, source temperature of 100°C, desolvation temperature of 500°C, cone gas at 100 liters/h, desolvation gas at 800 liters/h, and nebulizer bar at 6.0. Mobile phase A was 0.1% formic acid in water. Mobile phase B was 0.1% formic acid in acetonitrile. The mass accuracy of this system was ±5 Da.
CD and thermal denaturation.
Circular dichroism (CD) and thermal denaturation experiments were carried out in a Jasco (Easton, MD) J-815 spectrometer with a Peltier effect temperature controller. Quartz cells with a 0.1-cm path length were used for all experiments.
To assess the stability of KPC-2 and the K234R variant (10 μM) with the DBOs (100 μM), secondary structure changes were monitored for helical content by CD in the far-UV region between λ values of 200 and 300 nm. The spectra of the DBOs were recorded at the baseline and subtracted from the spectrum for the enzyme with the DBOs.
To determine the thermal stability, KPC-2 and the K234R variant (10 μM) were incubated with the DBOs (100 μM) and monitored for thermal denaturation by CD at λ values of 210 nm and 222 nm at between 20 and 80°C with a heating rate of 2°C/min. A two-state behavior was indicated by identical curves at each wavelength. Raw equilibrium denaturation data were normalized to the fraction of denatured protein (fU). With the assumption of a reversible two-state transition (N ↔ U, where N and U are the denatured and undenatured proteins, respectively) equilibrium constants (Keq) at any given temperature were calculated from equation 1:
| (1) |
With the assumption that the variation in enthalpy (ΔH) and entropy (ΔS) are temperature independent, the melting temperature (Tm) at the midpoint of equilibrium folding (where temperature [T] is equal to Tm) was determined using equation 2, where R is the gas constant:
| (2) |
Molecular modeling/docking.
Structural representations of KPC-2 and the K234R variant of KPC-2 β-lactamase were generated using the crystal coordinates of KPC-2 (PDB accession number 2OV5) and a Flexible Docking protocol in Discovery Studio 2016 (D.S. 2016 Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, San Diego, CA) molecular modeling software as previously described (33, 45). The minimized crystal structure of the KPC-2 enzyme was used to generate the K234R variant model. Both enzymes were minimized to a 0.01 root mean squared gradient, using a Conjugate Gradient protocol, with the Generalized Born with a simple SWitching (GBSW) implicit solvent model. The force field used was CHARMm with Particle Mesh Ewald electrostatics and SHAKE constraint. The intact nacubactam was built and minimized using the small molecules module. After the docking, the generated poses were analyzed and scored based on the interaction energy, and the most favorable ones were chosen to create the enzyme-nacubactam complexes. The Michaelis-Menten complexes were energetically minimized using the same Conjugate Gradient protocol with the GBSW implicit solvent model.
ACKNOWLEDGMENTS
F. Hoffmann-La Roche Ltd. provided inhibitors, antibiotics, and funding for the study. This research was supported by funds and/or facilities provided by the Louis Stokes Cleveland VA Medical Center to K.M.P.-W. and R.A.B. and by Veterans Affairs Merit Review Program award 1I01BX002872 to K.M.P.-W. and Veterans Affairs Merit Review Program award 1I01BX001974 to R.A.B. from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10. R.A.B. is also supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers R01AI100560, R01AI063517, R21AI114508, and R01AI072219.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, or the U.S. government.
REFERENCES
- 1.U.S. Department of Health and Human Services and Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. U.S. Department of Health and Human Services and Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc./drugresistance/threat-report-2013/index.html. [Google Scholar]
- 2.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority list Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 3.Humphries RM, Yang S, Hemarajata P, Ward KW, Hindler JA, Miller SA, Gregson A. 2015. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59:6605–6607. doi: 10.1128/AAC.01165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaibani P, Campoli C, Lewis RE, Volpe SL, Scaltriti E, Giannella M, Pongolini S, Berlingeri A, Cristini F, Bartoletti M, Tedeschi S, Ambretti S. 2018. In vivo evolution of resistant subpopulations of KPC-producing Klebsiella pneumoniae during ceftazidime/avibactam treatment. J Antimicrob Chemother 73:1525–1529. doi: 10.1093/jac/dky082. [DOI] [PubMed] [Google Scholar]
- 5.Both A, Buttner H, Huang J, Perbandt M, Belmar Campos C, Christner M, Maurer FP, Kluge S, Konig C, Aepfelbacher M, Wichmann D, Rohde H. 2017. Emergence of ceftazidime/avibactam non-susceptibility in an MDR Klebsiella pneumoniae isolate. J Antimicrob Chemother 72:2483–2488. doi: 10.1093/jac/dkx179. [DOI] [PubMed] [Google Scholar]
- 6.Giddins MJ, Macesic N, Annavajhala MK, Stump S, Khan S, McConville TH, Mehta M, Gomez-Simmonds A, Uhlemann AC. 2018. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC-2-harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother 62:e02101-17. doi: 10.1128/AAC.02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shields RK, Potoski BA, Haidar G, Hao B, Doi Y, Chen L, Press EG, Kreiswirth BN, Clancy CJ, Nguyen MH. 2016. Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis 63:1615–1618. doi: 10.1093/cid/ciw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson K, Hemarajata P, Sun D, Rubio-Aparicio D, Tsivkovski R, Yang S, Sebra R, Kasarskis A, Nguyen H, Hanson BM, Leopold S, Weinstock G, Lomovskaya O, Humphries RM. 2017. Resistance to ceftazidime-avibactam is due to transposition of KPC in a porin-deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrob Agents Chemother 61:e00989-17. doi: 10.1128/AAC.00989-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castanheira M, Huband MD, Mendes RE, Flamm RK. 2017. Meropenem-vaborbactam tested against contemporary Gram-negative isolates collected worldwide during 2014, including carbapenem-resistant, KPC-producing, multidrug-resistant, and extensively drug-resistant Enterobacteriaceae. Antimicrob Agents Chemother 61:e00567-17. doi: 10.1128/AAC.00567-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castanheira M, Rhomberg PR, Flamm RK, Jones RN. 2016. Effect of the β-lactamase inhibitor vaborbactam combined with meropenem against serine carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 60:5454–5458. doi: 10.1128/AAC.00711-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lapuebla A, Abdallah M, Olafisoye O, Cortes C, Urban C, Quale J, Landman D. 2015. Activity of meropenem combined with RPX7009, a novel β-lactamase inhibitor, against Gram-negative clinical isolates in New York City. Antimicrob Agents Chemother 59:4856–4860. doi: 10.1128/AAC.00843-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. 2008. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol 29:1099–1106. doi: 10.1086/592412. [DOI] [PubMed] [Google Scholar]
- 13.Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, Schlaeffer F, Sherf M. 2009. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect Control Hosp Epidemiol 30:972–976. doi: 10.1086/605922. [DOI] [PubMed] [Google Scholar]
- 14.Perez F, Endimiani A, Ray AJ, Decker BK, Wallace CJ, Hujer KM, Ecker DJ, Adams MD, Toltzis P, Dul MJ, Windau A, Bajaksouzian S, Jacobs MR, Salata RA, Bonomo RA. 2010. Carbapenem-resistant Acinetobacter baumannii and Klebsiella pneumoniae across a hospital system: impact of post-acute care facilities on dissemination. J Antimicrob Chemother 65:1807–1818. doi: 10.1093/jac/dkq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neuner EA, Yeh JY, Hall GS, Sekeres J, Endimiani A, Bonomo RA, Shrestha NK, Fraser TG, van Duin D. 2011. Treatment and outcomes in carbapenem-resistant Klebsiella pneumoniae bloodstream infections. Diagn Microbiol Infect Dis 69:357–362. doi: 10.1016/j.diagmicrobio.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, Marchese A, Spanu T, Ambretti S, Ginocchio F, Cristini F, Losito AR, Tedeschi S, Cauda R, Bassetti M. 2012. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin Infect Dis 55:943–950. doi: 10.1093/cid/cis588. [DOI] [PubMed] [Google Scholar]
- 17.Bush K. 2018. Past and present perspectives on β-lactamases. Antimicrob Agents Chemother 62:e01076-18. doi: 10.1128/AAC.01076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 20.Moya B, Barcelo IM, Bhagwat S, Patel M, Bou G, Papp-Wallace KM, Bonomo RA, Oliver A. 2017. WCK 5107 (zidebactam) and WCK 5153 are novel inhibitors of PBP2 showing potent “β-lactam enhancer” activity against Pseudomonas aeruginosa, including multidrug-resistant metallo-β-lactamase-producing high-risk clones. Antimicrob Agents Chemother 61:e02529-16. doi: 10.1128/AAC.02529-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durand-Reville TF, Guler S, Comita-Prevoir J, Chen B, Bifulco N, Huynh H, Lahiri S, Shapiro AB, McLeod SM, Carter NM, Moussa SH, Velez-Vega C, Olivier NB, McLaughlin R, Gao N, Thresher J, Palmer T, Andrews B, Giacobbe RA, Newman JV, Ehmann DE, de Jonge B, O'Donnell J, Mueller JP, Tommasi RA, Miller AA. 2017. ETX2514 is a broad-spectrum β-lactamase inhibitor for the treatment of drug-resistant Gram-negative bacteria including Acinetobacter baumannii. Nat Microbiol 2:17104. doi: 10.1038/nmicrobiol.2017.104. [DOI] [PubMed] [Google Scholar]
- 22.Morinaka A, Tsutsumi Y, Yamada M, Suzuki K, Watanabe T, Abe T, Furuuchi T, Inamura S, Sakamaki Y, Mitsuhashi N, Ida T, Livermore DM. 2015. OP0595, a new diazabicyclooctane: mode of action as a serine β-lactamase inhibitor, antibiotic and β-lactam 'enhancer.' J Antimicrob Chemother 70:2779–2786. doi: 10.1093/jac/dkv166. [DOI] [PubMed] [Google Scholar]
- 23.Livermore DM, Mushtaq S, Warner M, Woodford N. 2015. Activity of OP0595/β-lactam combinations against Gram-negative bacteria with extended-spectrum, AmpC and carbapenem-hydrolysing β-lactamases. J Antimicrob Chemother 70:3032–3041. doi: 10.1093/jac/dkv239. [DOI] [PubMed] [Google Scholar]
- 24.Mushtaq S, Vickers A, Woodford N, Haldimann A, Livermore DM. 2018. Activity of nacubactam (RG6080/OP0595) combinations against MBL-producing Enterobacteriaceae. J Antimicrob Chemother 74:953–960. doi: 10.1093/jac/dky522. [DOI] [PubMed] [Google Scholar]
- 25.Jin W, Wachino JI, Yamaguchi Y, Kimura K, Kumar A, Yamada M, Morinaka A, Sakamaki Y, Yonezawa M, Kurosaki H, Arakawa Y. 2017. Structural insights into the TLA-3 extended-spectrum β-lactamase and its inhibition by avibactam and OP0595. Antimicrob Agents Chemother 61:e00501-17. doi: 10.1128/AAC.00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morinaka A, Tsutsumi Y, Yamada K, Takayama Y, Sakakibara S, Takata T, Abe T, Furuuchi T, Inamura S, Sakamaki Y, Tsujii N, Ida T. 2017. In vitro and in vivo activities of the diazabicyclooctane OP0595 against AmpC-derepressed Pseudomonas aeruginosa. J Antibiot 70:246–250. doi: 10.1038/ja.2016.150. [DOI] [PubMed] [Google Scholar]
- 27.Morinaka A, Tsutsumi Y, Yamada K, Takayama Y, Sakakibara S, Takata T, Abe T, Furuuchi T, Inamura S, Sakamaki Y, Tsujii N, Ida T. 2016. In vitro and in vivo activities of OP0595, a new diazabicyclooctane, against CTX-M-15-positive Escherichia coli and KPC-positive Klebsiella pneumoniae. Antimicrob Agents Chemother 60:3001–3006. doi: 10.1128/AAC.02704-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaku N, Kosai K, Takeda K, Uno N, Morinaga Y, Hasegawa H, Miyazaki T, Izumikawa K, Mukae H, Yanagihara K. 2017. Efficacy and pharmacokinetics of the combination of OP0595 and cefepime in a mouse model of pneumonia caused by extended-spectrum-β-lactamase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 61:e00828-17. doi: 10.1128/AAC.00828-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monogue ML, Giovagnoli S, Bissantz C, Zampaloni C, Nicolau DP. 2018. In vivo efficacy of meropenem with a novel non-β-lactam–β-lactamase inhibitor, nacubactam, against Gram-negative organisms exhibiting various resistance mechanisms in a murine complicated urinary tract infection model. Antimicrob Agents Chemother 62:e02596-17. doi: 10.1128/AAC.02596-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing; twenty-seventh informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 31.The European Committee on Antimicrobial Susceptibility Testing. 2018. Breakpoint tables for interpretation of MICs and zone diameters, version 8.1. http://wwweucastorg.
- 32.Barnes MD, Winkler ML, Taracila MA, Page MG, Desarbre E, Kreiswirth BN, Shields RK, Nguyen MH, Clancy C, Spellberg B, Papp-Wallace KM, Bonomo RA. 2017. Klebsiella pneumoniae carbapenemase-2 (KPC-2), substitutions at Ambler position Asp179, and resistance to ceftazidime-avibactam: unique antibiotic-resistant phenotypes emerge from β-lactamase protein engineering. mBio 8:e00528-17. doi: 10.1128/mBio.00528-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levitt PS, Papp-Wallace KM, Taracila MA, Hujer AM, Winkler ML, Smith KM, Xu Y, Harris ME, Bonomo RA. 2012. Exploring the role of a conserved class A residue in the omega-loop of KPC-2 β-lactamase: a mechanism for ceftazidime hydrolysis. J Biol Chem 287:31783–31793. doi: 10.1074/jbc.M112.348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joris B, Ledent P, Dideberg O, Fonze E, Lamotte-Brasseur J, Kelly JA, Ghuysen JM, Frere JM. 1991. Comparison of the sequences of class A β-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother 35:2294–2301. doi: 10.1128/AAC.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winkler ML, Rodkey EA, Taracila MA, Drawz SM, Bethel CR, Papp-Wallace KM, Smith KM, Xu Y, Dwulit-Smith JR, Romagnoli C, Caselli E, Prati F, van den Akker F, Bonomo RA. 2013. Design and exploration of novel boronic acid inhibitors reveals important interactions with a clavulanic acid-resistant sulfhydryl-variable (SHV) β-lactamase. J Med Chem 56:1084–1097. doi: 10.1021/jm301490d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lahiri SD, Mangani S, Durand-Reville T, Benvenuti M, De Luca F, Sanyal G, Docquier JD. 2013. Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-lactamases. Antimicrob Agents Chemother 57:2496–2505. doi: 10.1128/AAC.02247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papp-Wallace KM, Winkler ML, Taracila MA, Bonomo RA. 2015. Variants of β-lactamase KPC-2 that are resistant to inhibition by avibactam. Antimicrob Agents Chemother 59:3710–3717. doi: 10.1128/AAC.04406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Durand-Reville TF, Lahiri S, Thresher J, Livchak S, Gao N, Palmer T, Walkup GK, Fisher SL. 2013. Kinetics of avibactam inhibition against class A, C, and D β-lactamases. J Biol Chem 288:27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drawz SM, Bonomo RA. 2010. Three decades of β-lactamase inhibitors. Clin Microbiol Rev 23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krishnan NP, Nguyen NQ, Papp-Wallace KM, Bonomo RA, van den Akker F. 2015. Inhibition of Klebsiella β-lactamases (SHV-1 and KPC-2) by avibactam: a structural study. PLoS One 10:e0136813. doi: 10.1371/journal.pone.0136813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Duin D, Perez F, Rudin SD, Cober E, Hanrahan J, Ziegler J, Webber R, Fox J, Mason P, Richter SS, Cline M, Hall GS, Kaye KS, Jacobs MR, Kalayjian RC, Salata RA, Segre JA, Conlan S, Evans S, Fowler VG Jr, Bonomo RA. 2014. Surveillance of carbapenem-resistant Klebsiella pneumoniae: tracking molecular epidemiology and outcomes through a regional network. Antimicrob Agents Chemother 58:4035–4041. doi: 10.1128/AAC.02636-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doi Y, Bonomo RA, Hooper DC, Kaye KS, Johnson JR, Clancy CJ, Thaden JT, Stryjewski ME, van Duin D, Gram-Negative Committee of the Antibacterial Resistance Leadership Group (ARLG)a. 2017. Gram-negative bacterial infections: research priorities, accomplishments, and future directions of the Antibacterial Resistance Leadership Group. Clin Infect Dis 64:S30–S35. doi: 10.1093/cid/ciw829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yigit H, Queenan AM, Rasheed JK, Biddle JW, Domenech-Sanchez A, Alberti S, Bush K, Tenover FC. 2003. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing β-lactamase KPC-2. Antimicrob Agents Chemother 47:3881–3889. doi: 10.1128/AAC.47.12.3881-3889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papp-Wallace KM, Winkler ML, Gatta JA, Taracila MA, Chilakala S, Xu Y, Johnson JK, Bonomo RA. 2014. Reclaiming the efficacy of β-lactam–β-lactamase inhibitor combinations: avibactam restores the susceptibility of CMY-2-producing Escherichia coli to ceftazidime. Antimicrob Agents Chemother 58:4290–4297. doi: 10.1128/AAC.02625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papp-Wallace KM, Taracila MA, Smith KM, Xu Y, Bonomo RA. 2012. Understanding the molecular determinants of substrate and inhibitor specificities in the carbapenemase KPC-2: exploring the roles of Arg220 and Glu276. Antimicrob Agents Chemother 56:4428–4438. doi: 10.1128/AAC.05769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]