Viral pathogens causing global disease burdens are often characterized by high rates of evolutionary changes. The extensive viral diversity at baseline can shorten the time to escape from therapeutic or immune selective pressure and alter mutational pathways. The impact of genotypic background on the barrier to resistance can be difficult to capture, particularly for agents in experimental stages or that are recently approved or expanded into new patient populations.
KEYWORDS: HIV-1, antiretroviral resistance, evolution, fitness, immunology, integrase, vaccine
ABSTRACT
Viral pathogens causing global disease burdens are often characterized by high rates of evolutionary changes. The extensive viral diversity at baseline can shorten the time to escape from therapeutic or immune selective pressure and alter mutational pathways. The impact of genotypic background on the barrier to resistance can be difficult to capture, particularly for agents in experimental stages or that are recently approved or expanded into new patient populations. We developed an evolutionary model-based counting method to quickly quantify the population genetic potential to resistance and assess population differences. We demonstrate its applicability to HIV-1 integrase inhibitors, as their increasing use globally contrasts with limited availability of non-B subtype resistant sequence data and corresponding knowledge gap. A large sequence data set encompassing most prevailing HIV-1 subtypes and resistance-associated mutations of currently approved integrase inhibitors was investigated. A complex interplay between codon predominance, polymorphisms, and associated evolutionary costs resulted in a subtype-dependent varied genetic potential for 15 resistance mutations against integrase inhibitors. While we confirm the lower genetic barrier of subtype B for G140S, we convincingly discard a similar effect previously suggested for G140C. A supplementary analysis for HIV-1 reverse transcriptase inhibitors identified a lower genetic barrier for K65R in subtype C through differential codon usage not reported before. To aid evolutionary interpretations of genomic differences for antiviral strategies, we advanced existing counting methods with increased sensitivity to identify subtype dependencies of resistance emergence. Future applications include novel HIV-1 drug classes or vaccines, as well as other viral pathogens.
INTRODUCTION
The advent of vaccines and antiviral treatment resulted globally in significant health gains and averted deaths by preventing viral infections and improving disease outcomes. Major human pathogens targeted by these strategies are fast-evolving genetically diverse viruses (1–3), allowing for rapid adaptation through the emergence of resistance-conferring mutations. A key concept in understanding the dynamics of resistance development is the genetic barrier, which ultimately quantifies the evolutionary time to viral escape from selective pressure (4–7). The virus makeup at baseline can shorten the evolutionary distance to resistance and, together with intervention- and patient-related factors, alter the mode and tempo of resistance emergence (1, 8). The imprint of the genotypic background on viral escape dynamics, however, can be difficult to capture, in particular for agents that only have been recently introduced. Resistance knowledge for these newer agents is often limited to in vitro selection experiments using genetically limited backbones or initially inferred from observations in well-controlled clinical studies and patient cohorts which underrepresent the full spectrum of viral diversity.
A notable example of success is the evolution of the management of human immunodeficiency virus type 1 (HIV-1) infection in the last 3 decades, with antivirals available from multiple drug classes that drastically reduced morbidity and mortality related to HIV-1. The recent class of integrase strand transfer inhibitors (INSTIs), directed against the integrase enzyme by blocking the strand transfer step of viral DNA integration, has considerably expanded treatment options and reduced the probability of virological failure, predominantly in resource-rich settings where INSTI use is widespread. To date, the INSTIs raltegravir (RAL), elvitegravir (EVG), dolutegravir (DTG), and bictegravir (BIC) are approved for HIV-1 treatment and often a preferred option for first-line regimens and dual regimens (9, 10). INSTIs are anticipated to become also widespread in low- and middle-income countries (LMICs), and the use of INSTIs has been shown to be cost-effective (11), although rates of acquired drug resistance are increasing in these settings. The impact of HIV-1 genetic diversity, classified into groups and subtypes, on resistance development has been well documented for the historical classes of protease and reverse transcriptase (RT) inhibitors, mainly resulting from preferential codon usage (12–14). The evolutionary mechanisms underlying viral escape from INSTI selective pressure, despite the identification of mutational pathways, are still unfolding, particularly for non-B subtypes, which are prevalent in LMICs (15). As short pathways toward INSTI resistance have been reported (16, 17), it is imperative to characterize the impact of HIV-1 diversity on the lowering of the genetic barrier to resistance.
Extensive subtype mappings of integrase diversity in treatment-naive patients revealed amino acid polymorphisms at resistance-associated positions (1, 18–20). Apart from a documented subtype B INSTI resistance pathway attributed to differential codon usage at position 140 (17, 21–23), the role of baseline nucleotide variation on INSTI resistance development has been less systematically investigated. The probabilistic models of resistance evolution that previously quantified the genetic barrier for the historical drug classes require sequence data of various subtypes from treatment-experienced patients (4–6), which are limited to date in the context of INSTIs (19, 24). Alternatively, the genetic barrier can be estimated by the number and type of required nucleotide substitutions to evolve from a wild-type virus to a resistant mutant (25). While this modality has been applied to HIV-1 integrase before (21, 22), these studies lacked resistance mutations only recently discovered, ignored subtype-specific effects, or analyzed limited subtype distributions and relied on arbitrary substitution cost assignments.
We present a novel, optimized, and evolutionary model-based methodology to quantify the genetic potential to resistance in fast-evolving viral pathogens, facilitating a priori identification of the impact of varied genetic background on resistance emergence. Here, we demonstrate our approach by the application to the HIV-1 INSTIs. With the rollout of INSTIs in LMICs, an increasing introduction of non-B subtypes in high-income countries (26–29), and the anticipated approval of additional INSTIs, this study addressed the need for an in-depth understanding of the dynamics underlying INSTI resistance development across HIV-1 subtypes. We derived an optimized genetic barrier score based on empirical substitution costs, which contrasts with previous approaches that used arbitrary costs differing from in vivo estimates (30). Furthermore, we advanced existing studies by considering a population-based estimate for the most prevailing subtypes globally and calculating subtype-tailored summary scores. The framework is easily applicable to novel HIV-1 drug classes (e.g., attachment or maturation inhibitors) and extended to vaccines or other pathogens (e.g., arboviruses, influenza virus, respiratory syncytial virus [RSV], herpesvirus, and varicella virus) which are currently targeted by antiviral development efforts.
RESULTS
Data set.
A total of 10,235 viral sequences coding for the HIV-1 integrase enzyme fulfilled the inclusion criteria, resulting in 410 (4%) sequences classified as subtype A, 5,174 (50.5%) sequences classified as subtype B, 1,837 (17.9%) sequences classified as subtype C, 1,630 (15.9%) sequences classified as CRF01_AE, 596 (5.8%) sequences classified as CRF02_AG, 170 (1.7%) sequences classified as subtype D, 257 (2.5%) sequences classified as subtype F, and 161 (1.6%) sequences classified as subtype G. The highest percentage of codons that did not fulfill the ambiguity criteria was 7.6% for position 125, and distributions of within-subtype pairwise diversity were unimodal and similar across subtypes (Fig. S5 and S6 provide data quality results).
Natural integrase variability.
Variation in genetic barrier across subtypes requires a combination of differences in wild-type triplet (WTt) frequencies and in cost scores. We first illustrate relevant integrase genetic variability using triplet entropy calculations to elucidate variation within a single subtype. Next, identifying predominant triplets can reveal major variation between subtypes. Triplets associated with INSTI resistance are particularly of interest because of their minimal cost.
Figure 1 shows within-subtype entropy values for each resistance position. Some of this within- subtype variation is translated into the presence of polymorphic mutations. Figure 2 shows the prevalence of resistance-associated mutations (RAMs) in the data set (Table S2 provides prevalence for all RAMs). The most prevalent mutations for subtype A were M50I (25.2%), L74I (22.4%), I203M (5.5%), and T97A (5.1%); for subtype B, K156N (17.6%), S230N (10.4%), M50I (9.4%), I203M (6.4%), S119R (5.5%), and V151I (5.2%); for subtype C, M50I (35.1%) and L74I (5.0%); for CRF01_AE, V165I (17.2%); for CRF02_AG, L74I (18.4%), M50I (10.6%), L74M (10.2%), E157Q (8.3%), and T97A (5.5%); for subtype D, I203M (15.3%), T97A (6.2%), and V165I (6.2%); for subtype F, V165I (30.2%), M50I (9.2%), G163R (6.2%), and G163K (5.4%); and for subtype G, M50I (15.1%), L74I (10.3%), and V165I (5.3%). These results illustrate both RAMs that prevalent across subtypes (e.g., M50I and L74I) and subtype-specific occurrences (e.g., K156N).
FIG 1.
Shannon entropy of nucleotide triplets as a measure of genetic diversity was calculated for each position to illustrate the extent of triplet variation within each of the 8 HIV-1 subtypes separately. In addition, for every position, we also calculated a single Shannon entropy value (All) of nucleotide triplets for all subtypes combined to increased variation between HIV-1 subtypes.
FIG 2.
We determined the subtype-specific prevalence (%) of each INSTI resistance-associated mutation in treatment-naive patients. The A49P, M50I, V54I, L68I/V, L74I/M, T97A, S119R, V151I, M154I, K156N, E157Q, K160N, G163K/R, V165I, I203M, S230N, and D232N mutations were observed with a frequency above 1% in at least one subtype.
Figure 1 also provides a triplet entropy value of all subtypes combined for each position. High values, compared to within-subtype values, indicate between-subtype variation and suggest differences in predominant codon usage. Of the 41 integrase positions investigated, 16 positions (50, 51, 66, 75, 97, 121, 138, 142, 143, 145, 146, 149, 154, 155, 203, and 230) showed a similar predominant triplet across all subtypes. Of the remaining 25 positions, 11 positions showed a consensus in predominant WTt among 7 subtypes (49, 54, 74, 92, 118, 148, 153, 157, 160, 170, and 263), 9 positions showed a consensus among 6 subtypes observed (68, 95, 119, 128, 140, 147, 156, 165, and 166), and, finally, 5 positions (114, 125, 151, 163, and 232) showed a consensus at 5 subtypes. These results illustrate well the extent of relevant integrase diversity within and between subtypes, encompassing varied entropy, resistance mutations, and codon predominance. Some positions (e.g., 114 and 140) that are fully conserved at the amino acid level showed high variability in codon usage, while other positions (e.g., 50 and 74) showed limited variation in codon predominance, but RAMs occurred at increased frequencies.
Calculated genetic barrier.
The observed baseline differences in triplet frequencies between subtypes will impact the genetic barrier to resistance only when they are also accompanied by variation in triplet scores. Subtype frequencies of wild-type nucleotide triplets, translated amino acids, and cumulative substitution costs to evolve into each triplet translating into the resistance mutation are available in Table S3.
Triplet score variability was subsequently investigated for patterns in entropy and codon usage, as described above. Limited variation in the evolutionary potential between subtypes is expected for the 35 RAMs at 16 integrase positions with a similar predominant triplet and, consequently, similar scores. Additionally, these positions are generally characterized by a low frequency of RAMs and a high level of consistent codon usage at baseline (Fig. S7 provides detailed codon information for each RAM), although some exceptions exist, with higher entropy values at positions 50, 138, and 203 (Fig. 1) or RAMs that are prevalent above 5% at low-entropy positions 97 and 230 (Fig. 2). Among the group of 25 positions, a larger heterogeneity in cost scores is expected due to varied predominance complementing variation in codon usage (Fig. 1) and RAM prevalence (Fig. 2). Subtype variation in predominant WTt resulted most strongly in a different cost score for the following mutations: V54I, L68I, L74I/M, E92A/G, Q95K, S119R, AT125K, A128T, G140S, S147G, Q148H/K/R, V151I, K156N, K160N, G163R, V165I, R166S, E170A, D232N, and R263K (Fig. S7).
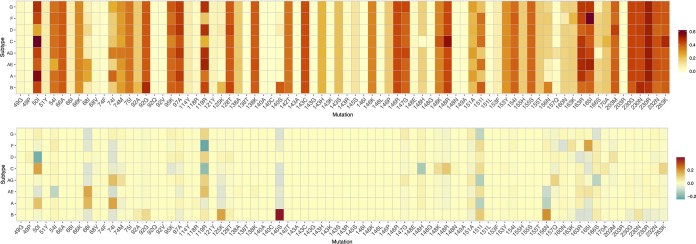
Finally, combining into a complex interplay, reported results on genetic diversity and evolutionary costs can be captured into a single value denoting a population-based genetic barrier (Fig. 3). Significant differences in the calculated genetic barrier were observed for 15 mutations at 12 positions (Fig. 3B), with higher genetic barriers for M50I (subtype D), V54I (subtype CRF01_AE), S119R (subtypes C and F), Q148H (subtype C), V151I (subtypes A, G, and CRF02_AG), K156N (subtypes A and CRF01_AE), and R166S (subtype CRF02_AG), and a lower genetic barrier for L68I (subtypes A and CRF01_AE), S119R (subtype CRF01_AE), AT125K (subtype B), G140S (subtype B), Q148R (subtype C), V151I (subtype B), and K156N (subtype B). Pairwise comparisons with subtype B additionally revealed a lower genetic barrier for M50I (subtypes A and C), L74I (subtypes A and AG), G163R (subtype CRF01_AE), and V165I (subtypes F and CRF01_AE). A similar analysis applied to the RT enzyme revealed a lower genetic barrier for 8 and a higher genetic barrier for 7 reverse transcriptase inhibitor (RTI) RAMs, including mutations at positions 106 and 65.
FIG 3.
Top, estimated population genetic barrier for each combination of a resistance-associated mutation and a subtype. A higher value (red) represents an increased potential for adaptation and is indicative of a lower genetic barrier to resistance. A subtype-specific value of this population genetic barrier is calculated by first assigning each wild-type triplet with a score that indicates the ease to evolve into the resistance amino acid (see Materials and Methods for a detailed description of how this cost score for a wild-type triplet is derived). Next, the sum of all triplet scores is weighted by the triplet prevalence so that the most frequently occurring triplets have a larger impact on the population genetic barrier estimate. Bottom, average difference in population genetic barrier of each subtype from the other subtypes. A lower genetic barrier to resistance for that subtype than for the other subtypes is shown in red, while a higher genetic barrier is shown in green.
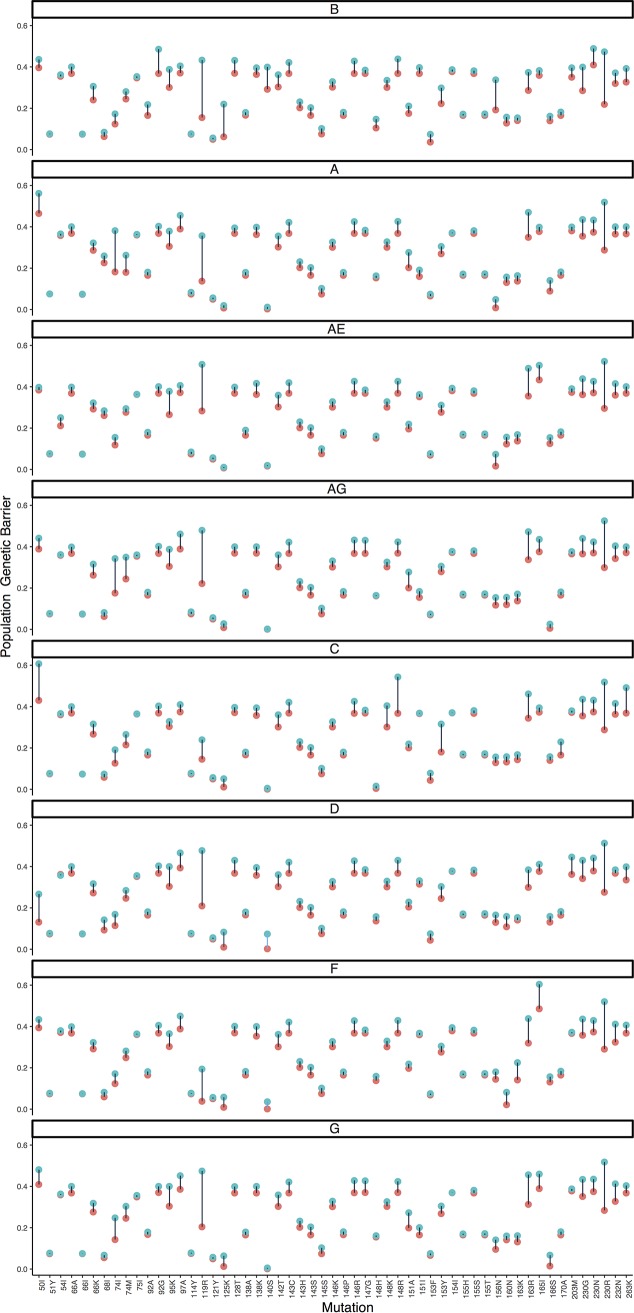
The calculation of a wild-type triplet score is based on the summation of all possible triplet cumulative costs, rather than considering the minimum cumulative cost (Table S3 shows all cumulative costs being considered). To illustrate the effect of this strategy, Fig. 4 provides a comparison of the evolutionary potentials calculated by our strategy and a simplified version which only considers the lowest cost for determining the score. Positions with high level of triplet variability (e.g., 119, 163, and 230) are primarily expected to be affected across all subtypes, but important subtype-specific effects were also observed (e.g., G140S in subtype B and Q148R and R263K in subtype C).
FIG 4.
Each of 8 panels shows the estimated population genetic barrier for a single subtype, with the estimate either calculated as described in Materials and Methods (blue) or calculated by a simpler but similar methodology (red). Instead of obtaining the cost score for each wild-type triplet by summing all possible cumulative costs (blue), thereby taking into account almost equally likely substitution pathways of a wild-type triplet to the resistance amino acid, a cost score can also be calculated by only using the minimum cumulative cost (red); therefore, this only considers the shortest substitution pathway to the resistance amino acid and ignores other but almost equally likely mutational pathways to resistance. To increase the comparability of the two measures, we also took the negative exponential of the minimum score. Only increases in values are possible due to the summation, and we restricted the figure to the subset of mutations with a difference larger than 0.01.
DISCUSSION
In this study, we developed a novel methodology to quantify the potential for viral escape from selective pressure in fast-evolving viruses and presented an application that evaluated the impact of the genetic background in HIV-1 integrase on the genetic barrier to INSTI resistance development. The integrase enzyme has proven to be a successful target for HIV-1 treatment, and the objective of maximizing the benefits of INSTIs is translated into their global rollout. As for other HIV-1 drug classes, the effectiveness of INSTIs can be challenged by the high evolutionary rates of HIV-1 when treatment conditions are nonoptimal (31–33). However, to date, mutational pathways that provide viral adaptation to INSTI pressure have not been fully characterized. Until now, the high effectiveness of INSTI-based regimens in high-income settings translated into low in vivo rates of RAM emergence, which resulted in a reduced need for in vitro phenotypic studies and, therefore, limited resistance data available. In particular, the role of natural nucleotide variation on resistance development is less well understood (17, 34). In addition to the presence of RAMs at baseline, differential codon usage can affect the mode and tempo of INSTI resistance pathways (22, 23). The study presented here provides new insights into the processes underlying INSTI resistance development by quantifying the genetic barrier to resistance, with a focus on the impact of genetic diversity on resistance evolvability between integrase positions and HIV-1 subtypes. Many factors govern the emergence of antiviral resistance, and the clinical implications of the presented results should be evaluated within this context. The genetic barrier estimates generated by our framework could be used to refine resistance interpretation algorithms (35, 36). Furthermore, our findings can support clinical decision making by virologists and clinicians when new patient populations are targeted by existing antivirals, such as the recommended introduction and scale-up of INSTIs in LMICs, which are characterized by a high prevalence of various HIV-1 non-B subtypes (19). Increasing rates of INSTI resistance in these countries can be anticipated, as the administration of INSTI-based treatments is likely to be confronted with high pretreatment resistance levels to backbone historical antivirals, lower patient adherence levels, and less frequent virological monitoring that will result in longer durations on suboptimal treatment (37). As the strength of selective pressure diminishes, resistance development becomes more likely and differences in genetic barriers more pronounced. Related to this, the transmission and persistence of transmitted drug resistance in drug-naive patients could be explored using our estimated cumulative substitution costs (38–40). Furthermore, our methodology is well suited to explore the effects of global virus diversity for new interventional agents, such as small-molecule integrase inhibitors (e.g., LEDGINS) with a new mechanism of action (41), for which drug resistance mutations are known based only upon in vitro drug resistance selection experiments using a limited number of laboratory or clinical strains (41), or even for ongoing drug development efforts guided by structural modeling that can identify new mutational interactions and hence for which the mutational sequence space related to the genetic barrier can be explored.
We investigated a large sequence data set of globally the most prevailing HIV-1 subtypes for their evolutionary potential for viral escape, considering all INSTIs currently available for HIV-1 treatment. This study confirms the nonpolymorphic amino acid nature of most INSTI resistance positions (19, 20), as primarily single-subtype occurrences of RAMs at low frequencies were detected. In contrast, varied codon predominance across subtypes was more pronounced, and consequently, 15 RAMs were detected that significantly varied in the estimated barrier across subtypes. For major INSTI mutations, modest variability in genetic barrier was observed, except for a potentially important lower genetic barrier of the RAM Q148H/R in subtype C, which causes high-level resistance to all INSTIs (42). Minor INSTI mutations, i.e., those having an impact on drug activity only in the presence of a major mutation or compensating for defects in replication capacity (42), were to a larger extent characterized by increased variability in diversity and associated scores. Most notable was the varied potential for G140S in subtype B viruses, confirming previous findings (21, 22), which is an accessory mutation often occurring in combination with Q148H. We and others have indeed previously observed the preference of the Q148H pathway in treatment-experienced patients infected with subtype B viruses (17, 23). This study also identified a higher estimated potential for the selection of R263K in subtype C due to distinct predominant codon usage, but a significant effect could not be established. The emergence of this infrequent INSTI RAM is associated with the use of DTG and is mostly observed in subtype B-infected patients (37), but it could become more common with the increased use of DTG in LMICs where subtype C prevails. R263K was observed following treatment failure in a subtype C-infected patient in the SAILING clinical trial (34) and recently in a subtype D-infected DTG-exposed patient (37).
Previous studies calculated the genetic barrier by distinguishing only between transitions and transversions, following an approach initially developed for the HIV-1 protease and reverse transcriptase enzymes (25). We improved these previous frameworks by applying an empirically derived and evolutionary model-based cost for each possible substitution and triplet position, which resulted in increased detection sensitivity. Particularly, transversions displayed substantial variation in costs compared to arbitrarily defined uniform values used in previous studies. Furthermore, our calculated genetic barrier incorporated cost scores from all possible pathways, compared to restricting it to the minimum score previously used, relying on a now outdated parsimony assumption. This approach allowed the detection of relevant increases in the estimated evolutionary potential (e.g., L74I), particularly for positions with a high level of codon variability. The improved sensitivity of our approach is well illustrated by the G140C mutation, for which we could not confirm the previously suggested subtype effect on its potential (21, 22), since this mutation requires a substitution at the first triplet position known to be evolutionarily costly. In contrast to G140S, the prevalence of G140C in patients failing INSTI-based treatment does not differ across subtypes in the Stanford HIV drug resistance database (19, 23), which strengthens our finding. A complete in-depth comparison of our findings with these studies is hindered by their limited number of HIV-1 subtypes and INSTI mutations included. As most genetic barrier information to date has been established for the historical drug class of RTIs, we applied our approach to the RT enzyme to validate our approach. Subtype dependencies in genetic barrier previously predicted and observed in clinical practice were confirmed (e.g., V106M in subtype C). However, compared to previous RTI genetic barrier counting approaches (25), the increased sensitivity of our method also identified a lower genetic barrier for K65R in subtype C. A higher selection rate of K65R in this subtype has been well established (13) and attributed to a mechanistic basis of template factors (43). However, we suggest an additional impact of codon usage when all possible paths are considered, a mechanism which was previously discarded (43). A similar analysis can be also performed for the RNase H region, which is also suggested to be implicated in RTI resistance development (44).
While our framework provides a population-based cost for every position, it only represents a simplified estimate of the actual in vivo genetic barrier, which results from a multimodal interplay that eventually defines the evolutionary time to drug resistance. The selection of a mutation and its rate of fixation depend both on non-virus-related factors, such as patient adherence and treatment potency, and on virus-related factors, such as the impact on viral fitness and (nonadditive) epistatic mutational interactions. Positions implicated in immune escape (e.g., 125) further influence the rate of resistance accumulation (45, 46). It is difficult to timely obtain information on all these influencing factors and hence to construct an accurate model capturing resistance evolutionary dynamics. As such an adequate model is lacking, our distance-based method offers a valuable alternative to assess the genetic barrier to resistance. Our methodology cannot predict novel RAMs or subtype-specific pathways. A cost matrix was used for all subtypes assuming that substitution rates are equal between subtypes; however, the use of group-specific cost matrices can be accommodated by our framework.
In conclusion, our findings are important in the context of up-scaled introduction of DTG and novel INSTIs in LMICs, where non-B subtypes prevail, but more studies are needed to further validate the clinical implications of our results. Future applications of this reproducible framework transcend novel HIV-1 drug classes or subtypes, as principles of HIV-1 drug resistance are generally shared with other pathogens known to escape selective pressure, and our methodology can be easily transferred to identify a role of genetic diversity of these pathogens, particularly for antivirals which are being evaluated in early development or clinical trials.
MATERIALS AND METHODS
HIV-1 data set and drug resistance mutations.
HIV-1 integrase sequences from INSTI-naive patients were obtained from the Stanford HIV drug resistance database (19), aligned codon-correct using VIRULIGN (47), and classified using the REGA subtyping tool v3 (48–50). One sequence per patient without a stop codon was retained. As the transmission of INSTI resistance is infrequently reported to date, we used the presence of INSTI signature mutations Y143C/H/R, Q148H/K/R, or N155H/S, which are nonpolymorphic in treatment-naive patients, as a proxy to identify possible incorrect INSTI-naive status (19). Integrase positions and mutations important for INSTI resistance were defined by an association with reduced susceptibility and virological response (16) or by inclusion in HIV-1 drug resistance interpretation systems (REGA v10, HIVdb v8.7, and ANRS v29) (51–53). A total of 41 codon positions and 77 amino acid mutations implicated in viral escape from currently approved INSTIs were investigated. In addition, the supplemental material presents the same approach applied to quantify the evolutionary potential to resistance of the RT enzyme (Fig. S1 to S4).
Genetic barrier to resistance.
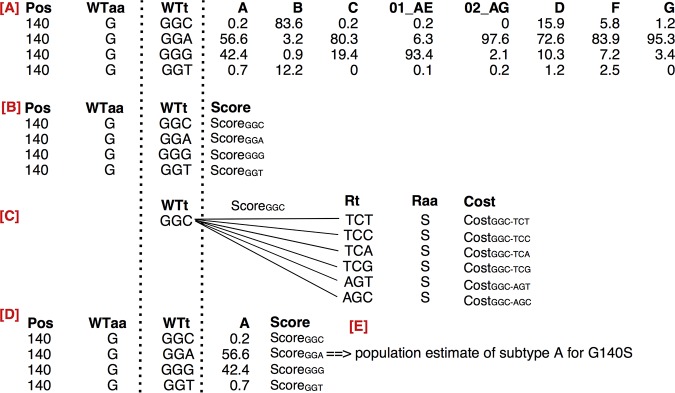
We define the genetic barrier in terms of a particular subepidemic, e.g., for HIV-1 with respect to all viruses that belong to a particular subtype, and of a resistance mutation defined by an amino acid and position. An overview of the procedure to obtain the genetic barrier is given in Fig. 5, using the resistance mutation G140S as an example. The calculation of the genetic barrier is a series of steps (see Fig. 5) which result in a subtype-specific estimate for G140S. Initially, we determine the extent of natural diversity at position 140 in the HIV-1 data set by identifying all wild-type triplets and their subtype prevalence. Next, we compute a score for each wild-type triplet that quantifies its evolvability into the resistance amino acid S at position 140, based on the costs associated with the possible nucleotide substitutions, as some substitutions are more costly for HIV-1 than others (54). Finally, a subtype-specific genetic barrier for G140S is obtained by summing all wild-type triplet scores weighted by the prevalence of respective wild-type triplets in the subtype of interest. In the following paragraphs, we provide a more detailed description of each of these steps.
FIG 5.
Methodology overview for resistance amino acid (Raa) S at position 140 (Pos). (A) Determination of the distribution wild-type triplets (WTt) naturally present, the translated amino acid WTaa, and their frequency across subtypes. (B) A score is assigned to each WTt to evolve into Raa. (C) This score is obtained by iteratively determining the cumulative substitution cost for the change of WTt into each resistance triplet (Rt) translating into the amino acid S, and subsequently by the summation of the different cumulative costs. (D) Each score is iteratively weighted by the subtype frequency of the WTt, here shown for subtype A. (E) A population estimate of the genetic barrier is obtained for subtype A. This procedure is repeated for each subtype, resulting in a subtype-specific population genetic barrier for G140S. When all resistance-associated mutations are considered, a matrix of genetic barrier values is created, where subtypes are shown as rows and mutations as columns. Figure 3A illustrates the visualization of this matrix.
Codon distributions.
At every position, variability at the nucleotide level was assessed by the prevalence of nucleotide triplets (codon) in each subtype (Fig. 5A). Codons with ambiguities consisting of >2 bases per nucleotide position or of two or more ambiguities per codon were not considered. A nucleotide ambiguity of exactly 2 bases was resolved in the two corresponding triplets, each counting for one-half of their respective frequencies. All triplets with a prevalence above 1% in at least one subtype were retained, and triplets with a prevalence above 50% were defined as predominant for that subtype. The prevalences of resistance mutations and codon entropy were inferred from the triplet distribution.
Substitution cost.
A penalty score is assigned to the different types of nucleotide substitution through the transformation of empirically estimated substitution matrices into corresponding cost matrices. For each codon index (ci) ∈ {1, 2, 3}, the cost matrix is given by the normalized and inverted substitution matrix following normalization, using the following equation:
| (Eq 1) |
This transformation assigns the substitution with the highest rate (e.g., G→A) a cost of 1, and the costs of the other substitutions were proportionally adapted to this baseline cost. A cost of zero is assigned when no change occurs. For our study, a substitution matrix for each codon position was derived from HIV-1 integrase nucleotide sequences independently collected from the Los Alamos database, with codon-based substitution patterns and rates estimated under the general time-reversible model (Table S1 provides information on cost matrices).
Genetic barrier to resistance.
Given a subtype ε, the genetic barrier to resistance mutation R is a cost function, GBε(R), that quantifies the number and type of nucleotide substitutions required for the virus to evolve from wild-type diversity in the virus population of ε to R (Fig. 5). This baseline diversity is defined by the set of wild-type triplets {WTt,i}, with the index i, ranging from 1 to the number of wild-type triplets. The prevalence of a given WTt is denoted as prev(WTt). Furthermore, as R is an amino acid, we enumerate all triplets that translate into R and refer to this set of resistance triplets as {Rt,j}, with the index j, ranging from 1 to the number of resistance triplets. For each WTt in {WTt,i}, we determine a score for the given WTt to evolve into R, thereby considering all triplets that translate into R (i.e., {Rt,j}):
| (Eq 2) |
where Cost (.) quantifies the cumulative substitution cost to mutate from the given WTt into a single resistance triplet Rt,j, as provided in equation 3. To obtain this cumulative substitution cost, we compute the sum of the substitution cost of each triplet position, using substitution matrices as defined by equation 1:
| (Eq 3) |
This calculation of a score incorporates all possible evolution paths from WTt to R and subsequently assigns lower cumulative costs (i.e., more likely mutation pathways) a higher contribution to the total triplet score. Taking into account different but almost equally likely evolution paths, compared to only considering the minimum cumulative substitution cost, results in an elevated sensitivity to detect subtype dependencies in the genetic barrier.
Finally, the genetic barrier GBε is defined as the sum of the scores (equation 2) of the different WTt in subtype ε, weighted according to their prevalence:
| (Eq 4) |
A GBε(R) is obtained for each resistance-associated mutation (RAM) and subtype. In order to visualize most pronounced differences between subtypes, we calculated the average distance in population genetic barriers for each subtype against other subtypes. The R code used in this study to calculate the population genetic barrier is made available at https://github.com/ktheyss/genetic-barrier/.
Statistics.
The nonparametric Mann-Whitney test was used to identify differences in the genetic barriers between HIV-1 subtypes and corrected for multiple-hypothesis testing using the Benjamini-Hochberg method (25). Each subtype was compared against a weighted sample (n = 500) of the other subtypes, repeated 1,000 times. In addition, pairwise comparisons of subtype B with each subtype separately were performed given that most knowledge on integrase resistance development is available for subtype B. Significant comparisons with a difference in the barrier score above 0.1 are retained. All analyses were done using the statistical software package R (55).
Supplementary Material
ACKNOWLEDGMENTS
Kristof Theys was funded by a postdoctoral grant from the Research Foundation—Flanders (FWO). Pieter J. K. Libin was funded by a Ph.D. grant from the FWO. This work was supported by the FWO grant G.0692.14N and grant PF/10/018 from the KU Leuven. This study was partly supported by European Funds through grant “Bio-Molecular and Epidemiological Surveillance of HIV Transmitted Drug Resistance, Hepatitis Co-Infections and Ongoing Transmission Patterns in Europe (BEST HOPE) (project funded through HIVERA: Harmonizing Integrating Vitalizing European Research on HIV/AIDS, grant 249697)”; by L’Oréal Portugal Medals of Honor for Women in Science 2012 (financed through L’Oréal Portugal, Comissão Nacional da Unesco and Fundação para a Ciência e Tecnologia [FCT; https://www.fct.pt/]); by FCT through funds to GHTM-UID/Multi/04413/2013, by the MigrantHIV project (financed by FCT: PTDC/DTP-EPI/7066/2014), and by the INTEGRIV project (financed by FCT: PTDC/SAU-INF/31990/2017).
K.T., A.B.A., and P.J.K.L. developed the methodology and designed the analyses. K.T. implemented and performed the analyses. All authors contributed to the interpretation of the results and writing of the manuscript.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00539-19.
REFERENCES
- 1.Li G, Piampongsant S, Faria NR, Voet A, Pineda-Pena AC, Khouri R, Lemey P, Vandamme AM, Theys K. 2015. An integrated map of HIV genome-wide variation from a population perspective. Retrovirology 12:18. doi: 10.1186/s12977-015-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuypers L, Li G, Libin P, Piampongsant S, Vandamme AM, Theys K. 2015. Genetic diversity and selective pressure in hepatitis C Virus genotypes 1–6: significance for direct-acting antiviral treatment and drug resistance. Viruses 7:5018–5039. doi: 10.3390/v7092857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kramvis A. 2014. Genotypes and genetic variability of hepatitis B virus. Intervirology 57:141–150. doi: 10.1159/000360947. [DOI] [PubMed] [Google Scholar]
- 4.Deforche K, Cozzi-Lepri A, Theys K, Clotet B, Camacho RJ, Kjaer J, Van Laethem K, Phillips A, Moreau Y, Lundgren JD, Vandamme AM, EuroSIDA Study Group. 2008. Modelled in vivo HIV fitness under drug selective pressure and estimated genetic barrier towards resistance are predictive for virological response. Antivir Ther 13:399–407. [PubMed] [Google Scholar]
- 5.Theys K, Deforche K, Beheydt G, Moreau Y, van Laethem K, Lemey P, Camacho RJ, Rhee SY, Shafer RW, Van Wijngaerden E, Vandamme AM. 2010. Estimating the individualized HIV-1 genetic barrier to resistance using a nelfinavir fitness landscape. BMC Bioinformatics 11:409. doi: 10.1186/1471-2105-11-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beerenwinkel N, Montazeri H, Schuhmacher H, Knupfer P, von Wyl V, Furrer H, Battegay M, Hirschel B, Cavassini M, Vernazza P, Bernasconi E, Yerly S, Boni J, Klimkait T, Cellerai C, Gunthard HF, Aubert V, Barth J, Bateau M, Bernasconi E, Boni J, Bucher HC, Burton-Jeangros C, Calmy A, Cavassini M, Egger M, Elzi L, Fellay J, Francioli P, Furrer H, Fux CA, Gorgievski M, Gunthard H, The SWISS HIV Cohort Study. 2013. The individualized genetic barrier predicts treatment response in a large cohort of HIV-1 infected patients. PLoS Comput Biol 9:e1003203. doi: 10.1371/journal.pcbi.1003203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Götte M. 2012. The distinct contributions of fitness and genetic barrier to the development of antiviral drug resistance. Curr Opin Virol 2:644–650. doi: 10.1016/j.coviro.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Megens S, Laethem KV. 2013. HIV-1 genetic variation and drug resistance development. Expert Rev Anti Infect Ther 11:1159–1178. doi: 10.1586/14787210.2013.844649. [DOI] [PubMed] [Google Scholar]
- 9.Panel on Antiretroviral Guidelines for Adults and Adolescents. 2018. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. U.S. Department of Health and Human Services, Washington, DC. [Google Scholar]
- 10.European AIDS Clinical Society. 2018. Guidelines. Version 9.1. European AIDS Clinical Society, Brussels, Belgium: http://www.eacsociety.org/files/2018_guidelines-9.1-english.pdf. [Google Scholar]
- 11.Phillips AN, Venter F, Havlir D, Pozniak A, Kuritzkes D, Wensing A, Lundgren JD, De Luca A, Pillay D, Mellors J, Cambiano V, Bansi-Matharu L, Nakagawa F, Kalua T, Jahn A, Apollo T, Mugurungi O, Clayden P, Gupta RK, Barnabas R, Revill P, Cohn J, Bertagnolio S, Calmy A. 2019. Risks and benefits of dolutegravir-based antiretroviral drug regimens in sub-Saharan Africa: a modelling study. Lancet HIV 6:e116–e127. doi: 10.1016/S2352-3018(18)30317-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abecasis AB, Deforche K, Bacheler LT, McKenna P, Carvalho AP, Gomes P, Vandamme AM, Camacho RJ. 2006. Investigation of baseline susceptibility to protease inhibitors in HIV-1 subtypes C, F, G and CRF02AG. Antivir Ther 11:581–589. [PubMed] [Google Scholar]
- 13.Theys K, Vercauteren J, Snoeck J, Zazzi M, Camacho RJ, Torti C, Schulter E, Clotet B, Sonnerborg A, De Luca A, Grossman Z, Struck D, Vandamme AM, Abecasis AB. 2013. HIV-1 subtype is an independent predictor of reverse transcriptase mutation K65R in HIV-1 patients treated with combination antiretroviral therapy including tenofovir. Antimicrob Agents Chemother 57:1053–1056. doi: 10.1128/AAC.01668-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner D, Brenner B, Moisi D, Detorio M, Cesaire R, Kurimura T, Mori H, Essex M, Maayan S, Wainberg MA. 2004. Nucleotide and amino acid polymorphisms at drug resistance sites in non-B-subtype variants of human immunodeficiency virus type 1. Antimicrob Agents Chemother 48:2993–2998. doi: 10.1128/AAC.48.8.2993-2998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charpentier C, Descamps D. 2018. Resistance to HIV integrase inhibitors: about R263K and E157Q mutations. Viruses 10:41. doi: 10.3390/v10010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanco JL, Varghese V, Rhee SY, Gatell JM, Shafer RW. 2011. HIV-1 integrase inhibitor resistance and its clinical implications. J Infect Dis 203:1204–1214. doi: 10.1093/infdis/jir025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han YS, Mesplede T, Wainberg MA. 2016. Differences among HIV-1 subtypes in drug resistance against integrase inhibitors. Infect Genet Evol 46:286–291. doi: 10.1016/j.meegid.2016.06.047. [DOI] [PubMed] [Google Scholar]
- 18.Varghese V, Liu TF, Rhee SY, Libiran P, Trevino C, Fessel WJ, Shafer RW. 2010. HIV-1 integrase sequence variability in antiretroviral naïve patients and in triple-class experienced patients subsequently treated with raltegravir. AIDS Res Hum Retroviruses 26:1323–1326. doi: 10.1089/aid.2010.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhee SY, Liu TF, Kiuchi M, Zioni R, Gifford RJ, Holmes SP, Shafer RW. 2008. Natural variation of HIV-1 group M integrase: implications for a new class of antiretroviral inhibitors. Retrovirology 5:74. doi: 10.1186/1742-4690-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers RE, Pillay D. 2008. Analysis of natural sequence variation and covariation in human immunodeficiency virus type 1 integrase. J Virol 82:9228–9235. doi: 10.1128/JVI.01535-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piralla A, Paolucci S, Gulminetti R, Comolli G, Baldanti F. 2011. HIV integrase variability and genetic barrier in antiretroviral naïve and experienced patients. Virol J 8:149. doi: 10.1186/1743-422X-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maïga AI, Malet I, Soulie C, Derache A, Koita V, Amellal B, Tchertanov L, Delelis O, Morand-Joubert L, Mouscadet JF, Murphy R, Cisse M, Katlama C, Calvez V, Marcelin AG. 2009. Genetic barriers for integrase inhibitor drug resistance in HIV type-1 B and CRF02_AG subtypes. Antivir Ther 14:123–129. [PubMed] [Google Scholar]
- 23.Theys K, Abecasis A, Libin P, Gomes PT, Cabanas J, Camacho RJ, Van Laethem K. 2015. Discordant predictions of residual activity could impact dolutegravir prescription upon raltegravir failure. J Clin Virol 70:120–127. doi: 10.1016/j.jcv.2015.07.311. [DOI] [PubMed] [Google Scholar]
- 24.Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res 31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van de Vijver DA, Wensing AM, Angarano G, Asjo B, Balotta C, Boeri E, Camacho R, Chaix ML, Costagliola D, De Luca A, Derdelinckx I, Grossman Z, Hamouda O, Hatzakis A, Hemmer R, Hoepelman A, Horban A, Korn K, Kucherer C, Leitner T, Loveday C, MacRae E, Maljkovic I, de Mendoza C, Meyer L, Nielsen C, Op de Coul EL, Ormaasen V, Paraskevis D, Perrin L, Puchhammer-Stockl E, Ruiz L, Salminen M, Schmit JC, Schneider F, Schuurman R, Soriano V, Stanczak G, Stanojevic M, Vandamme AM, Van Laethem K, Violin M, Wilbe K, Yerly S, Zazzi M, Boucher CA. 2006. The calculated genetic barrier for antiretroviral drug resistance substitutions is largely similar for different HIV-1 subtypes. J Acquir Immune Defic Syndr 41:352–360. doi: 10.1097/01.qai.0000209899.05126.e4. [DOI] [PubMed] [Google Scholar]
- 26.Dennis AM, Hue S, Learner E, Sebastian J, Miller WC, Eron JJ. 2017. Rising prevalence of non-B HIV-1 subtypes in North Carolina and evidence for local onward transmission. Virus Evol 3:vex013. doi: 10.1093/ve/vex013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hauser A, Hofmann A, Meixenberger K, Altmann B, Hanke K, Bremer V, Bartmeyer B, Bannert N. 2018. Increasing proportions of HIV-1 non-B subtypes and of NNRTI resistance between 2013 and 2016 in Germany: results from the national molecular surveillance of new HIV-diagnoses. PLoS One 13:e0206234. doi: 10.1371/journal.pone.0206234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oster AM, Switzer WM, Hernandez AL, Saduvala N, Wertheim JO, Nwangwu-Ike N, Ocfemia MC, Campbell E, Hall HI. 2017. Increasing HIV-1 subtype diversity in seven states, United States, 2006–2013. Ann Epidemiol 27:244–251. doi: 10.1016/j.annepidem.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Neogi U, Haggblom A, Santacatterina M, Bratt G, Gisslen M, Albert J, Sonnerborg A. 2014. Temporal trends in the Swedish HIV-1 epidemic: increase in non-B subtypes and recombinant forms over three decades. PLoS One 9:e99390. doi: 10.1371/journal.pone.0099390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zanini F, Puller V, Brodin J, Albert J, Neher RA. 2017. In vivo mutation rates and the landscape of fitness costs of HIV-1. Virus Evol 3:vex003. doi: 10.1093/ve/vex003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pena MJ, Chueca N, D’Avolio A, Zarzalejos JM, Garcia F. 2019. Virological failure in HIV to triple therapy with dolutegravir-based firstline treatment: rare but possible. Open Forum Infect Dis 6:ofy332. doi: 10.1093/ofid/ofy332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Achieng L, Riedel DJ. 2019. Dolutegravir resistance and failure in a Kenyan patient. J Infect Dis 219:165–167. doi: 10.1093/infdis/jiy436. [DOI] [PubMed] [Google Scholar]
- 33.Cardoso M, Baptista T, Diogo I, Aleixo MJ, Marques N, Mansinho K, Gomes P. 2018. Two cases of dolutegravir failure with R263K mutation. AIDS 32:2639–2640. doi: 10.1097/QAD.0000000000001978. [DOI] [PubMed] [Google Scholar]
- 34.Cahn P, Pozniak AL, Mingrone H, Shuldyakov A, Brites C, Andrade-Villanueva JF, Richmond G, Buendia CB, Fourie J, Ramgopal M, Hagins D, Felizarta F, Madruga J, Reuter T, Newman T, Small CB, Lombaard J, Grinsztejn B, Dorey D, Underwood M, GriZth S, Min S, Extended SAILING Study Team. 2013. Dolutegravir versus raltegravir in antiretroviral-experienced, integrase-inhibitor-naive adults with HIV: week 48 results from the randomised, double-blind, non-inferiority SAILING study. Lancet 382:700–708. doi: 10.1016/S0140-6736(13)61221-0. [DOI] [PubMed] [Google Scholar]
- 35.Frentz D, Boucher CA, Assel M, De Luca A, Fabbiani M, Incardona F, Libin P, Manca N, Muller V, O Nuallain B, Paredes R, Prosperi M, Quiros- Roldan E, Ruiz L, Sloot PM, Torti C, Vandamme AM, Van Laethem K, Zazzi M, van de Vijver DA. 2010. Comparison of HIV-1 genotypic resistance test interpretation systems in predicting virological outcomes over time. PLoS One 5:e11505. doi: 10.1371/journal.pone.0011505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vercauteren J, Beheydt G, Prosperi M, Libin P, Imbrechts S, Camacho R, Clotet B, De Luca A, Grossman Z, Kaiser R, Sonnerborg A, Torti C, Van Wijngaerden E, Schmit JC, Zazzi M, Geretti AM, Vandamme AM, Van Laethem K. 2013. Clinical evaluation of Rega 8: an updated genotypic interpretation system that signi1cantly predicts HIV-therapy response. PLoS One 8:e61436. doi: 10.1371/journal.pone.0061436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed N, Flavell S, Ferns B, Frampton D, Edwards SG, Miller RF, Grant P, Nastouli E, Gupta RK. 2019. Development of the R263K mutation to dolutegravir in an HIV-1 subtype D virus harboring 3 class-drug resistance. Open Forum Infect Dis 6:ofy329. doi: 10.1093/ofid/ofy329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castro H, Pillay D, Cane P, Asboe D, Cambiano V, Phillips A, Dunn DT, UK Collaborative Group on HIV Drug Resistance. 2013. Persistence of HIV-1 transmitted drug resistance mutations. J Infect Dis 208:1459–1463. doi: 10.1093/infdis/jit345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theys K, Deforche K, Vercauteren J, Libin P, van de Vijver DA, Albert J, Asjo B, Balotta C, Bruckova M, Camacho RJ, Clotet B, Coughlan S, Grossman Z, Hamouda O, Horban A, Korn K, Kostrikis LG, Kucherer C, Nielsen C, Paraskevis D, Poljak M, Puchhammer-Stockl E, Riva C, Ruiz L, Liitsola K, Schmit JC, Schuurman R, Sonnerborg A, Stanekova D, Stanojevic M, Struck D, Van Laethem K, Wensing AM, Boucher CA, Vandamme AM, SPREAD-Programme. 2012. Treatment-associated polymorphisms in protease are significantly associated with higher viral load and lower CD4 count in newly diagnosed drug-naive HIV-1 infected patients. Retrovirology 9:81. doi: 10.1186/1742-4690-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theys K, Libin P, Pineda-Pena AC, Nowe A, Vandamme AM, Abecasis AB. 2018. The impact of HIV-1 within-host evolution on transmission dynamics. Curr Opin Virol 28:92–101. doi: 10.1016/j.coviro.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Choi E, Mallareddy JR, Lu D, Kolluru S. 2018. Recent advances in the discovery of small-molecule inhibitors of HIV-1 integrase. Futur Sci OA 4:FSO338. doi: 10.4155/fsoa-2018-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shafer RW, Schapiro JM. 2008. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev 10:67–84. [PMC free article] [PubMed] [Google Scholar]
- 43.Brenner BG, Coutsinos D. 2009. The K65R mutation in HIV-1 reverse transcriptase: genetic barriers, resistance profile and clinical implications. HIV Ther 3:583–594. doi: 10.2217/hiv.09.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ngcapu S, Theys K, Libin P, Marconi VC, Sunpath H, Ndung’u T, Gordon ML. 2017. Characterization of nucleoside reverse transcriptase inhibitor-associated mutations in the RNase H region of HIV-1 subtype C infected individuals. Viruses 9:330. doi: 10.3390/v9110330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tschochner M, Chopra A, Maiden TM, Ahmad IF, James I, Furrer H, Gunthard HF, Mallal S, Rauch A, John M. 2009. Effects of HIV type-1 immune selection on susceptability to integrase inhibitor resistance. Antivir Ther 14:953–964. doi: 10.3851/IMP1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brockman MA, Chopera DR, Olvera A, Brumme CJ, Sela J, Markle TJ, Martin E, Carlson JM, Le AQ, McGovern R, Cheung PK, Kelleher AD, Jessen H, Markowitz M, Rosenberg E, Frahm N, Sanchez J, Mallal S, John M, Harrigan PR, Heckerman D, Brander C, Walker BD, Brumme ZL. 2012. Uncommon pathways of immune escape attenuate HIV-1 integrase replication capacity. J Virol 86:6913–6923. doi: 10.1128/JVI.07133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Libin P, Deforche K, Abecasis AB, Theys K. 2018. VIRULIGN: fast codon-correct alignment and annotation of viral genomes. Bioinformatics 35:1763–1765. doi: 10.1093/bioinformatics/bty851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pineda-Peña AC, Faria NR, Imbrechts S, Libin P, Abecasis AB, Deforche K, Gomez-Lopez A, Camacho RJ, de Oliveira T, Vandamme AM. 2013. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: performance evaluation of the new REGA version 3 and seven other tools. Infect Genet Evol 19:337–348. doi: 10.1016/j.meegid.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 49.Alcantara LC, Cassol S, Libin P, Deforche K, Pybus OG, Van Ranst M, Galvao-Castro B, Vandamme AM, de Oliveira T. 2009. A standardized framework for accurate, high-throughput genotyping of recombinant and non-recombinant viral sequences. Nucleic Acids Res 37:W634–W642. doi: 10.1093/nar/gkp455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Libin P, Beheydt G, Deforche K, Imbrechts S, Ferreira F, Van Laethem K, Theys K, Carvalho AP, Cavaco-Silva J, Lapadula G, Torti C, Assel M, Wesner S, Snoeck J, Ruelle J, De Bel A, Lacor P, De Munter P, Van Wijngaerden E, Zazzi M, Kaiser R, Ayouba A, Peeters M, de Oliveira T, Alcantara LC, Grossman Z, Sloot P, Otelea D, Paraschiv S, Boucher C, Camacho RJ, Vandamme AM. 2013. RegaDB: community-driven data management and analysis for infectious diseases. Bioinformatics 29:1477–1480. doi: 10.1093/bioinformatics/btt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Laethem K, De Luca A, Antinori A, Cingolani A, Perna CF, Vandamme AM. 2002. A genotypic drug resistance interpretation algorithm that significantly predicts therapy response in HIV-1-infected patients. Antivir Ther 7:123–129. [PubMed] [Google Scholar]
- 52.Betts BJ, Shafer RW. 2003. Algorithm specification interface for human immunodeficiency virus type 1 genotypic interpretation. J Clin Microbiol 41:2792–2794. doi: 10.1128/jcm.41.6.2792-2794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brun-Vézinet F, Descamps D, Ruffault A, Masquelier B, Calvez V, Peytavin G, Telles F, Morand-Joubert L, Meynard JL, Vray M, Costagliola D. 2003. Clinically relevant interpretation of genotype for resistance to abacavir. AIDS 17:1795–1802. doi: 10.1097/00002030-200308150-00008. [DOI] [PubMed] [Google Scholar]
- 54.Theys K, Feder AF, Gelbart M, Hartl M, Stern A, Pennings PS. 2018. Within patient mutation frequencies reveal fitness costs of CpG dinucleotides and drastic amino acid changes in HIV. PLoS Genet 14:e1007420. doi: 10.1371/journal.pgen.1007420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R Core Team. 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.r-project.org/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.