Quantitative assessment of antibiotic-responsive RNA transcripts holds promise for a rapid point-of-care (POC) diagnostic tool for antimicrobial susceptibility testing. These assays aim to distinguish susceptible and resistant isolates by transcriptional differences upon drug exposure. However, an often-overlooked dimension of designing these tests is that the genetic diversity within a species may yield differential transcriptional regulation independent of resistance phenotype.
KEYWORDS: AST, Neisseria gonorrhoeae, RNA-seq, antimicrobial susceptibility testing, azithromycin, ciprofloxacin, gene expression, gonorrhea, macrolide, transcriptome profiling
ABSTRACT
Quantitative assessment of antibiotic-responsive RNA transcripts holds promise for a rapid point-of-care (POC) diagnostic tool for antimicrobial susceptibility testing. These assays aim to distinguish susceptible and resistant isolates by transcriptional differences upon drug exposure. However, an often-overlooked dimension of designing these tests is that the genetic diversity within a species may yield differential transcriptional regulation independent of resistance phenotype. Here, we use a phylogenetically diverse panel of Neisseria gonorrhoeae and transcriptome profiling coupled with reverse transcription-quantitative PCR to test this hypothesis, to identify azithromycin responsive transcripts and evaluate their potential diagnostic value, and to evaluate previously reported diagnostic markers for ciprofloxacin resistance (porB and rpmB). Transcriptome profiling confirmed evidence of genetic distance and population structure impacting transcriptional response to azithromycin. Taking this into account, we found azithromycin-responsive transcripts overrepresented in susceptible strains compared to resistant strains and selected four candidate diagnostic transcripts (rpsO, rplN, omp3, and NGO1079) that were the most significantly differentially regulated between phenotypes across drug exposure. RNA signatures for these markers categorically predicted resistance in 19/20 cases, with the one incorrect categorical assignment for an isolate at the threshold of reduced susceptibility. Finally, we found that porB and rpmB expression were not uniformly diagnostic of ciprofloxacin resistance in a panel of isolates with unbiased phylogenetic sampling. Overall, our results suggest that RNA signatures as a diagnostic tool are promising for future POC diagnostics; however, development and testing should consider representative genetic diversity of the target pathogen.
INTRODUCTION
The rise of resistance to antimicrobial drugs presents a critical threat to the successful treatment of infectious diseases. Rapid point-of-care (POC) diagnostics for drug susceptibility could guide therapeutic decisions, thereby improving the time to appropriate therapy and limiting the extent of broad empirical antibiotic use. DNA-based susceptibility prediction tests are promising and may have niche applications but are subject to concerns about the comprehensiveness with which genotypic features accurately predict resistance (i.e., due to complex epistasis or the additive effects of other loci) and are also sensitive to the emergence of novel resistance alleles through mutation or horizontal gene transfer. In contrast, quantitative assessment of antibiotic-responsive RNA transcripts can rapidly differentiate pathogen strains by resistance phenotype independent of resistance mechanism or genetic background. Some of the earliest cellular responses to antibiotic exposure are mediated through transcriptional changes, which has recently been demonstrated in Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumannii, and Neisseria gonorrhoeae (1–4). These RNA-based antimicrobial susceptibility tests (AST) are particularly attractive as POC diagnostics as drug-susceptible and -resistant bacteria proceed down distinct physiological pathways indicative of cellular stress or normal cell growth in as little as 10 min (3)—much faster than the current “gold standard” for resistance phenotyping via MIC by agar dilution, Etest (bioMérieux), or disk diffusion, which can take days to weeks depending on bacterial growth rate.
Consideration of the impact of genetic diversity within the targeted pathogen species will be key to the success of effective RNA-based antimicrobial resistance diagnostics. Gene expression heterogeneity is known to occur between distinct lineages within bacterial species (5–7), and this strain-to-strain variation leaves the open possibility that transcripts may be antibiotic responsive in some strains but not others of the same phenotype. Thus, to identify antibiotic-responsive transcripts that define the resistance phenotype independent of genetic mechanism of resistance and genomic background, panels of clinical isolates must be surveyed to characterize the full transcriptional diversity of a species. However, while large panels of isolates have been used before to search for diagnostic transcripts (1–4), lack of consideration of phylogenetic structure and specific attention to diverse clade sampling within the assay design panel risks mistakenly attributing distinct RNA signatures to resistance phenotype when they instead derive from population structure (i.e., as a result of genetic drift or divergent selection).
Here, we investigate the impact of phylogenetic distance on strain variability in gene expression, and search for antibiotic-responsive RNA transcripts diagnostic of antimicrobial resistance in Neisseria gonorrhoeae, the Gram-negative pathogen that causes the sexually transmitted disease gonorrhea. Antimicrobial resistance in N. gonorrhoeae is one of the most critical contemporary threats to public health, with resistance emerging to every class of antimicrobials that have been used to empirically treat gonococcal infection (i.e., penicillins, sulfonamides, tetracyclines, fluoroquinolones, macrolides, and cephalosporins) (8, 9). With only a few novel antimicrobials and combination therapies currently in development (10–12) and rapidly rising incidence of infection (13), we face an imminent threat of gonorrhea that will become difficult to treat (9, 14). POC tests that move health care providers away from empirical treatment to data-driven prescription are urgently needed to support good antimicrobial stewardship by rapidly dispensing effective therapeutics to individuals and their sexual contacts and minimizing the selective pressures on bacterial bystanders from inappropriate treatment (15).
We focused on RNA-based diagnostic assay development for two clinically relevant drugs: azithromycin and ciprofloxacin. Azithromycin is a macrolide antibiotic that inhibits protein synthesis by binding to the 23S rRNA component of the 50S ribosome and is one of the two first-line drugs recommended as dual-antimicrobial therapy for uncomplicated cases of gonococcal infection by the U.S. Centers for Disease Control and Prevention (CDC) (16). Azithromycin resistance is especially attractive for phenotypic AST development, since there are multiple resistance mechanisms, including as-yet-unexplained pathways (17, 18). Resistance mechanisms that have been associated with or experimentally confirmed to be involved in reduced susceptibility to azithromycin include mutations in the 23S rRNA azithromycin binding sites (C2611T and A2059G) (17, 19, 20), mosaic multiple transferable resistance (mtr) efflux pump alleles acquired from commensal Neisseria species (17, 18, 21–23), mutations that enhance the expression of Mtr (24–26), mutations in rplD (17), rplV tandem duplications (17), and variants of the rRNA methylase genes ermC and ermB (27). Ciprofloxacin is a quinolone antibiotic that targets DNA gyrase (encoded by gyrA) and topoisomerase IV (encoded by parC) and was recommended as a first-line therapy for gonorrhea until 2007, at which point the prevalence of quinolone resistance overtook the 5% accepted threshold for suggested use (28). However, the development of a DNA-based diagnostic for quinolone susceptibility has led to the introduction of targeted quinolone use (29) and as such makes an attractive target for rapid POC diagnostics.
Here, we used transcriptome sequencing (RNA-seq) to profile the transcriptomes of a panel of N. gonorrhoeae isolates that are representative of the known diversity within the gonococcal population (17) in response to exposure to azithromycin. Using the transcriptomes from this panel, we sought to determine (i) whether there is a differential transcriptome-wide response to azithromycin between resistant and susceptible isolates, (ii) the impact of the evolutionary distance on transcriptome regulation, and (iii) whether there are RNA signatures of particular transcripts ubiquitously diagnostic of resistance phenotype across the species. After defining candidate diagnostic markers for azithromycin resistance, we then verified them via RT-qPCR in an expanded panel of isolates. We also assess the previously described RNA-based diagnostic markers for ciprofloxacin resistance in N. gonorrhoeae, porB and rpmB (3), in a phylogenetically representative panel of isolates.
RESULTS
Transcription profiling.
We used whole-transcriptome profiling to identify differentially expressed azithromycin-responsive RNA transcripts between resistant and susceptible isolates of N. gonorrhoeae after 60 and 180 min of below breakpoint azithromycin exposure (0.125 μg/ml; Fig. 1). Clinical isolates were sampled from across the phylogenetic diversity of the species, based on the collection reported in Grad et al. (17). This included paired resistant (MIC ≥ 1 μg/ml) and susceptible (MIC < 1 μg/ml) strains from four of the twelve substructured clades, termed Bayesian analysis of population structure (BAPS) groups (17) (Table 1; Fig. 2A). In total, we selected ten isolates: four susceptible isolates, two resistant isolates with mosaic mtr efflux pump alleles acquired from commensal Neisseria, three resistant isolates with the C2611T 23S rRNA mutation, and one resistant isolate with the A2059G 23S rRNA mutation (17, 18). A total of ∼258 million 50-bp paired-end reads were generated across 90 libraries. Each library had on average 3.17 ± 0.16 million reads, and an average of 1.82 ± 0.11 million reads per library were mapped to the FA1090 (AE004969.1) reference genome.
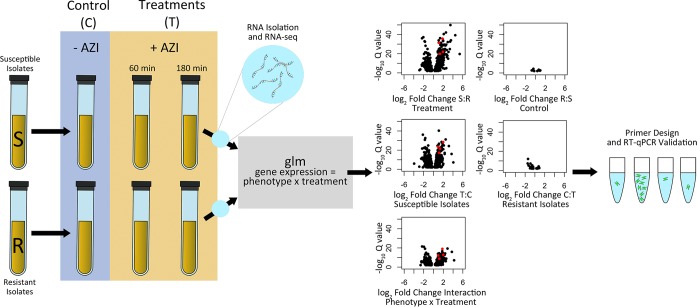
FIG 1.
Workflow for generating candidate diagnostic markers for azithromycin resistance. We sampled cultures of susceptible (MIC < 1 μg/ml) and resistant (MIC ≥1 μg/ml) isolates prior to the addition of azithromycin, after 60 min of exposure to 0.125 μg/ml of azithromycin, and after 180 min of exposure to 0.125 μg/ml of azithromycin. RNA-seq libraries were then constructed and sequenced, and a generalized linear model was fit to the resultant data with phenotype (resistant versus susceptible), treatment (control versus later time points), and the interaction between phenotype and treatment used as model terms. Volcano plots are shown for the control to 60-min exposure comparisons with an FDR cutoff of ≤0.01. Black circles indicate all significant transcripts for each model contrast, and red dots indicate the four candidate diagnostic markers ultimately chosen for RT-qPCR validation. Most of the observed responses occurred (i) in susceptible strains between the control and the drug exposed conditions, (ii) between resistant and susceptible isolates in the antibiotic exposure condition, and (iii) in isolates having a significant interaction term, indicating a differential response between susceptible and resistance isolates across treatments.
TABLE 1.
Properties of the clinical isolates used to develop diagnostic markers for azithromycin resistance (n = 20)
| Isolate | Location | Collection yr | BAPS | AZI MIC (μg/ml) | Resistance category | Resistance mechanism |
|---|---|---|---|---|---|---|
| Whole transcriptome | ||||||
| GCGS0524 | Albuquerque, NM | 2000 | 4 | 0.25 | Susceptible | |
| GCGS0276 | Kansas City, MO | 2000 | 4 | 1 | Resistant | mtr mosaic |
| GCGS0745 | Honolulu, HI | 2012 | 4 | 256 | Resistant | 23S A2059G |
| GCGS0996 | Seattle, WA | 2012 | 6 | 0.5 | Susceptible | |
| GCGS0834 | Los Angeles, CA | 2012 | 6 | 2 | Resistant | mtr mosaic |
| GCGS0641 | Orange County, CA | 2011 | 7 | 0.25 | Susceptible | |
| GCGS0354 | Dallas, TX | 2012 | 7 | 8 | Resistant | 23S C2599T |
| GCGS0550 | Birmingham, AL | 2012 | 7 | 8 | Resistant | 23S C2599T |
| GCGS0353 | Dallas, TX | 2011 | 8 | 0.03 | Susceptible | |
| GCGS0363 | Detroit, MI | 2011 | 8 | 4 | Resistant | 23S C2599T |
| RT-qPCR | ||||||
| GCGS1035 | Detroit, MI | 2007 | 1 | 0.5 | Susceptible | |
| GCGS0481 | Portland, OR | 2006 | 1 | 2 | Resistant | mtr mosaic |
| GCGS0926 | Minneapolis, MA | 2012 | 2 | 0.125 | Susceptible | |
| GCGS0298 | San Diego, CA | 2005 | 2 | 2 | Resistant | mtr mosaic |
| GCGS0605 | Miami, FL | 2002 | 3 | 0.25 | Susceptible | |
| GCGS1026 | Chicago, IL | 2008 | 5 | 16 | Resistant | rplV tandem duplication |
| GCGS0436 | Orange County, CA | 2010 | 7 | 2 | Resistant | 23S C2599T |
| GCGS0465 | Phoenix, AZ | 2011 | 10 | 0.06 | Susceptible | |
| GCGS0525 | Atlanta, GA | 2000 | 11 | 2 | Resistant | mtr mosaic |
| GCGS0313 | Baltimore, PA | 2012 | 12 | 0.06 | Susceptible |
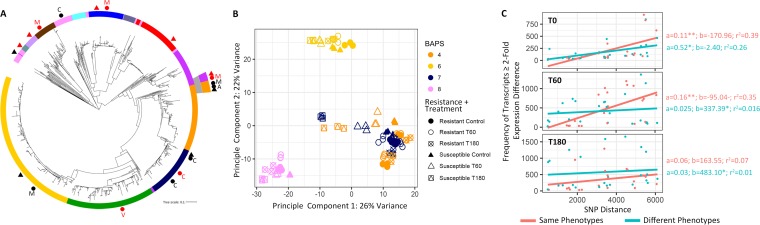
FIG 2.
Genetic distance impacts transcriptional regulation in phylogenetically diverse isolates. (A) A maximum-likelihood whole-genome-sequence phylogeny of 1102 Neisseria gonorrhoeae isolates from GISP, based on SNPs generated from mapping to the FA1090 reference genome (Grad et al. [17]). The annotation ring displays the Bayesian analysis of population structure (BAPS) groups. All annotated shapes represent the panel of isolates selected for identifying azithromycin-responsive RNA transcripts and evaluating their potential diagnostic value. Black circles represent resistant isolates selected for RNA-seq and RT-qPCR validation, black triangles represent susceptible isolates selected for RNA-seq and RT-qPCR validation, red circles show resistant isolates selected for RT-qPCR validation, and red triangles show susceptible isolates selected for RT-qPCR validation. Resistance mechanisms are indicated by A for the A2059G 23S rRNA mutation, C for the C2611T 23S rRNA mutation, M for the mosaic mtr mutation, and V for the rplV tandem duplication. (B) For all 90 RNA-seq libraries generated within this study, PCA revealed that the largest variance in gene expression can be attributed to genetic distance between isolates rather than drug exposure, resistance phenotype, or resistance mechanism. (C) We further tested the impact of evolutionary distance on transcriptional regulation, by comparing the number of pairwise SNP differences with the frequency of transcripts with a ≥2-fold expression difference across the ten isolates for which we had collected RNA-seq data at baseline (T0, no drug exposure), after 60 min of drug exposure (T60), and after 180 min of drug exposure (T180). Linear models were fit to either paired isolate comparisons that belonged to the same (resistant versus resistant or susceptible versus susceptible) or different (resistant versus susceptible) phenotypic classes. In all cases, we observed positive slope values.
Genetic distance and population structure impact transcriptome regulation.
First, we investigated the relationship between phylogenetic structure and transcriptome regulation. Principal-component analysis (PCA) revealed three distinct transcriptional clusters along the first two dimensions (PC1 and PC2), representing the greatest variability within the gene expression data set, which were primarily structured by genetic distance rather than resistance phenotype, resistance mechanism, or drug exposure condition (Fig. 2A and B). Isolates that belonged to BAPS groups 4 and 7, which were the most genetically similar as indicated by their grouping in adjacent phylogenetic clades, were also the most transcriptionally similar and clustered on the PCA plot (Fig. 2A and B). These two groups, in addition to containing susceptible isolates, harbored multiple mechanisms of resistance among resistant isolates including: the 23S rRNA C2611T, 23S rRNA A2059G, and mosaic mtr efflux pump mutations. Isolates from BAPS groups 6 and 8 clustered independently from each other, as well as the other clades sampled, and were also the most evolutionarily distant from other clades on the gonococcal tree (Fig. 2A and B).
The relationship between genetic distance and transcriptional regulation was also supported by a pairwise comparison of whole-genome single nucleotide polymorphism (SNP) distance versus pairwise fold change differences in gene expression across all ten isolates transcriptionally profiled in multiple treatment conditions. SNP distances for all isolate pairs ranged from 488 to 6,106 (mean = 3,410). For pairwise comparisons, we assessed isolate pairs with the same (resistant versus resistant or susceptible versus susceptible) or different (resistant versus susceptible) phenotypic classes. For both categories, paired isolates of increasing core genome SNP distances had an increasing number of differentially regulated transcripts (≥2-fold expression difference) between them at baseline (T0) and under treatment conditions (T60 and T180), as indicated by each linear model’s positive slope values (Fig. 2C). This pattern also held true for all transcripts regardless of their fold change in abundance between isolates (Fig. S1). Interestingly, at baseline (T0), the two markers previously reported as being diagnostic of ciprofloxacin resistance (3), porB and rpmB, diverged transcriptionally with increasing evolutionary distance between isolates as well (see Fig. S1 in the supplemental material).
Transcriptional response to azithromycin and candidate marker selection.
We next characterized the transcriptome-wide response to azithromycin across 60- and 180-min drug exposure treatments in our phylogenetically diverse isolate panel. To define the differential response patterns of resistant and susceptible isolates across treatment conditions, we fit a generalized linear model to the count data generated from all 90 RNA-seq libraries with phenotype (resistant versus susceptible), treatment (control versus later time points), and the interaction between phenotype and treatment used as model terms. Overall, 978 genes were significantly differentially expressed in at least one comparison involving resistance phenotype or condition from 0 to 60 min, and 1,177 genes were significantly differentially expressed from 0 to 180 min (Fig. 1; Fig. S2). For both drug exposure time points, the majority of response to condition was initiated only in susceptible isolates (Fig. 1; Fig. S2).
For susceptible isolates, we observed large-scale changes in gene expression in response to azithromycin in both treatments; thus, the 60-min time point was selected for designing candidate diagnostic markers. Candidate markers were required to be significantly differentially expressed between the control and drug exposure in the susceptible strains and between resistant and susceptible isolates under drug exposure and also to have a significant interaction term. From the 433 transcripts that fit these conditions, we subset those that were within the top 10% most significant false discovery rate (FDR)-corrected P values for each comparison yielding 28 transcripts. From these, we selected the transcripts that most clearly distinguished susceptible and resistant isolates (Fig. S2 and S3) to assess as markers via RT-qPCR. Genes encoding selected transcripts were present in all isolates within our panel, as well as the larger collection of 1,102 isolates reported in Grad et al. (17). For resistant isolates, there were no transcripts that significantly changed in expression between the control and drug exposure treatments that were also differentially expressed between resistant and susceptible isolates under drug exposure.
RT-qPCR validation.
(i) Azithromycin. The diagnostic potential of RNA-seq nominated markers to characterize azithromycin susceptibility was assessed in a larger panel (n = 20) of genetically diverse isolates with nonbiased phylogenetic sampling using RT-qPCR. In addition to the panel of isolates used for RNA-seq, ten more isolates were chosen from eight of the twelve BAPS groups for validation (Table 1 and Fig. 2A). Of these, five were susceptible, three were resistant and had mosaic mtr alleles, one was resistant and had the C2611T 23S rRNA mutation, and one was resistant and had a tandem duplication in rplV that has previously been associated with resistance (17).
We evaluated fold-change across control and 60-minute azithromycin exposure treatments for four markers (rpsO, NGO1079, omp3, and rplN) using the 2–ΔΔCT method (30, 31), normalizing to an empirically defined control gene (etfB) that did not significantly change in expression between any of our RNA-seq glm model contrasts (Fig. S3). Expression change was then compared between resistant and susceptible isolates. For both rpsO and NGO1079, change in RNA abundance differed significantly between resistant and susceptible isolates across treatments and there was no overlap in 2–ΔΔCT values between groups (Fig. 3A and B). While the 2–ΔΔCT values for omp3 and rplN also differed significantly between resistant and susceptible isolates, isolates with intermediate MIC values bounding the reduced susceptibly cutoff of 1 μg/ml displayed some overlap between phenotypes (Fig. 3C and D).
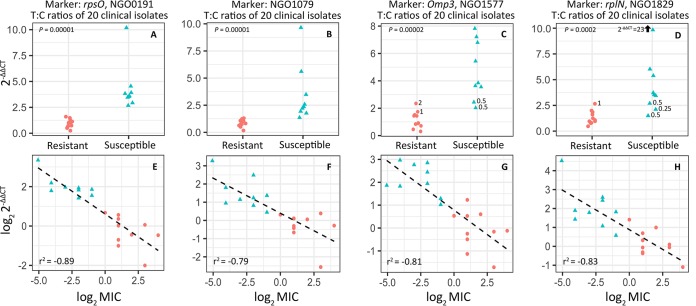
FIG 3.
Candidate diagnostic marker validation via RT-qPCR in a panel of 20 clinical isolates (9 susceptible and 11 resistant). (A to D) NGO0191 or rpsO (A), NGO1079 (B), NGO1577 or omp3 (C), and NGO1829 or rplN (D) fold changes from prior to the addition of azithromycin and 60 min after exposure (n = 3 or 4 biological replicates) were calculated using the 2–ΔΔCT method, normalizing to an empirically defined control transcript (NGO1935, etfB). All four markers showed significant differences in expression across conditions between resistant and susceptible isolate phenotypic classes; however, the fold change for omp3 and rplN did overlap between phenotypes for isolates with azithromycin MIC values close to the reduced susceptibility threshold (i.e., 0.25, 0.5, 1, and 2 μg/ml). (E to H) Linear models were fit to log transformed 2–ΔΔCT and MIC values for each marker for each isolate, omitting the GCGS0745 outlier (MIC > 256 μg/ml) and were used to assess the accuracy of 2–ΔΔCT values as a predictor of MIC (Table 2).
We examined the relationship between the transcriptional response of candidate markers to azithromycin exposure and MIC by fitting a linear model to the log2-transformed 2–ΔΔCT and MIC values. In the model fitting for this analysis, we omitted the outlier GCGS0745 (MIC > 256 μg/ml), since this isolate was the single isolate to depart from linear expectations between these two variables. We speculate that this behavior is attributable to the extent of resistance limiting azithromycin-induced changes in gene expression. For all other isolates across all tested markers, there was a strong negative relationship between log2-transformed 2–ΔΔCT and MIC values, with r2 values ranging between −0.79 and −0.89 (Fig. 3E to H). Fitted models were used to estimate the MICs of each isolate, including the GCGS0745 outlier given 2–ΔΔCT for each marker, which were then averaged across markers, and rounded to the nearest MIC on the discrete scale used by the Centers for Disease Control and Prevention’s Gonococcal Isolate Surveillance Project (GISP) clinical microbiology laboratories (32) (Table 2). Using this method, RNA signatures for these markers categorically predicted resistance in 19/20 cases, with the one susceptible isolate called as resistant predicted to be within one dilution of the actual MIC (predicted at 1 μg/ml, phenotyped at 0.5 μg/ml), and on the cusp of the reduced susceptibility phenotypic designation.
TABLE 2.
RNA response to 60 min of azithromycin exposure (2−ΔΔCT) as a predictor of MIC
| Isolate | Resistance category | AZI MIC (μg/ml) | MIC prediction (μg/ml) |
Avg MIC prediction (μg/ml) | MIC prediction discrete scale | Dilution differencea | |||
|---|---|---|---|---|---|---|---|---|---|
| NGO0191 | NGO1079 | NGO1577 | NGO1829 | ||||||
| GCGS0353 | Susceptible | 0.03 | 0.04 | 0.03 | 0.29 | 0.01 | 0.09 | 0.125 | +2 |
| GCGS0313 | Susceptible | 0.06 | 0.15 | 0.18 | 0.09 | 0.29 | 0.18 | 0.25 | +2 |
| GCGS0465 | Susceptible | 0.06 | 0.23 | 0.48 | 0.30 | 0.49 | 0.38 | 0.50 | +3 |
| GCGS0926 | Susceptible | 0.125 | 0.19 | 0.39 | 0.11 | 0.33 | 0.25 | 0.25 | +1 |
| GCGS0641 | Susceptible | 0.25 | 0.20 | 0.08 | 0.10 | 0.13 | 0.13 | 0.25 | 0 |
| GCGS0524 | Susceptible | 0.25 | 0.23 | 0.35 | 0.26 | 0.16 | 0.25 | 0.25 | 0 |
| GCGS0605 | Susceptible | 0.25 | 0.36 | 0.56 | 0.16 | 0.74 | 0.45 | 0.50 | +1 |
| GCGS1035 | Susceptible | 0.5 | 0.31 | 0.30 | 0.71 | 0.32 | 0.41 | 0.50 | 0 |
| GCGS0996 | Susceptible | 0.5 | 0.22 | 0.86 | 0.54 | 1.30 | 0.73 | 1 | +1 |
| GCGS0276 | Resistant | 1 | 0.88 | 1.00 | 1.20 | 0.51 | 0.89 | 1 | 0 |
| GCGS0834 | Resistant | 2 | 1.27 | 2.28 | 0.57 | 1.14 | 1.32 | 2 | 0 |
| GCGS0298 | Resistant | 2 | 6.39 | 2.01 | 2.77 | 1.74 | 3.23 | 4 | +1 |
| GCGS0436 | Resistant | 2 | 1.00 | 1.27 | 3.60 | 2.67 | 2.13 | 4 | +1 |
| GCGS0525 | Resistant | 2 | 4.47 | 3.01 | 1.21 | 3.54 | 3.05 | 4 | +1 |
| GCGS0481 | Resistant | 2 | 1.91 | 1.25 | 7.13 | 3.79 | 3.52 | 8 | +2 |
| GCGS0363 | Resistant | 4 | 3.30 | 1.33 | 1.11 | 0.81 | 1.64 | 2 | −1 |
| GCGS0354 | Resistant | 8 | 1.82 | 0.92 | 2.51 | 2.24 | 1.87 | 2 | −2 |
| GCGS0550 | Resistant | 8 | 20.83 | 25.84 | 13.29 | 2.65 | 15.65 | 16 | +1 |
| GCGS1026 | Resistant | 16 | 3.39 | 1.96 | 2.41 | 8.62 | 4.09 | 8 | −1 |
| GCGS0745 | Resistant | 256 | 1.59 | 4.38 | 0.90 | 0.91 | 1.94 | 2 | −7 |
That is, the predicted MIC – the measured MIC.
(ii) Ciprofloxacin. Khazaei et al. (3) previously proposed the markers porB and rpmB as candidates for an RNA-based ciprofloxacin AST in gonococcus, with both transcripts downregulated after 10 min of ciprofloxacin exposure in susceptible but not resistant isolates. However, our evaluation of the panel of isolates used by the study revealed a clade-biased ciprofloxacin susceptibility sampling pattern, with resistant isolates all belonging to a single clade and susceptible isolates sampled from elsewhere on the phylogeny (Fig. 4A). Here, we test the antibiotic-induced RNA signatures of these markers in more genetically diverse isolate panel to validate their diagnostic effectiveness.
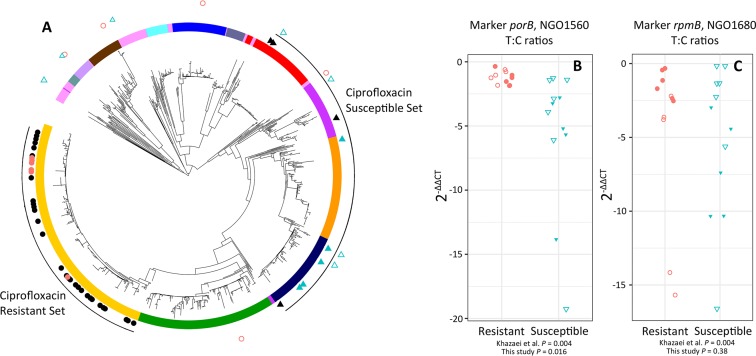
FIG 4.
Phylogenetic structure obscures diagnostic signatures of ciprofloxacin resistance. (A) The panel of isolates used to nominate porB and rpmB as diagnostic phenotypic markers for ciprofloxacin resistance had a highly structured distribution within the gonococcal population, with resistant isolates all sampled from a single clade (closed circles) and susceptible isolates sampled from distantly related clades (closed triangles). In this study, we sample a subset of the panel previously evaluated (3) (closed red circles and blue triangles), in addition to a panel of diverse resistant and susceptible isolates with a less biased sampling pattern (open red circles and blue triangles). RT-qPCR validation revealed the nominated markers porB (B) and rpmB (C) were diagnostic of resistance in the previously evaluated subset (closed red circles and blue triangles do not overlap); however, in the expanded panel the markers were not diagnostic (open red circles and blue triangles overlap).
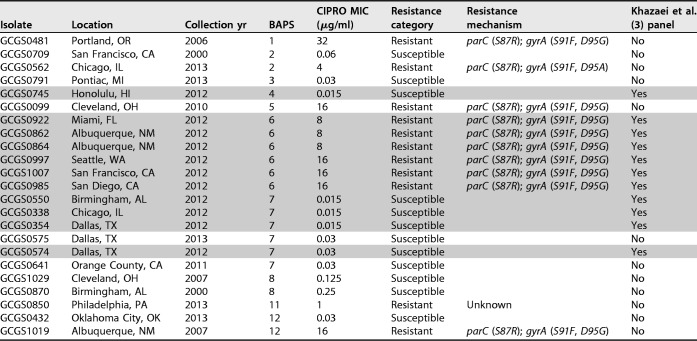
For our ciprofloxacin test panel, we selected a set of isolates from the panel that had previously been used to develop diagnostic markers for ciprofloxacin resistance (3) and also added isolates that represented a more phylogenetically comprehensive sampling. The set of isolates used for the prior diagnostic marker development included six resistant isolates (MIC ≥ 1 μg/ml) with parC (S87R) and gyrA (S91F, D95G) mutations from a single BAPS clade, and five susceptible isolates (MIC < 1 μg/ml) from BAPS clades located elsewhere on the tree (Table 3; Fig. 4A). The additional isolates tested in our study represented eight of the thirteen BAPS groups, and included one resistant isolate with an unknown mechanism of resistance; three resistant isolates with the ParC S87R, GyrA S91F/D95G haplotype; one resistant isolate with the ParC S87R, GyrA S91F/D95A haplotype; and seven susceptible isolates (Table 3; Fig. 4A).
TABLE 3.
Properties of the clinical isolates used to validate diagnostic markers for ciprofloxacin resistance (n = 23)a
The shaded rows indicate isolates used in the Khazaei et al. (3) panel.
Both porB and rpmB were diagnostic of reduced susceptibility to ciprofloxacin in the sample of isolates used by Khazaei et al. (3) that we tested, with no overlap in 2–ΔΔCT values (Fig. 4 and C). However, these markers displayed divergent transcription as a function of increasing genetic distance (Fig. S1), and in the expanded panel of isolates, including a more representative sampling of the known gonococcal genetic diversity, the 2–ΔΔCT values of porB and rpmB were not diagnostic of ciprofloxacin resistance phenotype (Fig. 4B and C).
DISCUSSION
RNA-based resistance diagnostics have the potential to revolutionize antimicrobial selection at the point of care. However, the design of diagnostic tests that rely on changes in RNA abundance after drug exposure can be vulnerable to erroneous candidate marker designation due to the potential for gene regulation divergence to reflect genetic distance and population structure rather than resistance phenotype. This danger can be amplified if the sampling of isolates for marker design is highly phylogenetically structured by resistance phenotype. Here, we used a combination of whole transcriptome profiling and RT-qPCR to test the importance of controlling for genetic diversity in the design of RNA-based tests, to identify azithromycin responsive transcripts and evaluate their potential diagnostic value, and to evaluate previously reported diagnostic markers for ciprofloxacin resistance (porB and rpmB).
We expected that if genetic distance was the main contributor to transcriptional variation between isolates, increasing genetic distance would also yield increasing differences in transcriptome regulation, regardless of the resistance phenotypes of the isolates being compared (see models in Fig. S5). Results from our principal-component analysis (PCA) demonstrated that for our panel of diverse gonococcal isolates, divergence in transcriptional profiles primarily reflected genetic distance and population structure, as indicated by the clustering of isolates along PC1 and PC2 by phylogenetic proximity (i.e., genetic relatedness; Fig. 2B). We also observed an interaction effect between resistance phenotype and drug exposure condition, where susceptible strains moved predictably along PC1 upon treatment, but resistant strains did not. Thus, while clustering position appears to be mostly dominated by phylogeny, supporting the overall importance of controlling for phenotype-specific clade sampling bias in the AST assay design panel isolates, the clustering of transcriptional profiles also supports conserved signatures of antibiotic response in susceptible isolates.
To further investigate the importance of controlling for phylogenetic structure in assay design panels, we quantified the pairwise interstrain whole-genome SNP distances, as a metric of evolutionary distance, and pairwise interstrain frequency of ≥2-fold expression differences by transcript across drug exposure conditions, as a metric of transcriptional regulatory evolution (Fig. 2C). Linear models fit to either same- or cross-phenotype comparisons indicated several conclusions. First, at baseline for both comparison types, regressions had significant and positive slopes, and y intercepts that were not significantly different than zero (Fig. 2C, T0), supporting genetic distance as the main contributor to gene expression variance between isolates under this condition. Second, after 60 min of azithromycin exposure, though genetic distance remained the main contributor to gene expression variance in the same phenotypic class comparison, for isolates of different phenotypic classes the slope was no longer significantly different than zero and the y intercept of 337.39 was significantly greater than zero. This suggests that differences in resistance phenotype had a greater impact on gene expression divergence under this condition than genetic distance (Fig. 2C, T60). Third, after 180 min of azithromycin exposure for isolates of the different phenotypic class comparison the slope was again not significantly different than zero and the y intercept of 483.10 was significantly greater than zero (Fig. 2C, T180). In contrast to the main result of our PCA supporting the domination of transcriptional signatures by phylogeny, these results suggest that gene expression variance driven by differences in phenotype will overwhelm any genetic distance effects, as isolates of different resistance phenotypes are diverted along distinct physiological trajectories in response to drug exposure. However, we suggest a measure of caution in this interpretation, since positive slope values in every treatment condition for cross-phenotype comparisons indicate that some variance in gene regulation is still likely driven by genetic distance effects rather than resistance phenotype alone.
To identify candidate diagnostic markers for azithromycin resistance, we broadly sampled isolates from the N. gonorrhoeae phylogenetic tree and selected four candidate diagnostic markers (rpsO, rplN, omp3, and NGO1079). For all candidate transcripts, RNA abundance increased after 60 min of azithromycin exposure in susceptible but not resistant isolates (Fig. 3), and upregulation was consistent with gene expression results from previous drug challenge studies. Macrolides induce translational stalling by binding the ribosome peptide exit tunnel and inhibiting the exit of nascent proteins, which has been shown to induce the upregulation of ribosomal protein-encoding transcripts (33–35). Upregulation of the 30S and 50S ribosomal protein-encoding transcripts rpsO and rplN in susceptible isolates of N. gonorrhoeae therefore likely serves as a good phenotypic signature of susceptibility. The omp3 gene encodes the major gonococcal outer membrane protein Rmp, which (i) has been implicated in the formation, stabilization, and operation of the trimeric porin structure of PorB; and (ii) contains a peptidoglycan-binding motif, suggesting a role in cross-linking the outer membrane proteins to the peptidoglycan and the maintenance of cell membrane integrity (36, 37). Interestingly, the Rmp protein is also upregulated after spectinomycin exposure in susceptible N. gonorrhoeae isolates (38). Finally, the putative oxidoreductase NGO1079 was upregulated upon drug exposure in our study. NGO1079 has previously been shown to be upregulated by the AraC-like regulator MpeR, and the expression of MpeR also indirectly modulates resistance to macrolides through transcriptional suppression of the repressor of the mtrCDE efflux pump (39).
Though all azithromycin-responsive transcripts were significantly upregulated in susceptible isolates compared to resistant isolates across treatment conditions, isolates with intermediate MIC values bounding the reduced susceptibly threshold displayed some overlap in 2–ΔΔCT values between phenotypic groups for some markers (Fig. 3C and D). For all markers we uncovered a strong negative relationship between the log2-transformed 2–ΔΔCT and MIC values (Fig. 3E to H). Using regression of these two variables across tested markers, we accurately predicted susceptibility in 19/20 cases (89% sensitivity and 92% specificity; Table 2), with the one susceptible isolate called as resistant on the boarder of the reduced susceptibility phenotypic designation (predicted at 1 μg/ml, phenotyped at 0.5 μg/ml). Though 2–ΔΔCT values could not perfectly predict resistance for isolates with intermediate MICs, phenotyping a susceptible isolate as resistant does not necessarily present a problem for treatment, since it only precludes the use of an antibiotic that may be effective. Conversely, diagnosing a resistant isolate as susceptible would lead to inappropriate treatment with an ineffective antibiotic. Thus, our results are promising and continued optimization of the assay conditions (markers, drug concentration, exposure time, etc.) may increase diagnostic accuracy and inform the development of a POC test for azithromycin susceptibility for gonococcal infections.
The sampling scheme for the assay design panel isolates used to identify the transcripts of porB and rpmB as diagnostic for ciprofloxacin resistance in the Khazaei et al. (3) study had a clade bias, with all resistant isolates from a single clade and all susceptible isolates from outside that clade (Fig. 4A). We confirmed that porB and rpmB could diagnosis resistance phenotype after 10 min of ciprofloxacin exposure in a set of the isolates used in the prior study (Fig. 4B and C). However, in evaluation of a panel of isolates representative of the broader diversity of the gonococcal population, the markers no longer showed diagnostic potential, displaying overlapping 2–ΔΔCT values between resistant and susceptible isolates (Fig. 4B and C). We also find that transcription of porB and rpmB is clearly impacted by evolutionary distance in control conditions (Fig. S1B). Together, these results suggest that the differential expression patterns of porB and rpmB found by Khazaei et al. (3) are largely independent of resistance phenotype and rather are more likely driven by gene expression evolution due to clade-specific selective pressures or genetic drift. Additional RNA-seq studies will be needed in a more genetically diverse isolate panel to nominate true RNA-based signatures diagnostic for ciprofloxacin resistance.
To our knowledge, our study is the first to comprehensively consider the genetic diversity of a species in development of RNA-based candidate diagnostic markers for resistance phenotype and is also the first study to nominate RNA signatures diagnostic of azithromycin susceptibility in N. gonorrhoeae. Overall, our results emphasize the power and potential of transcription-based phenotypic AST assays and also indicate the importance of selecting appropriate strains for the assay design panel in the research and development phase. Though our own strain panels were relatively small, we anticipate additional testing will confirm the importance of considering phylogenetic diversity in AST development. We suggest that strain panels for AST design must capture the genetic diversity of a species to accurately evaluate the factors underlying strain-to-strain gene expression heterogeneity and control for the impact of phylogenetic structure on phenotypic diagnostics.
MATERIALS AND METHODS
Bacterial strains and culturing.
N. gonorrhoeae isolates were acquired from the Centers for Disease Control’s Gonococcal Isolate Surveillance Project (GISP). GISP isolates are collected from the first 25 cases of men with gonococcal urethritis per month at 25 to 30 sentinel sexually transmitted disease clinics across the United States (32). Isolates were cultured on GCB agar medium supplemented with 1% IsoVitaleX (Becton, Dickinson Co., Franklin Lakes, NJ). After inoculation, plates were incubated at 37°C in a 5% CO2 atmosphere incubator for 16 to 18 h. All isolate stocks were stored at –80°C in Trypticase soy broth containing 20% glycerol. Isolates for both azithromycin and ciprofloxacin test panels were selected to be representative of the known diversity within the gonococcal population. Antimicrobial susceptibility testing was conducted using Etest strips (40) or the agar dilution method (32), and MICs were recorded after 24 h of growth. The European Committee on Antimicrobial Susceptibility Testing (EUCAST) defines azithromycin reduced susceptibility as MICs of ≥1 μg/ml (41), while GISP uses a breakpoint of ≥2 μg/ml (32). Therefore, we conservatively define azithromycin reduced susceptibility in this study by the EUCAST standard of ≥1 μg/ml. For ciprofloxacin resistance we adhere to the GISP and CLSI breakpoint of ≥1 μg/ml (32, 42), which was also the breakpoint used by Khazaei et al. (3).
Experimental design.
Cells harvested from overnight plates were suspended in GCP (7.5 g protease peptone #3, 0.5 g soluble starch, 2 g dibasic K2HPO4, 0.5 g monobasic KH2PO4, 2.5 g NaCl, ddH2O to 500 ml; Becton Dickinson) supplemented with 1% IsoVitaleX and 0.042% sodium bicarbonate, and incubated at 37°C for 2 h to mid-log phase. For azithromycin, samples were collected before exposure to azithromycin, 60 min after 0.125 μg/ml azithromycin exposure and 180 min after 0.125 μg/ml azithromycin exposure (Fig. 1). For ciprofloxacin, we replicated the sampling scheme of Khazaei et al. (3), and sampled paired 0.5 μg/ml ciprofloxacin exposed and unexposed cultures 10 min after drug treatment (Fig. S3). RNA was isolated using the Direct-Zol kit with DNase I treatment (Zymo Research, Irvine, CA).
RNA preparation and sequencing.
RNA libraries were prepared at the Broad Institute at the Microbial ‘Omics Core using a modified version of the RNAtag-seq protocol (43). Total RNA (500 ng) was fragmented, depleted of genomic DNA, dephosphorylated, and ligated to DNA adapters carrying 5′-AN8-3′ barcodes of known sequence with a 5′ phosphate and a 3′ blocking group. Barcoded RNAs were pooled and depleted of rRNA using the RiboZero rRNA depletion kit (Epicentre, Madison, WI). Pools of barcoded RNAs were converted to Illumina cDNA libraries in two main steps: (i) reverse transcription of the RNA using a primer designed to the constant region of the barcoded adaptor with addition of an adapter to the 3′ end of the cDNA by template switching using SMARTScribe (Clontech, Mountain View, CA) as described previously (44) and (ii) PCR amplification using primers whose 5′ ends target the constant regions of the 3′ or 5′ adaptors and whose 3′ ends contain the full Illumina P5 or P7 sequences. cDNA libraries were sequenced on the Illumina NextSeq 500 platform to generate 50-bp paired end reads.
RNA-seq analysis.
Barcode sequences were removed, and reads were then aligned to the FA1090 reference genome (AE004969.1) using BWA v.0.7.8 (45), mapping to the sense strand of the coding domain sequences (CDSs; n = 1,894). Differential expression analysis was conducted with DESeq2 v.1.10.1, which employs an empirical Bayes method to estimate gene-specific biological variation (46). A multifactor analysis was used to account for the effects of resistance phenotype (P), condition (C), or the interaction between both phenotype and condition on transcript expression (E), using the equation E = P + C + (P × C). Per-gene dispersions were estimated using the Cox-Reid profile-adjusted method and a negative binomial generalized log-linear model was fit to identify significantly differentially expressed transcripts. Subsequently, gene lists were corrected for multiple testing using an FDR (cutoff, ≤0.01). We then compared significant model factors to define expression profiles for each transcript.
Genomic read libraries were obtained from the NCBI’s Sequence Read Archive (project PRJEB2090) and the European Nucleotide Archive (projects PRJEB2999 and PRJEB7904). Pairwise SNP distances were generated by mapping read libraries to the FA1090 (AE004969.1) reference genome using BWA v.0.7.8, and calling variants with Pilon v1.16 (47) using mapping qualities of >40 and coverage depths of >5× set as minimum thresholds. SNP distance between each isolate pair was then calculated excluding gaps using custom scripts. Normalized transcript read counts were generated for coding domain regions annotated in the FA1090 reference for each isolate (see above), and the absolute log-fold change was calculated by comparing transcriptional counts per CDS between each isolate pair at baseline (no drug exposure). CDS regions with zero counts in any isolate within each pair were excluded.
RT-qPCR methods.
SuperScript IV reverse transcriptase (Thermo Fisher Scientific, Waltham, MA) was used to convert isolated RNA to cDNA using random hexamer primers. Annealing of primers to the template RNA was conducted in 13 μl reactions (3.8 μM primers, 1.3 mM deoxynucleoside triphosphate mix, and 500 ng of RNA) by heating the RNA-primer mix to 65°C for 5 min, followed by incubation on ice for 1 min. For reverse transcription, 4 μl of 5× SSIV buffer, 1 μl of 100 mM dithiothreitol, 1 μl of RNaseOUT, and 1 μl of SuperScript IV reverse transcriptase were added to the reaction mix, followed by incubation at 23°C for 10 min, followed in turn by 55°C for 10 min and 80°C for 10 min.
Primers for azithromycin susceptibility diagnostic markers were designed to amplify ∼100-bp fragments of both control and target genes (Table S1). The control gene etfB (NGO1935) was empirically defined from our RNA-seq results, and was required to have no significant change in expression across treatments for all isolates tested. Primers for ciprofloxacin diagnostic markers were obtained from Khazaei et al. (3) (Table S1). Real-time PCR was conducted with the Applied Biosystems PowerUp SYBR green Master Mix on the Applied Biosystems ViiA 7 real-time PCR system (Thermo Fisher Scientific), using the following cycling conditions: 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60 to 64°C for 1 min (see Table S1 for primer-specific details). The 6.25-μl PCR mix consisted of 1 μl of 1:100-diluted cDNA, 0.5× SYBR green master mix, and a 0.5 μM concentration of the forward and reverse primer pair. Reactions were performed in 384-well plates with three to four biological replicates per isolate tested per antibiotic exposure condition. CT values were calculated using the ViiA 7 software, and the fold change was calculated using the 2–ΔΔCT method (30, 31).
Data availability.
The transcriptome libraries are archived in the GenBank SRA (BioProject ID PRJNA518111). RT-qPCR data are available in Table S2 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jonathan Livny for technical assistance with transcriptome library preparation and Samantha Palace for aid in RT-qPCR assay design.
This study was supported by the Richard and Susan Smith Family Foundation (http://www.smithfamilyfoundation.net/) and the National Institutes of Health (R01 AI132606; https://www.nih.gov/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
R.P.B. is a coinventor on subject matter in U.S. provisional patent application no. 62/723,417 filed by the Broad Institute directed to RNA signatures for AST, similar to work described in the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00549-19.
REFERENCES
- 1.Barczak AK, Gomez JE, Kaufmann BB, Hinson ER, Cosimi L, Borowsky ML, Onderdonk AB, Stanley SA, Kaur D, Bryant KF, Knipe DM, Sloutsky A, Hung DT. 2012. RNA signatures allow rapid identification of pathogens and antibiotic susceptibilities. Proc Natl Acad Sci U S A 109:6217–6222. doi: 10.1073/pnas.1119540109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharyya RP, Liu J, Ma P, Bandyopadhyay N, Livny J, Hung DT. 2017. Rapid phenotypic antibiotic susceptibility testing through RNA detection. Open Forum Infect Dis 4:S33. doi: 10.1093/ofid/ofx162.082. [DOI] [Google Scholar]
- 3.Khazaei T, Barlow JT, Schoepp NG, Ismagilov RF. 2018. RNA markers enable phenotypic test of antibiotic susceptibility in Neisseria gonorrhoeae after 10 minutes of ciprofloxacin exposure. Sci Rep 8:11606. doi: 10.1038/s41598-018-29707-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y-H, Qin X-L, Yang J-Y, Liao Y-W, Wu X-Z, Zheng H-P. 2019. Identification and expression analysis of ceftriaxone resistance-related genes in Neisseria gonorrhoeae integrating RNA-Seq data and qRT-PCR validation. J Glob Antimicrob Resist 16:202–209. doi: 10.1016/j.jgar.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Gao Q, Kripke KE, Saldanha AJ, Yan W, Holmes S, Small PM. 2005. Gene expression diversity among Mycobacterium tuberculosis clinical isolates. Microbiology 151:5–14. doi: 10.1099/mic.0.27539-0. [DOI] [PubMed] [Google Scholar]
- 6.Homolka S, Niemann S, Russell DG, Rohde KH. 2010. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog 6:e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rose G, Cortes T, Comas I, Coscolla M, Gagneux S, Young DB. 2013. Mapping of genotype-phenotype diversity among clinical isolates of Mycobacterium tuberculosis by sequence-based transcriptional profiling. Genome Biol Evol 5:1849–1862. doi: 10.1093/gbe/evt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goire N, Lahra MM, Chen M, Donovan B, Fairley CK, Guy R, Kaldor J, Regan D, Ward J, Nissen MD, Sloots TP, Whiley DM. 2014. Molecular approaches to enhance surveillance of gonococcal antimicrobial resistance. Nat Rev Microbiol 12:223–229. doi: 10.1038/nrmicro3217. [DOI] [PubMed] [Google Scholar]
- 9.Unemo M, Shafer WM. 2014. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 27:587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unemo M. 2015. Current and future antimicrobial treatment of gonorrhoea: the rapidly evolving Neisseria gonorrhoeae continues to challenge. BMC Infect Dis 15:364. doi: 10.1186/s12879-015-1029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suay-García B, Pérez-Gracia M. 2018. Future prospects for Neisseria gonorrhoeae treatment. J Antibiot 7:49. doi: 10.3390/antibiotics7020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh V, Bala M, Bhargava A, Kakran M, Bhatnagar R. 2018. In vitro efficacy of 21 dual antimicrobial combinations comprising novel and currently recommended combinations for treatment of drug-resistant gonorrhea in future era. PLoS One 13:e0193678. doi: 10.1371/journal.pone.0193678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2018. New CDC analysis shows steep and sustained increases in STDs in recent years. Press release. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/nchhstp/newsroom/2018/press-release-2018-std-prevention-conference.html. Accessed 30 January 2019. [Google Scholar]
- 14.Eyre DW, Sanderson ND, Lord E, Regisford-Reimmer N, Chau K, Barker L, Morgan M, Newnham R, Golparian D, Unemo M, Crook DW, Peto TE, Hughes G, Cole MJ, Fifer H, Edwards A, Andersson MI. 2018. Gonorrhoea treatment failure caused by a Neisseria gonorrhoeae strain with combined ceftriaxone and high-level azithromycin resistance, England, February 2018. Eurosurveillance 23:364–366. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2018.23.27.1800323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tedijanto C, Olesen S, Grad Y, Lipsitch M. 2018. Estimating the proportion of bystander selection for antibiotic resistance among potentially pathogenic bacterial flora. Proc Natl Acad Sci U S A 115:E11988–E11995. doi: 10.1073/pnas.1810840115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. 2012. Update to CDC’s sexually transmitted diseases treatment guidelines, 2010: oral cephalosporins no longer a recommended treatment for gonococcal infections. MMWR Morb Mortal Wkly Rep 61:590. [PubMed] [Google Scholar]
- 17.Grad YH, Harris SR, Kirkcaldy RD, Green AG, Marks DS, Bentley SD, Trees D, Lipsitch M. 2016. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000–2013. J Infect Dis 214:1579–1587. doi: 10.1093/infdis/jiw420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wadsworth CB, Arnold BJ, Sater MRA, Grad YH. 2018. Azithromycin resistance through interspecific acquisition of an epistasis-dependent efflux pump component and transcriptional regulator in Neisseria gonorrhoeae. mBio 9:e01419-18. doi: 10.1128/mBio.01419-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng LK, Martin I, Liu G, Bryden L. 2002. Mutation in 23S rRNA associated with macrolide resistance in Neisseria gonorrhoeae. Antimicrob Agents Ch 46:3020–3025. doi: 10.1128/AAC.46.9.3020-3025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chisholm SA, Dave J, Ison CA. 2010. High-level azithromycin resistance occurs in Neisseria gonorrhoeae as a result of a single point mutation in the 23S rRNA genes. Antimicrob Agents Chemother 54:3812–3816. doi: 10.1128/AAC.00309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trembizki E, Doyle C, Jennison A, Smith H, Bates J, Lahra M, Whiley D. 2014. A Neisseria gonorrhoeae strain with a meningococcal mtrR sequence. J Med Microbiol 63:1113–1115. doi: 10.1099/jmm.0.074286-0. [DOI] [PubMed] [Google Scholar]
- 22.Whiley DM, Kundu RL, Jennison AV, Buckley C, Limnios A, Hogan T, Enriquez R, Nasser El J, George CR, Lahra MM. 2018. Azithromycin-resistant Neisseria gonorrhoeae spreading amongst men who have sex with men (MSM) and heterosexuals in New South Wales, Australia, 2017. J Antimicrob Chemother 73:1242–1246. doi: 10.1093/jac/dky017. [DOI] [PubMed] [Google Scholar]
- 23.Rouquette-Loughlin C, Reimche JL, Balthazar JT, Dhulipala V, Gernert K, Kersh E, Pham C, Pettus K, Abrams AJ, Trees DL, St Cyr S, Shafer WM. 2018. Mechanistic basis for decreased antimicrobial susceptibility in a clinical isolate of Neisseria gonorrhoeae possessing a mosaic-like mtr efflux pump locus. mBio 9:e02281-18. doi: 10.1128/mBio.02281-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagman KE, Shafer WM. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J Bacteriol 177:4162–4165. doi: 10.1128/jb.177.14.4162-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zarantonelli L, Borthagaray G, Lee EH, Veal W, Shafer WM. 2001. Decreased susceptibility to azithromycin and erythromycin mediated by a novel mtr(R) promoter mutation in Neisseria gonorrhoeae. J Antimicrob Chemother 47:651–654. doi: 10.1093/jac/47.5.651. [DOI] [PubMed] [Google Scholar]
- 26.Ohneck EA, Zalucki YM, Johnson PJT, Dhulipala V, Golparian D, Unemo M, Jerse AE, Shafer WM. 2011. A novel mechanism of high-level, broad-spectrum antibiotic resistance caused by a single base pair change in Neisseria gonorrhoeae. mBio 2:e00187-11. doi: 10.1128/mBio.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demczuk W, Martin I, Peterson S, Bharat A, Van Domselaar G, Graham M, Lefebvre B, Allen V, Hoang L, Tyrrell G, Horsman G, Wylie J, Haldane D, Archibald C, Wong T, Unemo M, Mulvey MR. 2016. genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant Neisseria gonorrhoeae in Canada from 1997 to 2014. J Clin Microbiol 54:1304–1313. doi: 10.1128/JCM.03195-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention. 2007. Update to CDC’s sexually transmitted diseases treatment guidelines, 2006: fluoroquinolones no longer recommended for treatment of gonococcal infections. MMWR Morb Mortal Wkly Rep 56:332. [PubMed] [Google Scholar]
- 29.Fifer H, Saunders J, Soni S, Sadiq ST, FitzGerald M. 2019. British Association for Sexual Health and HIV national guideline for the management of infection with Neisseria gonorrhoeae (2019). British Association for Sexual Health and HIV, London, United Kingdom. [Google Scholar]
- 30.Gibson UE, Heid CA, Williams PM. 1996. A novel method for real time quantitative RT-PCR. Genome Res 6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Centers for Disease Control and Prevention. 2016. Gonococcal isolate surveillance program (GISP) protocol. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/std/gisp/gisp-protocol-may-2016.pdf. Accessed 30 January 2019. [Google Scholar]
- 33.Ng WL, Kazmierczak KM, Robertson GT, Gilmour R, Winkler ME. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J Bacteriol 185:359–370. doi: 10.1128/jb.185.1.359-370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aakra A, Vebo H, Snipen L, Hirt H, Aastveit A, Kapur V, Dunny G, Murray B, Nes IF. 2005. transcriptional response of Enterococcus faecalis v583 to erythromycin. Antimicrob Agents Chemother 49:2246–2259. doi: 10.1128/AAC.49.6.2246-2259.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kannan K, Kanabar P, Schryer D, Florin T, Oh E, Bahroos N, Tenson T, Weissman JS, Mankin AS. 2014. The general mode of translation inhibition by macrolide antibiotics. Proc Natl Acad Sci U S A 111:15958–15963. P doi: 10.1073/pnas.1417334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Judd RC. 1989. Protein I: structure, function, and genetics. Clin Microbiol Rev 2:S41–S48. doi: 10.1128/CMR.2.Suppl.S41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koebnik R. 1995. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell surface proteins. Mol Microbiol 16:1269–1270. doi: 10.1111/j.1365-2958.1995.tb02348.x. [DOI] [PubMed] [Google Scholar]
- 38.Nabu S, Lawung R, Isarankura-Na-Ayudhya P, Isarankura-Na-Ayudhya C, Roytrakul S, Prachayasittikul V. 2014. Reference map and comparative proteomic analysis of Neisseria gonorrhoeae displaying high resistance against spectinomycin. J Med Microbiol 63:371–385. doi: 10.1099/jmm.0.067595-0. [DOI] [PubMed] [Google Scholar]
- 39.Mercante AD, Jackson L, Johnson PJT, Stringer VA, Dyer DW, Shafer WM. 2012. MpeR Regulates the mtr efflux locus in Neisseria gonorrhoeae and modulates antimicrobial resistance by an iron-responsive mechanism. Antimicrob. Antimicrob Agents Chemother 56:1491–1501. doi: 10.1128/AAC.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papp JR, Rowlinson M-C, O’Connor NP, Wholehan J, Razeq JH, Glennen A, Ware D, Iwen PC, Lee LV, Hagan C. 2018. Accuracy and reproducibility of the E-test to detect drug-resistant Neisseria gonorrhoeae to contemporary treatment. J Med Microbiol 67:68–73. doi: 10.1099/jmm.0.000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The European Committee on Antimicrobial Susceptibility Testing. 2019. Breakpoint tables for interpretation of MICs and zone diameters, version 9.0. EUCAST, Basel, Switzerland. [Google Scholar]
- 42.Clinical Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing, 29th ed CLSI supplement M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 43.Shishkin AA, Giannoukos G, Kucukural A, Ciulla D, Busby M, Surka C, Chen J, Bhattacharyya RP, Rudy RF, Patel MM, Novod N, Hung DT, Gnirke A, Garber M, Guttman M, Livny J. 2015. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat Methods 12:323–325. doi: 10.1038/nmeth.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. 2001. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques 30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:31–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome libraries are archived in the GenBank SRA (BioProject ID PRJNA518111). RT-qPCR data are available in Table S2 in the supplemental material.