LETTER
In our recent study of 203 sequential isolates evaluating the ability of whole-genome sequencing (WGS) to predict clarithromycin resistance in Mycobacterium abscessus (1), we demonstrated high sensitivity but poor specificity of mutations identified in a literature search. Most of the discrepancies occurred in isolates predicted to be inducibly resistant (they showed early time point susceptibility but became resistant after prolonged incubation).
WGS for nontuberculous mycobacteria has continued to be rolled out across England with all isolates from London included from January 2018, affording us the opportunity to examine the performance of the algorithm we described previously on an independent data set (none of the isolates reported here were included in our previous study). We prepared a customized Ariba (2) library to implement the algorithm outlined in our recent publication (1), extended to include the novel mutations which we described. Using this, we predicted clarithromycin susceptibility for 259 isolates (10 with intermediate susceptibility were excluded) and compared these results to those obtained from DST using RAPMYCO (Thermo Fisher) plates as previously described. Isolates were classified as resistant if they showed early or late time point (inducible) resistance.
This yielded a sensitivity of 194/197 (98%, 95% confidence interval [CI] = 96 to 100%) and a specificity of 61/62 (98%, 95% CI = 91 to 100%) (Fig. 1). For comparison, our previously reported values were a sensitivity of 95/100 (95%; 95% CI = 89 to 98%) and a specificity of 52/79 (66%; 95% CI = 54 to 76%). The very major error rate on this new data set was 3/194 (2%; 95% CI = 0 to 3%), and the major error rate was 1/62 (2%; 95% CI = −2 to 5%) (3). Of the novel rrl mutations which we described previously, T371C, G795A, and A1932G were also seen in this data set and again always associated with resistance. As in our original study, these occurred only in samples otherwise predicted inducibly resistant; the overall sensitivity/specificity was therefore the same regardless of whether they were included. For all isolates with discordant genotypic predictions or intermediate phenotypes, we considered the possibility of mixed infection or within patient subclones (4), using a previously described probabilistic method (5). This demonstrated that one such isolate predicted sensitive (due to being subspecies massiliense) but with a resistant phenotype was likely a mixed massiliense/abscessus infection. Two further isolates with intermediate phenotypes, one predicted sensitive and the other resistant, were found with high posterior probability (>99%) to have subclones with mutations at positions which confer macrolide resistance in the rrl gene.
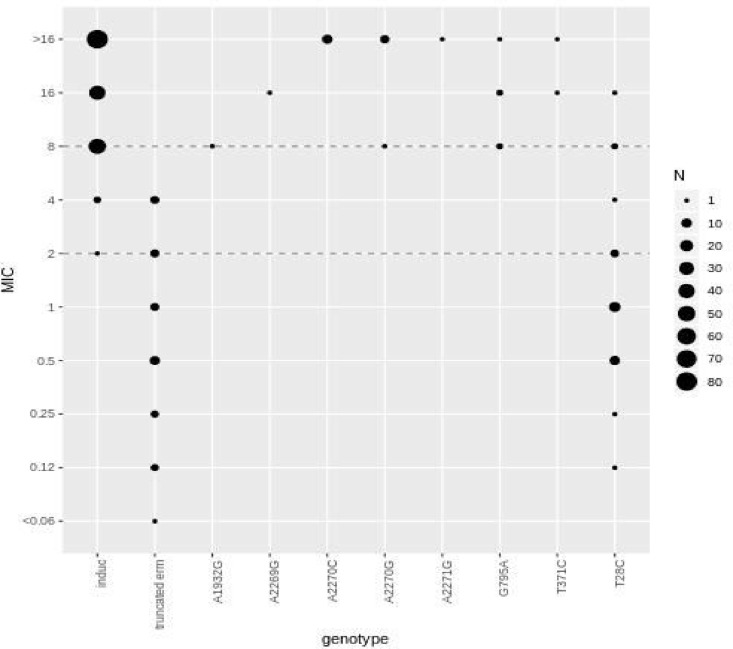
FIG 1.
MIC (μg/ml) frequency by genotype for 269 M. abscessus isolates (including those with intermediate phenotypes). Dotted lines represent the Clinical and Laboratory Standards Institute breakpoints for clarithromycin (≤2 μg/ml, sensitive; >2 to <8 μg/ml, intermediate; and ≥8 μg/ml, resistant). Induc, inducible resistance (the isolate is subspecies bolletii/abscessus with no other relevant mutation and thus genotypically predicted to show inducible resistance); truncated erm, massiliense isolate with truncated erm(41) gene and no other relevant mutations. All other mutations except T28C, which is a mutation in erm(41), represent nucleotide mutations in the rrl gene.
The improved performance of our algorithm may reflect the increased experience which has been gained in the RAPMYCO Sensititre method by laboratory staff over time. The performance of WGS for predicting clarithromycin susceptibility in M. abscessus is now approaching U.S. Food and Drug Administration standards; providing genotypic information alongside DST to clinicians is likely to be desirable. Considering mixed populations may add value to resistance prediction in M. abscessus. The sequencing data from this second data set is available under project accession number PRJNA420644 and our Ariba library (https://github.com/samlipworth/Mab_ariba) is freely available, which will allow others to easily apply our algorithm to their own data.
ACKNOWLEDGMENTS
The research was supported by the National Institute for Health Research (NIHR) Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance at University of Oxford in partnership with Public Health England and by Oxford NIHR Biomedical Research Centre. T.P. is an NIHR Senior Investigator. The report presents independent research funded by NIHR. The views expressed in this publication are those of the authors and not necessarily those of the NHS, NIHR, the Department of Health or Public Health England. Computation used the Oxford Biomedical Research Computing facility, a joint development between the Wellcome Centre for Human Genetics and the Big Data Institute supported by Health Data Research UK and the NIHR Oxford Biomedical Research Centre.
REFERENCES
- 1.Lipworth S, Hough N, Leach L, Morgan M, Jeffery K, Andersson M, Robinson E, Smith EG, Crook D, Peto T, Walker T. 2018. Whole-genome sequencing for predicting clarithromycin resistance in Mycobacterium abscessus. Antimicrob Agents Chemother 63:e01204-18. doi: 10.1128/AAC.01204-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, Keane JA, Harris SR. 2017. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom 3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuper KM, Boles DM, Mohr JF, Wanger A. 2009. Antimicrobial susceptibility testing: a primer for clinicians. Pharmacotherapy 29:1326–1343. doi: 10.1592/phco.29.11.1326. [DOI] [PubMed] [Google Scholar]
- 4.Shaw LP, Doyle RM, Kavaliunaite E, Spencer H, Balloux F, Dixon G, Harris KA. 2019. Children with cystic fibrosis are infected with multiple subpopulations of Mycobacterium abscessus with different antimicrobial resistance profiles. Clin Infect Dis doi: 10.1093/cid/ciz069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eyre DW, Cule ML, Griffiths D, Crook DW, Peto TEA, Walker AS, Wilson DJ. 2013. Detection of mixed infection from bacterial whole-genome sequence data allows assessment of its role in Clostridium difficile transmission. PLoS Comput Biol 9:e1003059. doi: 10.1371/journal.pcbi.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]