Abstract
Purpose
The goals of this study were to assess the performance of a novel lesion segmentation tool for longitudinal analyses, as well as to validate the generated lesion progression map between two time points using conventional and non-conventional MR sequences.
Material and methods
The lesion segmentation approach was evaluated with (LeMan-PV) and without (LeMan) the partial volume framework using “conventional” and “non-conventional” MR imaging in a two-year follow-up prospective study of 32 early RRMS patients. Manual segmentations of new, enlarged, shrunken, and stable lesions were used to evaluate the performance of the method variants. The true positive rate was estimated for those lesion evolutions in both white matter and cortex. The number of false positives was compared with two strategies for longitudinal analyses. New lesion tissue volume estimation was evaluated using Bland-Altman plots. Wilcoxon signed-rank test was used to evaluate the different setups.
Results
The best median of the true positive rate was obtained using LeMan-PV with non-conventional sequences (P < .05): 87%, 87%, 100%, 83%, for new, enlarged, shrunken, and stable WM lesions, and 50%, 60%, 50%, 80%, for new, enlarged, shrunken, and stable cortical lesions, respectively. Most of the missed lesions were below the mean lesion size in each category. Lesion progression maps presented a median of 0 false positives (range:0–9) and the partial volume framework improved the volume estimation of new lesion tissue.
Conclusion
LeMan-PV exhibited the best performance in the detection of new, enlarged, shrunken and stable WM lesions. The method showed lower performance in the detection of cortical lesions, likely due to their low occurrence, small size and low contrast with respect to surrounding tissues. The proposed lesion progression map might be useful in clinical trials or clinical routine.
Keywords: Cortical lesions, Multiple sclerosis, Longitudinal analysis, Lesion segmentation, Non-conventional MRI
Highlights
-
•
DIR and MP2RAGE sequences improve automated longitudinal assessment of MS lesions.
-
•
Partial volume estimation improves lesion segmentation in a longitudinal scenario.
-
•
Lesion progression maps simplify the assessment of disease activity and treatment response.
1. Introduction
Magnetic resonance imaging (MRI) is used for both Multiple Sclerosis (MS) diagnosis (Thompson et al., 2018), and longitudinal evaluation of disease progression and therapy response (Rovira et al., 2015).
Manual detection and segmentation of longitudinal changes in lesions, especially of new and enlarged lesions, including both white matter (WM) and cortical lesions, is challenging, time-consuming and often characterized by substantial intra- and inter-variability. Although several automated MS lesion segmentation methods have been proposed in the past 25 years, their application to time series analyses to study lesion evolution or disease progression is less frequent (Carass et al., 2017). Indeed, detection of new lesions, particularly of small size, or small enlarged areas is challenging, mainly due to partial volume (PV) effects unavoidable at conventional image spatial resolutions, as well as to low contrast-to-noise (Fartaria et al., 2017). Moreover, most of the automated methods were designed to detect lesions in the WM (Garcia-Lorenzo et al., 2013) and have rarely been applied to cortical lesions (Fartaria et al., 2016).
State-of-the-art methods on automated longitudinal analyses of MS progression can be classified in two main groups: i) lesion segmentation-based methods; ii) change detection-based methods (Lladó et al., 2012a). Lesion segmentation-based methods aim at segmenting MS lesions cross-sectionally in successive MR images and then to compute volumetric differences between the two time points (Ettinger et al., 1994; Guttmann Charles et al., 1999; Solomon and Sood, 2004; Aït-Ali et al., 2005; Köhler et al., 2019). Although any kind of cross-sectional approach is adaptable to segmentation-based methods, only few groups have evaluated them in longitudinal scenarios (Carass et al., 2017; Garcia-Lorenzo et al., 2013; Lladó et al., 2012b). This evaluation was mainly based on longitudinal volume correlation between times points within one subject (Carass et al., 2017), lacking the assessment on detection of new and enlarged lesions (Rovira et al., 2015). Change detection-based methods rely on capturing differences in serial MR images by either using subtraction images or deformation analyses from non-rigid registration between different time points (Lladó et al., 2012a). Subtraction techniques evaluate tissue transformations through intensity variations (Lladó et al., 2012a; Moraal et al., 2010; Ganiler et al., 2014; Marco et al., 2014; Bosc et al., 2003; Jain et al., 2016); deformation analyses, on the other hand, measure the mass effect on surrounding tissues due to the deformation caused by lesion enlargement or shrinkage (Rey et al., 2002). Although these methods ii) are sensitive to changes in the brain, they do not provide information about stable lesions and often show a considerable number of false positives due to noise, registration errors, or image artefacts.
In this work, we propose a strategy for longitudinal analysis of MS lesions based on a combination of lesion-segmentation and change-detection based approaches. Thus, the purpose of our study was to assess the performance of the partial-volume aware lesion segmentation tool, LeMan-PV, for longitudinal analyses (Fartaria et al., 2016; Fartaria et al., 2017; Fartaria et al., 2018), to evaluate the value of non-conventional MR sequences, and to propose a method for generation of a lesion progression map between two time points.
2. Material and methods
2.1. Subjects
Thirty-two RRMS patients underwent MR imaging at baseline (first time point, TP1, as previously reported in (Fartaria et al., 2016; Fartaria et al., 2018)) and after two years follow-up (second time point, TP2; follow-up time: 21.4 ± 2.5 months, range 16–27 months) in this prospective study. This patient cohort consisted of 18 females and 14 males with age 35 ± 9.9 years (mean ± standard deviation, range 20–60 years) at TP1. All patients were < 6 years from initial symptoms (32 ± 21.6 months, range 3–70 months) and disease diagnosis (26 ± 19.3 months, range 0–59 months) at TP1. Thirty patients has been under immunomodulatory treatment (high dosage interferon beta or fingolimod) for at least 3 months. No patient had received corticosteroid therapy within the 3 months preceding the enrolment. No significant evolution of the expanded disability status scale (EDSS) was measured over the two years (median EDSS: 1.5, range 1.5 to 2.5). MR images from twenty age-matched (33 ± 9.3 years, range 20–60 years) healthy controls, 12 females and 8 males, were also acquired at TP1 (see Table 1).
Table 1.
Study population: demographic and clinical information. SD: standard-deviation.
| Patients (time point 1) | Patients (time point 2) | Healthy controls (time point 1) | |
|---|---|---|---|
| Demographics | |||
| # Subjects | 32 | 32 | 20 |
| Age: mean ± SD | 35 ± 9.9 | 37 ± 10.0 | 33 ± 9.3 |
| (range) [years] | (20–60) | (22–62) | (20–60) |
| Sex (female/male) | 18/14 | 18/14 | 12/8 |
| Disease | |||
| 1st relapse: mean ± SD | 32 ± 21.6 | 53 ± 21.2 | --- |
| (range) [months] | (3–70) | (25–90) | |
| Diagnosis: mean ± SD | 26 ± 19.3 | 48 ± 18.9 | --- |
| (range) [months] | (0–59) | (20–81) | |
| EDSS: median (range) | 1.5 (1.5–2.5) | 1.5 (1.5–2.5) | --- |
| Follow-up | |||
| Time: mean ± SD | 21.4 ± 2.5 | – | |
| (range) [months] | (16–27) | ||
The imaging was performed between January 2012 and November 2014. The study was approved by the Ethics Committee of our institution, and all the subjects gave written informed consent prior to participation.
2.2. MRI
The MRI acquisitions were performed on a 3 T MRI (MAGNETOM Trio a Tim system, Siemens Healthcare, Erlangen, Germany) equipped with a 32-channel head coil. The imaging protocol consisted of MP-RAGE, MP2RAGE, 3D FLAIR, and 3D DIR with a spatial resolution of 1 × 1 × 1.2 mm3 (see Table 2). In this work, we considered MPRAGE and FLAIR as “conventional” sequences once they are currently established in clinical protocols. MP2RAGE and DIR were considered as “non-conventional” sequences since they are mainly used for research purposes. From the MP2RAGE acquisitions, we used only the homogeneous T1-weigthed contrast (“uniform image”) (Marques et al., 2010).
Table 2.
MRI protocol.
| MPRAGE | MP2RAGE | 3D FLAIR | DIR | |
|---|---|---|---|---|
| VS, mm3 | 1.0 × 1.0 × 1.2 | 1.0 × 1.0 × 1.2 | 1.0 × 1.0 × 1.2 | 1.0 × 1.0 × 1.2 |
| FOV, mm3 | 256 × 240 × 160 | 256 × 240 × 176 | 256 × 240 × 176 | 256 × 240 × 160 |
| AT, min | 5:12 | 8:22 | 6:27 | 12:52 |
| TR, ms | 2300 | 5000 | 5000 | 10,000 |
| TE, ms | 2.98 | 2.89 | 394 | 218 |
| TI, ms | 900 | 700/2500 | 1800 | 450/3650 |
2.3. Manual segmentation
Two experts - one radiologist and one neurologist, 10 and 6 years of experience, respectively - identified and marked fully blinded WM and cortical lesions on FLAIR, MP2RAGE, and DIR by consensus in two separate sessions (one for patients TP1 together with the healthy controls cohort, and one for patients TP2). Cortical lesions were identified according to the criteria defined by the recommendations by Geurts et al. (2011). A trained technician delineated each identified lesion in each image contrast. Lesions that were marked only by one rater were re-evaluated by both experts and excluded or included according to a final consensus. Finally, the delineations obtained from the different contrasts were registered to MP2RAGE space and merged into a single mask which we considered the most comprehensive representation of lesion load per patient in both time points (Kober et al., 2012; Bonnier et al., 2014; Bonnier et al., 2015). Based on this single mask per time point, both time points were compared and lesions classified, by the same technician, in one of the following four categories (based on criteria by (Moraal et al., 2010)):
-
•
New: lesion identifiable on the TP2 image but not on the TP1 image;
-
•
Enlarged: lesion diameter in TP2 increased by at least 50% with respect to TP1;
-
•
Shrunken: lesion diameter in TP2 decreased by at least 50% with respect to TP1;
-
•
Stable: any lesion that does not fall into any of the above criteria.
The lesion masks with longitudinal information (longitudinal lesion mask) were finally reviewed and corrected (if necessary) by consensus from the same experts and considered as reference. Lesion volume and count per each lesion type were computed for WM and cortical lesions based on the longitudinal lesion mask. Minimum lesion size was derived from the median lesion size found in the cohort of healthy controls (Grahl et al., 2017). Disease activity was measured according to the presence or absence of new or enlarged T2 hyperintense lesions (Giovannoni et al., 2017; Stangel et al., 2015). In this particular study, the disease was considered active in patients with at least one new or enlarged T2 hyperintense lesion. Patients with no new and/or enlarged lesion were considered patients with inactive disease.
2.4. Proposed methods
We assessed the following methods:
-
•
LeMan: a supervised approach based on the kNN classifier trained using a set of features obtained from the images and atlas-based prior probability maps of the two brain tissues (WM and GM), and CSF (Fartaria et al., 2016).
-
•
LeMan-PV: a Bayesian partial volume estimation algorithm (Fartaria et al., 2018), where spatial constraints for GM and lesions are included to drive the segmentation (Fartaria et al., 2017). The spatial constraint for GM is an atlas-based probability map, and the spatial constraint for lesions is derived a priori from the supervised approach described above.
2.5. Lesion progression map
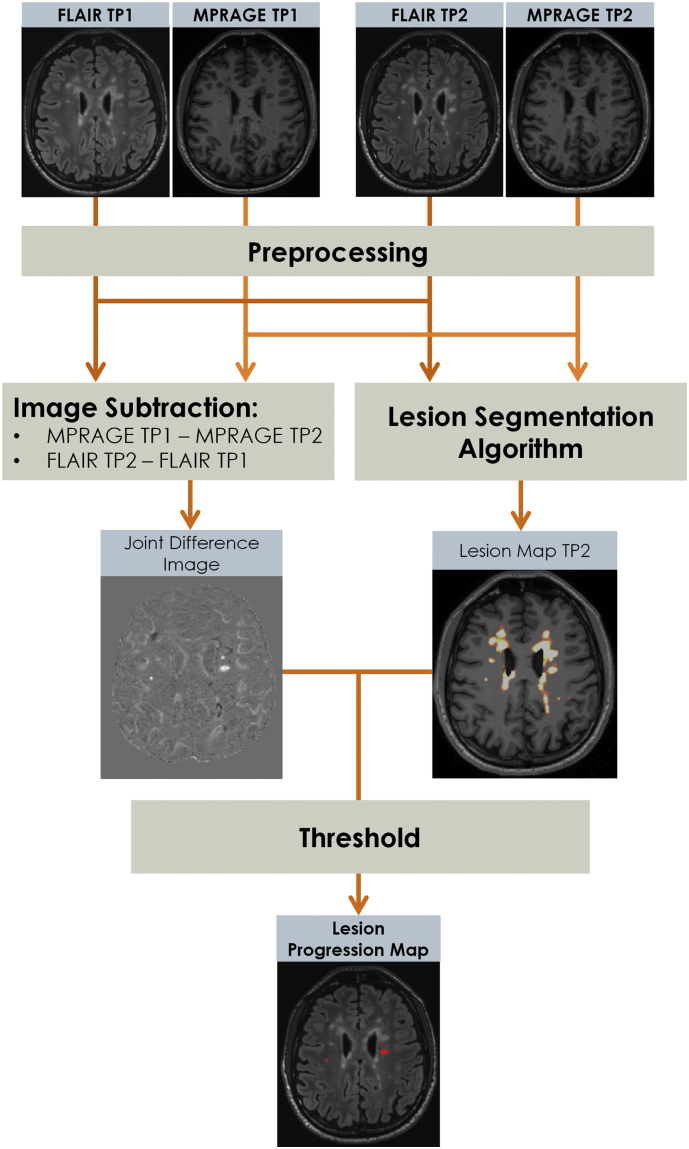
In addition, we proposed the lesion progression map, a binary mask that shows areas of new lesional tissue located in the WM and cortex, i.e. new WM and cortical lesions and enlarged parts of existing WM and cortical lesions. This longitudinal extension that can be applied to both LeMan and LeMan-PV, consisting of the following steps:
-
1.
TP1 and TP2 images previously submitted to pre-processing steps including N4 bias field correction and intensity normalization (see (Fartaria et al., 2016) for details) from each contrast were rigidly registered to MP-RAGE TP2 (Klein et al., 2010).
-
2.
[TP2] – [TP1] difference images were computed for FLAIR and DIR images. [TP1] – [TP2] difference images were computed for MPRAGE and MP2RAGE images.
-
3.
A “joint difference image” was obtained by summing up and normalizing by the maximum value the available difference images. In this study FLAIR/MPRAGE and DIR/FLAIR/MP2RAGE joint difference images were obtained for the “conventional” and “non-conventional” protocols, respectively.
-
4.
Lesion concentration maps (LeMan-PV output) or binary lesion masks (LeMan output) from TP2 were overlaid on the “joint difference image”. Voxels identified as lesions in the concentration maps or binary lesion masks that have an intensity in the joint difference image higher than a certain threshold were considered as new lesion tissue. The threshold was automatically set as the value that maximized the Dice similarity coefficient between the outcome of the thresholding operation and the manual segmentations.
-
5.
As a final step, voxels classified as new lesional tissue were binarized and a lesion progression map containing new and enlarged fractions of lesions was created.
An overview of this extension of the pipeline using the “conventional” imaging set is presented in Fig. 1. The total processing time per case, including the computation of the lesion progression map, was approximately seven minutes on a computer with a 3.40 GHz processor and 16.0 GB RAM.
Fig. 1.
Schematic diagram representing the computation of the lesion progression map (mask of new and enlarged fractions of lesions) using the “conventional” protocol: MPRAGE and 3D FLAIR. The pre-processing step relies on registration, bias field correction and intensity normalization as described in (Fartaria et al., 2016). The segmentation algorithm is our proposed partial volume aware algorithm that outputs the lesion concentration map (see (Fartaria et al., 2017; Fartaria et al., 2018) for details).
2.6. Evaluation of lesion detection
As lesion count is the main MRI biomarker used for MS diagnosis (Rovira et al., 2015), the performance of the algorithm was first evaluated in terms of lesion detection by computing the true positive rate (TPR) and the number of false positives (FP). TPR is thereby defined as the ratio of automatically detected lesions that overlap with lesions in the reference, and the total number of lesions in the reference (Styner et al., 2008). TPR was estimated for WM and cortical lesions for each longitudinal lesion category (new, enlarged, shrunken, and stable) for LeMan and LeMan-PV using “conventional” and “non-conventional” MRI contrasts. The number and the size of missed lesions were computed in each longitudinal lesion class.
In addition, we computed the number of FP in the lesion progression map to evaluate the misclassifications of new and enlarged lesions, since in clinical follow-up there is a low tolerance for misclassifications of this particular type of lesions.
2.7. Comparison of lesion progression map and other quantification approaches
To evaluate the gain of using the proposed extension of the algorithm, we compared the number of FP from the lesion progression map with two commonly used strategies to compare two different time points. The first strategy consisted of a simple comparison of LeMan-PV concentration maps in TP1 and TP2 (“segmentation-based strategy”), computing new and enlarged lesions according to lesion volume changes between the two time points (Ettinger et al., 1994). The second comparison strategy was inspired by Ganiler's work (Ganiler et al., 2014) and consisted of applying a threshold on the joint difference image to detect new and enlarged fractions of enlarged lesions. It should be noted that both strategies use LeMan-PV as the underlying segmentation algorithm, but derive the new/enlarged lesion masks differently. Doing so allowed us to evaluate the added benefit of the proposed extension of the LeMan-PV algorithm.
2.8. Evaluation of volume estimation
Lesion volume estimation was evaluated for new and enlarged fractions of lesions by a Bland-Altman plot. This plot represents the agreement between manual and automatic segmentations of new lesional tissue, showing the performance of the proposed approach for the assessment of disease activity.
2.9. Statistical analyses
TPRs for each longitudinal lesion category were compared for LeMan and LeMan-PV and for the “conventional” and “non-conventional” sets of sequences using the Wilcoxon signed-rank test. The goal of this comparison was to evaluate the advantage of modelling partial volume and using non-conventional MRI on WM and cortical lesion detection in a longitudinal scenario.
Wilcoxon signed-rank test was also used to compare the FP obtained from the proposed approach and from the other strategies used in the literature to compute new and enlarged lesions.
The significant variables were identified with P values below 0.05.
3. Results
3.1. Manual segmentation
Only WM lesions were found in the healthy control cohort. The median of WM lesion volume in this cohort was 0.006 ml (range 0.0012 to 0.1032 ml), which was used to define the minimum lesion size in the patient cohort for both WM and cortical lesions. Consequently, lesions with volumes lower than 0.006 ml (approximately 5 voxels with the image resolution used here) were excluded from the masks and not considered for evaluation.
Fig. 2 shows the percentage (y-axis) and the number of new, enlarged, shrunken, and stable WM (orange numbers) and cortical (blue numbers) lesions in our follow-up cohort of patients. Overall, stable WM lesions were more frequent (total number of lesions/average volume: 691/0.1898 ml) followed by stable cortical lesions, enlarged and new WM lesions (281/0.1506 ml, 169/0.1733 ml, and 60/0.0737 ml, respectively). Enlarged and new cortical lesions (40/0.2661 ml, 16/0.0416 ml), and shrunken lesions (41/0.0821 ml, and 2/0.102 ml for WM and cortical lesions, respectively) were less frequent in our cohort.
Fig. 2.
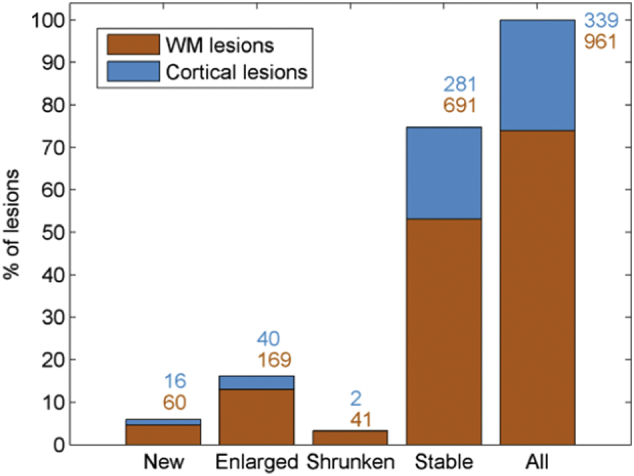
Stacked bar plot of longitudinal manual segmentation results of WM (in orange) and cortical lesion (in blue) load, for all types of lesions and for each lesion category: new, enlarged, shrunken and stable. Numbers next to the bars represent the number of WM (button number) and cortical (top number) lesions in our cohort for each lesion category. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Twenty-three patients appeared to show a positive disease activity with a median ± median absolute deviation of 0.78 ± 0.67 ml and 6 ± 5 for volume and count of new and enlarged lesions, respectively.
3.2. Lesion detection and false positive rate
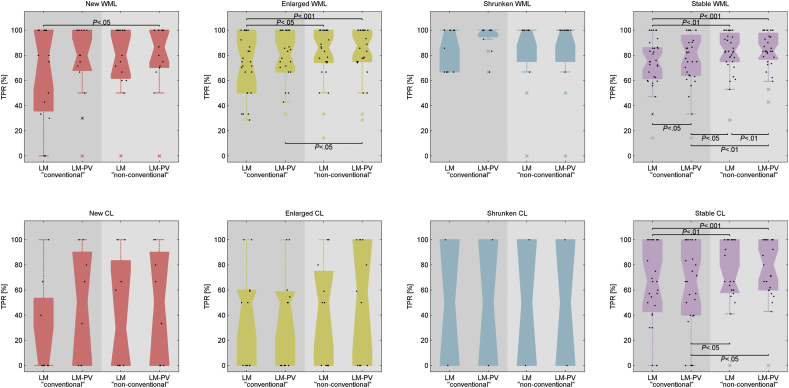
Except for the shrunken category, detection of WM lesions improved significantly when the partial volume estimation algorithm (LeMan-PV) was used with the “non-conventional” protocol (P < .001, Fig. 3) reaching the best TPRs in each longitudinal lesion category (median TPR of 87%, 87%, 100%, and 83% for new, enlarged, shrunken, and stable lesions, respectively).
Fig. 3.
Boxplots of the TPR for all patients for white matter (WML) and cortical (CL) lesions across different longitudinal categories of lesions: new (red), enlarged (yellow), shrunken (blue), and stable (pink). The x-axis labelled different configurations of the lesion segmentation approach: LeMan (LM, approach without partial volume estimation); LeMan-PV (LM-PV, approach with partial volume estimation); Results using the “conventional”, and “non-conventional” imaging sets are present in the darker and lighter grey background, respectively. The crosses in the plots represent outliers in our cohort. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Although the TPR improved when partial volume was estimated, the results were not significantly different for new and enlarged lesions (P = .25).
Except for stable cortical lesions, TPR did not appear significantly different for new, enlarged and shrunken lesions in the different configurations of the method (P ≥ .25). However, the detection of stable cortical lesions improved (P < .05) when the “non-conventional” protocol was used. As for WM lesions, the best results in cortical lesion detection were obtained using the LeMan-PV algorithm with the “non-conventional protocol (median TPR of 50%, 60%, 50%, 80%, for new, enlarged, shrunken, and stable lesions, respectively, see Fig. 3).
Overall, the configuration that provided the best TPR (LeMan-PV with “non-conventional” protocol) showed that missed lesions are much smaller with respect to the mean lesion size for each longitudinal category (see Fig. 4). The mean lesion size for missed WM lesions were 0.0240 ml, 0.0282 ml, 0.0300 ml, and 0.0253 ml for new, enlarged, shrunken, and stable lesions, respectively. Missed cortical lesions were of a bigger mean size: 0.0365 ml, 0.0531 ml, 0.0174 ml, and 0.0323 ml for new, enlarged, shrunken, and stable lesions, respectively.
Fig. 4.
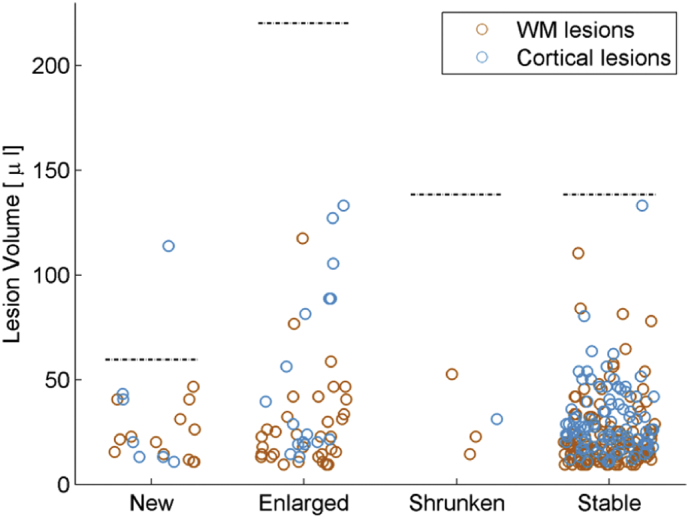
Scatter plot of the volume of missed white matter (WM, orange) and cortical (blue) lesions for each longitudinal lesion category: new, enlarged, shrunken and stable. The dashed lines represent the mean lesion volume in each category according to the ground truth. These results were computed using the configuration that provided the best true positive rate: LeMan-PV with “non-conventional” protocol. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
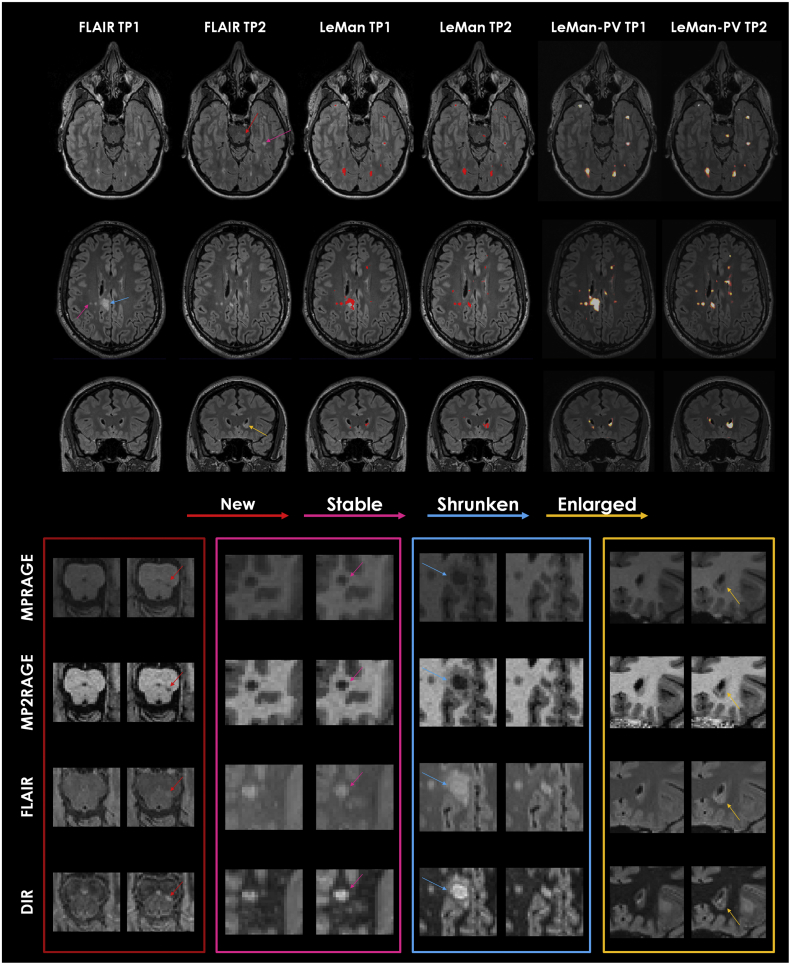
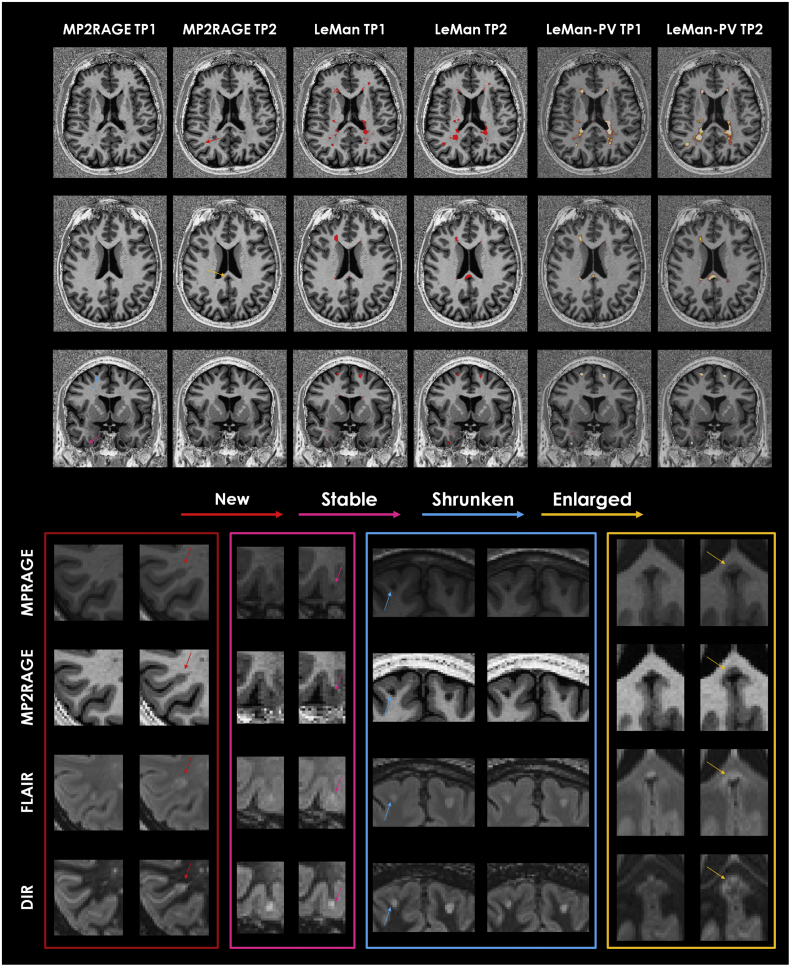
Examples of WM and cortical lesion segmentations from LeMan and LeMan-PV obtained using the “conventional” and “non-conventional” protocols for the different longitudinal lesion categories are presented in Fig. 5, Fig. 6.
Fig. 5.
From top to bottom: Axial FLAIR image in a 34-year-old woman; Axial FLAIR image in a 32-year-old man; Coronal FLAIR image in a 39-year-old woman. All cases are relapsing-remitting MS. FLAIR images are shown for the registered time point 1 (TP1) and the respective time point 2 (TP2). Results from algorithm before (LeMan) and after (LeMan-PV) applying the partial volume model using the conventional protocol are shown in the 3rd-4th and 5th–6th columns, respectively. The output of Leman is a binary mask and the output of LeMan-PV is a concentration map. Examples of new, stable, shrunken, and enlarged WM lesions are indicated with red, pink, blue, and yellow arrows, respectively. Patches of the zoomed area of the different lesion categories in each image contrast (MPRAGE, MP2RAGE, FLAIR, and DIR) are shown at the bottom. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
From top to bottom: Axial MP2RAGE image in a 24-year-old woman; Axial MP2RAGE image in a 35-year-old man; Coronal MP2RAGE image in a 33-year-old man. All cases are relapsing-remitting MS. MP2RAGE images are shown for the registered time point 1 (TP1) and the respective time point 2 (TP2). Results from algorithm before (LeMan) and after (LeMan-PV) applying the partial volume model using the non-conventional protocol are shown in the 3rd-4th and 5th–6th columns, respectively. The output of Leman is a binary mask and the output of LeMan-PV is a concentration map. Examples of new, stable, shrunken, and enlarged cortical lesions are indicated with red, pink, blue, and yellow arrows, respectively. Patches of the zoomed area of the different lesion categories in each image contrast (MPRAGE, MP2RAGE, FLAIR, and DIR) are shown at the bottom. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
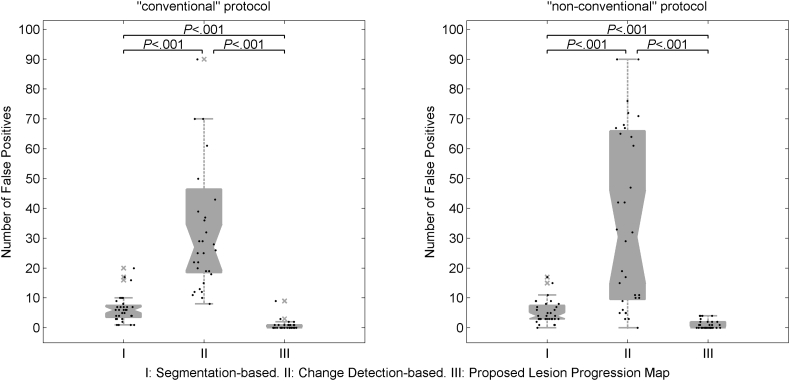
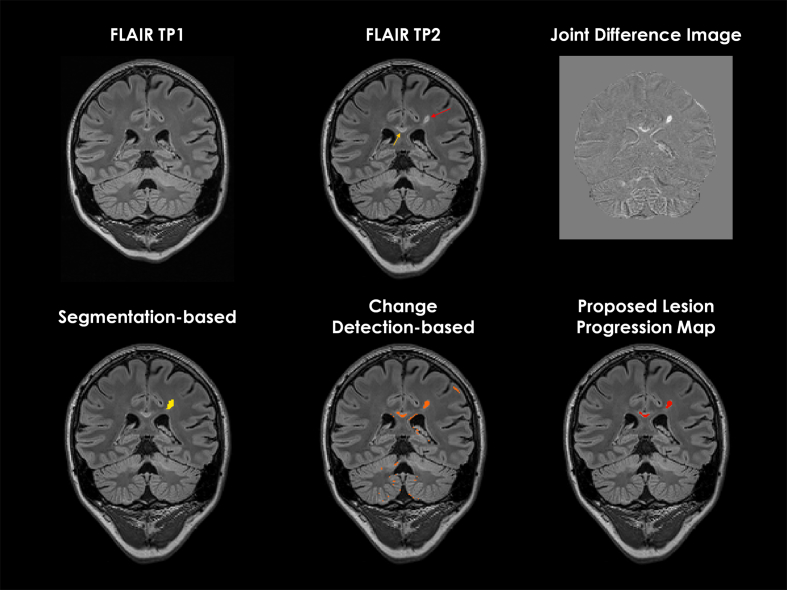
We found that the optimal threshold for the generation of the lesion progression maps correspond to intensity 0.3 in the normalized joint difference image. Using the “non-conventional” imaging set, the proposed lesion progression map showed the lowest number of false positives with a median of 0 (range 0 to 9), against 6 (range 1 to 20), and 20 (range 8 to 90) for segmentation-based, and change detection-based strategies, respectively. Similar results were obtained when the “conventional” protocol was used (see Fig. 7). An example of the lesion progression map obtained using the “conventional” imaging set is shown for comparison with the outputs from the segmentation-based, and change detection-based strategies in Fig. 8.
Fig. 7.
Boxplots of number of new and enlarged false positives using the “conventional” and “non-conventional” protocols from: I) segmentation-based and II) change detection-based methods, and III) proposed lesion progression map.
Fig. 8.
Examples of the proposed lesion progression map, and output from segmentation-based, and change detection-based methods. Top row shows the coronal slices of FLAIR images in a 26-year-old woman with relapsing-remitting MS at time point 1 (TP1) and time point 2 (TP2), and skull-stripped joint difference image obtained using the “conventional” set of images: FLAIR and MPRAGE. Red and yellow arrows indicate examples of new and enlarged lesions, respectively, according to the ground truth. The bottom row shows the output of new and enlarged fractions of lesions in yellow, orange, and red from segmentation-based, change detection based strategies, and the proposed lesion progression map. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Lesion volume difference estimation
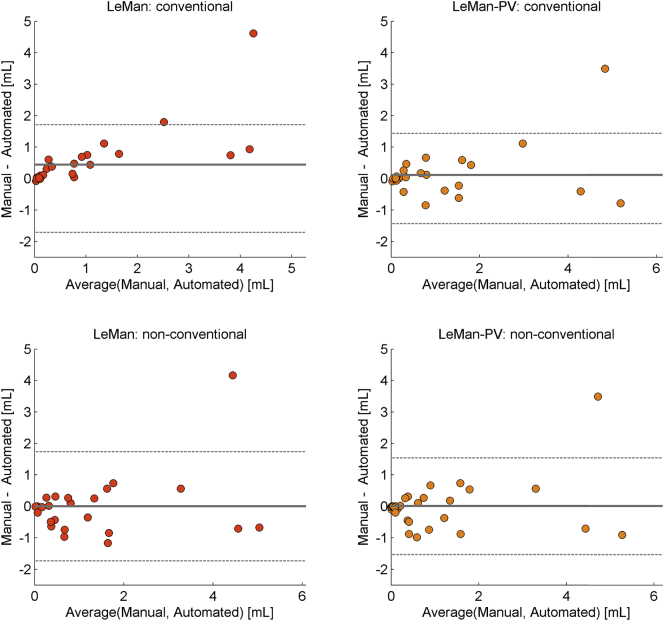
Fig. 9 shows the results of new lesion tissue volume agreement between the manual and automatic segmentations by Bland-Altman plots. The agreement was computed for the different configurations of the algorithm. Except for one patient, the quantification of new lesion tissue lies within the deviations, indicating a good agreement with the ground truth. The best agreement was achieved by LeMan-PV using either the “non-conventional” (0.01 ± 1.5 ml), or “conventional” protocol (0.01 ± 1.4 ml), followed by LeMan with the “non-conventional” protocol (0.01 ± 1.8 ml) and LeMan with the “conventional” (0.4 ± 1.7 ml) protocol.
Fig. 9.
Bland-Altman plot for total new lesional tissue volume between manual and automated segmentations. Results without (LeMan) and with (LeMan-PV) partial volume model computed using the “conventional”, and the “non-conventional” imaging datasets.
4. Discussion
In this work, we assessed the performance of the LeMan and LeMan-PV algorithms on a longitudinal MRI dataset. We also extended the methods to evaluate disease activity automatically by introducing a “lesion progression map”, aimed at directly visualising the clinically relevant information. We performed automatic detection and segmentation of new, enlarged, shrunken and stable lesions located in both WM and the cortex; in this context it is worth noting that cortical lesions were recently included in the MS diagnostic criteria (Thompson et al., 2018) and proved to be an independent predictor of disease progression (Treaba et al., 2019).
We also evaluated the advantage of modelling PV to detect new small lesions and to improve the lesion segmentation as well as the detection of lesion volume changes. Indeed, new lesions may be very small and therefore susceptible to PV effects. In addition, a precise segmentation of the lesion boarders is essential to detect volume changes, i.e. lesion enlargement or shrinkage.
Our results demonstrate that new, enlarged and stable WM lesion detection improved when the PV framework and the “non-conventional” MRI, such DIR and MP2RAGE were used. However, the method showed lower performance when applied to cortical lesion detection in all lesion categories. Similar to the observations in WM lesions, the TPR was higher when LeMan-PV with the “non-conventional” protocol was used, confirming our findings in previous work (Fartaria et al., 2016). However, this improvement was only significant for stable cortical lesions, most probably due to the relatively small number of samples in the other categories (the number of new cortical lesions in the entire cohort was 16).
The minimum lesion size has a strong influence on detection performance (Fartaria et al., 2016; Fartaria et al., 2018). Yet, there is still no clear definition of minimum MS lesion size in the literature. Current criteria are based on 2D sequences suggesting an in-plane diameter of 3 mm as the minimum for WM (Polman et al., 2005) and cortical (Geurts et al., 2011) lesions. This is mainly due to the low resolution in these – now considered deprecated – 2D protocols and would amount to ≈0.015 ml, i.e. 13 voxels in our 3D acquisitions. However, clinically relevant new lesions may be smaller than that. Here, we considered a minimum lesion volume of 0.006 ml obtained by evaluating the median lesion size in a cohort of healthy controls. It should be noted that our definition of minimum lesion size is considerably lower than in most of the automated methods for MS lesion segmentation in the literature (Cabezas et al., 2014; Datta and Narayana, 2013; Guizard et al., 2015; Sajja et al., 2006; Yamamoto et al., 2010).
All the missed lesions in each longitudinal lesion category were below the mean lesion size estimated using the ground truth lesion masks. In our cohort of patients, the mean lesion size for new lesions was approximately 0.070 ml and the mean lesion size for missed new WM and cortical lesions was 0.024 ml and 0.037 ml, respectively.
Beyond lesion detection and segmentation, we propose a lesion progression map as a binary mask showing areas of new lesional tissue, i.e. new lesions and the enlarged part of existing lesions. These maps showed a very low number of false positives when compared to segmentation-based and detection-based evaluation strategies. The remaining false positives were found on the pons region, due to flow artefacts common in 3D FLAIR images (Naganawa et al., 2004). The proposed approach combines segmentation-based and detection based techniques to reduce the dependency on baseline lesion segmentation and the sensitivity to noise, registration errors and artefacts.
Furthermore, the Bland-Altman analyses showed the advantage of using PV estimation, where volumes of new and enlarged fractions of lesions were close to the ground truth when LeMan-PV was used. The proposed method showed better results for longitudinal WM lesion assessment than for cortical lesions regardless whether the “conventional” or the “non-conventional” protocol was used; this is mainly due to the particular size and shape of cortical lesions and their resulting susceptibility to PV effects on tissue borders, as well as, their low contrast-to-noise ratio (Fartaria et al., 2017; Fartaria et al., 2016; Fartaria et al., 2019).
One of the main limitations of this study is the low number of lesion samples, especially for cortical lesions. As reported in our previous study (Fartaria et al., 2016), our cohort of early MS patients is characterized by low lesion loads compared to studies reported in the literature (Garcia-Lorenzo et al., 2013; Datta and Narayana, 2013; Schmidt et al., 2012; Shiee et al., 2010). This small number of samples is a disadvantage for the supervised step of our approach (since there is less training data), which is likely to compromise the performance of lesion detection. In addition, the number of missed lesions has an artificially large penalizing effect in metrics like TPR when applied to patients with low lesion loads. Another limitation is linked to the type of the data used for the validation of the proposed method. We used the same consistent protocol and the same scanner for the follow-up acquisitions. It should be noted that this ideal setting cannot always be achieved in routine clinical practice.
In conclusion, we evaluated the longitudinal performance of LeMan and LeMan-PV, which are methods designed to detect and segment WM and cortical lesions. LeMan-PV showed the best performance in detecting new, enlarged, shrunken and stable lesions in the WM. To the best of our knowledge, our method is the first attempt to segment cortical lesions, although the lower performance on detection of this type of lesions leaves room for improvement. We have also developed a lesion progression map that might be useful in clinical trials or clinical routine to evaluate disease activity and treatment response.
Future work should aim at including patients in more non-conventional stages, with higher number of samples of cortical lesions in order to increase the number of examples in the training set, which will improve the detection of lesions located in the cortex. Sophisticated statistical change techniques (Bosc et al., 2003), and methods based on deformation fields (Rey et al., 2002) should also be investigated and combined to LeMan-PV to evaluate their improvements in sensitivity to new and enlarged lesion detection. We also aim at augmenting the information from the proposed lesion progression map by adding the quantification of resolved lesional tissue. Quantification of resolved lesional tissue is not yet suggested in MS follow-up guidelines (Rovira et al., 2015), but can provide additional insights on treatment response in clinical routine. Lastly, an evaluation of the applicability of the method to images from different scanners and protocols is currently ongoing. Another work in progress, not yet explored, is to use the quantitative T1 maps from MP2RAGE (Kober et al., 2012; Marques et al., 2010). This could improve the robustness of the segmentation results, especially for longitudinal assessment and multicenter trials due to the increased independence of quantitative maps from the used hardware and reconstruction.
Acknowledgments
Acknowledgements
This study was supported by the Swiss National Science Foundation PZ00P3 131914/1 and PP00P3_176984, the Swiss MS Society and the Societé Académique Vaudoise. The funding sources had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report or in the decision to submit the paper for publication.
References
- Aït-Ali L.S., Prima S., Hellier P., Carsin B., Edan G., Barillot C., editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2005. Springer Berlin Heidelberg; Berlin, Heidelberg: 2005. STREM: a robust multidimensional parametric method to segment MS lesions in MRI. [DOI] [PubMed] [Google Scholar]
- Bonnier G., Roche A., Romascano D., Simioni S., Meskaldji D., Rotzinger D. Advanced MRI unravels the nature of tissue alterations in early multiple sclerosis. Ann. Clin. Transl. Neurol. 2014;1(6):423–432. doi: 10.1002/acn3.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnier G., Roche A., Romascano D., Simioni S., Meskaldji D.E., Rotzinger D. Multicontrast MRI quantification of focal inflammation and degeneration in multiple sclerosis. Biomed. Res. Int. 2015;2015 doi: 10.1155/2015/569123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosc M., Heitz F., Armspach J.-P., Namer I., Gounot D., Rumbach L. Automatic change detection in multimodal serial MRI: application to multiple sclerosis lesion evolution. NeuroImage. 2003;20(2):643–656. doi: 10.1016/S1053-8119(03)00406-3. [DOI] [PubMed] [Google Scholar]
- Cabezas M., Oliver A., Roura E., Freixenet J., Vilanova J.C., Ramio-Torrenta L. Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding. Comput. Methods Prog. Biomed. 2014;115(3):147–161. doi: 10.1016/j.cmpb.2014.04.006. PubMed PMID: 24813718. [DOI] [PubMed] [Google Scholar]
- Carass A., Roy S., Jog A., Cuzzocreo J.L., Magrath E., Gherman A. Longitudinal multiple sclerosis lesion segmentation: resource and challenge. NeuroImage. 2017;148:77–102. doi: 10.1016/j.neuroimage.2016.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Narayana P.A. A comprehensive approach to the segmentation of multichannel three-dimensional MR brain images in multiple sclerosis. Neuroimage Clin. 2013;2:184–196. doi: 10.1016/j.nicl.2012.12.007. PubMed PMID: 24179773; PubMed Central PMCID: PMC3777770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger G.J., Grimson W.E.L., Lozano-Perez T., Wells W.M., White S.J., Kikinis R., editors. Proceedings of IEEE Workshop on Biomedical Image Analysis. 1994. Automatic registration for multiple sclerosis change detection. 24-25 Jun 1994. [Google Scholar]
- Fartaria M.J., Bonnier G., Roche A., Kober T., Meuli R., Rotzinger D. Automated detection of white matter and cortical lesions in early stages of multiple sclerosis. J. Magn. Reson. Imaging. 2016;43(6):1445–1454. doi: 10.1002/jmri.25095. [DOI] [PubMed] [Google Scholar]
- Fartaria M.J., O'Brien K., Sorega A., Bonnier G., Roche A., Falkovskiy P. An ultra-high field study of cerebellar pathology in early relapsing-remitting multiple sclerosis using MP2RAGE. Investig. Radiol. 2017;52(5):265–273. doi: 10.1097/RLI.0000000000000338. (PubMed PMID: 00004424-201705000-00002) [DOI] [PubMed] [Google Scholar]
- Fartaria Mário João, Todea Alexandra, Kober Tobias, O'Brien Kieran, Krueger Gunnar, Meuli Reto. Partial volume-aware assessment of multiple sclerosis lesions. NeuroImage. 2018;18:245–253. doi: 10.1016/j.nicl.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fartaria M.J., Sati P., Todea A., Radue E.-W., Rahmanzadeh R., O'Brien K. Automated detection and segmentation of multiple sclerosis lesions using ultra–high-field MP2RAGE. Investig. Radiol. 2019;54(6):356–364. doi: 10.1097/RLI.0000000000000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fartaria M.J., Roche A., Meuli R., Granziera C., Kober T., Bach Cuadra M., editors. Segmentation of Cortical and Subcortical Multiple Sclerosis Lesions Based on Constrained Partial Volume Modeling. Springer International Publishing; Cham: 2017. [Google Scholar]
- Ganiler O., Oliver A., Diez Y., Freixenet J., Vilanova J.C., Beltran B. A subtraction pipeline for automatic detection of new appearing multiple sclerosis lesions in longitudinal studies. Neuroradiology. 2014;56(5):363–374. doi: 10.1007/s00234-014-1343-1. [DOI] [PubMed] [Google Scholar]
- Garcia-Lorenzo D., Francis S., Narayanan S., Arnold D.L., Collins D.L. Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med. Image Anal. 2013;17(1):1–18. doi: 10.1016/j.media.2012.09.004. PubMed PMID: 23084503. [DOI] [PubMed] [Google Scholar]
- Geurts J.J., Roosendaal S.D., Calabrese M., Ciccarelli O., Agosta F., Chard D.T. Consensus recommendations for MS cortical lesion scoring using double inversion recovery MRI. Neurology. 2011;76(5):418–424. doi: 10.1212/WNL.0b013e31820a0cc4. PubMed PMID: 21209373. [DOI] [PubMed] [Google Scholar]
- Giovannoni G., Tomic D., Bright J.R., Havrdová E. “No evident disease activity”: the use of combined assessments in the management of patients with multiple sclerosis. Mult. Scler. J. 2017;23(9):1179–1187. doi: 10.1177/1352458517703193. PubMed PMID: 28381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahl S., Pongratz née Biberacher V., Schmidt P., Engl C., Bussas M., Radetz A. European Committee for Treatment and Research in Multiple Sclerosis - ECTRIMS; 2017. Defining a Minimal Meaningful Lesion Size in Multiple Sclerosis. [Google Scholar]
- Guizard N., Coupé P., Fonov V.S., Manjón J.V., Arnold D.L., Collins D.L. Rotation-invariant multi-contrast non-local means for MS lesion segmentation. NeuroImage. 2015;8:376–389. doi: 10.1016/j.nicl.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttmann Charles R.G., Kikinis R., Anderson Mark C., Jakab M., Warfield Simon K., Killiany Ron J. Quantitative follow-up of patients with multiple sclerosis using MRI: reproducibility. J. Magn. Reson. Imaging. 1999;9(4):509–518. doi: 10.1002/(sici)1522-2586(199904)9:4<509::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Jain S., Ribbens A., Sima D.M., Cambron M., De Keyser J., Wang C. Two time point MS lesion segmentation in brain MRI: an expectation-maximization framework. Front. Neurosci. 2016;10(576) doi: 10.3389/fnins.2016.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Staring M., Murphy K., Viergever M.A., Pluim J.P. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging. 2010;29(1):196–205. doi: 10.1109/TMI.2009.2035616. PubMed PMID: 19923044. [DOI] [PubMed] [Google Scholar]
- Kober T., Granziera C., Ribes D., Browaeys P., Schluep M., Meuli R. MP2RAGE multiple sclerosis magnetic resonance imaging at 3 T. Investig. Radiol. 2012;47(6):346–352. doi: 10.1097/RLI.0b013e31824600e9. PubMed PMID: 22543966. [DOI] [PubMed] [Google Scholar]
- Köhler Caroline, Wahl Hannes, Ziemssen Tjalf, Linn Jennifer, Kitzler Hagen H. Exploring individual multiple sclerosis lesion volume change over time: development of an algorithm for the analyses of longitudinal quantitative MRI measures. NeuroImage: Clinical. 2019;21 doi: 10.1016/j.nicl.2018.101623. 101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lladó X., Ganiler O., Oliver A., Martí R., Freixenet J., Valls L. Automated detection of multiple sclerosis lesions in serial brain MRI. Neuroradiology. 2012;54(8):787–807. doi: 10.1007/s00234-011-0992-6. [DOI] [PubMed] [Google Scholar]
- Lladó X., Oliver A., Cabezas M., Freixenet J., Vilanova J.C., Quiles A. Segmentation of multiple sclerosis lesions in brain MRI: a review of automated approaches. Inf. Sci. 2012;186(1):164–185. [Google Scholar]
- Marco B., Francesca R., GR A., Laura S.M., Brandon W., MP M. Automated identification of brain new lesions in multiple sclerosis using subtraction images. J. Magn. Reson. Imaging. 2014;39(6):1543–1549. doi: 10.1002/jmri.24293. [DOI] [PubMed] [Google Scholar]
- Marques J.P., Kober T., Krueger G., van der Zwaag W., Van de Moortele P.F., Gruetter R. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage. 2010;49(2):1271–1281. doi: 10.1016/j.neuroimage.2009.10.002. PubMed PMID: 19819338. [DOI] [PubMed] [Google Scholar]
- Moraal B., Wattjes M.P., Geurts J.J., Knol D.L., van Schijndel R.A., Pouwels P.J. Improved detection of active multiple sclerosis lesions: 3D subtraction imaging. Radiology. 2010;(1) doi: 10.1148/radiol.09090814. [DOI] [PubMed] [Google Scholar]
- Naganawa S., Koshikawa T., Nakamura T., Kawai H., Fukatsu H., Ishigaki T. Comparison of flow artifacts between 2D-FLAIR and 3D-FLAIR sequences at 3 T. Eur. Radiol. 2004;14(10):1901–1908. doi: 10.1007/s00330-004-2372-7. [DOI] [PubMed] [Google Scholar]
- Polman C.H., Reingold S.C., Edan G., Filippi M., Hartung H.-P., Kappos L. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald criteria”. Ann. Neurol. 2005;58(6):840–846. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- Rey D., Subsol G., Delingette H., Ayache N. Automatic detection and segmentation of evolving processes in 3D medical images: application to multiple sclerosis. Med. Image Anal. 2002;6(2):163–179. doi: 10.1016/s1361-8415(02)00056-7. [DOI] [PubMed] [Google Scholar]
- Rovira A., Wattjes M.P., Tintore M., Tur C., Yousry T.A., Sormani M.P. Evidence-based guidelines: MAGNIMS consensus guidelines on the use of MRI in multiple sclerosis[mdash]clinical implementation in the diagnostic process. Nat. Rev. Neurol. 2015 doi: 10.1038/nrneurol.2015.106. advance online publication. [DOI] [PubMed] [Google Scholar]
- Sajja B.R., Datta S., He R.J., Mehta M., Gupta R.K., Wolinsky J.S. Unified approach for multiple sclerosis lesion segmentation on brain MRI. Ann. Biomed. Eng. 2006;34(1):142–151. doi: 10.1007/s10439-005-9009-0. PubMed PMID: WOS:000236354800013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P., Gaser C., Arsic M., Buck D., Forschler A., Berthele A. An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. Neuroimage. 2012;59(4):3774–3783. doi: 10.1016/j.neuroimage.2011.11.032. PubMed PMID: 22119648. [DOI] [PubMed] [Google Scholar]
- Shiee N., Bazin P.L., Ozturk A., Reich D.S., Calabresi P.A., Pham D.L. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. Neuroimage. 2010;49(2):1524–1535. doi: 10.1016/j.neuroimage.2009.09.005. PubMed PMID: 19766196; PubMed Central PMCID: PMC2806481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon J., Sood A., editors. 2004 2nd IEEE International Symposium on Biomedical Imaging: Nano to Macro (IEEE Cat No 04EX821) 2004. 4-D lesion detection using expectation-maximization and hidden Markov model. 15-18 April 2004. [Google Scholar]
- Stangel M., Penner I.K., Kallmann B.A., Lukas C., Kieseier B.C. Towards the implementation of 'no evidence of disease activity' in multiple sclerosis treatment: the multiple sclerosis decision model. Ther. Adv. Neurol. Disord. 2015;8:3–13. doi: 10.1177/1756285614560733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styner M., Lee J., Chin B., Chin M., Commowick O., Tran H. 3D segmentation in the clinic: a grand challenge II: MS lesion segmentation. MIDAS J. 2008;2008:1–6. [Google Scholar]
- Thompson A.J., Banwell B.L., Barkhof F., Carroll W.M., Coetzee T., Comi G. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17(2):162–173. doi: 10.1016/S1474-4422(17)30470-2. [DOI] [PubMed] [Google Scholar]
- Treaba C.A., Granberg T.E., Sormani M.P., Herranz E., Ouellette R.A., Louapre C. Longitudinal characterization of cortical lesion development and evolution in multiple sclerosis with 7.0-T MRI. Radiology. 2019;291:740–749. doi: 10.1148/radiol.2019181719. 181719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto D., Arimura H., Kakeda S., Magome T., Yamashita Y., Toyofuku F. Computer-aided detection of multiple sclerosis lesions in brain magnetic resonance images: false positive reduction scheme consisted of rule-based, level set method, and support vector machine. Comput. Med. Imaging Graph. 2010;34(5):404–413. doi: 10.1016/j.compmedimag.2010.02.001. [DOI] [PubMed] [Google Scholar]