Ceftazidime-avibactam (CAZ/AVI) combines ceftazidime with a diazabicyclooctane non-β-lactam β-lactamase inhibitor. This has potent inhibitory activity against KPC-type enzymes. We studied activity of clinically relevant regimens of CAZ/AVI against two KPC-2-bearing Klebsiella pneumoniae isolates (sequence type 258 recovered sequentially from the same patient) with and without ompK36 mutations in a hollow fiber infection model. The baseline total bacterial burden exceeded 109 CFU.
KEYWORDS: KPC beta-lactamase, hollow fiber infection model, pharmacodynamics
ABSTRACT
Ceftazidime-avibactam (CAZ/AVI) combines ceftazidime with a diazabicyclooctane non-β-lactam β-lactamase inhibitor. This has potent inhibitory activity against KPC-type enzymes. We studied activity of clinically relevant regimens of CAZ/AVI against two KPC-2-bearing Klebsiella pneumoniae isolates (sequence type 258 recovered sequentially from the same patient) with and without ompK36 mutations in a hollow fiber infection model. The baseline total bacterial burden exceeded 109 CFU. For both isolates, there was early multi-log CFU/ml reductions in the bacterial burden for all regimens. Bacterial subpopulations with reduced susceptibilities to CAZ/AVI were isolated only from the no-treatment control arms. All CAZ/AVI regimens resulted in undetectable colony counts between days 6 and 8. At day 10, the total volume of each CAZ/AVI arm was plated, with no organisms recovered from any regimen, documenting complete eradication. A population model was fit to avibactam concentrations and total colony count outputs. The model fit was acceptable and demonstrated a large kill rate constant (Kkill = 6.29 h−1) and a relatively low avibactam concentration at which kill rate was half maximal (C50 = 2.19 mg/liter), concordant with the observed bacterial burden decline. A threshold analysis identified time > 4 mg/liter of avibactam as the index most closely linked to bacterial burden decline. Given the clinical outcomes seen with KPC-bearing organisms and the toxicities that occur when patients are treated with currently available polymyxins, drugs such as CAZ/AVI should have a prominent place in early therapy.
INTRODUCTION
The advent of β-lactamases that hydrolyze carbapenem antibiotics was a major event threatening our ability to adequately treat serious infections in hospitalized patients infected with organisms carrying these enzymes. Examination of outcomes from patients in prospective cohort studies (1, 2) and from a case-control study (3) demonstrated that mortality from carbapenemase-producing Klebsiella pneumoniae were substantial, ranging from 32 to 36%. Of note, in one study (3), these isolates were associated with a 61% mortality in patients with pneumonia. Few antimicrobials are available to appropriately treat these pathogens. Polymyxin analogues often have an MIC of ≤2 mg/liter but have a rapid emergence of resistance during monotherapy and are associated with a substantial rate of acute kidney injury (4, 5).
Ceftazidime-avibactam (CAZ/AVI) has been approved for use (6) in complicated urinary tract infections, complicated intra-abdominal infections, and hospital-acquired and ventilator-associated bacterial pneumonia (HABP/VABP). Avibactam is a diazabicyclooctane, non-β-lactam β-lactamase inhibitor with an inhibitory profile that includes (among others) KPC-2 and KPC-3 β-lactamases.
We report here the effects of the combination of CAZ/AVI in a fixed regimen of 2 g ceftazidime plus 500 mg avibactam against two isolates of Klebsiella pneumoniae (7) carrying KPC-2 β-lactamases, as determined in a 10-day hollow fiber infection model (HFIM), where the organisms were exposed to the plasma concentration-time profile of CAZ/AVI. The isolates studied were recovered from the same patient and differed only by an IS5 insertion in the ompK36 promoter. We also wanted to look at different administration profiles of drug to determine their impacts on bacterial cell kill. Finally, we modeled the effect of different infusion times and clinically relevant exposures of CAZ/AVI on the total population of both Klebsiella pneumoniae KPC-2-bearing organisms. By studying this isogenic pair (except for the ompK36 promoter mutation), we could identify the impact of the porin deletion on CAZ/AVI activity.
RESULTS
Organism MIC and mutational frequency to resistance.
Two KPC-2-producing Klebsiella pneumoniae isolates were evaluated. The isolate designated 20KPN had an IS5 insertion in the ompK36 promoter, while 86KPN was a wild-type strain. Both carry a KPC-2 enzyme. The broth microdilution MIC for serial 2-fold dilutions of ceftazidime tested with a fixed concentration of 4 mg/liter of avibactam (CAZ/AVI) were 0.5/4.0 mg/liter and 0.125/4.0 mg/liter, respectively. The mutational frequencies to resistance were 1 in 7.62 log10(CFU) (or 1 in 4.17 × 107 CFU) for 20KPN and 1 in 7.94 Log10(CFU/) (or 1 in 8.71 × 107 CFU) for 86KPN when tested with CAZ/AVI at 3× the baseline MIC for ceftazidime in the presence of 4 mg/liter of avibactam incorporated into the selecting agar. The CAZ/AVI MIC values for colonies which grew on the antibiotic-supplemented agars for 20KPN were 1 to 2 mg of ceftazidime when tested with 4 mg/liter of avibactam. For 86KPN, colonies which grew on drug-containing agar had ceftazidime MIC values of 4 to 8 mg/liter when tested in combination with 4 mg/liter of avibactam.
Klebsiella pneumoniae bacterial kill and resistance emergence.
The plasma pharmacokinetics reported in adult humans for clinically relevant CAZ/AVI regimens were simulated in the HFIM. There was a no-treatment control and six different exposures to CAZ/AVI. These six exposures are listed below.
(i) The CAZ/AVI exposure was the –1 standard deviation (–SD) concentration-time profile for a regimen of 2 g of ceftazidime administered intravenously (i.v.) every 8 h in combination with avibactam 500 mg i.v. as a 2-h infusion every 8 h.
(ii) The CAZ/AVI exposure was the mean concentration-time profile for a regimen of 2 g of ceftazidime i.v. every 8 h in combination with avibactam at 500 mg i.v. as a 2-h infusion every 8 h.
(iii) The CAZ/AVI exposure was the +1 standard deviation (+SD) concentration-time profile for a regimen of 2 g of ceftazidime i.v. every 8 h in combination with avibactam 500 mg i.v. as a 2-h infusion every 8 h.
(iv) The CAZ/AVI exposure was the mean concentration-time profile for a regimen of 2 g of ceftazidime i.v. every 8 h in combination with avibactam at 500 mg i.v. as a 4-h infusion every 8 h.
(v) The CAZ/AVI exposure was the mean concentration-time profile for a regimen of 2 g of ceftazidime i.v. every 6 h in combination with avibactam at 500 mg i.v. as a 2-h infusion every 6 h.
(vi) The CAZ/AVI exposure was 6 g of ceftazidime plus 1.5 g of avibactam as a continuous infusion (both drugs) after a small loading dose.
For 20KPN, the baseline bacterial burden was 12 ml as 8.45 Log10(CFU/ml) [9.53 Log10(CFU) total burden]. For 86KPN, the baseline bacterial burden was 12 ml as 8.19 Log10(CFU/ml) [9.27 Log10(CFU) total burden].
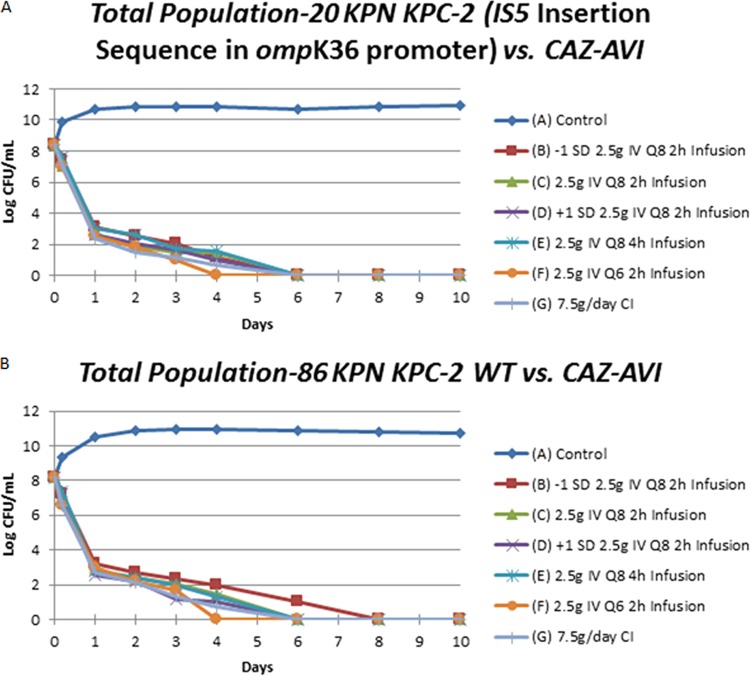
The microbiological effect plots for the different regimens of CAZ/AVI for the two isolates are shown in Fig. 1A (20KPN) and Fig. 1B (86KPN). All of the CAZ/AVI regimens rapidly killed the bacterium inoculated into the hollow fiber cartridge by day 1, with a lower rate of killing thereafter. No colonies were detected after 4 to 8 days of treatment. On day 10, the total volume of each hollow fiber cartridge was removed, washed, and cultured in its entirety to check for organism eradication. These end-of-study cultures were sterile.
FIG 1.
Bacterial cell kill for six regimens of ceftazidime/avibactam plus a no-treatment control for two isolates of KPC-2-bearing Klebsiella pneumoniae. (A) Total populations of 20KPN; (B) total populations of 86KPN.
Less-susceptible organisms were not recovered from any active CAZ/AVI regimen for either 20KPN or 86KPN. For the no-treatment controls, no less-susceptible isolates were recovered at baseline (t = 0 h) for either isolate. However, at the 4-h time point, 20KPN had less-susceptible isolates recovered. For 86KPN, less-susceptible isolates were not seen in its respective control arm until 24 h. The majority of the less-susceptible isolates isolated from the control arms had CAZ MICs of 2 to 4 mg/liter when tested in combination with a fixed concentration of 4 mg/liter of avibactam. A few isolates had CAZ/AVI MICs of 8/4 mg/liter, and one isolate had an MIC of 32/4 mg/liter.
Mathematical analysis.
Multiple analyses were performed. We performed an analysis where only data from 20KPN alone and 86KPN alone were employed. The analysis included two system outputs: drug concentration of avibactam and total colony counts. Since only the no-treatment control had less-susceptible isolates recovered and there was no selective pressure, these data were not employed. It should be noted that there were no resistant clones isolated from the no-treatment control at baseline for either isolate even though the total bacterial burden slightly exceeded the inverse of the mutational frequency to resistance (see above). The explanation is that with the volume plated for resistance selection (100 μl) is such that finding a less-susceptible isolate is a function of a Poisson distribution. In the case of 20KPN, there were slightly fewer than 7 colonies/ml that would be expected, given the estimate of the total bacterial burden and mutational frequency to resistance. With the sample volume for the less-susceptible population, it is not surprising that there were no less-susceptible colonies found at baseline. They were recovered at hour 4, where the total bacterial burden was 9.89 log10(CFU/ml) and the number of less-susceptible isolates was approximately 186 colonies/ml.
We modeled only the avibactam concentrations as drug exposures. We chose to do this because the two ceftazidime MIC values in the presence of 4.0 mg/liter of avibactam (0.5/4.0 mg/liter and 0.125/4.0 mg/liter for 86KPN and 20KPN, respectively) were always exceeded by the actual measured exposures to ceftazidime. We posited that the variability in cell kill observed would be explained by the variability in the avibactam concentrations.
For both isolates, the bacterial burden was reduced to approximately zero between days 4 and 6 for 20KPN and between days 4 and 8 for 86KPN. The individual analyses showed similar parameter estimates (data not shown). This caused us to comodel both data sets simultaneously. We show the mean, median and standard deviation of the parameter estimates in Table 1. The mean and median values are similar with a small coefficient of variation for most of the parameter values. The exception is for the C50. This is to be expected, as the regimens were set up to be different, with the first three evaluations representing the −1 SD, mean, and +1 SD for the exposure to avibactam for a regimen of 2 g ceftazidime/500 mg avibactam administered as a 2-h infusion. The fourth was the mean value administered as a 4-h infusion, the fifth was a regimen employing the mean parameters (but with a 6-hourly administration of 2 g/500 mg), and the 6th was a rapid load, followed by a continuous infusion of 6 g/1,500 mg per day. The fit of the model to the data was acceptable. The predicted-observed regressions for both outputs (avibactam concentrations and total colony counts) are displayed in Table 2.
TABLE 1.
Means, medians, and standard deviations of population model parameter estimates for comodeling both strains 20KPN and 86KPNa
| Parameter | V | CL | Kg | Kkill | C50 | H | POPMAX | IC2 |
|---|---|---|---|---|---|---|---|---|
| Units | Liters | Liters/h | h−1 | h−1 | mg/liter | CFU/ml | CFU/ml | |
| Mean | 28.6 | 9.55 | 5.04 | 6.29 | 2.17 | 13.7 | 6.16 × 1010 | 2.58 × 108 |
| Median | 28.6 | 10.3 | 5.30 | 5.59 | 0.196 | 14.5 | 6.02 × 1010 | 1.44 × 108 |
| SD (%CV) | 3.52 (12.3) | 1.72 (18.0) | 0.609 (12.1) | 1.39 (22.1) | 3.02 (139) | 6.75 (49.1) | 4.00 × 109 (6.49) | 2.19 × 108 (85.1) |
V, volume of distribution; CL, clearance; Kg, first-order growth rate constant; Kkill, first order bacterial kill rate constant; C50, avibactam concentration at which the Kkill is half maximal; H, Hill’s constant (unitless); POPMAX, maximal total bacterial burden; IC2, initial bacterial burden.
TABLE 2.
Pre-Bayesian (population) and Bayesian (individual) regressions for avibactam concentrations and total bacterial burden
| Regression | Avibactam concn (mg/liter) | Total bacterial burden, log10(CFU/ml) |
|---|---|---|
| Pre-Bayesian | Observed = 0.986 × predicted + 0.586; r2 = 0.853 | Observed = 1.13 × predicted + 0.772; r2 = 0.755 |
| Bayesian | Observed = 1.01 × predicted + 0.251; r2 = 0.949 | Observed = 0.891 × predicted + 0.146; r2 = 0.808 |
Bayesian parameter estimates.
The parameter estimates derived from the mean parameter vector for the parameters related to drug effect are shown in Table 3. The parameter estimates show reproducibility between strains
TABLE 3.
Bayesian parameter estimates for drug concentration-time profile and effect parameters as derived from the mean parameter vectora
| Regimenb | Isolate | V | CL | Kg | Kkill | C50 | H |
|---|---|---|---|---|---|---|---|
| –1SD | 20KPN | 27.2 | 11.5 | 5.29 | 5.57 | 0.350 | 2.17 |
| 86KPN | 25.3 | 11.6 | 5.29 | 5.57 | 0.350 | 2.01 | |
| Mean | 20KPN | 30.0 | 9.35 | 5.12 | 6.02 | 0.993 | 19.2 |
| 86KPN | 33.2 | 9.01 | 4.45 | 7.81 | 4.16 | 19.5 | |
| +1SD | 20KPN | 28.1 | 6.30 | 5.18 | 8.78 | 7.38 | 14.3 |
| 86KPN | 29.8 | 6.30 | 5.18 | 8.78 | 7.38 | 14.3 | |
| 4hrinfusion | 20KPN | 28.5 | 9.21 | 5.30 | 5.50 | 0.101 | 19.0 |
| 86KPN | 31.0 | 10.3 | 5.35 | 5.56 | 0.102 | 19.5 | |
| Q6h | 20KPN | 28.8 | 9.00 | 5.30 | 5.57 | 0.103 | 6.74 |
| 86KPN | 30.7 | 10.4 | 5.35 | 5.57 | 0.123 | 17.8 | |
| CI | 20KPN | 20.3 | 11.2 | 3.34 | 5.40 | 5.26 | 10.9 |
| 86KPN | 31.0 | 10.3 | 5.35 | 5.57 | 0.101 | 19.6 |
Definitions for parameters are given in Table 1.
–1SD, regimen for 2 g/500 mg as a 2-h infusion every 8 h for parameters that produce a concentration time curve one standard deviation below the mean; mean, parameters produce the mean concentration-time profile for 2 g/500 mg every 8 h; +1SD, regimen for 2 g/500 mg as a 2-h infusion every 8 h for parameters that produce a concentration time curve one standard deviation above the mean; 4hrinfusion, the mean concentration-time curve is produced, but with a 4-h infusion; Q6h, the 2-g/500-mg dose is administered as a 2-h infusion every 6 h; CI, 6 g/1.5 g are administered as a continuous infusion.
Pharmacodynamic driver.
Examination of Fig. 1 shows that both isolates have the maximal variability in response occurring at day 4. This is because the bacterial burden is bounded from above because over time, in the absence of drug, organisms will enter stationary phase. The bacterial burden is also bounded from below by reaching an extinction event in all the drug-containing regimens.
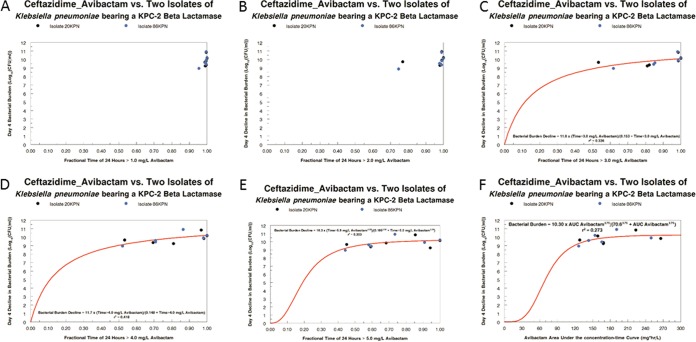
We fit only the avibactam concentration-time profiles to allow calculation of individual regimen AUC values, as well as the time that avibactam concentrations remained above a threshold, which we ranged from 1 through 5 mg/liter in increments of 1 mg/liter. The results are displayed in Fig. 2. The threshold avibactam concentrations of 1 and 2 mg/liter did not produce enough range to explain the variability in bacterial cell kill. Threshold values of 3, 4, and 5 mg/liter of avibactam demonstrated reasonable relationships with a sigmoid-Emax effect model, with the threshold of 4 mg/liter explaining the largest amount of overall variance and was statistically significant (P = 0.023). The AUC of avibactam was also evaluated but did not explain a larger amount of variance compared to fractional (of the dosing interval) time above an avibactam concentration of 4 mg/liter, and the regression was not statistically significant. Note that the decline in bacterial burden is relative to the no-treatment controls for the two isolates on day 4.
FIG 2.
Linkage of avibactam concentrations in the hollow fiber model to bacterial cell kill employing a sigmoid Emax effect model.
DISCUSSION
KPC β-lactamases have destabilized the practice of modern medicine. The carbapenem class of antimicrobials was the last bulwark of defense for seriously infected patients, particularly those with immune dysfunction, such as neutropenic patients, patients with a transplant, or patients taking biologics that alter immune function (e.g., anti-tumor necrosis factor monoclonal antibodies).
For a number of years, clinicians have had to employ polymyxin antimicrobials to treat patients infected with KPC-bearing organisms. Unfortunately, these agents (colistin and polymyxin B) frequently cause acute kidney injury (5). They also display a central nervous system adverse event profile (8). Further, resistance emergence during therapy is a frequent occurrence. Finally, between-patient variability in clearance is a major problem for identifying a drug dose that is highly likely to provide efficacious therapy. This is particularly the case for colistin, as it is administered as the prodrug: colistin methanesulfonate.
Ceftazidime/avibactam (CAZ/AVI) is the first new β-lactam/β-lactamase inhibitor combination approved with significant activity against KPC-2- and KPC-3-bearing β-lactamases. In this set of experiments, we have studied two isolates of KPC-2-bearing Klebsiella pneumoniae in the hollow fiber infection model. This model provides a tool for examining the antibacterial effect of a new agent. In addition, because of the bacterial burden it is capable of supporting, the evaluation for resistant subpopulation amplification is straightforward.
As seen in Fig. 1, both isolates were studied at initial bacterial burdens that exceeded 8 log10(CFU/ml) [and exceeded a 9-log10(CFU) total bacterial burden]. Nonetheless, all the regimens of CAZ/AVI evaluated sterilized the system over the period of 10 days. All units actually had bacterial counts that were undetectable starting at days 4 to 8. In no instance were less-susceptible subpopulations identified in the active therapy arms for either isolate.
Resistance emergence during CAZ/AVI has been described during therapy (9–11) in Klebsiella pneumoniae due to alterations to the β-lactamase (KPC-2 and KPC-3), leading to high-level resistance (11). Mutations in ompK36 have also been described to shift CAZ/AVI MIC’s in Klebsiella. The isolates studied are isogenic and differ only in having an IS5 insertion in the ompK36 promoter. Therefore, studying both gives insight into the impact of this mutation on the activity of the CAZ/AVI regimens studied.
In this set of experiments, we were unable to recover any less susceptible isolates. In the experimental design, we evaluated a concentration-time profile of CAZ/AVI that was 1 SD below the mean concentration-time curve. This still prevented amplification of a less-susceptible subpopulation. We speculate the inability to select resistant clones was due to a combination of relatively low MICs (0.5 and 0.125 mg/liter), ceftazidime concentrations where even the resistant isolates had long ceftazidime concentrations time > MIC, and not going low enough on the AVI exposure.
The bacterial cell kill was impressive for both isolates, achieving an inability to recover any colonies at days 6 and 8, with an initial burden of >8 log10(CFU/ml). For the day 4 colony counts, a 2-log10(CFU/ml) decline from the baseline was calculated to require approximately 10% of the dosing interval to have avibactam concentrations in excess of 4 mg/liter.
An independent issue is the question of the pharmacodynamic driver for bacterial cell kill. As stated above, because the ceftazidime concentrations were always above the MIC (determined in the presence of 4 mg/liter of avibactam) for both strains, we hypothesized that variability in response would be attributable to the avibactam concentrations. We looked at the avibactam AUC as a driver, as well as the time greater than avibactam concentrations between 1 and 5 mg/liter, rising in an increment of 1 mg/liter. As can be seen in Fig. 2, low avibactam concentrations (1 and 2 mg/liter) had very high fractions of the dosing interval with coverage above these numbers. When we looked at 3, 4, and 5 mg/liter, the time greater than a threshold of 4 mg/liter explained the largest amount of the variance and was statistically significant (P = 0.023). The avibactam AUC was also examined, but this driver explained substantially less of the variance than time > 4 mg/liter of avibactam, and the relationship was not statistically significant.
Consequently, prolonging the infusion of CAZ/AVI may result in improving bacterial cell kill. Many patients receiving CAZ/AVI are very seriously infected and reside in the intensive care unit (12) Such patients will have issues with intravascular catheter management. A prolonged infusion would have the attribute of allowing other i.v. medications to be delivered without the problem of line incompatibilities. Further, prolonged infusions have been demonstrated to be clinically advantageous, especially in patients with higher APACHE II scores (13).
When optimizing outcomes, it is important to look both at bacterial cell kill and at resistance suppression. Because no resistance emergence was identified, we were not able to identify an exposure threshold that would allow resistance to become a very low likelihood event. Given that we went as low as 1 SD of exposure below the mean, it may be that even lower exposures would need to be investigated or isolates with a higher mutational frequency to resistance would require study. The current data indicate that for the approved dose and schedule of CAZ/AVI, resistance emergence in KPC-2-bearing isolates would be infrequent, at least in isolates like 20KPN with an IS5 insertion in ompK36.
KPC-bearing organisms continue to be a major problem. With help from governmental organizations like BARDA, DTRA, and NIAID, the pharmaceutical industry has risen to the challenge and has gained U.S. Food and Drug Administration (FDA)-approved indications for three new molecules with substantial activity against these organisms. Indeed, in each instance, there are data that indicate that these agents (CAZ/AVI, plazomicin, and meropenem/vaborbactam) have superior outcomes compared to polymyxin-based regimens (14–16). We note that eravacycline has good in vitro activity against KPC-bearing isolates, but we were unable to find any clinical data similar to that seen for the other three agents. For the sake of our patients, stewardship groups need to reevaluate the use of polymyxin agents in the circumstance where the presence of KPC-bearing organisms is strongly suspected. The lack of optimal activity, the large between-patient variability, and the substantial toxicity profile all mitigate against the continued use of these agents. CAZ/AVI and similar molecules should be agents of choice in this clinical circumstance.
MATERIALS AND METHODS
Microorganisms.
The two clinical isolates of KPC-2-bearing Klebsiella pneumoniae were obtained from a patient treated at the University of Pittsburgh Medical Center. Both isolates harbored blaKPC-2, blaSHV-11, blaSHV-12, and blaOXA-9, as well as a mutation in ompK35 resulting in a premature stop codon at amino acid position 89. Isolate 20KPN harbored an IS5 insertion in the ompK36 promoter, while isolate 86KPN harbored wild-type ompK36.
Drugs.
Ceftazidime/avibactam was kindly supplied by Allergan, Inc.
In vitro susceptibility testing.
The in vitro susceptibility to CAZ/AVI was determined using the microdilution broth and the agar dilution method described by the Clinical and Laboratory Standards Institute (17) in cation-adjusted Mueller-Hinton broth (CA-MHB) and Mueller-Hinton agar (MHA), respectively. For the respective method, the susceptibility testing was performed using serial 2-fold dilutions of ceftazidime in combination with a fixed concentration of 4 mg/liter of avibactam. The MICs were read after the cultures were incubated for 16 to 20 h at 35°C in ambient air.
Mutation frequency.
Overnight broth cultures of 20KPN and 86KPN were quantitatively cultured onto drug-free MHA plates to estimate the total bacterial burden and also onto agar supplemented with 3× baseline MIC values of ceftazidime of the respective isolate in the presence of 4 mg/liter of avibactam. After 48 h of incubation, the colonies on drug-free and antibiotic-supplemented agars were enumerated. The mutation frequency value was calculated by dividing the total number of colonies on drug-supplemented agar by the number of colonies which grew on drug-free agar. To validate that the colonies which grew on antibiotic-supplemented agars had reduced susceptibilities to the test antibiotics, MICs were determined for several of the colonies collected from the drug-containing plates.
Hollow fiber infection model.
An HFIM was used to investigate the pharmacodynamics of CAZ/AVI against 20KPN and 86KPN. A peristaltic pump circulated CA-MHB between the central compartment of the hollow fiber cartridges (FiberCell Systems, Frederick, MD) and the central compartment. CAZ/AVI was administered into the central compartment by a programmable syringe pump. Fresh CA-MHB was pumped from a reservoir into the central compartment, and the same volume of drug-containing medium was removed as waste. The rate was controlled to simulate pharmacokinetic profiles for CAZ/AVI (18). The extracapillary space of each HFIM was inoculated with 12 ml of bacterial suspension. The desired inoculum was confirmed with quantitative cultures. The HFIM was incubated at 35°C in ambient air. Over the 10-day experiments, 0.4-ml portions of bacterial suspension were collected from the extracapillary space. Serial dilutions in 0.1-ml volumes were then quantitatively cultured onto both drug-free agar and agar supplemented with 3× the baseline MIC of ceftazidime plus 4 mg/liter of avibactam to enumerate the impact of each antibiotic regimen on the total and less-susceptible bacterial populations, respectively.
Study design.
Each hollow fiber experiment consisted of six active treatment arms and a no-treatment control. As outlined in Results, the CAZ/AVI concentration-time profiles for the mean pharmacokinetic (PK) values, as well as the PK profiles that were −1 SD and +1 SD relative to the mean, were simulated for ceftazidime at 2 g in combination with avibactam at 500 mg administered i.v. every 8 h (q8h) as 2- and 4-h prolonged infusions. A higher, non-FDA-approved dose of CAZ/AVI of 2 g/500 mg given every 6 h as 2-h infusions and CAZ/AVI as 6 g/1.5 g per day as a continuous infusion after a small loading dose (to rapidly attain steady state) were evaluated. Each hollow fiber arm was sampled for quantitative cultures of the bacterial densities at baseline (time = 0 h) and at 0.17, 1, 2, 3, 4, 6, 8, and 10 days after therapy initiation for microbiological endpoints (total and less-susceptible K. pneumoniae populations). To validate that the intended PK profiles were simulated, serial samples of medium were collected from each antibiotic treatment arm over the first 48 h of an experiment and frozen at –80°C until assayed for ceftazidime and avibactam concentrations using a validated liquid chromatography-tandem mass spectrometry (LC-MS/MS) method (see below).
Mathematical modeling.
The system of differential equations employed for modeling the interaction of antimicrobial agents has been previously described (19). The model was implemented in the program BigNPAG (20). We simultaneously modeled two system outputs for the analysis of the data. These were (i) concentration of avibactam and (ii) total K. pneumoniae colony counts.
BigNPAG partitions model error into assay (fixed) and residual (random). The assay error is calculated as an output dependent SD, SD = A0 + A1[C] + A2[C]2 + A4[C]3, where [C] is the measured output of drug concentration or the log10-transformed colony count. There is one set of coefficients, A, for each of the two output equations. In addition, we used a fitted multiplicative term, γ, such that each observation in the fitting process was weighted by the Fisher information, i.e., 1/(γ × SD2).
Pre-Bayesian (population) regressions were generated employing the population mean parameter vector for generating the predicted output values. Bayesian (individual) regressions were generated using the mean Bayesian posterior parameter values for each of the HFIM experiment tubes.
Sigmoid Emax effect modeling was performed using the ADAPT 5 package of programs (21). We used the maximum-likelihood estimator in the package to fit the model to the data.
Ceftazidime/avibactam UPLC-MS/MS assay for Mueller-Hinton II broth.
Samples in Mueller-Hinton II broth were removed from storage at –80°C and allowed to thaw at room temperature. Using 1.5-ml microcentrifuge tubes, 10.0 μl of each sample and 10 μl of the internal standard (cefepime, 50.0 μg/ml in water) was added, followed by 0.500 ml of LC-MS-grade water. Each sample was then capped, vortexed well for 30 s, and centrifuged for 10 min at 16,168 × g. After centrifugation, 100 μl of each sample supernatant and 100 μl of LC-MS-grade water was transferred to a 96-well plate (or vial) for analysis by LC-MS/MS.
Determination of ceftazidime and avibactam concentrations was performed using LC-MS/MS consisting of an Acquity I-Class UPLC (Waters) and an API5000 triple quadrupole mass spectrometer (AB Sciex). Separation was achieved using a Kinetex C18 (100 by 3.0 mm, 2.6 μm; Phenomenex) high-pressure liquid chromatography column at 40°C with a run time of 3.50 min. Mobile phases consisted of 0.1% formic acid (FA) in water (A) and 0.1% FA in acetonitrile (B) at a flow rate of 0.500 ml/min. The gradient profile was as follows: 0 to 0.5 min, 5% B; 0.5 to 1.5 min, 5 to 40% B; 1.51 to 2.50 min, 95% B; and 2.51 to 3.5 min, 95 to 5% B. A 1-μl injection volume was used for analysis.
The mass spectrometer was operated in both positive and negative ion mode using the turbo ion spray (TIS) probe interface. Multiple reaction monitoring (MRM) m/z 264/95.8 (quantifier) and m/z 264/79.8 (qualifier) was used for Avibactam, m/z 547.1/468 (quantifier) and m/z 547.1/166.9 (qualifier) was used for ceftazidime, and m/z 481.2/396.2 for the internal standard cefepime. API5000 parameters (in arbitrary units) were as follows: CAD 6, CUR 30, GS1 60, GS2 60, IS 5500(CAZ)-4500(AVI), TEM 650°C; MRM 264/95.8: DP –120, CE –42, CXP –11, dwell 200 ms; MRM 264/79.8: DP –120, CE –56, CXP –9, dwell 200 ms; MRM 547.1/468: DP 111, CE 19, CXP 18, dwell 200 ms; MRM 547.1/166.9: DP 76, CE 37, CXP 24, dwell 200 ms; MRM 481.2/396.2: DP 81, CE 19, CXP 14, dwell 200 ms. Calculations were performed using Analyst software v1.6.2 (AB Sciex). Parameter abbreviations are as follows: CAD, collision cell gas setting; CUR, curtain plate gas setting; GS1 (gas 1), nebulizer gas setting; GS2 (gas 2), auxiliary gas setting; IS, ion spray voltage; TEM, temperature of heater gas; DP, declustering potential; CE, collision cell energy; CXP, collision cell exit potential.
Linearity for Avibactam in Mueller-Hinton II broth with a range of 0.250 to 50.0 μg/ml was demonstrated over four separate runs with a correlation coefficient (R) of ≥0.9979 and a linear regression (R2) of ≥0.9958. Intra- and inter-run accuracies for each calibration curve were within ±5.7 and ±3% of nominal concentrations, respectively. The calibration curve precision intrarun ranged from 0.1 to 9.8%, and the inter-run precision ranged from 2.6 to 4.9%. The quality control (QC) samples intra- and inter-run accuracies were within ±9 and ±5.9% of nominal concentrations, respectively. The QC sample precision intrarun ranged from 1.1 to 8.1%, and inter-run precision ranged from 3.5 to 6%.
Ceftazidime linearity in Mueller-Hinton II broth with a range of 1.00 to 200 μg/ml was demonstrated over four separate runs with a correlation coefficient (R) of ≥0.9987 and linear regression (R2) of ≥0.9974. Intra- and inter-run accuracies for each calibration curve were within ±7 and ±5.5% of nominal concentrations, respectively. The calibration curve precision intrarun ranged from 0.7 to 9.6%, and the inter-run precision ranged from 2.2 to 6.3%. The QC sample intra- and inter-run accuracies were within ±10.8 and ±5.4% of nominal concentrations, respectively. The QC sample precision intrarun ranged from 0.8 to 9%, and the inter-run precision ranged from 2.8 to 8%.
ACKNOWLEDGMENT
This study was supported, in part, by Allergan, Inc.
REFERENCES
- 1.de Maio Carrilho CMD, Marques de Oliverira L, Gaudereto J, Perozin JS, Urbano MR, Camargo CH, Grion CM, Levin AS, Costa SF. 2016. A prospective study of treatment of carbapenem-resistant Enterobacteriaceae infections and risk factors associated with outcome. BMC Infect Dis 16:629. doi: 10.1186/s12879-016-1979-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamma PD, Goodman KE, Harris AD, Tekle T, Roberts A, Taiwo A, Simner PJ. 2017. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin Infect Dis 64:257–264. doi: 10.1093/cid/ciw741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papadimitriou-Olivgeris M, Fligou F, Bartzavali C, Zotou A, Spyropoulou A, Koutsileou K, Vamvakopoulou S, Sioulas N, Karamouzos V, Anastassiou ED, Spiliopoulou I, Christofidou M, Marangos M. 2017. Carbapenemase-producing Klebsiella pneumoniae bloodstream infection in critically ill patients: risk factors and predictors of mortality. Eur J Clin Microbiol Infect Dis 36:1125–1131. doi: 10.1007/s10096-017-2899-6. [DOI] [PubMed] [Google Scholar]
- 4.Zavascki AP, Girardello R, Magagnin CM, Antochevis LC, Maciel RA, Palmeiro JK, Gales AC. 2018. Emergence of polymyxin B resistance in a polymyxin B-susceptible KPC-producing Klebsiella pneumoniae causing bloodstream infection in a neutropenic patient during polymyxin B therapy. Diagn Microbiol Infect Dis 90:134–138. doi: 10.1016/j.diagmicrobio.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Forrest A, Garonzik SM, Thamlikitkul V, Giamarellos-Bourboulis EJ, Paterson DL, Li J, Silveira FP, Nation RL. 2017. Pharmacokinetic/toxicodynamic analysis of colistin-associated acute kidney injury in critically ill patients. Antimicrob Agents Chemother 61:e01367-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allergan. 2019. AVYCAZ package insert. Allergan, Madison, NJ: https://www.accessdata.fda.gov/drugsatfda_docs/label/2015/206494s000lbl.pdf. Accessed 24 January 2019. [Google Scholar]
- 7.Shields RK, Clancy CJ, Hao B, Chen L, Press EG, Iovine NM, Kreiswirth BN, Nguyen MH. 2015. Effects of Klebsiella pneumoniae carbapenemase subtypes, extended spectrum β-lactamases, and porin mutations on the in vitro activity of ceftazidime-avibactam against carbapenem-resistant K. pneumoniae. Antimicrob Agents Chemother 59:5793–5797. doi: 10.1128/AAC.00548-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai C, Xiao X, Li J, Ciccotosto GD, Cappai R, Tang S, Schneider-Futschik EK, Hoyer D, Velkov T, Shen J. 2019. Molecular mechanisms of neurotoxicity induced by polymyxins and chemoprevention. ACS Chem Neurosci 10:120–131. doi: 10.1021/acschemneuro.8b00300. [DOI] [PubMed] [Google Scholar]
- 9.Giddins MJ, Macesic N, Annavajhala MK, Stump S, Khan S, McConville TH, Mehta M, Gomez-Simmonds A, Uhlemann ACl. 2018. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC-2-harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother 62:e02101-17. doi: 10.1128/AAC.02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnes MD, Winkler ML, Taracila MA, Page MG, Desarbre E, Kreiswirth BN, Shields RK, Nguyen MH, Clancy C, Spellberg B, Papp-Wallace KM, Bonomo RA. 2017. Klebsiella pneumoniae carbapenemase-2 (KPC-2), substitutions at Ambler position Asp179, and resistance to ceftazidime-avibactam: unique antibiotic resistance phenotypes emerge from β-lactamase protein engineering. mBio 8:e00528-17. doi: 10.1128/mBio.00528-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen MH, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. doi: 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shields RK, Nguyen MH, Chen L, Press EG, Kreiswirth BN, Clancy CJ. 2018. Pneumonia and renal replacement therapy are risk factors for ceftazidime-avibactam treatment failures and resistance among patients with carbapenem-resistant Enterobacteriaceae infections. Antimicrob Agents Chemother 62:e02497-17. doi: 10.1128/AAC.02497-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodise T, Lomaestro B, Drusano GL. 2007. Piperacillin/tazobactam for Pseudomonas aeruginosa infections: clinical implications of an extended infusion dosing strategy. Clin Infect Dis 44:357–363. doi: 10.1086/510590. [DOI] [PubMed] [Google Scholar]
- 14.McKinnell JA, Connollt LE, Pushkin R, Jubb AM, O’Keeffe B, Serio AW, Smith A, Gall J, Riddle V, Krause KM. 2017. Improved outcomes with plazomicin compared with colistin in patients with bloodstream infections caused by carbapenem-resistant Enterobacteriaceae: results from the CARE study. IDWeek, San Diego, CA. [Google Scholar]
- 15.Wunderink RG, Giamarellos-Bourboulis EJ, Rahav G, Mathers AJ, Bassetti M, Vazquez J, Cornely OA, Solomkin J, Bhowmick T, Bishara J, Daikos GL, Felton T, Furst MJL, Kwak EJ, Menichetti F, Oren I, Alexander EL, Griffith D, Lomovskaya O, Loutit J, Zhang S, Dudley MN, Kaye KS. 2018. Effect and safety of meropenem-vaborbactam versus best-available therapy in patients with carbapenem-resistant Enterobacteriaceae infections: the TANGO II randomized clinical trial. Infect Dis Ther 7:439–455. doi: 10.1007/s40121-018-0214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shields RK, Nguyen MH, Chen L, Press EG, Potoski BA, Marini RV, Doi Y, Kreiswirth BN, Clancy CJ. 2017. Ceftazidime-avibactam is superior to other treatment regimens against carbapenem-resistant Klebsiella pneumoniae bacteremia. Antimicrob Agents Chemother 61:e00883-17. doi: 10.1128/AAC.00883-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed CLSI document M07-A9 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 18.Li J, Zhou D, Das S, Lovern MR, Green ML, Chiu JS, Riccobene TA, Carrothers TJ, Al-Huniti N. 2018. Population PK modeling for ceftazidime-avibactam in patients with complicated intra-abdominal infection and complicated urinary tract infection. American Association of Pharmaceutical Scientists Annual Meeting and Exposition, Orlando, FL. [Google Scholar]
- 19.Heinrichs MT, Drusano GL, Brown DL, Maynard MS, Sy SKB, Rand KH, Peloquin CA, Louie A, Derendorf H. 2019. Dose optimization of moxifloxacin and linezolid against tuberculosis using mathematical modeling and simulation. Int J Antimicrob Agents 53:275–283. doi: 10.1016/j.ijantimicag.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Leary RH, Jelliffe R, Schumitzky A, Van Guilder M. 2001. An adaptive grid nonparametric approach to population pharmacokinetic/dynamic (PK/PD) population models, p 389–394. In Proceedings of the 14th IEEE Symposium on Computer-Based Medical Systems. IEEE, New York, NY. [Google Scholar]
- 21.D’Argenio DZ, Schumitzky A, Wang X. 2009. ADAPT 5 user’s guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical Simulations Resource, Los Angeles, CA. [Google Scholar]