We studied the molecular mechanisms involved in the postantibiotic effect of the fluoroquinolones levofloxacin and moxifloxacin in Streptococcus pneumoniae. Wild-type strain R6 had postantibiotic effects of 2.05 ± 0.10 h (mean ± standard deviation [SD]) and 3.23 ± 0.45 h at 2.5× and 10× MIC of levofloxacin, respectively. Moxifloxacin exhibited lower effects of 0.87 ± 0.1 and 2.41 ± 0.29 h at 2.5× and 10× MIC, respectively.
KEYWORDS: DNA cleavage, fluoroquinolones, levofloxacin, moxifloxacin, postantibiotic effect, reactive oxygen species, Streptococcus pneumoniae, transcriptional regulation
ABSTRACT
We studied the molecular mechanisms involved in the postantibiotic effect of the fluoroquinolones levofloxacin and moxifloxacin in Streptococcus pneumoniae. Wild-type strain R6 had postantibiotic effects of 2.05 ± 0.10 h (mean ± standard deviation [SD]) and 3.23 ± 0.45 h at 2.5× and 10× MIC of levofloxacin, respectively. Moxifloxacin exhibited lower effects of 0.87 ± 0.1 and 2.41 ± 0.29 h at 2.5× and 10× MIC, respectively. Fluoroquinolone-induced chromosome fragmentation was measured at equivalent postantibiotic effects for levofloxacin (2.5× MIC) and moxifloxacin (10× MIC). After 2 h of drug removal, reductions were approximately 7-fold for levofloxacin and 3-fold for moxifloxacin, without further decreases at later times. Variations in reactive oxygen species production were detected after 4 to 6 h of drug withdrawals, with decreases ≥400-fold for levofloxacin and ≥800-fold for moxifloxacin at 6 h. In accordance, after 4 to 6 h of drug withdrawal, the levofloxacin-induced upregulation of the fatCDEB operon, introducing iron in the bacteria, decreased up to 2- to 3-fold, and the moxifloxacin-induced upregulation of several genes involved in the production of pyruvate was reduced 3- to 7-fold. In accordance, lower postantibiotic effects (up to 1 h) were observed in strain R6 ΔspxB, lacking the main enzyme involved in oxygen peroxide production, than in R6. Although no change in the recovery of chromosome fragmentation was observed between R6 and R6 ΔspxB, 3.5 × 103-fold lower reactive oxygen species production was observed in R6 ΔspxB, without changes after drug removal. These results show that reactive oxygen species are the main factors directing the postantibiotic effect of levofloxacin and moxifloxacin in S. pneumoniae.
INTRODUCTION
Streptococcus pneumoniae, a main human pathogen, is the primary cause of community-acquired pneumonia, meningitis, bacteremia, and otitis media in children. Worldwide, one million children ≤5 years old die annually of pneumococcal infections (1). Given the spread of pneumococcal isolates resistant to beta-lactams and macrolides (2), the fluoroquinolones (FQs) levofloxacin (LVX) and moxifloxacin (MXF) are currently recommended for the treatment of adult patients with pneumonia (3). FQ resistance in S. pneumoniae has a low prevalence (<3%) in Europe (4, 5), although it is higher in Canada (7.3%) (6) and in some locations of Asia (10.5%) (7). However, an increase in resistance may occur if the use of FQs is increased (8). An important pharmacodynamic parameter of antibiotics, which has clinical impact on the antibiotic dosing regimens, is the postantibiotic effect (PAE). PAE is defined as the delayed regrowth of bacteria following short exposure to suprainhibitory concentrations of an antibiotic (9). PAEs of FQS in S. pneumoniae were previously determined (10–14). However, the mechanisms involved in this FQ-induced PAE have not been studied. FQs target DNA gyrase and DNA topoisomerase IV, essential enzymes for the maintenance of DNA topology and, subsequently, of replication and transcription (15). They interact with these topoisomerases and stabilize an intermediate of their reaction, DNA-FQ-topoisomerase complexes, which lead to the generation of harmful double-stranded DNA breaks (16). In addition, the inhibition of gyrase triggers the generation of a hydroxyl radical via the Fenton reaction, contributing to the lethality of FQs (17–19). We previously detected the production of reactive oxygen species (ROS; chemical species formed upon incomplete reduction of oxygen, including the superoxide anion, hydrogen peroxide, and hydroxyl radical) in S. pneumoniae after FQ treatment. The inhibition of topoisomerase IV by LVX (20) or of both topoisomerase IV and gyrase by MXF (21) triggered global changes in the transcriptome. We proposed these changes to be mediated by local changes in supercoiling. The differences in the transcriptome triggered by either drug would be due to differences in sequence recognition by the topoisomerases, a process which is itself affected by DNA supercoiling. LVX treatment upregulates the fatDCEB operon, which is responsible for iron intake. This upregulation leads to an increase of intracellular iron and, in turn, to the shift in the Fenton reaction (Fe2+ + H2O2 → Fe3+ + OH− + OH·) toward the production of hydroxyl radicals. Treatment with MXF upregulates pathways affecting pyruvate levels, which lead to a parallel increase in intracellular H2O2 mediated by the pyruvate oxidase enzyme SpxB (EC 1.2.3.3). Then, both LVX and MXF stimulate the Fenton reaction by causing an increase in the concentrations of Fe2+ and H2O2, respectively. In consequence, an increase in the amount of hydroxyl radicals occurs, contributing to the lethality of these FQs.
We present here a study whose aim was to understand the mechanism involved in the PAEs of LVX and MXF in S. pneumoniae. Chromosomal DNA fragmentation, ROS production, and transcriptional alterations were monitored in the recovery phase.
RESULTS
The length of PAE depends on the presence of an active SpxB enzyme.
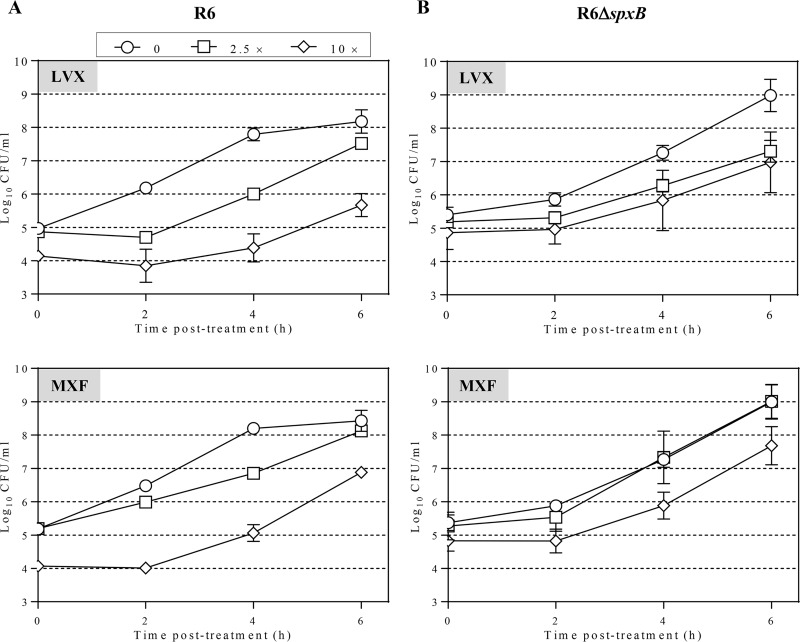
In strain R6, the PAE of LVX was dependent on its concentration, being 1.6-fold higher at 10× MIC than at 2.5× MIC (P = 0.01). Likewise, the PAE of MXF was 2.8-fold higher at 10× MIC than at 2.5× MIC (P = 0.001). These proportions were maintained in the R6 ΔspxB strain; LVX PAE was 1.8-fold higher at 10× MIC than at 2.5× MIC (P = 0.001), and the MXF PAE was 2.2-fold higher at 10× MIC than at 2.5× MIC (P = 0.0007). However, R6 ΔspxB had shorter PAEs than R6 at 2.5× MIC of LVX (2.05 ± 0.10 h versus 1.18 ± 0.13 h; mean ± standard deviation [SD]) and 10× MIC of MXF (2.41 ± 0.29 h versus 1.53 ± 0.12 h) (Fig. 1; Table 1). Strain R6 ΔspxB is isogenic with R6 except for a deletion in the spxB gene, which codes for the pyruvate oxidase enzyme, responsible for the production of oxygen peroxide (22, 23). Then, it can be concluded that the duration of FQ PAEs is related to the presence of a pyruvate oxidase enzyme encoded by spxB.
FIG 1.
PAE in S. pneumoniae R6 (A) and R6 ΔspxB (B). Cultures containing approximately 108 CFU/ml were subjected to 1 h treatment with either LVX or MXF at the indicated concentrations. Cultures were diluted 1,000-fold in medium without drug and growth was followed for 6 h. Viable cells were determined by plating on blood agar plates. Log10 of CFU/ml (means ± SDs) from three independent replicates are presented.
TABLE 1.
PAE in S. pneumoniae R6 and R6 ΔspxBa
| Strain | PAE (h) (P value) for:b
|
|||
|---|---|---|---|---|
| LVX (×MIC) |
MXF (×MIC) |
|||
| 2.5 | 10 | 2.5 | 10 | |
| R6 | 2.05 ± 0.10 | 3.23 ± 0.45 | 0.87 ± 0.1 | 2.41 ± 0.29 |
| R6 ΔspxB | 1.18 ± 0.13 (0.0008) | 1.53 ± 0.06 (0.003) | 0.43 ± 0.15 (0.015) | 1.53 ± 0.12 (0.008) |
Bacteria grown to 108 CFU/ml were exposed to the drugs for 1 h. Treatment finished by 1/1,000 dilution with fresh medium. R6 ΔspxB has a deletion in spxB (20).
PAE was measured by viable count method. Results are the averages ± SDs from three independent replicates. P values are for the differences between R6 and R6 ΔspxB. The MICs of LVX and MXF for both strains were 1 μg/ml and 0.06 μg/ml, respectively.
Fragmentation of chromosomal DNA reverted soon after drug removal.
The effects of LVX and MXF on R6 were tested at concentrations in which the bacteria exhibited similar PAEs, i.e., 2.5× MIC of LVX (2.05 ± 0.10 h) and 10× MIC of MXF (2.41 ± 0.29 h). For R6 ΔspxB, 10× MICs of both FQs were used, given they conferred the highest PAEs to the strain (2.17 ± 0.15 h for LVX and 1.53 ± 0.12 h for MXF). In R6, the reduction in DNA fragmentation was observed at 2 h, with no further decrease. This was detected in the LVX treatment both in the decrease of the fragmented DNA (compression zone [CZ], the band in which the nicked fragments of chromosomal DNAs that are unresolved in the gel migrate) and in the increase of nonfragmented DNA in the well. However, in the MXF treatment, the decrease was only observed in the fragmented DNA (Fig. 2). This difficultly in reproducibly measuring nonfragmented DNA trapped in the well and the lack of correlation between the reduction in the CZ band and the recovery of nonfragmented DNA suggest that, in the case of MXF treatment, this approach is semiquantitative.
FIG 2.
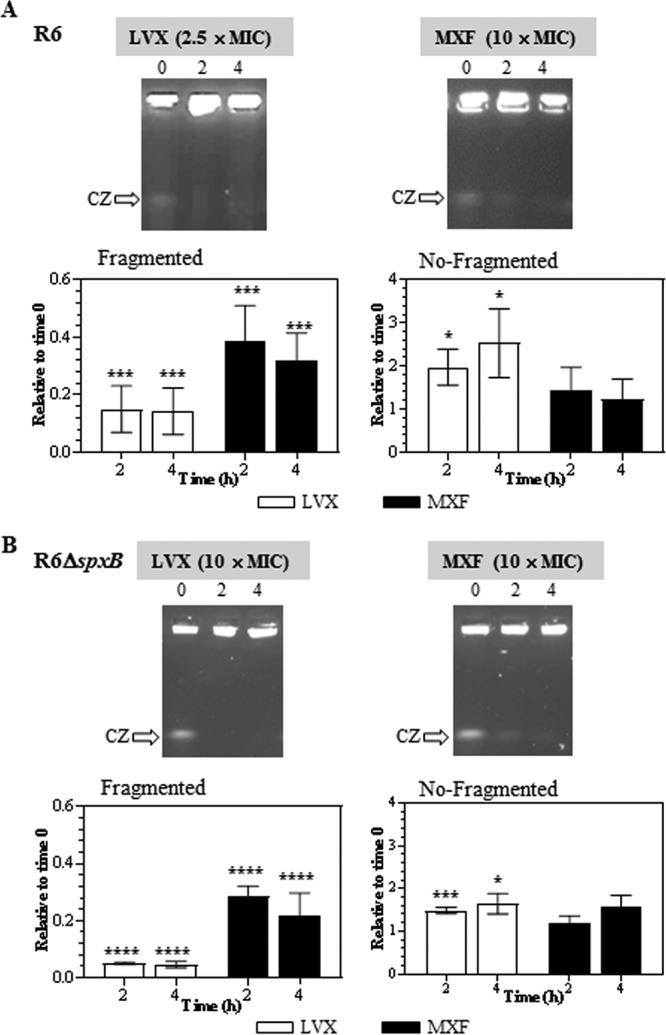
PAE and chromosome fragmentation. Cultures of S. pneumoniae R6 (A) or R6 ΔspxB (B) containing 108 CFU/ml were treated with antibiotics as described for Fig. 1. DNAs from cells taken at different times in the PAE phase were subjected to pulsed-field gel electrophoresis. CZ, compression zone, the band in which the nicked fragments of chromosomal DNAs that are unresolved in the gel migrate. Quantification of the fragmented chromosome (signal in the CZ relative to the combined signal of the lane plus well) and of the nonfragmented chromosome (signal in the well relative to the combined signal of the lane plus well) relative to time zero is shown. Fold variations (means ± SDs) from three independent replicates are presented. Student t test significances are with respect to time zero. *, P < 0.05; ***, P < 0.0001; ****, P < 0.00001.
For LVX, reductions of the CZ signal were up to 7-fold at 2 and 4 h, and the increase in the nonfragmented DNA was 2-fold (2 h) and up to 3-fold (4 h). For MXF, reductions in the CZ signals of up to 3-fold were detected at 2 and 4 h. In R6 ΔspxB, reductions in the CZ signal were up to 20-fold (LVX) and up to 3-fold (MXF) after 2 and 4 h. The increase in the nonfragmented DNA was up to 1.5-fold after 2 and 4 h of MXF treatment. Although the recovery in R6 ΔspxB after LVX treatment was higher, no differences were observed between R6 and R6 ΔspxB after MXF treatment. In both strains, recoveries after 2 h and 4 h treatment were equivalent.
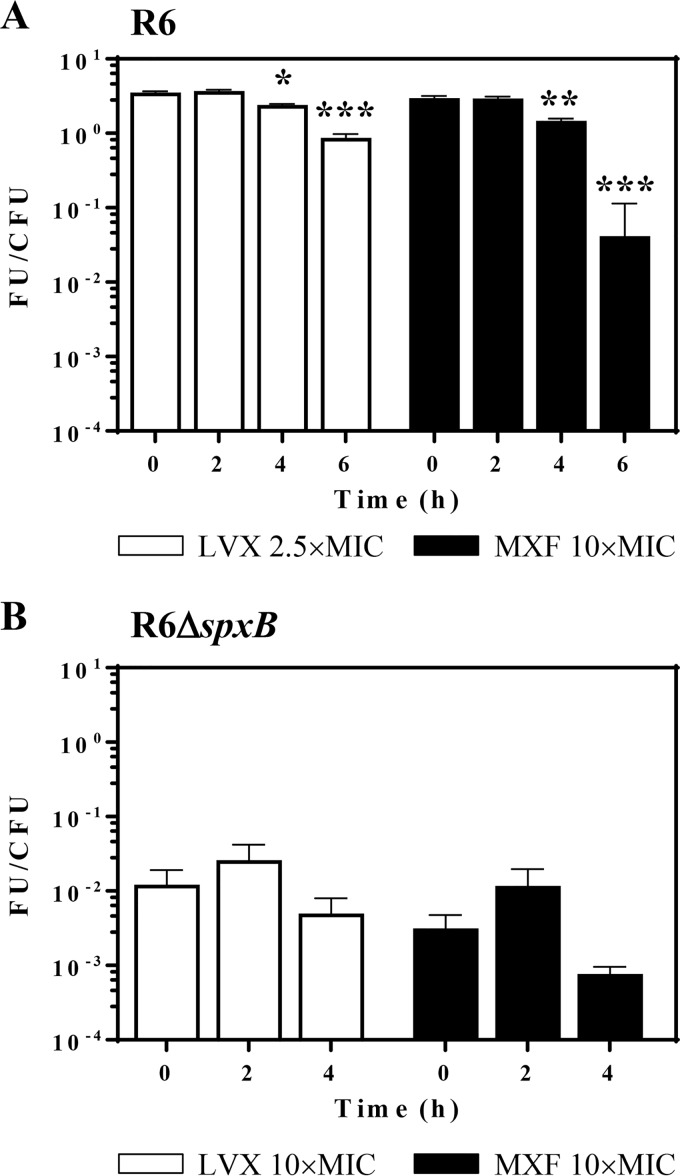
ROS production reverted at 4 and 6 h in the PAE phase according to the transcriptional alterations induced by LVX and MXF.
The effects of LVX and MXF on R6 were tested at the same concentrations used in the fragmentation assays to maintain similar PAEs for both drugs. The amounts of ROS production after 1 h treatment (time zero in Fig. 3) were similar with both drugs. While no changes were observed 2 h after drug withdrawal, decreases in ROS production were detected after 4 to 6 h of drug withdrawals. Similar decreases were observed for both FQs. After 6 h of drug withdrawal, decreases ≥400-fold for LVX and ≥800-fold for MXF were detected (Fig. 3A). In R6 ΔspxB, the intrinsic production of ROS was 3.5 × 103-fold lower than in R6, and no significant changes were observed under FQ treatment (Fig. 3A).
FIG 3.
ROS production in PAE phases. Cultures of S. pneumoniae R6 (A) or R6 ΔspxB (B) containing 108 CFU/ml were treated for 1 h with either LVX or MXF at the indicated concentrations, diluted 1,000-fold, and incubated in medium without drug. Samples were taken at the indicated times, and the total ROS content of the bacteria was measured as described in Materials and Methods. FU, fluorescence units; values were divided by the number of viable cells (CFU). Results (means ± SDs) from three independent replicates are presented. Student t test significances are with respect time zero. *, P < 0.05; **, P < 0.001; ***, P < 0.0001.
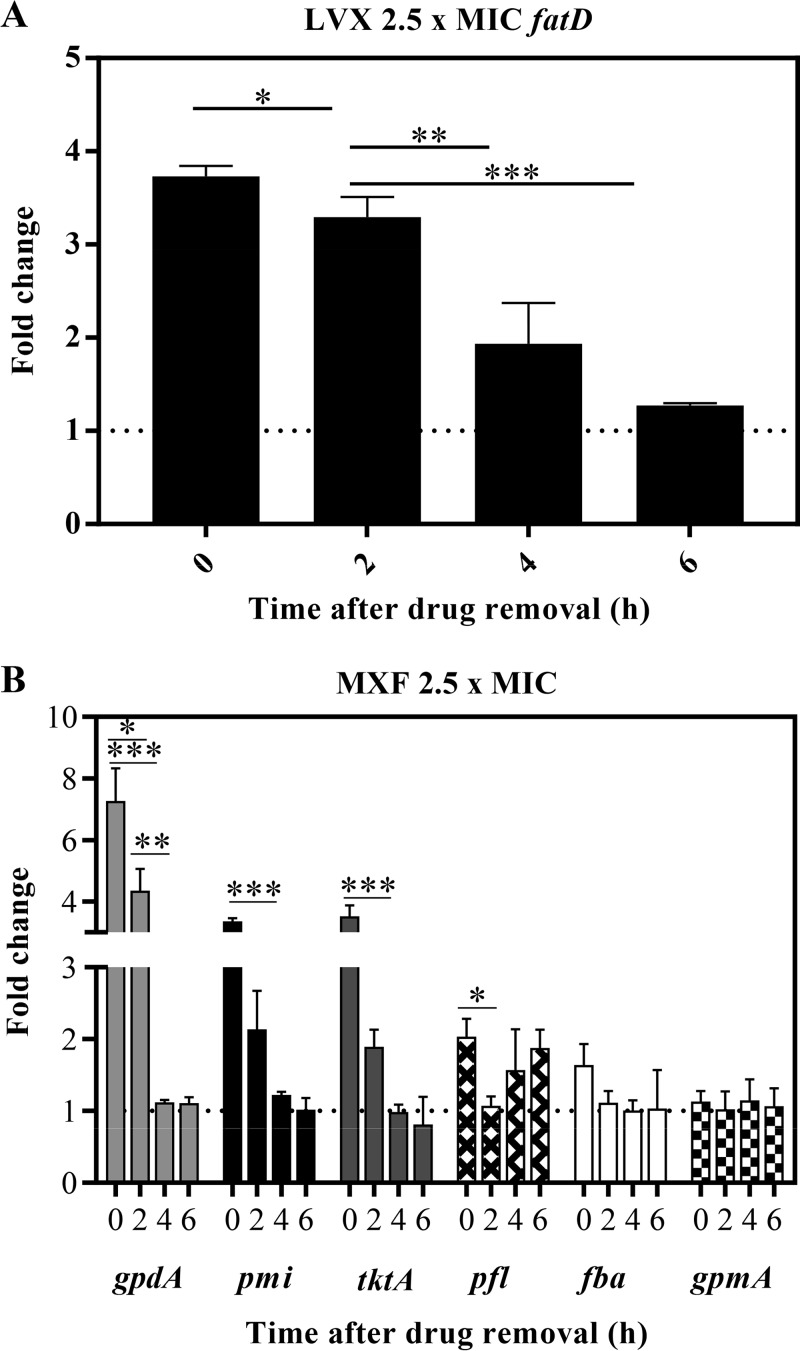
Accordingly, after 4 to 6 h of LVX withdrawal, the induced upregulation of the transcription of the fatCDEB operon, involved in the introduction of iron in the bacteria, was reduced up to 2- to 3-fold (Fig. 4A), and the MXF-induced upregulation of several genes encoding enzymes involved in the production of pyruvate was reduced 3- to 7-fold in the PAE phase (Fig. 4B). In the MXF treatment, 6 genes were chosen to test transcription by reverse transcription-quantitative PCR (qRT-PCR). These genes were previously shown to be upregulated by MXF (21). These included tktA and pmi, whose products convert mannose-6P to fructose-6P and ribulose-5P to glucose-6P, respectively. Glucose-6P is further converted to fructose-6P, the first substrate of glycolysis. After an initial increase in the transcription of these genes, a reversion was observed in the PAE phase (Fig. 4B). There were also three genes coding glycolytic enzymes (fba, gpdA, and gpmA), with a significant increase only for gpdA, and subsequent recovery was observed. Although no significant variations were observed for fba, this gene showed a behavior consistent with upregulation and recovery. The same behavior was observed for pfl, which encodes formate acetyltransferase that converts acetyl coenzyme A (acetyl-CoA) to pyruvate. The differences observed in the transcription of the chosen genes and that previously reported (21) could be due to the different MXF concentrations and times used. Nevertheless, 4 of the 6 genes analyzed showed an upregulation after 1 h treatment and further recovery during the PAE phases; the recovery was mostly observed at 4 to 6 h after FQ withdrawal.
FIG 4.
Transcriptional alterations in the PAE phases. Cultures of S. pneumoniae R6 containing 108 CFU/ml were subjected to 1 h treatment with either LVX (A) or MXF 4 (B) at 2.5× MIC. Cultures were diluted as described for Fig. 1 Samples were taken, total RNA was extracted, and cDNAs were synthetized and subjected to qRT-PCR determinations. Values were made relative to 16S rRNA genes. Fold change variations (means ± SDs) with respect to the nontreated cultures from three independent replicates are presented. *, P < 0.05; **, P < 0.01; ***, P < 0.0001 by Student t tests.
DISCUSSION
Postantibiotic effect induced by FQs is fundamental to determine optimal dosage regimens that achieve therapeutic success and avoid the emergence of resistance. In addition, knowledge of the postantibiotic recovery period is critical to understand persistence phenotypes. However, there are few studies on its mechanism and the factors on which it depends. This study related the recovery of DNA cleavage and ROS levels with its duration.
By measuring the PAE of LVX and MXF in S. pneumoniae R6 strain and its isogenic R6 ΔspxB derivative, we showed PAEs of up to 1 h longer in R6. In both strains, the resealing of DNA breaks after drug withdrawals was equivalent in time, occurring after 2 h without further decrease at later times. At equivalent LVX PAEs, 2.05 ± 0.10 h and 2.17 ± 0.15 for R6 and R6 ΔspxB, respectively, reductions in fragmented DNA, estimated as the amount of CZ species, were higher for R6 ΔspxB than for R6 (20-fold versus 7-fold). In the case of MXF, PAEs of 3.23 ± 0.45 h for R6 and 1.53 ± 0.12 h for R6 ΔspxB did not produce differences in the reductions of fragmented DNA (3-fold in both strains). These results indicate that the main factor determining the duration of PAE was not the recovery from chromosome fragmentation mediated by the FQs. However, big differences were observed in ROS production. The decrease in ROS production was detected only in R6 and after 4 and 6 h of drug withdrawal. No changes in ROS production after FQ removal were detected in R6 ΔspxB. The delayed decrease of ROS, with respect to the reduction in DNA fragmentation, could be related to the stability of this species. Production of ROS was also recently suggested as the mechanism of antibacterial and antibiofilm activity of other drugs, such as thymoquinone (24), ciprofloxacin, and meropenem (25). Previous studies also related ROS increases with longer PAEs induced by aminoglycosides in Pseudomonas aeruginosa under hyperoxic conditions (26). The photolysis of hydrogen peroxide induced significantly longer PAE in Staphylococcus aureus and Streptococcus salivarius due to the generation of hydroxyl radicals (27). Another factor to consider is the lower growth rate of R6 ΔspxB, which might also confer some protection against FQs and thereby reduce the PAE. The role of ROS in the lethality of antibiotics under anaerobic conditions has been a matter of controversy (28). Some studies denied this role (29, 30), although others clearly show a connection, especially for FQs (17, 19). The model was proposed in Escherichia coli, an aerobic bacterium. It was shown that the primary drug-target interactions stimulate the tricarboxylic acid cycle, this in turn leads to the hyperactivation of the electron transport chain that stimulates H2O2 formation. Superoxide damages Fe-S clusters of enzymes, making Fe2+ available for oxidation by the Fenton reaction. There are several differences between this model and the one we propose for S. pneumoniae (31), a facultative anaerobe that obtains energy from carbohydrate fermentation via glycolysis. The genome of this bacterium does not code for enzymes of the tricarboxylic acid cycle nor for cytochromes and heme-containing proteins involved in aerobic respiration. In addition, its F0F1-ATPase does not synthesize ATP; it instead uses ATP to pump protons out of the cell, being the main regulator of the intracellular pH (32). In our model, both LVX and MXF, by means of transcriptional alterations, stimulated the Fenton reaction by increasing the concentrations of Fe2+ and H2O2, respectively. We had shown that these effects have a low contribution to the lethality from the FQs (20, 21). In this study, we show that ROS production is the main factor determining the length of PAE. The efficacy of FQs could be increased either by elevating the levels of intracellular ferrous iron or by increasing the accumulation of ROS. This could also prolong PAE, determining longer dosing intervals, reducing adverse effects, and lowering costs while formulating a daily administration dosage.
MATERIALS AND METHODS
Postantibiotic effect.
S. pneumoniae was grown in a casein hydrolysate based medium (AGCH) with 0.2% yeast extract and 0.3% sucrose (33). MICs for R6 and R6 ΔspxB strains were 1 μg/ml for LVX and 0.06 μg/ml for MXF. PAE was calculated by measuring bacterial regrowth after antibiotic treatment by the viable plate count method (34). Cultures were grown in AGCH broth to approximately 108 CFU/ml and treated for 1 h with diverse drug concentrations in 24-well (flat bottom) polystyrene microtiter dishes. Growth controls without antibiotic were included with each experiment. After 1/1,000 dilution with fresh medium, cultures were incubated for 6 h and viable bacteria were counted every 2 h by plating on Mueller-Hinton blood agar plates. PAE was quantified with the formula PAE = T − C. It measures the time required for the viable bacteria counts to increase by 1 log10 above the counts observed immediately after washing in exposed cultures (T) compared to that in culture not exposed to antibiotic (C). A PAE of ≥0.5 h was considered significant (35).
Analysis of chromosomal fragmentation.
Pulse-field gel electrophoresis (PFGE) was used to detect chromosomal fragmentation as described previously (36). Approximately 3 × 106 cells were lysed in solid agarose inserts in a buffer containing 10 mM Tris HCl (pH 8), 1 M NaCl, 0.1 M EDTA, 0.5% Brij 58, 0.2% deoxycholate, 0.5% sarkosyl, 20 μg/ml RNaseE, and 100 μg/ml lysozyme. Inserts were treated with 1 mg/ml proteinase K and washed before placing them in a 1% low-gelling agarose (Pronadisa) gel in 0.5% TBE buffer (45 mM Tris-borate (pH 8), 1 mM EDTA). Gels were electrophoresed in a Chef-DR III system (Bio-Rad) for 20 h at 5.8 V/cm with a 0.1- to 40-s switch-time ramp at 14°C. Gels were stained with 0.5 μg/ml of ethidium bromide, and bands visualized under UV light.
Detection of ROS.
The intracellular oxidation levels were measured using dihydrorhodamine 123 dye (Sigma-Aldrich). This compound is nonfluorescent, but oxidation converts it to the fluorescent product rhodamine 123; therefore, measured fluorescence is proportional to the level of oxidation (37). Cells were grown exponentially to an optical density at 620 nm (OD620) of 0.4 before fluoroquinolones were added. Samples (1 ml) were collected and processed as previously described (20). Briefly, cells were washed with phosphate-buffered saline (PBS; pH 7.2). They were incubated in PBS with 2.5 μg/ml of dihydrorhodamine 123 for 30 min at 37°C in the dark. Cells were then washed, and fluorescence was measured using a Tecan Infinite 2000 device with a filter for excitation and emission wavelengths of 485 nm and 535 nm, respectively. Results, expressed as relative fluorescence units (RFU), were normalized according to the number of live cells at each time point.
RNA extraction and qRT-PCR experiments.
Total RNA was extracted with an RNeasy Midi kit (Qiagen) according to the manufacturer’s instructions. Three DNase treatments were performed to avoid any DNA contamination. Synthesis of cDNA was carried out in 20-μl reaction mixtures containing 3 μg of RNA, 0.5 mM each deoxynucleotide triphosphate, 2 pmol of random hexanucleotides, 40 U of RNaseOUT RNase inhibitor (Invitrogen), 200 U of SuperScript IV reverse transcriptase (Invitrogen), and 1 U of RNase H (Invitrogen) in the buffer recommended by the manufacturer. These cDNAs were subjected to qRT-PCR (chromo 4; Bio-Rad) in 20-μl reaction mixtures containing 2 μl of cDNA, 0.4 μM each specific primer, and 10 μl of LigthCycler FastStart Universal A SYBR green master mix (Roche). Amplification was achieved with 42 cycles of a three-segment program: denaturation (30 s at 94°C), annealing (30 s at 45 to 56°C), and elongation (30 s at 68°C). To normalize the three independent cDNA replicates, values were divided by those obtained from the amplification of internal fragments of 16S rRNA genes. Oligonucleotide pairs used were as follows: 5′-CCATCGGCTAGTCTGACCCAAAA-3′ and 5′-ATCCCAATCAGAGGCAACATCCAC-3′ (fatD); 5′-TTGGCTAAAGAAGTTGTTGAAAAA-3′ and 5′-AAGGGCCGTGGATGTTACC-3′ (fba); 5′-GAAAAACAAACCGTCGCCGTCTT-3′ and 5′-ACAACAAACAAAATCGCATCCACA-3′ (gpdA); 5′-AGGGAACAGATCTTGCTACTTTG-3′ and 5′-CTTCTTTTGACTTGGCATTGTGAC-3′ (pmi); 5′-GAAACGTGCTATCAAAACAACTAA-3′ and 5′-TGCTCATCATCACGGTCCATA-3′ (gpmA); 5′-GGGCAGAGGCTCCGAAGGTA-3′ and 5′-TGGCTAGTCAAGGCGAAAAA-3′ (tktA); 5′-ACAGAGCGTTCACTTCACATCA-3′ and 5′-AGCTGGGTCTGGTTCGTATCC-3′ (pfl).
ACKNOWLEDGMENT
This study was supported by grant BIO2017-82951-R from Plan Nacional de I+D+I of the Ministry of Economy and Competitiveness.
REFERENCES
- 1.World Health Organization. 2007. Pneumococcal conjugate vaccine for childhood immunization–WHO position paper. Wkly Epidemiol Rec 82:93–104. (In French.) [PubMed] [Google Scholar]
- 2.Jacobs MR, Felmingham D, Appelbaum PC, Grüneberg RN, Alexander Project Group. 2003. The Alexander project 1998–2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J Antimicrob Chemother 52:229–246. doi: 10.1093/jac/dkg321. [DOI] [PubMed] [Google Scholar]
- 3.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, Dowell SF, File TM Jr, Musher DM, Niederman MS, Torres A, Whitney CG. 2007. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 44 Suppl 2:S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riedel S, Beekmann SE, Heilmann KP, Richter SS, García-de-Lomas J, Ferech M, Goosens H, Doern GV. 2007. Antimicrobial use in Europe and antimicrobial resistance in Streptococcus pneumoniae. Eur J Clin Microbiol Infect Dis 26:485–490. doi: 10.1007/s10096-007-0321-5. [DOI] [PubMed] [Google Scholar]
- 5.Domenech A, Tirado-Vélez JM, Fenoll A, Ardanuy C, Yuste J, Liñares J, de la Campa AG. 2014. Fluoroquinolone-resistant pneumococci: dynamics of serotypes and clones in Spain in 2012 compared with those from 2002 and 2006. Antimicrob Agents Chemother 58:2393–2399. doi: 10.1128/AAC.02669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adam HJ, Hoban DJ, Gin AS, Zhanel GG. 2009. Association between fluoroquinolone usage and a dramatic rise in ciprofloxacin-resistant Streptococcus pneumoniae in Canada, 1997–2006. Int J Antimicrob Agents 34:82–85. doi: 10.1016/j.ijantimicag.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Ip M, Chau SL, Chi F, Cheuk ES, Ma H, Lai RW, Chan PK. 2007. Longitudinally tracking of fluoroquinolone resistance and its determinants in penicillin-susceptible and -nonsusceptible Streptococcus pneumoniae isolates in Hong Kong, 2000 to 2005. Antimicrob Agents Chemother 51:2192–2194. doi: 10.1128/AAC.00139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen DK, McGeer A, de Azavedo JC, Low DE. 1999. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N Engl J Med 341:233–239. doi: 10.1056/NEJM199907223410403. [DOI] [PubMed] [Google Scholar]
- 9.MacKenzie FM, Gould IM. 1993. The post-antibiotic effect. J Antimicrob Chemother 32:519–537. doi: 10.1093/jac/32.4.519. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs PC, Barry AL, Brown SD. 1997. Streptococcus pneumoniae killing rate and post-antibiotic effect of levofloxacin and ciprofloxacin. J Chemother 9:391–393. doi: 10.1179/joc.1997.9.6.391. [DOI] [PubMed] [Google Scholar]
- 11.Licata L, Smith CE, Goldschmidt RM, Barrett JF, Frosco M. 1997. Comparison of the postantibiotic and postantibiotic sub-MIC effects of levofloxacin and ciprofloxacin on Staphylococcus aureus and Streptococcus pneumoniae. Antimicrob Agents Chemother 41:950–955. doi: 10.1128/AAC.41.5.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boswell FJ, Andrews JM, Wise R, Dalhoff A. 1999. Bactericidal properties of moxifloxacin and post-antibiotic effect. J Antimicrob Chemother 43 Suppl B:43–49. doi: 10.1093/jac/43.suppl_2.43. [DOI] [PubMed] [Google Scholar]
- 13.Spangler SK, Lin G, Jacobs MR, Appelbaum PC. 1998. Postantibiotic effect and postantibiotic sub-MIC effect of levofloxacin compared to those of ofloxacin, ciprofloxacin, erythromycin, azithromycin, and clarithromycin against 20 pneumococci. Antimicrob Agents Chemother 42:1253–1255. doi: 10.1128/AAC.42.5.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies TA, Kelly LM, Pankuch GA, Credito KL, Jacobs MR, Appelbaum PC. 2000. Antipneumococcal activities of gemifloxacin compared to those of nine other agents. Antimicrob Agents Chemother 44:304–310. doi: 10.1128/AAC.44.2.304-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Champoux JJ. 2001. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 16.Drlica K, Malik M, Kerns RJ, Zhao X. 2008. Quinolone-mediated bacterial death. Antimicrob Agents Chemother 52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. 2007. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol 3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 20.Ferrándiz MJ, de la Campa AG. 2014. The fluoroquinolone levofloxacin triggers the transcriptional activation of iron transport genes that contribute to cell death in Streptococcus pneumoniae. Antimicrob Agents Chemother 58:247–257. doi: 10.1128/AAC.01706-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrándiz MJ, Martín-Galiano AJ, Arnanz C, Zimmerman T, de la Campa AG. 2016. Reactive oxygen species contribute to the bactericidal effects of the fluoroquinolone moxifloxacin in Streptococcus pneumoniae. Antimicrob Agents Chemother 60:409–417. doi: 10.1128/AAC.02299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idanpaan-Heikkila I, Rosenow C, Masure HR. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol 19:803–813. doi: 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 23.Pericone CD, Overweg K, Hermans PWM, Weiser JN. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun 68:3990–3997. doi: 10.1128/IAI.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goel S, Mishra P. 2018. Thymoquinone inhibits biofilm formation and has selective antibacterial activity due to ROS generation. Appl Microbiol Biotechnol 102:1955–1967. doi: 10.1007/s00253-018-8736-8. [DOI] [PubMed] [Google Scholar]
- 25.Van Acker H, Gielis J, Acke M, Cools F, Cos P, Coenye T. 2016. The role of reactive oxygen species in antibiotic-induced cell death in Burkholderia cepacia complex bacteria. PLoS One 11:e0159837. doi: 10.1371/journal.pone.0159837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park MK, Myers RA, Marzella L. 1993. Hyperoxia and prolongation of aminoglycoside-induced postantibiotic effect in Pseudomonas aeruginosa: role of reactive oxygen species. Antimicrob Agents Chemother 37:120–122. doi: 10.1128/aac.37.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Odashima Y, Nakamura K, Ikai H, Kanno T, Meirelles L, Sasaki K, Niwano Y. 2014. Postantibiotic effect of disinfection treatment by photolysis of hydrogen peroxide. J Chemother 26:92–100. doi: 10.1179/1973947813Y.0000000114. [DOI] [PubMed] [Google Scholar]
- 28.Dwyer DJ, Collins JJ, Walker GC. 2015. Unraveling the physiological complexities of antibiotic lethality. Annu Rev Pharmacol Toxicol 55:313–332. doi: 10.1146/annurev-pharmtox-010814-124712. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 31.de la Campa AG, Ferrándiz MJ, Martín-Galiano AJ, García MT, Tirado-Vélez JM. 2017. The transcriptome of Streptococcus pneumoniae induced by local and global changes in supercoiling. Front Microbiol 8:1447. doi: 10.3389/fmicb.2017.01447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martín-Galiano AJ, Ferrándiz MJ, de la Campa AG. 2001. The promoter of the operon encoding the F0F1 ATPase of Streptococcus pneumoniae is inducible by pH. Mol Microbiol 41:1327–1338. doi: 10.1046/j.1365-2958.2001.02597.x. [DOI] [PubMed] [Google Scholar]
- 33.Lacks SA, López P, Greenberg B, Espinosa M. 1986. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J Mol Biol 192:753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- 34.Lowdin E, Odenholt-Tornqvist I, Bengtsson S, Cars O. 1993. A new method to determine postantibiotic effect and effects of subinhibitory antibiotic concentrations. Antimicrob Agents Chemother 37:2200–2205. doi: 10.1128/AAC.37.10.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.García Y, Gómez MJ, Ramos MC, Gómez-Lus ML, Prieto J. 1998. The postantibiotic effect of amoxicillin/clavulanic acid on Streptococcus pneumoniae strains of different serotypes and penicillin sensitivity. Rev Esp Quimioter 11:157–160. (In Spanish.) [PubMed] [Google Scholar]
- 36.Khan SR, Kuzminov A. 2013. Trapping and breaking of in vivo nicked DNA during pulse field gel electrophoresis. Anal Biochem 443:269–281. doi: 10.1016/j.ab.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeom J, Imlay JA, Park W. 2010. Iron homeostasis affects antibiotic-mediated cell death in Pseudomonas species. J Biol Chem 285:22689–22695. doi: 10.1074/jbc.M110.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]