Combination antiretroviral therapy has achieved dramatic reductions in the mortality and morbidity in people with HIV-1 infection. Darunavir (DRV) represents a most efficacious and well-tolerated protease inhibitor (PI) with a high genetic barrier to the emergence of drug-resistant HIV-1. However, highly DRV-resistant variants have been reported in patients receiving long-term DRV-containing regimens.
KEYWORDS: AIDS, human immunodeficiency virus, multidrug resistance, protease inhibitors
ABSTRACT
Combination antiretroviral therapy has achieved dramatic reductions in the mortality and morbidity in people with HIV-1 infection. Darunavir (DRV) represents a most efficacious and well-tolerated protease inhibitor (PI) with a high genetic barrier to the emergence of drug-resistant HIV-1. However, highly DRV-resistant variants have been reported in patients receiving long-term DRV-containing regimens. Here, we report three novel HIV-1 PIs (GRL-057-14, GRL-058-14, and GRL-059-14), all of which contain a P2-amino-substituted-bis-tetrahydrofuranylurethane (bis-THF) and a P2′-cyclopropyl-amino-benzothiazole (Cp-Abt). These PIs not only potently inhibit the replication of wild-type HIV-1 (50% effective concentration [EC50], 0.22 nM to 10.4 nM) but also inhibit multi-PI-resistant HIV-1 variants, including highly DRV-resistant HIVDRVRP51 (EC50, 1.6 nM to 30.7 nM). The emergence of HIV-1 variants resistant to the three compounds was much delayed in selection experiments compared to resistance to DRV, using a mixture of 11 highly multi-PI-resistant HIV-1 isolates as a starting HIV-1 population. GRL-057-14 showed the most potent anti-HIV-1 activity and greatest thermal stability with wild-type protease, and potently inhibited HIV-1 protease’s proteolytic activity (Ki value, 0.10 nM) among the three PIs. Structural models indicate that the C-5-isopropylamino-bis-THF moiety of GRL-057-14 forms additional polar interactions with the active site of HIV-1 protease. Moreover, GRL-057-14’s P1-bis-fluoro-methylbenzene forms strong hydrogen bonding and effective van der Waals interactions. The present data suggest that the combination of C-5-aminoalkyl-bis-THF, P1-bis-fluoro-methylbenzene, and P2ʹ-Cp-Abt confers highly potent activity against wild-type and multi-PI-resistant HIV strains and warrant further development of the three PIs, in particular, that of GRL-057-14, as potential therapeutic for HIV-1 infection and AIDS.
INTRODUCTION
The treatment of human immunodeficiency virus type 1 (HIV-1) infection and AIDS has drastically improved with the use of combined antiretroviral therapy (cART), and the mortality and morbidity related to HIV-1 infection have reduced substantially (1). Protease inhibitors (PIs) are considered key drugs of cART regimens that competitively inhibit the cleavage of the Gag-Pol polyproteins during viral maturation, resulting in the production of noninfectious virions. In general, PIs have a relatively high genetic barrier to the emergence of drug-resistant viruses compared to nonnucleoside reverse transcriptase inhibitors (NNRTIs) and most integrase strand transfer inhibitors (INSTIs), even in patients with poor adherence (2–4). Darunavir (DRV), a bis-tetrahydrofuranylurethane (bis-THF) containing nonpeptidic PI (5), is preferred for cART-naive HIV-1 infected patients due to its tolerance and a high genetic barrier to the development of drug-resistant viruses (6). However, the emergence of DRV resistance has been reported in patients failing DRV-containing regimens in multiple clinical trials (7–11). We previously selected highly DRV-resistant HIV-1 variants in vitro (HIVDRVRP10, HIVDRVRP30, and HIVDRVRP51) by employing a mixture of eight multidrug-resistant (MDR) HIV-1 variants as a starting HIV-1 population, and reported a combination of four amino acid substitutions (V32I, L33F, I54M, and I84V) in the protease-encoding gene were responsible for the high-level DRV resistance in HIVDRVRP51 (12). We also reported that the V32I substitution serves as one of the key mutations for the development of DRV resistance (13). In this regard, the development of novel antiviral agents, which are active against wild-type and existing drug-resistant HIV-1s, possess a high genetic barrier against the emergence of drug-resistant HIV-1, and have favorable safety features, is urgently needed.
Here, we report three novel nonpeptidic PIs, GRL-057-14, GRL-058-14, and GRL-059-14, which contain P2-C-5-aminoalkyl-bis-THF and P2ʹ-cyclopropyl-amino-benzothiazole (Cp-Abt) as the common structural features, exhibit potent antiviral activity against not only wild-type HIV but also against various drug-resistant variants, including highly DRV-resistant HIVDRVRP51. We also found that these compounds bind to the HIV-1 protease specifically and inhibit protease enzymatic function, and do not allow the emergence of highly resistant viruses even after 50 weeks of selection in vitro.
RESULTS
GRL-057-14, GRL-058-14, and GRL-059-14 exert potent antiviral activity against wild-type HIV-1NL4-3.
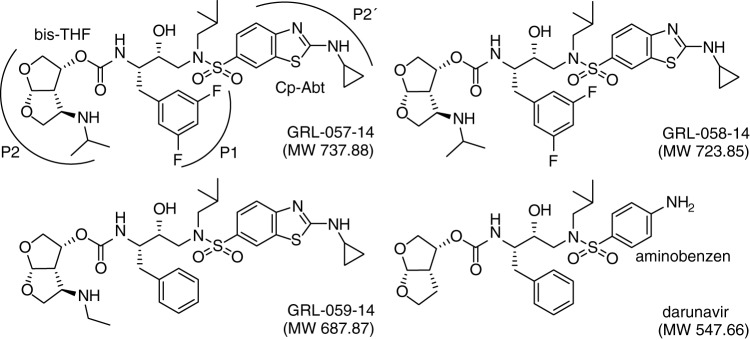
We designed, synthesized, and identified three novel PIs, GRL-057-14, GRL-058-14, and GRL-059-14. These three PIs have in common C-5-modified bis-THF and Cp-Abt moieties in the P2 and P2ʹ sites, respectively (Fig. 1). The C-5 substituents are isopropylamine for GRL-057-14 and ethyl-amine for GRL-058-14 and GRL-059-14, respectively. GRL-057-14 and GRL-058-14 have a meta-bis-fluoro-methylbenzene as the P1 site moiety, while GRL-059-14 and DRV contain a methylbenzene at the P1 site (Fig. 1). As shown in Table 1, all three PIs (GRL-057-14, GRL-058-14, and GRL-059-14) showed potent antiviral activity against wild-type HIV-1NL4-3, with 50% effective concentration (EC50) values of 0.34, 2.7, and 3.6 nM, respectively. Notably, GRL-057-14 was the most potent agent among the three novel PIs and the currently available FDA-approved PIs examined, including DRV (Table 1). These compounds were also active against HIV-2EHO, while amprenavir (APV) and tipranavir (TPV) were much less potent, in line with others’ data (14, 15). Interestingly, GRL-057-14 and GRL-058-14 showed more potent activity against HIV-2EHO (EC50 values of 0.0045 and 0.58 nM, respectively) than against HIV-1NL4-3 (EC50 values of 0.34 and 2.7 nM, respectively). Moreover, all three PIs had favorable cytotoxicity profiles, with 50% cytotoxicity concentration (CC50) values of 45.4, 42.8, and 40.5 μM, giving selectivity index (SI) values of 133,530, 15,850, and 11,250, respectively (Table 1).
FIG 1.
The structures of GRL-057-14, GRL-058-14, GRL-059-14, and darunavir (DRV).
TABLE 1.
Antiviral activity and cytotoxicity of GRL-057-14, GRL-058-14, and GRL-059-14 against wild-type HIVsa
| Compound | EC50b
(nM) against |
CC50c (μM) in MT-4 cells | SId with HIV-1NL4-3 (CC50/EC50) | |
|---|---|---|---|---|
| HIV-1NL4-3 | HIV-2EHO | |||
| GRL-057-14 | 0.34 ± 0.3 | 0.0045 ± 0.0026 | 45.4 ± 6.4 | 133,530 |
| GRL-058-14 | 2.7 ± 0.3 | 0.58 ± 0.29 | 42.8 ± 3.9 | 15,850 |
| GRL-059-14 | 3.6 ± 0.1 | 21.9 ± 6.4 | 40.5 ± 0.5 | 11,250 |
| APV | 28.3 ± 1.7 | 354.7 ± 9.3 | >100 | >3,500 |
| LPV | 1.3 ± 1.1 | 12.9 ± 4.1 | 46.9 ± 1.9 | 36,000 |
| TPV | 25.4 ± 1.1 | 285.8 ± 27.3 | 42.8 ± 2.8 | 1,685 |
| SQV | 2.8 ± 0.4 | 5.8 ± 3.3 | 41.2 ± 0.6 | 14,710 |
| DRV | 3.1 ± 1.0 | 24.8 ± 6.5 | 284.7 ± 37.5 | 91,840 |
Anti-HIV-1 activity was determined using MT-4 cells by MTT assay. Each assay was conducted in triplicate. The values represent mean values (±1 SD) of two or three independent experiments.
EC50, 50% effective concentration.
CC50, 50% cytotoxic concentration.
SI, selectivity index (CC50/EC50).
GRL-057-14, GRL-058-14, and GRL-059-14 exert potent antiviral activity against various PI-resistant HIV-1 variants, including highly DRV-resistant variants.
We then attempted to examine whether GRL-057-14, GRL-058-14, and GRL-059-14 were active against a variety of HIV-1 variants that had been selected in vitro with each of four FDA-approved PIs, amprenavir (APV), lopinavir (LPV), saquinavir (SQV), and tipranavir (TPV) (Table 2). All of the resulting variants (HIVAPV-5μM, HIVLPV-5μM, HIVSQV-5μM, and HIVTPV-15μM) had acquired multiple amino acid substitutions in the protease-encoding gene, which have reportedly been associated with viral resistance to PIs (Fig. S1A) (12, 16, 17). The four PI-resistant variants were highly resistant to each corresponding PI, with which the variant was selected. The EC50 differences were >30-fold to >415-fold compared to those values against HIV-1NL4-3. The variants also showed some cross-resistance against PIs, with which such variants were not selected (Table 2). All three novel PIs were active against all the four variants, with EC50 values of 0.00001 nM (10 fM) to 17.6 nM. It was of note that GRL-057-14 showed extremely potent activity against all the four variants examined, presenting EC50 values ranging from 0.00001 to 0.00063 nM (10 fM to 630 fM), which were significantly more potent than that against HIV-1NL4-3. We also determined the antiviral activity of the three novel PIs and selected FDA-approved PIs, nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs), and INSTI against HIV11MIX and in vitro-selected DRV-resistant HIV-1 (HIVDRVRs; HIVDRVRP10, HIVDRVRP30, and HIVDRVRP51) (Table 2). HIVDRVRs were selected in the presence of increasing concentrations of DRV with propagation of the mixture of 8 highly multi-PI-resistant, DRV-susceptible clinical HIV-1 variants (12). Although the four NRTIs (zidovudine [AZT], tenofovir disoproxil fumarate [TDF], abacavir [ABC], and 4ʹ-ethynyl-2-fluoro-2-deoxyadenosine [EFdA/MK-8591]) were active against HIV-1NL4-3, they had become much less active against HIV11MIX and HIVDRVRs, since these viruses were derived from a mixed population of clinical HIV-1 strains isolated from heavily NRTI-experienced AIDS patients. As expected, an INSTI, dolutegravir (DTG), showed highly potent activity against all of the variants, since none of these patients had received INSTI (Table 2). All three novel PIs were active against all the HIV-1 variants tested, with EC50 values of 0.000044 nM (44 fM) to 30.7 nM (Table 2). It was noted that these three novel PIs had potent activity against the two in vitro DRV-selected variants (HIVDRVRP10 and HIVDRVRP30), with EC50 values ranging from 0.000044 nM (44 fM) to 2.5 nM, which were more potent than that against HIV-1NL4-3 (Table 2). All three novel PIs also remained substantially active against the most highly DRV-resistant HIVDRVRP51, with the difference in EC50 values ranging from only 1- to 7-fold (Table 2).
TABLE 2.
Antiviral activity of selected compounds against PI-resistant HIV-1 variantsa
| Compound | EC50 (nM) againstb
: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| HIV-1NL4-3 | HIVAPV-5μM | HIVLPV-5μM | HIVSQV-5μM | HIVTPV-15μM | HIV11MIX | HIVDRVRP10 | HIVDRVRP30 | HIVDRVRP51 | |
| GRL-057-14 | 0.22 ± 0.09 | 0.00001 ± 0.00001 (0.00005) | 0.00001 ± 0.000005 (0.000005) | 0.00063 ± 0.00047 (0.003) | 0.0001 ± 0.00001 (0.005) | 0.0027 ± 0.0012 (0.01) | 0.000044 ± 0.00002 (0.0002) | 0.00067 ± 0.00093 (0.003) | 1.6 ± 1.9 (7) |
| GRL-058-14 | 9.2 ± 5.3 | 0.3 ± 0.1 (0.04) | 0.00002 ± 0.000005 (0.000002) | 0.0066 ± 0.0035 (0.0007) | 3.3 ± 1.2 (0.4) | 5.6 ± 2.4 (0.6) | 0.25 ± 0.28 (0.03) | 0.09 ± 0.08 (0.01) | 9.4 ± 11.5 (1) |
| GRL-059-14 | 10.4 ± 4.4 | 2.6 ± 0.4 (0.25) | 0.005 ± 0.004 (0.0005) | 0.023 ± 0.02 (0.002) | 17.6 ± 4.0 (1.7) | 19.8 ± 9.8 (1.9) | 2.5 ± 3.1 (0.2) | 0.51 ± 0.31 (0.05) | 30.7 ± 15.8 (3) |
| APV | 29.4 ± 9.0 | >1,000 (>34) | 615.1 ± 109.8 (21) | 48.6 ± 11.3 (1.7) | 339.5 ± 33.0 (11.5) | 263.0 ± 72.0 (9) | 327 ± 21.2 (11) | >1,000 (>35) | >1,000 (>35) |
| LPV | 2.4 ± 2.6 | 20.0 ± 4.0 (8.3) | >1,000 (>415) | 21.8 ± 16.4 (9) | 444.1 ± 185.5 (185) | 291.8 ± 59.9 (122) | 306.0 ± 130.6 (128) | >1,000 (>415) | >1,000 (>415) |
| SQV | 2.5 ± 1.0 | 1.2 ± 0.1 (0.5) | 0.08 ± 0.05 (0.03) | >1,000 (>400) | 455.4 ± 138.1 (180) | 22.3 ± 6.5 (9) | 15.5 ± 3.5 (6) | 197.7 ± 11.2 (80) | 271.3 ± 14.9 (108) |
| TPV | 33.2 ± 2.1 | 6.5 ± 3.7 (0.2) | 17.0 ± 3.3 (0.5) | 27.4 ± 6.5 (0.8) | >1,000 (>30) | 40.0 ± 4.8 (1.2) | 145.1 ± 18.1 (4) | 298.0 ± 23.3 (9) | 354.3 ± 51.8 (10) |
| NFV | 32.1 ± 3.2 | 10.4 ± 3.5 (0.3) | 243.3 ± 46.5 (8) | 114.5 ± 95.4 (3.5) | >1,000 (>30) | 563.7 ± 138.5 (18) | 632.7 ± 78.5 (20) | >1,000 (>30) | >1,000 (>30) |
| IDV | 25.4 ± 9.8 | 19.0 ± 1.2 (0.7) | 306.6 ± 90.9 (12) | 53.8 ± 47.6 (2) | >1,000 (>40) | >1,000 (>40) | >1,000 (>40) | >1,000 (>40) | >1,000 (>40) |
| DRV | 3.0 ± 1.0 | 20.6 ± 0.7 (6.9) | 20.4 ± 2.2 (6.8) | 2.1 ± 0.3 (0.7) | 25.4 ± 2.5 (8.5) | 3.5 ± 0.4 (1) | 27.6 ± 24.1 (9) | 220.5 ± 24.4 (73) | 4,530.7 ± 1,275.6 (1,510) |
| AZT | 32.5 ± 6.1 | ND | ND | ND | ND | >1,000 (>30) | >1,000 (>30) | >1,000 (>30) | >1,000 (>30) |
| TDF | 305.7 ± 69.9 | ND | ND | ND | ND | 7,041.6 ± 1,965.8 (23) | >10,000 (>30) | >10,000 (>30) | >10,000 (>30) |
| ABC | 1514.1 ± 841.8 | ND | ND | ND | ND | >10,000 (>6) | >10,000 (>6) | >10,000 (>6) | >10,000 (>6) |
| EFdA | 0.98 ± 0.23 | ND | ND | ND | ND | 39.1 ± 11.4 (48) | 29.4 ± 8.0 (30) | 27.7 ± 11.5 (28) | 59.1 ± 16.2 (60) |
| DTG | 0.7 ± 0.7 | ND | ND | ND | ND | 0.5 ± 0.3 (0.7) | 0.2 ± 0.2 (0.3) | 0.4 ± 0.2 (0.6) | 0.9 ± 1.1 (1) |
Anti-HIV-1 activity was determined with a p24 assay employing MT-4 cells. Each assay was conducted in triplicate. The values represent mean values (±1 SD) of two or three independent experiments. Numbers in parentheses represent fold changes in EC50s for each isolate compared to the EC50s for wild-type HIV-1NL4-3.
EC50, 50% effective concentration; ND, not determined.
GRL-057-14, GRL-058-14, and GRL-059-14 exert potent antiviral activity against various recombinant MDR HIV-1 strains and non-B subtype.
We also attempted to evaluate the anti-HIV-1 activity of GRL-057-14, GRL-058-14, and GRL-059-14 against five recombinant clinical HIV-1 isolates (rCLHIVs), which were originally isolated from patients who had received antiretroviral therapy containing multiple PIs (the median number of PIs received was 5, excluding the use of ritonavir as a boosting agent) over a median duration of 7.5 years. These viruses acquired 16 to 23 PI-resistance associated amino acid substitutions in their protease-encoding gene (Fig. S1A) (18, 19). APV, LPV, and SQV had anti-HIV-1 activity against rCLHIVs, with approximately 100-fold reductions of EC50 values compared to that against HIV-1NL4-3; even the highest concentration (1 μM) of the PIs tested failed to achieve the 50% virus replication reduction in multiple variants (Table 3). Overall, TPV, NFV, and IDV had smaller fold changes (0.7- to 30-fold change) compared to those of APV, LPV, and SQV, although the absolute EC50 values of those drugs were comparable (Table 3). DRV had also been compromised in its activity, with approximately ∼100-fold changes in EC50 values compared to that against HIV-1NL4-3. However, the three current PIs still retained their anti-HIV-1 activity against all of the rCLHIVs examined, and the fold changes were as low as 0.002 to 10 (Table 3). It is noteworthy that these PIs were significantly more potent against some of the rCLHIVs, especially against rCLHIVF16 and rCLHIVT48 (Table 3). It has been reported that non-B subtype HIV-1s, especially subtype-C HIV-1, which causes about half of HIV-1 infections worldwide, mostly in sub-Saharan Africa (20), have less susceptibility against PIs (21–23) due to its amino acid polymorphisms that have been associated with drug resistance in the subtype B viruses (24). In this regard, we determined the antiviral activity of PIs against an X4-tropic subtype A strain (HIV-192/UG/029) and an R5-tropic subtype C strain (HIV-197/ZA/003) using phytohemagglutinin (PHA)-stimulated peripheral blood mononuclear cells (PBMCs) as target cells. As shown in Table S1, all three novel PIs (GRL-057-14, GRL-058-14, and GRL-059-14) effectively inhibited the replication of the strains examined. In these data, GRL-057-14 was as potent as DRV; while the other two PIs were less potent than DRV (Table S1).
TABLE 3.
Antiviral activity of PIs against recombinant clinical HIV-1 variantsa
| Compound | EC50 (nM) againstb
: |
|||||
|---|---|---|---|---|---|---|
| HIV-1NL4-3c | rCLHIVF16 | rCLHIVT48 | rCLHIVV40 | rCLHIVT44 | rCLHIVM45 | |
| GRL-057-14 | 0.22 ± 0.09 | 0.003 ± 0.001 (0.01) | 0.0005 ± 0.0002 (0.002) | 2.7 ± 1.0 (10) | 0.47 ± 0.47 (2) | 0.07 ± 0.06 (0.3) |
| GRL-058-14 | 9.2 ± 5.3 | 0.01 ± 0.01 (0.001) | 0.18 ± 0.11 (0.02) | 2.1 ± 1.3 (0.2) | 0.6 ± 0.5 (0.06) | 0.52 ± 0.16 (0.05) |
| GRL-059-14 | 10.4 ± 4.4 | 0.47 ± 0.45 (0.05) | 1.8 ± 1.7 (0.2) | 27.3 ± 3.7 (2.7) | 20.5 ± 12.9 (2) | 12.8 ± 3.2 (1.2) |
| APV | 29.4 ± 9.0 | >1,000 (>35) | >1,000 (>35) | 476.4 ± 64.0 (16) | >1,000 (>35) | >1,000 (>35) |
| LPV | 2.4 ± 2.6 | >1,000 (>415) | 290.2 ± 117.9 (120) | 396.0 ± 84.5 (165) | 590.1 ± 38.8 (245) | 426.2 ± 81.7 (175) |
| SQV | 2.5 ± 1.0 | 192.0 ± 205.4 (75) | 210.9 ± 138.0 (85) | >1,000 (>400) | 268.5 ± 40.3 (105) | 252.1 ± 83.7 (100) |
| TPV | 33.2 ± 2.1 | 153.6 ± 3.8 (5) | 235.7 ± 120.3 (7) | 21.7 ± 7.1 (0.7) | 181.5 ± 11.7 (5) | 464.8 ± 100.3 (14) |
| NFV | 32.1 ± 3.2 | 359.4 ± 194.2 (10) | 547.9 ± 134.5 (17) | 254.3 ± 67.4 (8) | 809.3 ± 138.4 (25) | >1,000 (>30) |
| IDV | 25.4 ± 9.8 | 162.0 ± 128.9 (6) | 376.8 ± 40.5 (15) | 438.2 ± 27.1 (17) | 416.9 ± 59.8 (6) | 524.8 ± 80.0 (20) |
| DRV | 3.0 ± 1.0 | 338.4 ± 152.5 (110) | 438.0 ± 239.9 (146) | 11.9 ± 7.8 (4) | 181.5 ± 22.3 (60) | 215.8 ± 91.0 (70) |
Anti-HIV-1 activity was determined with a p24 assay employing MT-4 cells. Each assay was conducted in triplicate. The values represent mean values (±1 SD) of two or three independent experiments. Numbers in parentheses represent fold changes in EC50s for each isolate compared to the EC50s for wild-type HIV-1NL4-3.
EC50, 50% effective concentration.
Data from Table 2 (HIV-1NL4-3).
GRL-057-14, GRL-058-14, and GRL-059-14 had significantly increased thermal stability with HIV-1 protease.
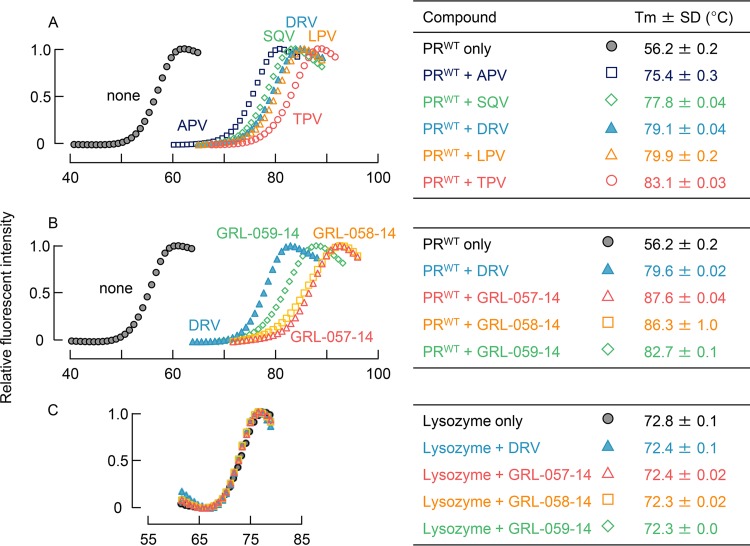
In an attempt to corroborate and interrogate the mechanism of the potent anti-HIV-1 activity of the three novel PIs, we examined the thermal stability with wild-type HIV-1 protease (PRWT) in the presence and absence of each PI, using differential scanning fluorimetry (DSF) (25). As illustrated in Fig. 2A and B, as HIV-1 PRWT was assumed to be denatured as the temperature increased and hydrophobic patches of the protein became exposed, the fluorescent dye (SYPRO orange) is thought to bind to the patches, and strong fluorescent light is emitted (26). The fluorescence intensity curves were analyzed, and the melting temperature (Tm) values were determined. When the PRWT was denatured in the absence of PIs, the Tm value was 56.18°C, which drastically increased when the denaturation occurred in the presence of PIs, giving Tm values ranging from 75.42°C (for APV) to 83.08°C (for TPV) and ΔTm values ranging from 19.24°C to 26.90°C (Fig. 2A). There was also a Tm value increment with DRV, giving a ΔTm value of 22.88°C, suggesting that the thermal stability of PRWT increased when all of the PIs tested bound to PRWT. Of note, GRL-057-14, GRL-058-14, and GRL-059-14 had significantly increased Tm values of 87.61, 86.27, and 82.68°C, respectively (Fig. 2B). These values were much higher than those of other PIs tested, giving much greater ΔTm values and strongly suggesting that these three novel PIs more strongly bind to PRWT than the FDA-approved PIs examined here. On the other hand, the stability curves of lysozyme did not shift at all with DRV and the three PIs, strongly suggesting that the binding of DRV and the three PIs to PRWT was highly specific to PRWT (Fig. 2C).
FIG 2.
Thermal stability of wild-type HIV-1 protease (PRWT) and lysozyme in the presence and absence of each compound using differential scanning fluorimetry (DSF). The relative fluorescence intensity was plotted at each temperature. Melting temperature (Tm) values were calculated from the top of the peak in the derivative plot. (A) Melting curves of PRWT in the presence and absence of selected FDA-approved PIs. Thermal stability curves with all the compounds shifted to higher temperatures than in the absence of each compound with Tm values ranging from 75.42 (amprenavir [APV]) to 83.08°C (tipranavir [TPV]), with a ΔTm value of 19.24°C to 26.90°C. (B) Melting curves of PRWT in the presence and absence of GRL-057-14, GRL-058-14, and GRL-059-14. The curve of DRV is shown as a reference. All three compounds significantly shifted the thermal stability curves to higher temperatures than in the absence of each compound. Their Tm values were much higher than those of all the FDA-approved PIs tested (A). (C) Melting curves of lysozyme in the presence and absence of GRL-057-14, GRL-058-14, GRL-059-14, and DRV. None of the PIs examined affected the thermal stability curves, indicating that all the PIs examined were highly specific to PRWT and had no significant interactions with lysozyme.
GRL-057-14 inhibited the HIV-1 protease activity in vitro.
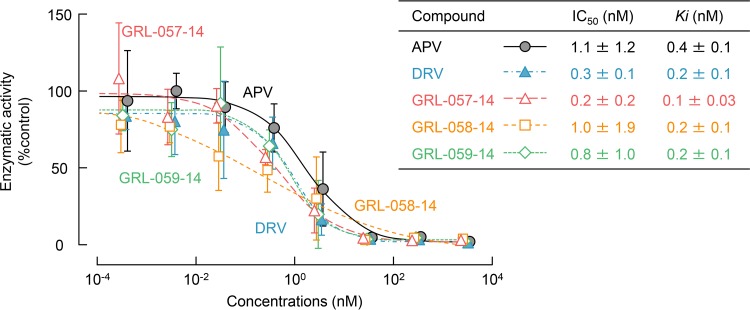
We also determined the inhibition constants (Ki) of PIs in HIV-1 protease enzyme inhibition assays. First, we conducted the enzyme reaction assay in the absence of PIs at several different concentrations (up to 100 μM) of the substrate to determine the catalytic rate constant (kcat) and the Michaelis constant (Km). The change in fluorescence was measured, and the relative fluorescent units (RFUs) was converted into the concentrations of 5-((2-aminoethyl)amino)naphthalene-1-sulfonic acid (EDANS). The kcat and Km values were 42.59 s−1 and 33.32 μM, respectively, and the specific constant (kcat/Km) value was 1.28 × 106 s−1 · M−1. Then, we conducted the enzyme inhibition assays in the presence of selected PIs and the three novel PIs under the same conditions. All of the compounds tested virtually completely blocked the PRWT activity at the concentration of 100 nM and above. APV inhibited the PRWT activity, with a 50% inhibitory concentration (IC50) value of 1.05 nM and Morrison Ki value of 0.44 nM. DRV inhibited the PRWT activity much more effectively, with lower IC50 and Morrison Ki values of 0.32 nM and 0.23 nM, respectively. Among all of the tested compounds, GRL-057-14 inhibited the PRWT activity most effectively, with IC50 and Morrison Ki values of 0.22 nM and 0.10 nM, respectively (Fig. 3). These data further corroborated and confirmed that the potent activity of the three PIs derived from their protease-specific activity.
FIG 3.
Inhibition of wild-type HIV-1 protease (PRWT) enzymatic activity by GRL-057-14, GRL-058-14, GRL-059-14, APV, and DRV. All the compounds tested completely blocked PRWT activity at 100 nM and beyond. GRL-057-14 inhibited PRWT activity with an IC50 of 0.22 nM and a Morrison Ki of 0.10 nM.
In vitro selection of HIV-1 variants resistant to GRL-057-14, GRL-058-14, and GRL-059-14.
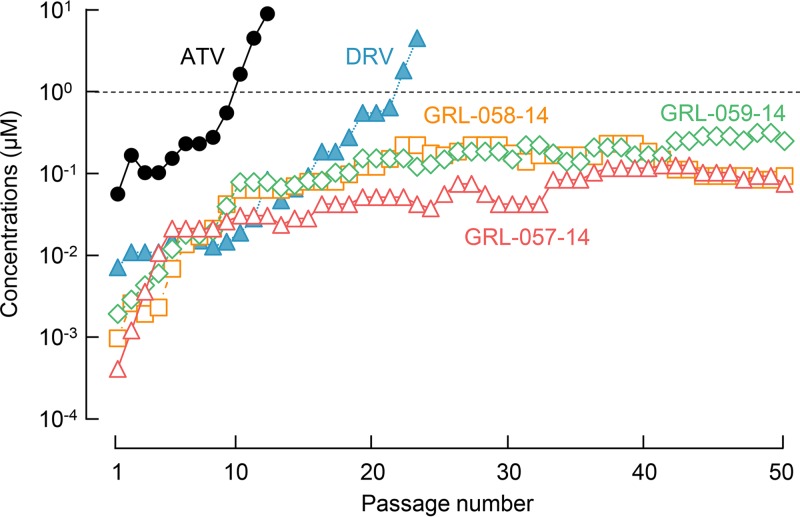
We also attempted to select HIV-1 variants resistant to atazanavir (ATV), DRV, GRL-057-14, GRL-058-14, and GRL-059-14. We previously reported that the use of a mixture of multi-PI-resistant viruses as a starting population for in vitro selection makes it highly possible to select highly drug-resistant variants through homologous recombination among the mixed population of drug-resistant HIV-1 variants (12, 17, 27). Thus, we employed HIV11MIX as a starting HIV-1 population for the selection (Fig. S1B). As expected, the viruses relatively quickly acquired resistance against ATV by passage 10, and finally, by passage 12, they became capable of propagating even in the presence of 10 μM ATV (HIV11MIXATV-P12) (Fig. 4, solid circles in black). This HIV11MIXATV-P12 population apparently originated from HIVC but additionally contained V32I, I50L, A71V, and I84V substitutions (Fig. 5A). When selected with DRV, the viral population was capable of replicating in the presence of 1 μM DRV by the end of 22 passages (Fig. 4, solid triangles in blue). The viral population that replicated in the presence of 5 μM DRV in passage 23 (HIV11MIXDRV-P23) also apparently originated from HIVC and additionally contained L23I, L24M, A28S, I50V, and I84V substitutions (Fig. 5B).
FIG 4.
In vitro selection of HIV-1variants resistant against GRL-057-14, GRL-058-14, GRL-059-14, ATV, and DRV. A mixture of 11 multi-PI-resistant viruses (HIV11MIX) was propagated in MT-4 cells in the presence of increasing concentrations of each compound. When exposed to ATV (closed circles), the virus became resistant to the drug by passage 10, and the virus propagated even in the presence of 10 μM ATV by the end of passage 12. The development of DRV-resistant viruses was delayed compared to that of ATV, but the virus propagated even in the presence of 5 μM DRV by the end of passage 23 (closed triangles). HIV11MIX exposed to GRL-057-14, GRL-058-14, and GRL-059-14 replicated only poorly around 0.05 μM and beyond, and drug-resistant variants did not emerge throughout the selection periods.
FIG 5.
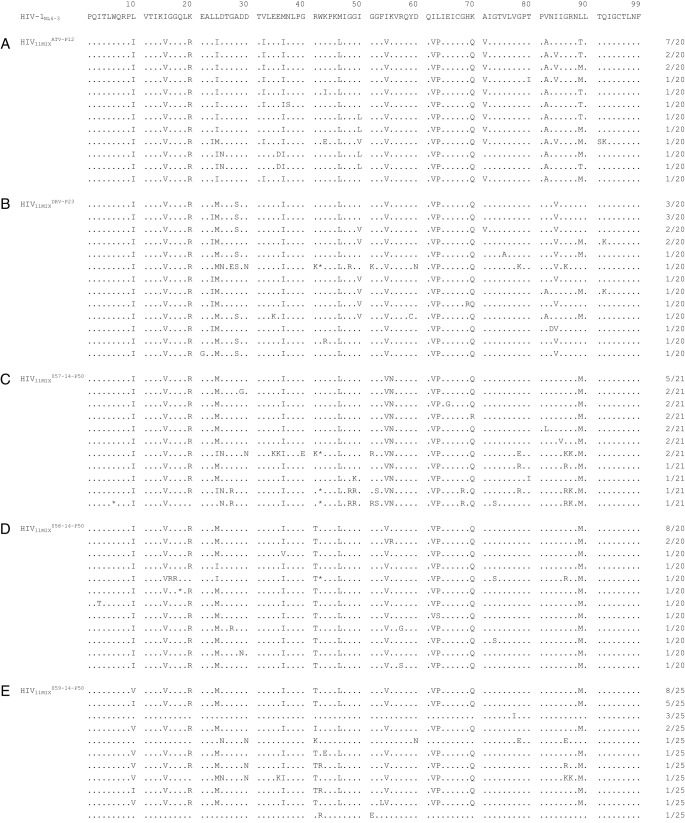
Amino acid sequences of the protease-encoding region of viruses selected with increasing concentrations of (A) ATV, (B) DRV, (C) GRL-057-14, (D) GRL-058-14, and (E) GRL-059-14. The HIV-1NL4-3 sequence is displayed at the top of the figures as a reference. Identity with the sequence at individual amino acid positions is indicated by dots. The fractions on the right are the number of viruses from which each clone is presumed to have originated, divided by the number of clones examined. The stop codon is indicated with an asterisk (*). The selection procedures are described in the legend to Fig. 4.
In contrast, when exposed to GRL-057-14, GRL-058-14, or GRL-059-14, HIV11MIX failed to propagate in the presence of 0.4 μM and beyond until the conclusion of 50 passages of selection (HIV11MIX057-14-P50, HIV11MIX058-14-P50, and HIV11MIX059-14-P50) (Fig. 4). However, when the amino acid sequences of HIV-1 protease-encoding gene, as selected with each compound, were determined, a variety of substitutions were identified. All HIV11MIX exposed to the three PIs had newly acquired L24M by 50 passages (Fig. 5C to E). Additionally, HIV11MIX exposed to GRL-057-14 had newly acquired K55N by 50 passages (HIV11MIX057-14-P50), which was previously seen in HIV-1 variants selected by DRV and its derivatives (13, 28) (Fig. 5C). Moreover, GRL-058-14- and GRL-059-14-exposed HIV11MIX (HIV11MIX058-14-P50 and HIV11MIX059-14-P50) had acquired R41T substitution, which had been only seen in HIV-1 variants selected by DRV (13, 29) (Fig. 5D and E), although R41K substitution is one of the common polymorphisms (24) and has not been associated with PI resistance (30). Interestingly, in most of the HIV-1 populations exposed to GRL-057-14, GRL-058-14, GRL-059-14, and DRV, the V82A/T substitution, which was seen in most constituents of HIV11MIX (Fig. S1B), reverted to the wild-type valine (Fig. 5B to E), while ATV-selected virus (HIV11MIXATV-P12) did not undergo this reversion (Fig. 5A). To evaluate the impact of the three substitutions (L24M, R41T, and K55N) on the susceptibility of HIV-1 to the three PIs, we generated recombinant infectious clones based on HIV-1NL4-3, rHIVL24M, rHIVR41T, and rHIVK55N, using the site-directed mutagenesis technique. All of these clones replicated well compared to HIV-1NL4-3 (Fig. S2). Both GRL-059-14 and DRV were comparably active against all of these clones. GRL-058-14, which has an additional two fluorine atoms in the P1-methylbenzene, was more potent to rHIVL24M than GRL-059-14 and DRV. Interestingly, GRL-057-14 exerted extremely potent activity against all of these clones at EC50 values of 350 aM to 150 fM (Table 4). The locations of the relatively unique amino acid substitutions identified in the present study (L24M, R41T, and K55N) are illustrated in Fig. S3, together with amino acid substitutions often associated with HIV-1 resistance to PIs.
TABLE 4.
Antiviral activity of PIs against HIV-1 variants carrying single amino acid substitutionsa
| Compound | EC50 (nM) againstb
: |
|||
|---|---|---|---|---|
| HIV-1NL4-3c | rHIVL24M | rHIVR41T | rHIVK55N | |
| GRL-057-14 | 0.22 ± 0.09 | 0.00000035 ± 0.00000021 (0.000002) | 0.00015 ± 0.00021 (0.0007) | 0.000006 ± 0.000002 (0.00003) |
| GRL-058-14 | 9.2 ± 5.3 | 0.2 ± 0.03 (0.02) | 2.0 ± 1.1 (0.2) | 6.3 ± 1.8 (0.7) |
| GRL-059-14 | 10.4 ± 4.4 | 18.5 ± 2.1 (1.8) | 2.0 ± 0.5 (0.2) | 4.5 ± 2.7 (0.4) |
| DRV | 3.0 ± 1.0 | 3.0 ± 1.0 (1.0) | 2.7 ± 1.5 (0.9) | 1.3 ± 0.01 (0.4) |
Anti-HIV-1 activity was determined with a p24 assay employing MT-4 cells. Each assay was conducted in triplicate. The values represent mean values (±1 SD) of two or three independent experiments. Numbers in parentheses represent fold changes in EC50s for each isolate compared to the EC50s for wild-type HIV-1NL4-3.
EC50, 50% effective concentration.
Data from Table 2 (HIV-1NL4-3).
Additionally, HIV11MIX selected with each PI had acquired multiple amino acid substitutions in the various parts of the Gag-encoding region by the end of the selection (Fig. S4). Such substitutions included L68F and N124D in the matrix (p17) region; H219Q and D284E in the capsid (p24) region; K403R, R406K, D425E, and A431V in the nucleocapsid (p7) region; I437T/V in the spacer peptide (SP2; p1) region; and L449F, P478T, A487S, S488A, R490K, and S495N in the p6 region (Fig. S4). H219Q, located at a Gag noncleavage site, has been reported as a part of multiple Gag mutations that appear in common among HIV-1 variants selected with APV, JE-2147, KNI-272, and UIC-94003 (31). It has been shown that H219Q gives greater replication advantage to HIV-1 by interacting with the viral cyclophilin A (CypA), especially in CypA-rich MT-2 cells compared to that in PBMCs that contain less CypA (32). A431V and I437T/V are located at the Gag cleavage site (p7/p1) and have been reportedly associated with PI treatment- and/or PI resistance-associated mutations (33, 34). L449F located at the (p1/p6) cleavage site has been reported in a population-based analysis to be invariably linked to I50V mutation (35). Viruses with V82A and V82A/L90M substitutions in protease in combination with a mutation at Gag L449 reportedly have lower susceptibility against PIs than viruses without the L449 mutation (36), although V82A and V82A/L90M substitutions in the protease-encoding region were not observed in the HIV-1s selected with GRL-057-14, GRL-058-14, or GRL-059-14.
Structural analysis.
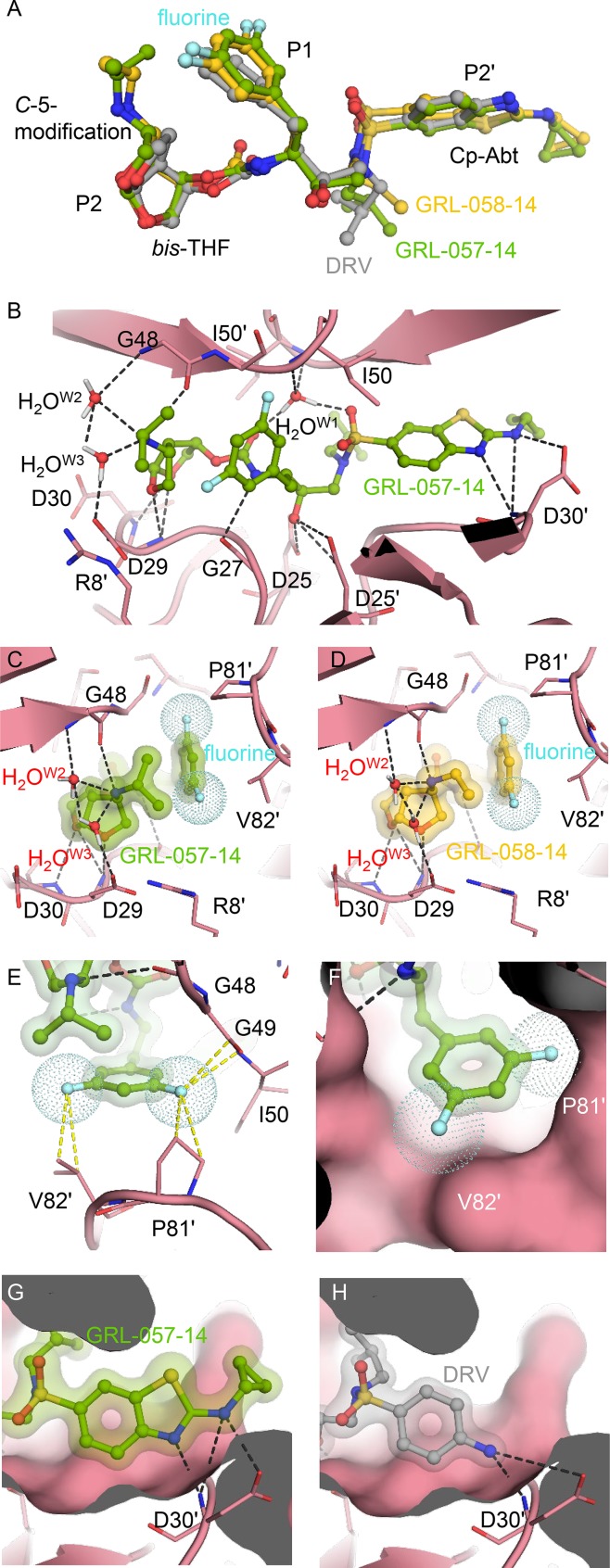
Finally, we attempted to examine the molecular interactions of GRL-057-14 and GRL-058-14 with PRWT to delineate the molecular basis of their potent anti-HIV-1 activity compared to that of DRV (Tables 1–3). Both inhibitors contain extended moieties attached at the C-5 position of bis-THF, a larger Cp-Abt group, and bis-fluorine substitution at the two meta-positions of the P1-benzene ring (Fig. 1). When the structural models of the three inhibitors, GRL-057-14, GRL-058-14, and DRV, were superimposed, no significant differences were seen except for the presence of two fluorine atoms, the C-5-substituted aminoalkyl moieties, and Cp-Abt moiety (Fig. 6A). Notably, inside the S2 subpocket, one direct and two water-mediated hydrogen bonds take place between one flap main chain (Gly48) and the basic amine at position C-5 (Fig. 6B). Such interactions between the inhibitor and the flap main chain are not seen in the interactions of DRV with protease (5). Of note, the present structural model describing the interaction of the C-5-aminoalkyl substituent originated from a crystal structure (PDB identifier [ID] 5BRY), which revealed the presence of water-mediated hydrogen bonds between the protease inhibitor containing THF amine derivative and the Gly48 (37). After careful evaluation of the PDB coordinates (PDB ID 5BRY), we noticed the presence of two water molecules involved in the hydrogen bonding between the C-5-aminoalkyl group and the side chain of Asp29 (Fig. 6B to D). These water molecules presumably strongly interconnect the flap residue of Gly48 with Asp29 on the opposite site (Fig. 6B to D). Moreover, the C-5-isopropylamine group located in close proximity to the meta-bis-fluorobenzene should be associated with favorable CH-π interactions (Fig. 6C). Of note, close interactions of the two fluorine atoms with the hydrogens of isopropyl (Fig. 6C) and ethyl (Fig. 6D) groups are seen, which should function as a glue to link the P1 and P2 moieties, thereby stabilizing the structures of both inhibitors (Fig. 6C and D). As shown in Tables 1 to 3, it is strongly suggested that the presence of two fluorine atoms in the P1-benzene in GRL-058-14 confers greater potency than that of nonfluorinated GRL-059-14. This greater potency is well explained by the finding that the two fluorine atoms engage in halogen bond interactions with surrounding residues, such as Pro81ʹ and Val82ʹ (Fig. 6E and F). Additionally, the P2ʹ-Cp-Abt group suitably fills the S2ʹ site and forms additional polar and van der Waals interactions (Fig. 6G and H).
FIG 6.
Structural analysis of PRWT complexed with DRV, GRL-057-14, and GRL-058-14. (A) Superimposed structural profile of DRV (gray), GRL-057-14 (green), and GRL-058-14 (yellow). Note that three compounds are well aligned around the scaffold of DRV. (B) GRL-057-14 (green) forming hydrogen bond interactions inside the HIV-1 protease binding pocket. The catalytic water (W1) and bridging water molecules (W2 and W3) near the S2 site are indicated. Although the hydrogen-bonding pattern is well-conserved with the scaffold molecule of DRV, additional hydrogen bonds with Asp30′, as well as with Gly48, are observed. (C and D) The P2 moieties of GRL-057-14 (green) and GRL-058-14 (yellow). The C-5-aminoalkyl group of both GRL-057-14 and GRL-058-14 forms a direct hydrogen bond with the carbonyl group of Gly48 and a water-mediated hydrogen bond interaction with the amide of Gly48. (E) Two fluorine atoms (cyan dots) form interactions with neighboring residues of Pro81ʹ and Val82ʹ but also engage halogen interactions with the isopropyl-amine and ethyl-amine groups linked to the C-5 position of THF (C and D). (F) The P1-meta-bis-fluoro-methylbenzene also forms good van der Waals interactions. Note that the Cp-Abt group of GRL-057-14 forms three hydrogen bonds with Asp30ʹ (G), whereas the aniline group of DRV forms only two hydrogen bonds with the same residue (H), suggesting that GRL-057-14 more tightly binds to Asp30ʹ. Moreover, the cyclopropyl group attached to the amino-benzothiazole of GRL-057-14 protrudes from the periphery of the binding pocket (G), forming additional interactions.
DISCUSSION
We have previously reported PIs with a P2-bis-THF moiety (5, 38–42) that are potent against not only wild-type HIV-1 but also against HIV-2 and multi-PI-resistant laboratory and clinical isolates. We have also reported PIs with P2ʹ-Cp-Abt moiety (28, 43–45), including GRL-142, which has shown unprecedented potent activity against a variety of HIV strains (44). The extreme potency of P2ʹ-Cp-Abt-containing PIs is thought to stem from the favorable van der Waals interactions, in which the Cp-Abt is involved. The formation of hydrogen bonds by the P2 and P2ʹ ligands of PIs with polar groups in S2 and S2ʹ regions of the HIV-1 protease homodimer is thought to be one of the key factors for the much-delayed emergence of drug-resistant HIV-1 variants (37, 43). Thus, we further designed, synthesized, and identified three novel PIs (GRL-057-14, GRL-058-14, and GRL-059-14) that contain both a P2-bis-THF moiety carrying the C-5-modification and a P2ʹ-Cp-Abt moiety (Fig. 1). Among them, GRL-057-14, which contains P2-C-5-isopropylamino-bis-THF, P1-meta-bis-fluoro-methylbenzene, and P2ʹ-Cp-Abt moieties, showed the greatest antiviral potency, with EC50 values of 0.34 nM against HIV-1NL4-3 (Table 1). GRL-057-14 also maintained its potent antiviral activity against a series of MDR HIV-1s, including highly DRV-resistant viruses (HIVDRVRs), with EC50 values ranging from 0.000044 nM (44 fM) to 1.6 nM, while other tested FDA-approved drugs totally failed to suppress viral propagation or required much higher concentration (Table 2). Furthermore, the three compounds we developed were found to have potent antiviral activity against a series of clinically isolated strains of HIV-1, with EC50 values ranging from 0.0005 nM (500 fM) to 27.3 nM (Table 3).
In the selection with the three PIs, L24M substitution, which was not seen in any of the HIV11MIX constituents, was newly emerged by selection passage 50. This substitution has also been seen in DRV-selected HIV11MIXDRV-P23 and is reported to be significantly associated with PI exposure (46, 47), although its clinical features are still unclear (48). Of note, recombinant HIV-1 clone rHIVL24M, carrying an L24M amino acid substitution, proved to be hypersusceptible to GRL-057-14 (Table 4), likely making HIV-1 with L24M unable to replicate in the presence of GRL-057-14. Similarly, rHIVR41T and rHIVK55N were also hypersusceptible to GRL-057-14. However, HIV11MIX057-14-P50, which harbored both L24M and K55N substitutions, continued to replicate in the presence of GRL-057-14. Interestingly, HIV11MIX058-14-P50 and HIV11MIX059-14-P50, which contained L24M and R41T substitutions, also continued to replicate in the presence of GRL-058-14 and GRL-059-14. The mechanism of these “paradoxical” features of HIV11MIX057-14-P50, HIV11MIX058-14-P50, and HIV11MIX059-14-P50 remains to be elucidated, while the presence of the C-5-modified moiety rather than the presence of two fluorine atoms at the P1 site should be at least in part responsible for the “paradoxical” features.
The V82A/T substitution, which has been documented as one of the key amino acid substitutions in the emergence of HIV-1 variants resistant to most FDA-approved PIs except DRV (49, 50). In fact, in HIV11MIX selected with ATV, the V82A/T substitution, which was seen in 10 of the 11 virus constituents (Fig. S1B), persisted even when the virus had acquired a high-level resistance to ATV (Fig. 5A). However, in most of the HIV-1 populations exposed to GRL-057-14, GRL-058-14, and GRL-059-14, the V82A/T substitution reverted to the wild-type valine (Fig. 5B to E), suggesting that the drug resistance profiles of GRL-057-14, GRL-058-14, and GRL-059-14 should be different from those of FDA-approved PIs. This reversion has also been reported to have occurred when HIV11MIX was selected with two novel C-5-modified PIs, GRL-058 and GRL-079 (28). Thus, it is assumed that the C-5 modification in part prevents HIV-1 from acquiring V82A/T substitution, which is likely to be associated with the high genetic barrier to the emergence of HIV-1 variants resistant to C-5-modified PIs, including GRL-057-14, GRL-058-14, and GRL-059-14.
It is noteworthy that, when HIV-1NL4-3 was selected with DRV over 23 passages, HIV11MXDRV-P23 had acquired the A28S substitution, which is known to result in a >1,500-fold decrease in kcat/Km in the proteolytic activity of protease (51) and leads to highly poor replication fitness of HIV-1 (38, 52). Although A28S substitution was seen in the selection of HIV-1 with a few PIs, including TMC-126 (38), GRL-98065 (39), brecanavir (52), and GRL-1398 (27), the substitution had never appeared in the selection of HIV-1 with DRV. It is of note that the presence of the P2′-aminobenzene moiety of DRV was thought to prevent HIV-1 from acquiring the A28S substitution (27, 38, 39); however, the appearance of A28S substitution with DRV in the present study suggests that the development of A28S with DRV was possible, although the probability of its appearance is very low. In fact, database analysis by Wu et al. has demonstrated that substitutions at A28 position occurs only rarely (0.9%) in patients receiving PIs (53).
In conclusion, our efforts to maximize the antiviral potency of the prototypic protease inhibitor, DRV, with an extremely high genetic barrier, have successfully led to the discovery of GRL-142 (44). Furthermore, the additional modification at the C-5 position of P2 bis-THF of DRV, together with bis-fluorination of the P1-benzene and P2′-Cp-Abt, resulted in PIs with extremely potent anti-HIV-1 activity, particularly against multi-PI-resistant HIV-1 variants. In particular, GRL-057-14 should warrant further development for potential clinical application.
MATERIALS AND METHODS
Cells and viruses.
MT-4 cells, originated from adult male human, were grown in RPMI 1640-based culture medium (Lonza, Basel, Switzerland), while HEK293T cells, originated from fetal female human, were cultured in DMEM (Dulbecco's modified Eagle medium; Lonza) supplemented with 10% fetal calf serum (Gemini Bio-Products, West Sacramento, CA) plus 100 U/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml amphotericin B (Gibco antibiotic-antimycotic; Thermo Fisher Scientific, Waltham, MA). The cells were authenticated using short tandem repeat (STR) profiling (ATCC, Manassas, VA), and maintained in a humidified atmosphere containing 5% CO2 at 37°C. The following HIV-1 and HIV-2 strains were used for the drug susceptibility assays and in vitro selection experiments; wild-type laboratory HIV isolate HIV-1NL4-3, HIV-2EHO, four laboratory-selected PI-resistant HIV-1 variants (HIVAPV-5μM, HIVLPV-5μM, HIVSQV-5μM, and HIVTPV-15μM) (Fig. S1A), a mixture of eleven HIV-1 clinical strains (HIV11MIX; consisting of HIVA, HIVB, HIVC, HIVG, HIVTM, HIVMM, HIVJSL, HIVSS, HIVES, HIVEV, and HIV13-52) (Fig. S1B), three highly DRV-resistant viruses (HIVDRVRs; HIVDRVRP10, HIVDRVRP30, and HIVDRVRP51) (Fig. S1A), and five recombinant clinical HIV-1 isolates (rCLHIVs; rCLHIVF16, rCLHIVT48, rCLHIVV40, rCLHIVT44, and rCLHIVM45) (Fig. S1A). Four PI-resistant HIV-1s were selected in vitro in the presence of increasing concentrations of APV, LPV, and SQV with propagation of a wild-type HIV-1NL4-3 (5) and TPV with HIV11MIX (17). The mixture of 11 different MDR clinical HIV-1 strains (HIV11MIX) was employed in the present study. Seven such isolates were originally isolated from the patients who were enrolled into a clinical study of APV and ABC at the Clinical Center, National Institutes of Health, and failed the APV-plus-ABC therapy (54); HIVA, HIVB, HIVC, HIVG, HIVTM, HIVMM, HIVJSL were isolated from patients 3, 1, 2, 4, 6, 5, and 7, respectively (55). Three other isolates (HIVSS, HIVES, and HIVEV) were isolated from patients 8 (38), ES, and EV (56), respectively, who participated in the same clinical trial. HIV13-52 was originally isolated from a patient who was enrolled in a phase I/II study of TDF (57) and described as PtERS2 (58). These patients had failed existing anti-HIV-1 regimens after receiving 7 to 11 anti-HIV-1 drugs over the previous 24 to 83 months in the late 1990s. These clinical strains contained numbers of amino acid substitutions in the protease- and reverse transcriptase-encoding genes and have been genotypically and phenotypically characterized as multi-PI- and multi-NRTI-resistant HIV-1 (Fig. S1A) (17, 59). Three highly DRV-resistant viruses were selected in vitro in the presence of increasing concentrations of DRV with propagation of a mixture of eight highly multi-PI-resistant HIV-1 strains (Fig. S1A) (12). Five clinical HIV-1 isolates were kindly provided by Robert Shafer of Stanford University, and recombinant HIV-1 variants were produced using HIV-1NL4-3-based molecular clones and generated by ligating patient-derived amplicons encompassing approximately 200 nucleotides of Gag (beginning at the unique ApaI restriction site), the entire protease, and the first 72 nucleotides of reverse transcriptase, using the expression vector pNLPFB (a generous gift from Tomozumi Imamichi of the National Institute of Allergy and Infectious Diseases) (Fig. S1A) (18, 19). The clinical HIV-1 isolates were chosen from 32 isolates that had been obtained from multi-PI-treated patients whose protease genotype contained prototypical patterns of PI resistance. The median duration of continuous PI treatment was 7.5 years (range, 6 to 10 years), and the median number of PIs such patients received was 5 (range, 4 to 8 [excluding the use of ritonavir for pharmacokinetic boosting]). X4-tropic subtype A virus (HIV-192/UG/029) and R5-tropic subtype C virus (HIV-197/ZA/003) were obtained through the NIH AIDS Reagent Program.
Antiviral agents.
Three novel C-5-modified P2-bis-tetrahydrofuranylurethane (bis-THF) and P2ʹ-cyclopropyl-amino-benzothiazole (Cp-Abt) moieties containing nonpeptidic PIs, GRL-057-14, GRL-058-14, and GRL-059-14 (Fig. 1) (molecular weights of 737.88, 723.85, and 687.87, respectively) were designed and synthesized. The synthetic methods of these PIs will be published elsewhere. 4ʹ-Ethynyl-2-fluoro-2-deoxyadenosine (EFdA/MK-8591) was synthesized as previously described (60). Darunavir (DRV), amprenavir (APV), lopinavir (LPV), saquinavir mesylate (SQV), nelfinavir mesylate hydrate (NFV), 3ʹ-azido-3ʹ-deoxythymidine (zidovudine; AZT), and abacavir (ABC) were purchased from Sigma-Aldrich (St. Louis, MO). Indinavir sulfate (IDV) and tipranavir (TPV) were purchased from United States Biological (Swampscott, MA). Tenofovir disoproxil fumarate (TDF) was purchased from BioVision (Milpitas, CA).
Anti-HIV-1 and cytotoxicity assays.
The susceptibility of HIV-1NL4-3 and HIV-2EHO to various drugs was determined as previously described (55), using the methyl thiazol tetrazolium (MTT) assay with minor modifications (61). In brief, MT-4 cells were exposed to 50 × 50% tissue culture infectious dose (TCID50) of a selected HIV strain. After viral exposure, the cells (5 × 103 cells in 100 μl) were plated into each well of a 96-well flat microtiter culture plate containing various concentrations of the compound with a total volume of 200 μl per well. After incubation for 7 days at 37°C, the number of viable cells in each well was measured using Cell Counting Kit-8 (Dojindo, Kumamoto, Japan). An anti-HIV-1 assay against isolates resistant to various drugs was also conducted using the p24 assay as previously described (28, 44). In brief, 50 μl of the culture medium was harvested at the end of the incubation, and the amount of p24 Gag protein was determined by using a fully automated chemiluminescent enzyme immunoassay system (Lumipulse G1200; Fujirebio Inc., Tokyo, Japan). The potency of HIV-1 inhibition of a compound was determined based on its inhibitory effect on virally induced cytopathicity in MT-4 cells [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay] or on its suppressive effect of the production of p24 Gag protein in HIV-1-exposed MT-4 cells compared with that in drug-free control cell cultures (p24 assay), and EC50 values were determined. Cytotoxicity of a compound in MT-4 cells was also determined (61). Cells were plated into a 96-well plate at a density of 5 × 103 cells in 100 μl culture medium in each well and were continuously exposed to various concentrations of the compound throughout the period of the 7 days of culture. The number of viable cells in each well was determined using Cell Counting Kit-8, and CC50 values were determined. Each assay was conducted in triplicate on two or three different occasions.
Thermal stability analysis using differential scanning fluorimetry.
Wild-type HIV-1 protease (PRWT) was purchased from ProSpec-Tany TechnoGene (Ness-Ziona, Israel). Chicken lysozyme was purchased from Affymetrix (Santa Clara, CA). The final concentrations of each protein and compounds were 5 μM. Differential scanning fluorimetry (DSF) assays were conducted as previously described (25, 26), using a Protein Thermal Shift dye kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. All of the preparation procedures were conducted on ice and samples were kept on ice until the instrument run. Twenty microliters of PRWT or lysozyme solution were successively heated from 25 to 95°C, and the changes in the fluorescence intensity were measured by using a StepOne real-time PCR system (Thermo Fisher Scientific). The data were analyzed using StepOne software version 2.3 and Protein Thermal Shift software version 1.0 (Thermo Fisher Scientific), and melting temperature (Tm) values were obtained. All of the assays were conducted in duplicate on two different occasions.
Enzyme inhibition assay.
The inhibitory activity of selected PIs was determined as previously described, with modifications (62, 63). The change of the fluorescence intensity associated with the PRWT (ProSpec-Tany TechnoGene)-elicited cleavage of the fluorogenic substrate Arg-Glu(EDANS)-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-Lys(DABCYL)-Arg (HIV protease substrate 1; Sigma-Aldrich) was measured. All preparation procedures were done on ice, and the enzymatic reaction was processed at 37°C. Enzymatic assays were performed in 0.1 M sodium acetate, 1.0 M sodium chloride, 1.0 mM ethylenediaminetetraacetic acid (EDTA), 1.0 mM dithiothreitol (DTT), and 1 mg/ml bovine serum albumin (BSA) pH 4.7 buffer containing various concentrations of a test compound with 30 μM substrate and 25 nM PRWT. Fluorescence was measured using a SpectraMax M5e multimode microplate reader (Molecular Devices, Sunnyvale, CA) with an excitation wavelength of 340 nm and emission wavelength of 490 nm, and the relative fluorescent units (RFU) were obtained. Maximum velocity (Vmax) was calculated on SoftMax Pro (Molecular Devices), and the potency of HIV-1 protease activity inhibition by each compound was determined based on its reduced effectiveness on Vmax. Enzymatic assays for determination of the catalytic rate constant (kcat) and the Michaelis constant (Km) were also conducted under the same conditions. To determine the number of fluorophores out of RFU values, a standard curve was generated with 10-fold serial dilutions of EDANS (AnaSpec, Fremont, CA), and the RFU values for each sample were converted to the numbers of EDANS molecules. IC50 values and Morrison Ki and Km values were calculated on Prism 7 (GraphPad Software, La Jolla, CA).
In vitro selection of highly GRL-057-14-resistant HIV-1 variants.
We also attempted to select HIV-1 variants resistant to GRL-057-14, GRL-058-14, and GRL-059-14, as previously described (12, 16, 19, 28, 64). In brief, 30 × TCID50 of each of eleven highly multi-PI-resistant HIV-1 isolates (HIVA, HIVB, HIVC, HIVG, HIVTM, HIVMM, HIVJSL, HIVSS, HIVES, HIVEV, and HIV13-52) were mixed and propagated in a mixture of an equal number of PHA-stimulated PBMCs and MT-4 cells (5 × 105), in an attempt to adapt the mixed viral population for their replication in MT-4 cells. The cell-free supernatant was harvested on day 7 of coculture, and the mixed HIV-1 preparation in the supernatant obtained was designated as HIV11MIX. On the first passage, MT-4 cells (5 × 105) were exposed to HIV11MIX-containing supernatant and cultured in the presence of each compound at an initial concentration to the EC50 dose for approximately 7 days. On the last day of each passage, 2 ml of the cell-free supernatant was harvested and transferred to a culture of fresh, uninfected MT-4 cells in the presence of increased concentrations of the compound for the following round of culture. In this round of culture, three drug concentrations (increased by 1-, 2-, and 3-fold compared to the previous concentration) were employed. When the replication of HIV-1 in the presence of the compounds was confirmed by p24 Gag protein production (greater than 200 ng/ml), we assumed that the HIV-1 isolate substantially replicated, and the highest drug concentration among the three concentrations was used to continue the selection (for the next round of culture). This protocol was repeatedly used until the drug concentration reached the targeted concentration (regularly 5 μM). Proviral DNA preparations obtained from the lysates of infected cells at indicated passages were subjected to nucleotide sequencing.
Determination of nucleotide sequences.
Molecular cloning and determination of the nucleotide sequences of HIV-1 strains passaged in the presence of each compound was performed as previously described (17, 19, 65). In brief, high-molecular-weight DNA was extracted from HIV-1-exposed MT-4 cells by using InstaGene matrix (Bio-Rad Laboratories, Hercules, CA) and was subjected to molecular cloning, followed by nucleotide sequence determination. The extracted proviral DNA was amplified using TaKaRa Ex Taq (TaKaRa Bio Inc., Shiga, Japan) with the following primers: KAPA-1 (5ʹ-GCA GGG CCC CTA GGA AAA AGG GCT GTT GG-3ʹ) and Ksma2.1 (5ʹ-CCA TCC CGG GCT TTA ATT TTA CTG GTA C-3ʹ) for the protease-encoding region, while LTR F2 (5ʹ-GAG ACT CTG GTA ACT AGA GAT C-3ʹ) and PR12 (5ʹ-CTC GTG ACA AAT TTC TAC TAA TGC-3ʹ) for first round PCR and LTR F2 and Ksma2.1 for second round PCR for the entire Gag- and the protease-encoding region (65). PCR preparations were incubated at 95°C for 1 min, followed by 30 cycles of 95°C for 15 s, 55°C for 20 s, and 72°C for 1 min, with a final extension at 72°C for 7 min. The PCR products were purified using illustra MicroSpin S-400 HR columns (GE Healthcare Life Science, Pittsburgh, PA). The purified PCR products were directly cloned into pGEM-T Easy Vector (Promega, Madison, WI), and transformed into One Shot MAX Efficiency DH5α-T1R competent cells (Thermo Fisher Scientific). Approximately 20 colonies were selected by pick-up PCR, and plasmids were isolated using a QIAprep spin miniprep kit (Qiagen, Hilden, Germany). Purified plasmids were subjected to sequencing using the BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems). The Sanger sequencing was conducted at the CCR Genomics Core at the National Cancer Institute.
Viral replication assays.
Viral replication assays were performed using recombinant HIV-1 mutants harboring single amino acid substitutions, as previously described (59, 66). In brief, recombinant HIV-1NL4-3-based infectious clones were generated using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) and transfected into HEK293T cells by using Attractene transfection reagent (Qiagen). The viruses were harvested at 72 h after the transfection, and viral p24 antigen was measured using a Lumipulse G1200 instrument. Replication of HIV-1 mutants was determined as described previously (59, 66), with minor modifications. Briefly, MT-4 cells (density, 1.5 × 106 cells in 20 ml RPMI medium) were exposed to the HIV-1 mutants with 20 ng/ml of the p24 antigen and were cultured without the antiretroviral agents for 10 days. Fresh MT-4 cells (1.0 × 106) were added on days 4 and 8. The amount of the p24 antigen was measured every 2 days, and the average value of each data point was determined from two independent assays.
Structural analysis.
We used PDB ID 5DGW as the starting HIV-1 protease structure from the protein data bank (67). The initial conformation of GRL-057-14 was built in the active site by a structural overlay of similar chemical moieties of GRL-057-14 with the crystal structures of DRV (PDB ID 4HLA) (41), GRL-105-11A (PDB ID 5DGW) (67), and GRL-142 (PDB ID 5TYS) (44). After an initial minimization of GRL-057-14-HIV-1 protease complex using an OPLS3 force field, we used the electron density map of HIV-1 protease structure in complex with GRL-011-11A (PDB ID 5BRY) to further refine the conformation of the C-5 substituent of bis-THF (37). The PDB coordinates and topology file of GRL-057-14 were generated using PRODRG server (68) and manually fitted into an electron density map of GRL-011-11A using Coot (69). A final round of minimization was performed using the OPLS3 force field as implemented in Maestro version 10.7.015 (Schrödinger, LLC, New York, NY). Structural interactions of GRL-058-14 with HIV-1 protease were obtained by chemical modification of GRL-057-14 followed by minimization. Figures of the structural interactions of the inhibitors with protease were generated using PyMOL version 1.7.6.6.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the Intramural Research Program of Center for Cancer Research, National Cancer Institute, National Institutes of Health (grant BC011105 and SC006738 to H.M.); by grants for the promotion of AIDS research from the Ministry of Health, Welfare, and Labor of Japan; by grants from the Japan Agency for Medical Research and Development (AMED; grants JP15fk0410001 and JP18fk0410001 to H.M.); by grants from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT; grant number 17H04222 to H.M.); by the Platform Project for Supporting Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science), funded by MEXT and AMED (grant 17K19577 to H.M.); by grants from the National Institutes of Health (grant R01GM53386 to A.K.G.); and by a grant from the National Center for Global Health and Medicine (NCGM) Research Institute. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH, Bethesda, MD (http://hpc.nih.gov).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00372-19.
REFERENCES
- 1.HIV-CAUSAL Collaboration. 2010. The effect of combined antiretroviral therapy on the overall mortality of HIV-infected individuals. AIDS 24:123–137. doi: 10.1097/QAD.0b013e3283324283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner EM, Hullsiek KH, Telzak EE, Sharma S, Peng G, Burman WJ, MacArthur RD, Chesney M, Friedland G, Mannheimer SB, Terry Beirn Community Programs for Clinical Research on AIDS and the International Network for Strategic Initiatives in Global HIV Trials. 2010. Antiretroviral medication adherence and class-specific resistance in a large prospective clinical trial. AIDS 24:395–403. doi: 10.1097/QAD.0b013e328335cd8a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powderly WG. 2010. Integrase inhibitors in the treatment of HIV-1 infection. J Antimicrob Chemother 65:2485–2488. doi: 10.1093/jac/dkq350. [DOI] [PubMed] [Google Scholar]
- 4.Blanco JL, Varghese V, Rhee SY, Gatell JM, Shafer RW. 2011. HIV-1 integrase inhibitor resistance and its clinical implications. J Infect Dis 203:1204–1214. doi: 10.1093/infdis/jir025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koh Y, Nakata H, Maeda K, Ogata H, Bilcer G, Devasamudram T, Kincaid JF, Boross P, Wang YF, Tie Y, Volarath P, Gaddis L, Harrison RW, Weber IT, Ghosh AK, Mitsuya H. 2003. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob Agents Chemother 47:3123–3129. doi: 10.1128/AAC.47.10.3123-3129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saag MS, Benson CA, Gandhi RT, Hoy JF, Landovitz RJ, Mugavero MJ, Sax PE, Smith DM, Thompson MA, Buchbinder SP, Del Rio C, Eron JJ Jr, Fätkenheuer G, Günthard HF, Molina JM, Jacobsen DM, Volberding PA. 2018. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2018 recommendations of the International Antiviral Society—USA Panel. JAMA 320:379–396. doi: 10.1001/jama.2018.8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitsuya Y, Liu TF, Rhee SY, Fessel WJ, Shafer RW. 2007. Prevalence of darunavir resistance-associated mutations: patterns of occurrence and association with past treatment. J Infect Dis 196:1177–1179. doi: 10.1086/521624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert-Niclot S, Flandre P, Canestri A, Peytavin G, Blanc C, Agher R, Soulie C, Wirden M, Katlama C, Calvez V, Marcelin AG. 2008. Factors associated with the selection of mutations conferring resistance to protease inhibitors (PIs) in PI-experienced patients displaying treatment failure on darunavir. Antimicrob Agents Chemother 52:491–496. doi: 10.1128/AAC.00909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Meyer S, Vangeneugden T, van Baelen B, de Paepe E, van Marck H, Picchio G, Lefebvre E, de Béthune M-P. 2008. Resistance profile of darunavir: combined 24-week results from the POWER trials. AIDS Res Hum Retroviruses 24:379–388. doi: 10.1089/aid.2007.0173. [DOI] [PubMed] [Google Scholar]
- 10.Delaugerre C, Pavie J, Palmer P, Ghosn J, Blanche S, Roudiere L, Dominguez S, Mortier E, Molina JM, de Truchis P. 2008. Pattern and impact of emerging resistance mutations in treatment experienced patients failing darunavir-containing regimen. AIDS 22:1809–1813. doi: 10.1097/QAD.0b013e328307f24a. [DOI] [PubMed] [Google Scholar]
- 11.De Meyer S, Lathouwers E, Dierynck I, De Paepe E, Van Baelen B, Vangeneugden T, Spinosa-Guzman S, Lefebvre E, Picchio G, de Béthune M-P. 2009. Characterization of virologic failure patients on darunavir/ritonavir in treatment-experienced patients. AIDS 23:1829–1840. doi: 10.1097/QAD.0b013e32832cbcec. [DOI] [PubMed] [Google Scholar]
- 12.Koh Y, Amano M, Towata T, Danish M, Leshchenko-Yashchuk S, Das D, Nakayama M, Tojo Y, Ghosh AK, Mitsuya H. 2010. In vitro selection of highly darunavir-resistant and replication-competent HIV-1 variants by using a mixture of clinical HIV-1 isolates resistant to multiple conventional protease inhibitors. J Virol 84:11961–11969. doi: 10.1128/JVI.00967-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoki M, Das D, Hayashi H, Aoki-Ogata H, Takamatsu Y, Ghosh AK, Mitsuya H. 2018. Mechanism of darunavir (DRV)’s high genetic barrier to HIV-1 resistance: a key V32I substitution in protease rarely occurs, but once it occurs, it predisposes HIV-1 to develop DRV resistance. mBio 9:e02425-17. doi: 10.1128/mBio.02425-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Witvrouw M, Pannecouque C, Switzer WM, Folks TM, De Clercq E, Heneine W. 2004. Susceptibility of HIV-2, SIV and SHIV to various anti-HIV-1 compounds: implications for treatment and postexposure prophylaxis. Antivir Ther 9:57–65. [PubMed] [Google Scholar]
- 15.Desbois D, Roquebert B, Peytavin G, Damond F, Collin G, Benard A, Campa P, Matheron S, Chene G, Brun-Vezinet F, Descamps D, French ANRS HIV-2 Cohort (ANRS CO 05 VIH-2). 2008. In vitro phenotypic susceptibility of human immunodeficiency virus type 2 clinical isolates to protease inhibitors. Antimicrob Agents Chemother 52:1545–1548. doi: 10.1128/AAC.01284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aoki M, Venzon DJ, Koh Y, Aoki-Ogata H, Miyakawa T, Yoshimura K, Maeda K, Mitsuya H. 2009. Non-cleavage site gag mutations in amprenavir-resistant human immunodeficiency virus type 1 (HIV-1) predispose HIV-1 to rapid acquisition of amprenavir resistance but delay development of resistance to other protease inhibitors. J Virol 83:3059–3068. doi: 10.1128/JVI.02539-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aoki M, Danish ML, Aoki-Ogata H, Amano M, Ide K, Das D, Koh Y, Mitsuya H. 2012. Loss of the protease dimerization inhibition activity of tipranavir (TPV) and its association with the acquisition of resistance to TPV by HIV-1. J Virol 86:13384–13396. doi: 10.1128/JVI.07234-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh Y, Aoki M, Danish ML, Aoki-Ogata H, Amano M, Das D, Shafer RW, Ghosh AK, Mitsuya H. 2011. Loss of protease dimerization inhibition activity of darunavir is associated with the acquisition of resistance to darunavir by HIV-1. J Virol 85:10079–10089. doi: 10.1128/JVI.05121-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aoki M, Hayashi H, Yedidi RS, Martyr CD, Takamatsu Y, Aoki-Ogata H, Nakamura T, Nakata H, Das D, Yamagata Y, Ghosh AK, Mitsuya H. 2016. C-5-modified tetrahydropyrano-tetrahydofuran-derived protease inhibitors (PIs) exert potent inhibition of the replication of HIV-1 variants highly resistant to various PIs, including darunavir. J Virol 90:2180–2194. doi: 10.1128/JVI.01829-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hemelaar J, Gouws E, Ghys PD, Osmanov S, WHO-UNAIDS Network for HIV Isolation and Characterisation. 2011. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kantor R, Smeaton L, Vardhanabhuti S, Hudelson SE, Wallis CL, Tripathy S, Morgado MG, Saravanan S, Balakrishnan P, Reitsma M, Hart S, Mellors JW, Halvas E, Grinsztejn B, Hosseinipour MC, Kumwenda J, La Rosa A, Lalloo UG, Lama JR, Rassool M, Santos BR, Supparatpinyo K, Hakim J, Flanigan T, Kumarasamy N, Campbell TB, Eshleman SH, AIDS Clinical Trials Group (ACTG) A5175 Study Team. 2015. Pretreatment HIV drug resistance and HIV-1 subtype C are independently associated with virologic failure: results from the multinational PEARLS (ACTG A5175) clinical trial. Clin Infect Dis 60:1541–1549. doi: 10.1093/cid/civ102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Häggblom A, Svedhem V, Singh K, Sönnerborg A, Neogi U. 2016. Virological failure in patients with HIV-1 subtype C receiving antiretroviral therapy: an analysis of a prospective national cohort in Sweden. Lancet HIV 3:e166–74. doi: 10.1016/S2352-3018(16)00023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutherland KA, Collier DA, Claiborne DT, Prince JL, Deymier MJ, Goldstein RA, Hunter E, Gupta RK. 2016. Wide variation in susceptibility of transmitted/founder HIV-1 subtype C isolates to protease inhibitors and association with in vitro replication efficiency. Sci Rep 6:38153. doi: 10.1038/srep38153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velazquez-Campoy A, Todd MJ, Vega S, Freire E. 2001. Catalytic efficiency and vitality of HIV-1 proteases from African viral subtypes. Proc Natl Acad Sci U S A 98:6062–6067. doi: 10.1073/pnas.111152698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi H, Takamune N, Nirasawa T, Aoki M, Morishita Y, Das D, Koh Y, Ghosh AK, Misumi S, Mitsuya H. 2014. Dimerization of HIV-1 protease occurs through two steps relating to the mechanism of protease dimerization inhibition by darunavir. Proc Natl Acad Sci U S A 111:12234–12239. doi: 10.1073/pnas.1400027111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niesen FH, Berglund H, Vedadi M. 2007. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 27.Ide K, Aoki M, Amano M, Koh Y, Yedidi RS, Das D, Leschenko S, Chapsal B, Ghosh AK, Mitsuya H. 2011. Novel HIV-1 protease inhibitors (PIs) containing a bicyclic P2 functional moiety, tetrahydropyrano-tetrahydrofuran, that are potent against multi-PI-resistant HIV-1 variants. Antimicrob Agents Chemother 55:1717–1727. doi: 10.1128/AAC.01540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delino NS, Aoki M, Hayashi H, Hattori SI, Chang SB, Takamatsu Y, Martyr CD, Das D, Ghosh AK, Mitsuya H. 2018. GRL-079, a novel HIV-1 protease inhibitor, is extremely potent against multidrug-resistant HIV-1 variants and has a high genetic barrier against the emergence of resistant variants. Antimicrob Agents Chemother 62:e02060-17. doi: 10.1128/AAC.02060-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dierynck I, Van Marck H, Van Ginderen M, Jonckers TH, Nalam MN, Schiffer CA, Raoof A, Kraus G, Picchio G. 2011. TMC310911, a novel human immunodeficiency virus type 1 protease inhibitor, shows in vitro an improved resistance profile and higher genetic barrier to resistance compared with current protease inhibitors. Antimicrob Agents Chemother 55:5723–5731. doi: 10.1128/AAC.00748-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee SY, Sankaran K, Varghese V, Winters MA, Hurt CB, Eron JJ, Parkin N, Holmes SP, Holodniy M, Shafer RW. 2016. HIV-1 protease, reverse transcriptase, and integrase variation. J Virol 90:6058–6070. doi: 10.1128/JVI.00495-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gatanaga H, Suzuki Y, Tsang H, Yoshimura K, Kavlick MF, Nagashima K, Gorelick RJ, Mardy S, Tang C, Summers MF, Mitsuya H. 2002. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J Biol Chem 277:5952–5961. doi: 10.1074/jbc.M108005200. [DOI] [PubMed] [Google Scholar]
- 32.Gatanaga H, Das D, Suzuki Y, Yeh DD, Hussain KA, Ghosh AK, Mitsuya H. 2006. Altered HIV-1 Gag protein interactions with cyclophilin A (CypA) on the acquisition of H219Q and H219P substitutions in the CypA binding loop. J Biol Chem 281:1241–1250. doi: 10.1074/jbc.M505920200. [DOI] [PubMed] [Google Scholar]
- 33.Nijhuis M, van Maarseveen NM, Lastere S, Schipper P, Coakley E, Glass B, Rovenska M, de Jong D, Chappey C, Goedegebuure IW, Heilek-Snyder G, Dulude D, Cammack N, Brakier-Gingras L, Konvalinka J, Parkin N, Kräusslich HG, Brun-Vezinet F, Boucher CA. 2007. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med 4:e36. doi: 10.1371/journal.pmed.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kletenkov K, Hoffmann D, Böni J, Yerly S, Aubert V, Schöni-Affolter F, Struck D, Verheyen J, Klimkait T, Swiss HIV Cohort Study. 2017. Role of Gag mutations in PI resistance in the Swiss HIV cohort study: bystanders or contributors? J Antimicrob Chemother 72:866–875. doi: 10.1093/jac/dkw493. [DOI] [PubMed] [Google Scholar]
- 35.Maguire MF, Guinea R, Griffin P, Macmanus S, Elston RC, Wolfram J, Richards N, Hanlon MH, Porter DJ, Wrin T, Parkin N, Tisdale M, Furfine E, Petropoulos C, Snowden BW, Kleim JP. 2002. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to I50V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J Virol 76:7398–7406. doi: 10.1128/jvi.76.15.7398-7406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolli M, Stawiski E, Chappey C, Schiffer CA. 2009. Human immunodeficiency virus type 1 protease-correlated cleavage site mutations enhance inhibitor resistance. J Virol 83:11027–11042. doi: 10.1128/JVI.00628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh AK, Martyr CD, Osswald HL, Sheri VR, Kassekert LA, Chen S, Agniswamy J, Wang YF, Hayashi H, Aoki M, Weber IT, Mitsuya H. 2015. Design of HIV-1 protease inhibitors with amino- bis-tetrahydrofuran derivatives as P2-ligands to enhance backbone-binding interactions: synthesis, biological evaluation, and protein-ligand X-ray studies. J Med Chem 58:6994–7006. doi: 10.1021/acs.jmedchem.5b00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshimura K, Kato R, Kavlick MF, Nguyen A, Maroun V, Maeda K, Hussain KA, Ghosh AK, Gulnik SV, Erickson JW, Mitsuya H. 2002. A potent human immunodeficiency virus type 1 protease inhibitor, UIC-94003 (TMC-126), and selection of a novel (A28S) mutation in the protease active site. J Virol 76:1349–1358. doi: 10.1128/JVI.76.3.1349-1358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amano M, Koh Y, Das D, Li J, Leschenko S, Wang YF, Boross PI, Weber IT, Ghosh AK, Mitsuya H. 2007. A novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI), GRL-98065, is potent against multiple-PI-resistant human immunodeficiency virus in vitro. Antimicrob Agents Chemother 51:2143–2155. doi: 10.1128/AAC.01413-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tojo Y, Koh Y, Amano M, Aoki M, Das D, Kulkarni S, Anderson DD, Ghosh AK, Mitsuya H. 2010. Novel protease inhibitors (PIs) containing macrocyclic components and 3(R), 3a(S), 6a(R)- bis-tetrahydrofuranylurethane that are potent against multi-PI-resistant HIV-1 variants in vitro. Antimicrob Agents Chemother 54:3460–3470. doi: 10.1128/AAC.01766-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yedidi RS, Maeda K, Fyvie WS, Steffey M, Davis DA, Palmer I, Aoki M, Kaufman JD, Stahl SJ, Garimella H, Das D, Wingfield PT, Ghosh AK, Mitsuya H. 2013. P2′ benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel nonpeptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety. Antimicrob Agents Chemother 57:4920–4927. doi: 10.1128/AAC.00868-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amano M, Salcedo-Gomez PM, Zhao R, Yedidi RS, Das D, Bulut H, Delino NS, Sheri VR, Ghosh AK, Mitsuya H. 2016. A modified P1 moiety enhances in vitro antiviral activity against various multidrug-resistant HIV-1 variants and in vitro central nervous system penetration properties of a novel nonpeptidic protease inhibitor, GRL-10413. Antimicrob Agents Chemother 60:7046–7059. doi: 10.1128/AAC.01428-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh AK, Rao KV, Nyalapatla PR, Osswald HL, Martyr CD, Aoki M, Hayashi H, Agniswamy J, Wang YF, Bulut H, Das D, Weber IT, Mitsuya H. 2017. Design and development of highly potent HIV-1 protease inhibitors with a crown-like oxotricyclic core as the P2-ligand to combat multidrug-resistant HIV variants. J Med Chem 60:4267–4278. doi: 10.1021/acs.jmedchem.7b00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aoki M, Hayashi H, Rao KV, Das D, Higashi-Kuwata N, Bulut H, Aoki-Ogata H, Takamatsu Y, Yedidi RS, Davis DA, Hattori SI, Nishida N, Hasegawa K, Takamune N, Nyalapatla PR, Osswald HL, Jono H, Saito H, Yarchoan R, Misumi S, Ghosh AK, Mitsuya H. 2017. A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency. elife 6:e28020. doi: 10.7554/eLife.28020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh AK, Rao KV, Nyalapatla PR, Kovela S, Brindisi M, Osswald HL, Sekhara Reddy B, Agniswamy J, Wang YF, Aoki M, Hattori SI, Weber IT, Mitsuya H. 2018. Design of highly potent, dual-acting and central-nervous-system-penetrating HIV-1 protease inhibitors with excellent potency against multidrug-resistant HIV-1 variants. ChemMedChem 13:803–815. doi: 10.1002/cmdc.201700824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baxter JD, Schapiro JM, Boucher CA, Kohlbrenner VM, Hall DB, Scherer JR, Mayers DL. 2006. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J Virol 80:10794–10801. doi: 10.1128/JVI.00712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shahriar R, Rhee SY, Liu TF, Fessel WJ, Scarsella A, Towner W, Holmes SP, Zolopa AR, Shafer RW. 2009. Nonpolymorphic human immunodeficiency virus type 1 protease and reverse transcriptase treatment-selected mutations. Antimicrob Agents Chemother 53:4869–4878. doi: 10.1128/AAC.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Westen GJ, Hendriks A, Wegner JK, Ijzerman AP, van Vlijmen HW, Bender A. 2013. Significantly improved HIV inhibitor efficacy prediction employing proteochemometric models generated from antivirogram data. PLoS Comput Biol 9:e1002899. doi: 10.1371/journal.pcbi.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wensing AM, Calvez V, Günthard HF, Johnson VA, Paredes R, Pillay D, Shafer RW, Richman DD. 2017. 2017 Update of the drug resistance mutations in HIV-1. Top Antivir Med 24:132–133. [PMC free article] [PubMed] [Google Scholar]
- 50.Rhee SY, Clutter D, Fessel WJ, Klein D, Slome S, Pinsky BA, Marcus JL, Hurley L, Silverberg MJ, Kosakovsky Pond SL, Shafer RW. 2019. Trends in the molecular epidemiology and genetic mechanisms of transmitted human immunodeficiency virus type 1 drug resistance in a large US clinic population. Clin Infect Dis 68:213–221. doi: 10.1093/cid/ciy453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong L, Hartsuck JA, Foundling S, Ermoliefe J, Tang J. 2008. Active-site mobility in human immunodeficiency virus, type 1, protease as demonstrated by crystal structure of A28S mutant. Protein Sci 7:300–305. doi: 10.1002/pro.5560070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yates PJ, Hazen R, St Clair M, Boone L, Tisdale M, Elston RC. 2006. In vitro development of resistance to human immunodeficiency virus protease inhibitor GW640385. Antimicrob Agents Chemother 50:1092–1095. doi: 10.1128/AAC.50.3.1092-1095.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu TD, Schiffer CA, Gonzales MJ, Taylor J, Kantor R, Chou S, Israelski D, Zolopa AR, Fessel WJ, Shafer RW. 2003. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J Virol 77:4836–4847. doi: 10.1128/jvi.77.8.4836-4847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Falloon J, Piscitelli S, Vogel S, Sadler B, Mitsuya H, Kavlick MF, Yoshimura K, Rogers M, LaFon S, Manion DJ, Lane HC, Masur H. 2000. Combination therapy with amprenavir, abacavir, and efavirenz in human immunodeficiency virus (HIV)-infected patients failing a protease-inhibitor regimen: pharmacokinetic drug interactions and antiviral activity. Clin Infect Dis 30:313–318. doi: 10.1086/313667. [DOI] [PubMed] [Google Scholar]
- 55.Yoshimura K, Kato R, Yusa K, Kavlick MF, Maroun V, Nguyen A, Mimoto T, Ueno T, Shintani M, Falloon J, Masur H, Hayashi H, Erickson J, Mitsuya H. 1999. JE-2147: a dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc Natl Acad Sci U S A 96:8675–8680. doi: 10.1073/pnas.96.15.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamiya S, Mardy S, Kavlick MF, Yoshimura K, Mistuya H. 2004. Amino acid insertions near Gag cleavage sites restore the otherwise compromised replication of human immunodeficiency virus type 1 variants resistant to protease inhibitors. J Virol 78:12030–12040. doi: 10.1128/JVI.78.21.12030-12040.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hazra R, Gafni RI, Maldarelli F, Balis FM, Tullio AN, DeCarlo E, Worrell CJ, Steinberg SM, Flaherty J, Yale K, Kearney BP, Zeichner SL. 2005. Tenofovir disoproxil fumarate and an optimized background regimen of antiretroviral agents as salvage therapy for pediatric HIV infection. Pediatrics 116:e846–854. doi: 10.1542/peds.2005-0975. [DOI] [PubMed] [Google Scholar]
- 58.Harada S, Hazra R, Tamiya S, Zeichner SL, Mitsuya H. 2007. Emergence of human immunodeficiency virus type 1 variants containing the Q151M complex in children receiving long-term antiretroviral chemotherapy. Antiviral Res 75:159–166. doi: 10.1016/j.antiviral.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Maeda K, Desai DV, Aoki M, Nakata H, Kodama EN, Mitsuya H. 2013. Delayed emergence of HIV-1 variants resistant to 4′-ethynyl-2-fluoro-2′-deoxyadenosine: comparative sequential passage study with lamivudine, tenofovir, emtricitabine and BMS-986001. Antivir Ther 19:179–189. doi: 10.3851/IMP2697. [DOI] [PubMed] [Google Scholar]
- 60.Nakata H, Amano M, Koh Y, Kodama E, Yang G, Bailey CM, Kohgo S, Hayakawa H, Matsuoka M, Anderson KS, Cheng YC, Mitsuya H. 2007. Activity against human immunodeficiency virus type 1, intracellular metabolism, and effects on human DNA polymerases of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Antimicrob Agents Chemother 51:2701–2708. doi: 10.1128/AAC.00277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takamatsu Y, Tanaka Y, Kohgo S, Murakami S, Singh K, Das D, Venzon DJ, Amano M, Higashi-Kuwata N, Aoki M, Delino NS, Hayashi S, Takahashi S, Sukenaga Y, Haraguchi K, Sarafianos SG, Maeda K, Mitsuya H. 2015. 4′-Modified nucleoside analogs: potent inhibitors active against entecavir-resistant hepatitis B virus. Hepatology 62:1024–1036. doi: 10.1002/hep.27962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matayoshi ED, Wang GT, Krafft GA, Erickson J. 1990. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science 247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 63.Muzammil S, Armstrong AA, Kang LW, Jakalian A, Bonneau PR, Schmelmer V, Amzel LM, Freire E. 2007. Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations. J Virol 81:5144–5154. doi: 10.1128/JVI.02706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amano M, Tojo Y, Salcedo-Gómez PM, Parham GL, Nyalapatla PR, Das D, Ghosh AK, Mitsuya H. 2015. A novel tricyclic ligand-containing nonpeptidic HIV-1 protease inhibitor, GRL-0739, effectively inhibits the replication of multidrug-resistant HIV-1 variants and has a desirable central nervous system penetration property in vitro. Antimicrob Agents Chemother 59:2625–2635. doi: 10.1128/AAC.04757-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amano M, Miguel Salcedo-Gómez P, Yedidi RS, Delino NS, Nakata H, Venkateswara Rao K, Ghosh AK, Mitsuya H. 2017. GRL-09510, a unique P2-crown-tetrahydrofuranylurethane -containing HIV-1 protease inhibitor, maintains its favorable antiviral activity against highly-drug-resistant HIV-1 variants in vitro. Sci Rep 7:12235. doi: 10.1038/s41598-017-12052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takamatsu Y, Das D, Kohgo S, Hayashi H, Delino NS, Sarafianos SG, Mitsuya H, Maeda K. 2018. The high genetic barrier of EFdA/MK-8591 stems from strong interactions with the active site of drug-resistant HIV-1 reverse transcriptase. Cell Chem Biol 25:1268–1278.e3. doi: 10.1016/j.chembiol.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ghosh AK, Martyr CD, Kassekert LA, Nyalapatla PR, Steffey M, Agniswamy J, Wang YF, Weber IT, Amano M, Mitsuya H. 2015. Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands. Org Biomol Chem 13:11607–11621. doi: 10.1039/C5OB01930C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schuttelkopf AW, van Aalten DM. 2004. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 69.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.