Abstract
Mycobacterium tuberculosis is one of the most dangerous pathogens in humans. It acts as an etiological agent of tuberculosis (TB), infecting almost one-third of the world's population. Owing to the high incidence of multidrug-resistant TB and extensively drug-resistant TB, there is an urgent need for novel and effective alternative therapies. Peptide-based therapy has several advantages, such as diverse mechanisms of action, low immunogenicity, and selective affinity to bacterial cell envelopes. However, the identification of anti-tubercular peptides (AtbPs) via experimentation is laborious and expensive; hence, the development of an efficient computational method is necessary for the prediction of AtbPs prior to both in vitro and in vivo experiments. To this end, we developed a two-layer machine learning (ML)-based predictor called AtbPpred for the identification of AtbPs. In the first layer, we applied a two-step feature selection procedure and identified the optimal feature set individually for nine different feature encodings, whose corresponding models were developed using extremely randomized tree (ERT). In the second-layer, the predicted probability of AtbPs from the above nine models were considered as input features to ERT and developed the final predictor. AtbPpred respectively achieved average accuracies of 88.3% and 87.3% during cross-validation and an independent evaluation, which were ~8.7% and 10.0% higher than the state-of-the-art method. Furthermore, we established a user-friendly webserver which is currently available at http://thegleelab.org/AtbPpred. We anticipate that this predictor could be useful in the high-throughput prediction of AtbPs and also provide mechanistic insights into its functions.
Keywords: Extremely randomized tree, Two-step feature selection, Antitubercular peptide, Tuberculosis, Cross-validation, Two-layer framework
Graphical Abstract
Highlights
-
•
We developed a novel computational framework for the identification of anti-tubercular peptides using Extremely randomized tree.
-
•
AtbPpred displayed superior performance compared to the existing method on both benchmark and independent datasets.
-
•
We constructed a user-friendly web server that implements the proposed AtbPpred method.
1. Introduction
Tuberculosis (TB) is a major deadly disease caused by an infection from the bacterium Mycobacterium tuberculosis [1,2]. The World Health Organization (WHO) recently reported that around 10 million people were infected with TB and 1.6 million people died from the disease in 2017 alone (https://www.who.int/en/news-room/fact-sheets/detail/tuberculosis). Despite evolving treatment strategies, its prevalence is increasing owing to inadequate drug use, poor-quality drugs, and the premature discontinuation of treatment by patients, leading to amplified drug-resistant strains of Mycobacterium tuberculosis [3]. Multidrug-resistant tuberculosis (MDR-TB) is a form of TB caused by bacteria that do not respond to first-line anti-TB drugs, like rifampicin and isoniazid [4]. Owing to its high prevalence, MDR-TB is considered a public health crisis and a health security threat (https://www.who.int/en/news-room/fact-sheets/detail/tuberculosis). Another form of MDR-TB, extensively drug-resistant tuberculosis, does not respond to second-line anti-TB drugs and accordingly is much more serious. Furthermore, the adverse side effects of anti-tubercular drugs and prolonged treatment durations are major issues. Therefore, it is necessary to find alternative effective therapeutics with new mechanisms of anti-tubercular action or methods to potentiate existing drugs with minimized treatment periods and costs.
Since the serendipitous finding of penicillin in the 1920s, peptide-based therapy has gained momentum in the area of drug discovery. One of the most influential characteristics of peptides is their pleiotropic effects towards a wide range of biological targets, thus making them better drug candidates with lower toxicity than that of small molecules. Several studies have shown the anti-tubercular properties of peptides and their unique mechanism of actions, suggesting that they are an ideal approach for TB management [5]. Peptides derived from human immune and non-immune cells, venom, fungi, bacteria, and several other sources have been shown to act as effective anti-tubercular agents [3]. Some aspects of anti-tubercular peptides (AtbPs), like their low immunogenicity, selective affinity to negatively charged bacterial cell envelopes, and targeted immune responses against bacterium, make them attractive alternatives to conventional TB drugs [3,6].
Owing to the growing interest in peptide-based therapy for the identification of effective anti-tubercular agents, their design and discovery process should be accelerated. However, the experimental identification and development of AtbPs is time-consuming and expensive. To assist and expedite the discovery of AtbPs, in silico methods are needed prior to their synthesis [5]. A trending computational method, machine learning (ML), could provide an excellent platform for the prediction and design of AtbPs. To the best of our knowledge, only one ML-based method has been developed for AtbP prediction [7]. The authors utilized five different classifiers (support vector machine (SVM), random forest (RF), SMO, J48, Naïve-Bayes) and four different feature encodings (amino acid composition (AAC), dipeptide composition (DPC), binary profiles (NC5) and terminal composition). Two prediction models, such as SVM-based ensemble model and SVM using hybrid features achieved better performances [7]. Despite encouraging results, the method has certain limitations in terms of efficiency and accuracy. Furthermore, feature selection techniques for optimization of features and exploration of other feature encodings were not implemented.
Henceforth, in this study, we proposed a novel sequence-based two-layer predictor, i.e. AtbPpred, for the classification of AtbPs or non-AtbPs from given peptide sequences. To develop a prediction model, we explored nine different feature encodings that covers various aspects of sequence information, including AAC, DPC, NC5, composition-transition-distribution (CTD), quasi-sequence-order (QSO), amino acid index (AAI), conjoint triad (CTF), grouped tripeptide composition (GTPC), and grouped dipeptide composition (GDPC). Subsequently, a two-step feature selection protocol was applied on each encoding and identified their corresponding optimal feature set. In the first-layer, nine optimal feature set-based prediction models were developed using extremely randomized tree (ERT), whose predicted probability scores were further considered as input features to ERT in the second-layer and developed the final model. Comparative results for AtbPpred and the state-of-the-art method using benchmark and independent datasets showed remarkable improvements. Therefore, we foresee that our work will pave way for the development of novel computational methods and facilitate experimentalists in the discovery of novel AtbPs.
2. Materials and Methods
2.1. Proposed Predictor Framework
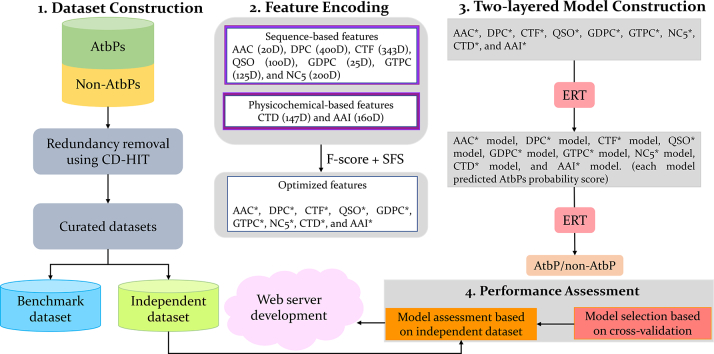
The overall procedure for the proposed peptide sequence-based predictor is shown in Fig. 1. It consists of four steps: (i) construction of benchmark and independent datasets; (ii) feature extraction and optimization using two-step feature selection (F-score algorithm was applied for feature ranking, followed by a sequential forward search (SFS)) protocol; and (iii) using ERT, the optimal feature set of nine different encodings based prediction models were developed individually in the first layer, whose predicted probability of AtbPs were integrated and further considered as input features to ERT for the development of the final prediction model in the second layer. (iv) performance assessment and webserver development. In our predictor, the predicted outcome for each sequence is 0 or 1, where 0 denotes non-AtbP and 1 denotes AtbP.
Fig. 1.
Framework of the proposed algorithm. It consists of four steps: (i) dataset construction, (ii) feature extraction and their optimization using two-step feature selection protocol, (iii) construction of two-layer prediction model, and (iv) assessment of performance and development of webserver.
3. Construction of Benchmark and Independent Datasets
Recently, Usmani et al. [7] constructed antitubercular peptide datasets and developed prediction models. Basically, two datasets were constructed, namely AntiTb_MD and AntiTb_RD, where both datasets were comprised of identical positive samples (246 AtbPs), but different negative samples (246 non-AtbPs). AntiTb_RD and AntiTb_MD respectively contained random peptides generated from Swiss-Prot and anti-bacterial peptides. In this study, we utilized both the datasets and developed a prediction model separately. All positive samples contained true experimentally validated ATbPs derived from the public database, AntiTbpdb [8]. Table S1 shows the distribution of ATbPs from different species, where major contribution is from Mycobacterium tuberculosis (66%), moderate contribution is from Mycobacterium smegmatis (24%), and other species contribution is marginal. Moreover, the sequence lengths in two separate datasets were in the range of 5 to 61 amino acids. We utilized the same dataset for the following reasons: (i) comprised of most recently updated datasets; (ii) contains nonredundant data; and (iii) allowance of a fair comparison between our proposed and the existing methods. For each dataset, 80% of the samples (199 AtbPs and 199 non-AtbPs) were randomly selected for the development of a prediction model and the remaining 20% of samples (47 AtbPs and 47 non-AtbPs) were utilized for model evaluation.
4. Feature Extraction
Generally, feature extraction is one of the important steps in designing well-performed classifiers. It generates a fixed length of feature vectors from the given peptide sequences that have varied lengths. Features used in this work can be categorized into two major groups: sequence-based features and physicochemical-based features.
5. Sequence-Based Features
The differences between positive and negative samples can be reflected by amino acid sequences bearing various factors, including profiles, composition, permutation and combination modes of amino acids, and physicochemical properties. Here, we extracted seven types of sequence-based features: AAC, DPC, QSO, CTF, GTPC, GDPC, and NC5.
-
(i)
Amino acid composition (AAC)
AAC reflects the occurrences of standard amino acids in a given peptide normalized by the sequence length and is widely applied in bioinformatics [[9], [10], [11]]. It has a fixed length of 20 features, which can be formulated as follows:
| (1) |
where fvi= (i = 1,2,3…,20) is the normalized frequency of the ith amino acid in a given peptide. Ri is the quantity of type i observed in a peptide.
-
(ii)
Dipeptide composition (DPC)
DPC reflects the composition of a residue pair (e.g. Ala-Ala, Ala-Cys) occurring in a given peptide, further describing the fraction of amino acids and their local order [7,12]. It has a fixed length of 400 vectors, which can be formulated as follows:
| (2) |
where fvj represents the frequency of jth amino acid pair in {AA, AC, AD, AE,…,YY}.
-
(iii)
Binary profile (NC5)
In the NC5, each amino acid is encoded as a 20-dimensional 0/1 vector. For instance, the amino acids of type A (b(A)) and type C (b(C)) are encoded as (1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0) and (0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0), respectively. For a given peptide P, its N- or C– terminus with a length of m amino acids was translated as follows:
| (3) |
where the BPF(m) dimension is 20 × m, where m is assigned a value of 5 at both termini to obtain BPFN5 and BPFC5. Furthermore, these two termini (NC5) were combined to generate 200-dimensional feature vector.
-
(iv)
Grouped tripeptide composition (GTPC)
In GTPC encoding, amino acids are divided into five categories according to their physicochemical properties: aliphatic group (G, A, V, L, M, and I), aromatic group (F, Y, and W), positive charge group (K, R, and H), negative charged group (G, D, and E) and uncharged group (S, T, C, P, N, and Q). The tripeptide composition of these five categories generates a fixed length of 125-dimensional feature vector.
-
(v)
Grouped dipeptide composition (GDPC)
In GDPC encoding, the dipeptide composition of five categories of amino acid physicochemical properties (listed in GTPC) generates a fixed length of 25-dimensional feature vector.
-
(vi)
Quasi-sequence-order (QSO)
QSO encoding of the given peptide sequence results in a fixed length of a 100-dimensional feature vector, by measuring the physicochemical distance between the amino acids within the sequence. A detailed description of QSO feature encoding along with a set of equations has been provided in previous studies [13,14].
-
(vii)
Conjoint triad (CTF)
CTF encoding generates a 343-dimensional feature vector for a given peptide sequence by clustering amino acids into seven classes according to their dipoles and side chain volumes. A detailed description of CTF with a set of equation has been reported previously [15].
5.1. Physicochemical Properties-Based Features
-
(i)
Composition-Transition-Distribution (CTD)
In the CTD feature, composition (C) indicates the composition of amino acids, transition (T) signifies the percentage of amino acid residues with certain characteristic that are followed by other amino acids, and distribution (D) measures the sequence length within which 1%, 25%, 50%, 75%, and 100% of the amino acids with certain characteristics are located. In CTD, composition, transition, and distribution are respectively encoded as a 21, 21, 105-dimensional feature vector.
-
(ii)
Amino acid index (AAI)
Previously, eight high-quality AAIs (accessions LIFS790101, TSAJ990101, MAXF760101, BIOV880101, CEDJ970104, BLAM930101, MIYS990104, and NAKH920108) were identified from 566 total AAIs in the AAIndex database [16,17] by applying a clustering technique [18]. AAI generates a 160 (=20 amino acids × 8 properties) dimensional vector, which has been widely applied in numerous sequence-based prediction tasks [19].
6. Feature Optimization
Feature optimization is one of the important steps in ML [20] that has been used in the improvisation of classification performance. In this study, an F-score algorithm with a SFS protocol was used to filter out noisy and irrelevant features, after which a subset containing optimal features was selected. This two-step protocol has been successfully applied in various predictions [[21], [22], [23]]. In the first step, an F-score algorithm is used to rank the actual features, and to sort these features in a descending order, thereby generating a ranked feature list. The F-score of the ith feature is defined as:
| (4) |
where , and represent mean values of the ith feature in the combined (both positive and negative), positive, and negative datasets, respectively. n+ and n− represent the number of positive and negative samples, respectively. and represent the ith feature of jth positive instance and ith feature of jth negative instance, respectively.
In the second step, two features were chosen from the ranked features list, and added sequentially as an input feature to ERT which were further utilized for training and developing the corresponding prediction models. Ultimately, the features corresponding to the model with highest accuracy were recognized as optimal features for the respective ML classifier.
7. Machine Learning Algorithms
We explored the commonly used six different ML algorithms, including ERT, gradient boosting (GB), k-nearest neighbor (KNN), logistic regression (LR), random forest (RF), and SVM. Another five ML-based algorithms have been discussed previously [19,[24], [25], [26], [27], [28], [29], [30]], whose parameter search range is given in supplementary Table S2. Since ERT implemented in AtbPpred exhibited better performance than other ML methods, a brief description of this method and its utilization is detailed below.
8. Extremely Randomized Tree (ERT)
ERT, another powerful decision tree based method developed by Geurts et al. [31], has been widely used in various sequence-based prediction problems [19,32]. ERT incorporates a stronger randomization technique that reduces the variance of the model. The ERT algorithm is similar to that of RF, except for two main differences: (i) ERT uses all training samples to construct each tree with varying parameters, rather than the bagging procedure used in RF; and (ii) ERT randomly chooses the node split upon construction of each tree, rather than the best split used in RF. In this study, the ERT algorithm was implemented using the scikit-learn (v 0.18.1) library in Python [33]. The grid search approach is used for optimizing the number of trees (ntree), number of randomly selected features (mtry), and minimum number of samples required to split an internal node (nsplit) of the ERT algorithm. The search ranges for the three parameters were 50 ≤ntree≤ 2000 with a step size of 25, 1 ≤mtry≤ 15 with a step size of 1, and 1 ≤nsplit≤ 12 with a step size of 1, respectively.
8.1. 10-Fold Cross-Validation (CV)
Three CV methods, i.e. an independent dataset test, a sub-sampling (or k-fold CV) test, and a leave-one-out CV (LOOCV) test, are frequently used to calculate the expected success rate of a developed predictor [34,35]. Among the three methods, the LOOCV test is deemed the least arbitrary and most objective, as demonstrated by Eqs. 28–32 in ref. [36]. Although it is widely used to examine the quality of various predictors [[37], [38], [39], [40], [41], [42], [43]], it is time- and resource-intensive. Thus, 10-fold CV was used to examine the proposed models. In 10-fold CV, the training dataset was randomly partitioned into 10 subsets. One subset was used as a test set and the remaining nine subsets were used as the training sets. This procedure is repeated 10 times, where each subset is treated as a test set at least once. Results were averaged to obtain the performance of the classifier.
8.2. Performance Evaluation
To evaluate the performance of the constructed models, four common measurements in binary classification tasks were used [23,29,[44], [45], [46], [47], [48]], i.e. sensitivity, specificity, accuracy, and Matthews correlation coefficients (MCC). They were calculated as follows:
| (5) |
where TP is the number of true positives (i.e. AtbPs classified correctly as AtbPs) and TN is the number of true negatives (i.e. non-AtbPs classified correctly as non-AtbPs). FP is the number of false positives (i.e. AtbPs classified incorrectly as non-AtbPs) and FN is the number of false negatives (i.e. non-AtbPs classified incorrectly as AtbPs). Additionally, the receiver operating characteristic (ROC) curve, was generated to visually evaluate the comprehensive performance of different classifiers.
9. Results and Discussion
9.1. Evaluation of Various ML-Classifiers Using Nine Different Feature Encodings
Generally, exploring different classifiers using the same dataset is essential, rather than selecting a specific classifier [20,22,49]. Hence, we explored commonly used six different ML algorithms (ERT, RF, SVM, GB, LR, and KNN) [[50], [51], [52], [53], [54]] to evaluate the effectiveness of the ML method in AtbP prediction. For a fair comparison, all ML classifiers were trained and evaluated using 10-fold CV on benchmark datasets, whose corresponding ML parameters were tuned and the optimal parameters were identified using a grid search procedure. To generate a robust prediction model, 10-fold CV was repeated 10 times for each classifier by random partitioning of the benchmark datasets, which lead to ten optimized ML parameters for each classifier. However, we considered the median ML parameter estimates to develop a final prediction model for each feature encoding.
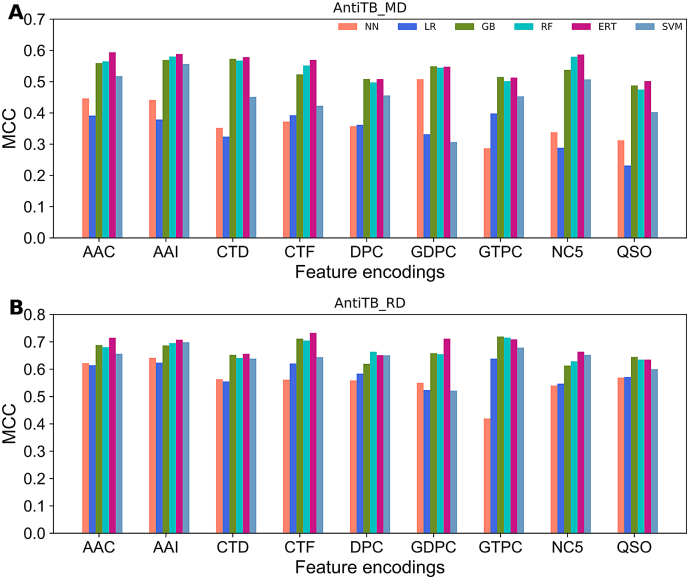
Fig. 2A and B respectively represent the performance of six different classifiers on AntiTb_MD and AntiTb_RD datasets. To get an overview of each classifier performance, we computed the average MCC from nine different feature encodings (Fig. 2A), where ERT, GB, KNN, LR, RF, and SVM respectively achieved an average MCC of 0.550, 0.535, 0.379, 0.344, 0.540, and 0.453. Particularly, ERT showed 1.0–20.6% higher MCC scores than that of other five classifiers, demonstrating its superiority in AtbPs prediction. From Fig. 2B, ERT, GB, KNN, LR, RF, and SVM respectively achieved an average MCC of 0.687, 0.666, 0.558, 0.586, 0.666, and 0.638. Specifically, ERT showed 2.1–12.9% higher MCC scores when compared to other classifiers. Overall, ERT classifier attained better performance when compared to other five classifiers regardless of the datasets. Hence, we considered only ERT classifier for further analysis.
Fig. 2.
Performance of various classifiers in distinguishing between AtbPs and non-AtbPs with respect to nine feature descriptors. (A) and (B) respectively represent performances based on AntiTb_MD and AntiTb_RD benchmark datasets.
10. Selection of Optimal Features for Each Encoding
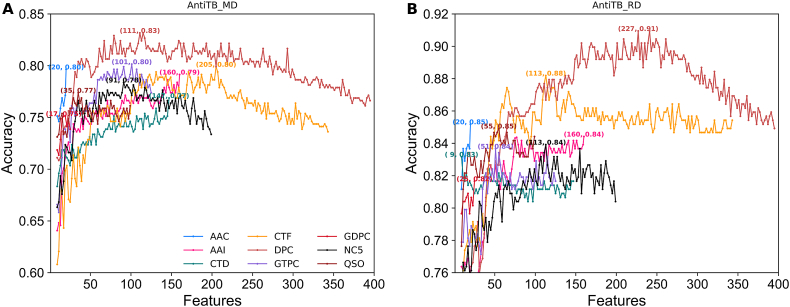
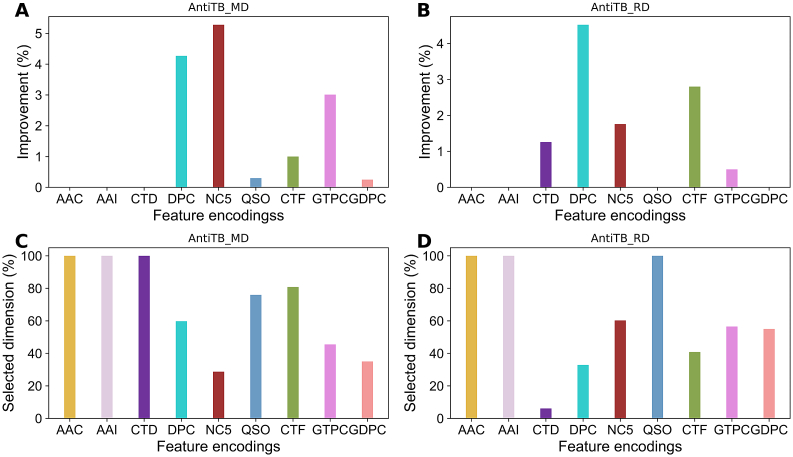
To examine whether the feature selection protocol could improve each encoding-based prediction model performance, we applied a commonly used two-step feature selection protocol (i.e. F-score based ranking, followed by SFS) on each encoding. Fig. 3A shows the accuracy curves with gradual addition of features from the ranked feature list for the ERT classifier based on nine different encodings using AntiTb_MD dataset. Results showed that the six feature encodings (DPC, NC5, QSO, CTF, GTPC, and GDPC), whose accuracy curve gradually improved, reached its maximum point and subsequently declined upon the addition of ranked features. Conversely, three encodings (AAC, AAI, and CTD) reached its maximum point using all features, further indicating the equal significance of all features. Using AntiTb_RD dataset, the accuracy curve gradually improved for six feature encodings (CTD, DPC, NC5, QSO, CTF, and GTPC) and reached its maximum point, which then declined upon the addition of ranked features (Fig. 3B). However, a maximum point was observed for three encodings (AAC, AAI, and GDPC) with all features. To check the improvement, we computed the difference in accuracy between the optimal feature set and control (using all features). Models developed using AntiTB_MD dataset (Fig. 4A) showed a major improvement (>3%) for three encodings (GTPC, NC5, and DPC), a marginal improvement (<1%) for three encodings (QSO, CTF, and GDPC), and a tie for three encodings (AAC, AAI, and CTD). Using AntiTB_RD dataset, a major improvement was observed only for DPC encoding (Fig. 4B), a slight improvement (< 3%) for four encodings (CTF, NC5, CTF, and GTPC), and a tie for four encodings (AAC, AAI, QSO, and GDPC). Furthermore, the selected optimal features are significantly reduced for both datasets, where AntiTB_MD (Fig. 4C) and AntiTB_RD (Fig. 4D) optimal features respectively contained 59.1% and 51.7% from the total nine feature encodings. Overall, feature selection protocol improved majority of feature encoding performances and significantly reduced the feature dimension on both AntiTB_MD and AntiTB_RD datasets.
Fig. 3.
Sequential forward search (SFS) for discriminating between AtbPs and non-AtbPs. The maximum accuracy (SFS peak) obtained from 10-fold cross-validation is shown for each feature encoding. (A) and (B) respectively represent performances based on AntiTb_MD and AntiTb_RD benchmark datasets.
Fig. 4.
Comparison between original features and optimal features in terms of performance and feature dimension. The percentage of improvement in accuracy calculated between the control and the optimal feature set is shown in (A) and (B), respectively represent AntiTb_MD and AntiTb_RD. The percentage of selected features (optimal features) from the original features shown in (C) and (B), respectively represent AntiTb_MD and AntiTb_RD.
11. Construction of AtbPpred
Two-layer prediction model has been successfully applied to address various biological problems [20,22,27,28,55,56]. Inspired by these studies, we implemented a similar approach for AtbPs prediction. The optimal models obtained for each encoding from the above step are considered as the first layer models (Table 1). Since the performance pattern is similar between two datasets (AntiTB_MD and AntiTB_RD), we focused only on AntiTB_MD. Using AntiTB_MD dataset, DPC encoding appeared to be the most powerful encoding and it achieved the MCC and accuracy of 0.664 and 0.832, respectively (Table 1). Whereas, the remaining eight encodings also achieved a reasonable performance with accuracies ranging from 75 to 80%, further indicating its practicality in AtbPs prediction due to their complementary feature representation from a different perspective. Instead of selecting the best model from Table 1, we considered all these models to generate a robust and final prediction in the second layer. Basically, we considered the predicted probability of AtbPs (values ranging from 0.0 to 1.0) from nine individual optimal models as input features to ERT and developed a final prediction model called AtbPpred.
Table 1.
Performance of various classifiers on the benchmark dataset.
| Dataset | Encoding | MCC | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|---|
| AntiTb_MD | AAC | 0.594 | 0.797 | 0.764 | 0.829 | 0.853 |
| AAI | 0.588 | 0.792 | 0.724 | 0.859 | 0.857 | |
| CTD | 0.559 | 0.769 | 0.633 | 0.905 | 0.809 | |
| CTF | 0.599 | 0.799 | 0.774 | 0.824 | 0.849 | |
| DPC | 0.664 | 0.832 | 0.809 | 0.854 | 0.886 | |
| GDPC | 0.506 | 0.751 | 0.694 | 0.809 | 0.798 | |
| GTPC | 0.604 | 0.802 | 0.774 | 0.829 | 0.837 | |
| NC5 | 0.568 | 0.784 | 0.779 | 0.789 | 0.826 | |
| QSO | 0.548 | 0.774 | 0.769 | 0.779 | 0.845 | |
| AntiTb_RD | AAC | 0.715 | 0.852 | 0.764 | 0.940 | 0.909 |
| AAI | 0.708 | 0.844 | 0.729 | 0.960 | 0.906 | |
| CTD | 0.665 | 0.832 | 0.799 | 0.864 | 0.883 | |
| CTF | 0.765 | 0.882 | 0.859 | 0.905 | 0.908 | |
| DPC | 0.820 | 0.910 | 0.889 | 0.930 | 0.945 | |
| GDPC | 0.635 | 0.817 | 0.779 | 0.853 | 0.883 | |
| GTPC | 0.674 | 0.837 | 0.814 | 0.859 | 0.889 | |
| NC5 | 0.684 | 0.839 | 0.774 | 0.905 | 0.878 | |
| QSO | 0.708 | 0.849 | 0.769 | 0.930 | 0.881 |
The first and the second column represent the dataset and the feature encoding employed in this study. The third, fourth, fifth, sixth, and the seventh columns, respectively represent the MCC, accuracy, sensitivity, specificity, and AUC.
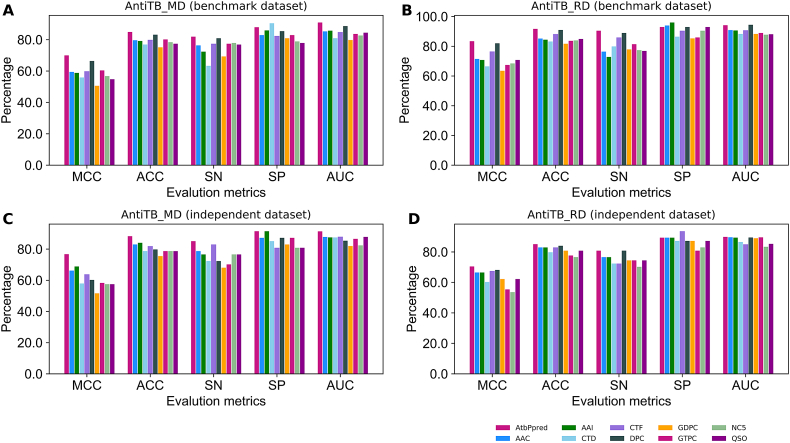
AtbPpred based on AntiTB_MD dataset achieved the best performance with the MCC, accuracy, sensitivity, specificity, and area under the curve of 0.700, 0.849, 0.819, 0.879, and 0.909, respectively. To show the effectiveness of AtbPpred, we compared its performance with nine feature encoding predictors (Fig. 5A). Specifically, the MCC and accuracy of the proposed predictor was 3.57–19.4% and 1.7–9.8% higher than the individual predictors, thus indicating the effectiveness of our approach by integrating various feature encodings, leading to an improved performance. Similarly, AtbPpred based on AntiTB_RD dataset achieved the best performance with the MCC, accuracy, sensitivity, specificity, and area under the curve of 0.834, 0.917, 0.905, 0.930, and 0.942, respectively. Specifically, the MCC and accuracy of the proposed predictor was 0.7–19.9% and 0.8–1.0% higher than the individual predictors (Fig. 5B), thus indicating the effectiveness of our approach.
Fig. 5.
Performance comparison of AtbPpred and nine different feature encodings based methods. (A) and (B) respectively represent the performances based on AntiTb_MD and AntiTb_RD benchmark datasets. (C) and (D) respectively represent performances based on AntiTb_MD and AntiTb_RD independent datasets.
12. Performance of AtbPpred on Independent Dataset
To assess the generalization, robustness, and practical application of our proposed method, we evaluated its performance using the independent dataset and compared the results with those obtained using first-layer models. AtbPpred based on AntiTB_MD achieved the best performance with the MCC, accuracy, sensitivity, specificity and area under the curve of 0.793, 0.894, 0.830, 0.957, and 0.934, respectively (Fig. 5C). Specifically, the MCC and accuracy of the proposed predictor was 7.9–25.1% and 4.3–12.8% higher than the first layer models. Similarly, AtbPpred based on AntiTB_MD achieved the best performance with the MCC, accuracy, sensitivity, specificity, and area under the curve of 0.704, 0.851, 0.809, 0.894, and 0.899, respectively (Fig. 5D). Specifically, the MCC and accuracy of the proposed predictor was 2.2–16.8% and 1.0–8.5% higher than the first layer models, thus indicating the utility and robustness of our two-layer approach. To summarize, the experiments based on the independent dataset highlights the importance and requisite to employ more comprehensive and discriminative feature encodings and integrate them into a consolidated framework to further enhance the model design and performance.
13. Comparison of AtbPpred With the Existing Method
We developed two prediction models using the AntiTb_MD and AntiTb_RD datasets and compared their performances with the state-of-the art method, Antitbpred. Notably, Antitbpred also contains two prediction models using two different datasets. The rationale for considering this method in our analysis is as follows: (i) the authors trained and validated their prediction models using the same training dataset presented in this study and (ii) this method has been reported to demonstrate excellent performance in AtbP identification.
First, we compared the performance of our proposed AtbPpred method on two benchmarking datasets. As shown in Table 2, AtbPpred achieved an overall better performance than Antitbpred in terms of MCC, accuracy, sensitivity, and specificity on two benchmarking datasets. Using a P-value threshold of 0.01, our proposed method significantly outperformed Antitbpred. Secondly, we compared the performance of AtbPpred on two independent datasets. AtbPpred achieved an overall better performance than Antitbpred in terms of MCC, accuracy, sensitivity, and specificity on two independent datasets (Table 3). Inclusively, AtbPpred shows the best and consistent performance on both benchmark and independent datasets, further suggesting its ability to perform well with unknown peptides when compared to the existing method.
Table 2.
Performance of various classifiers on the benchmark dataset.
| Dataset | Methods | MCC | Accuracy | Sensitivity | Specificity | AUC | P-value |
|---|---|---|---|---|---|---|---|
| AntiTb_MD | AtbPpred | 0.700 | 0.849 | 0.819 | 0.879 | 0.909 | – |
| Antitbpred | 0.550 | 0.775 | 0.768 | 0.773 | 0.820 | 0.000656 | |
| AntiTb_RD | AtbPpred | 0.834 | 0.917 | 0.905 | 0.930 | 0.942 | – |
| Antitbpred | 0.640 | 0.817 | 0.787 | 0.846 | 0.870 | 0.001013 |
The first and the second column represent the dataset and the classifier name employed in this study. The third, fourth, fifth, sixth, and the seventh columns respectively represent the MCC, accuracy, sensitivity, specificity, and AUC. For comparison, we have included Antitbpred metrics reported in the literature [7]. The last column represents the pairwise comparison of ROC area under curves (AUCs) between AtbPpred and the Antitbpred using a two-tailed t-test. P < .01 indicates a statistically meaningful difference between AtbPpred and the selected method (shown in bold).
Table 3.
Performance of various classifiers on the independent dataset.
| Dataset | Methods | MCC | Accuracy | Sensitivity | Specificity | AUC | P-value |
|---|---|---|---|---|---|---|---|
| AntiTb_MD | AtbPpred | 0.793 | 0.894 | 0.830 | 0.957 | 0.934 | – |
| Antitbpred | 0.520 | 0.759 | 0.750 | 0.767 | 0.830 | 0.020 | |
| AntiTb_RD | AtbPpred | 0.705 | 0.851 | 0.809 | 0.894 | 0.899 | – |
| Antitbpred | 0.570 | 0.785 | 0.733 | 0.838 | 0.860 | 0.4470 |
The first and the second column represent the dataset and the classifier name employed in this study. The third, fourth, fifth, sixth, and the seventh columns respectively represent the MCC, accuracy, sensitivity, specificity, and AUC. For comparison, we have included Antitbpred metrics reported in the literature [7]. The last column represents the pairwise comparison of ROC area under curves (AUCs) between AtbPpred and Antitbpred using a two-tailed t-test. P < .05 indicates a statistically meaningful difference between AtbPpred and the selected method (shown in bold).
14. Feature Selection Analysis
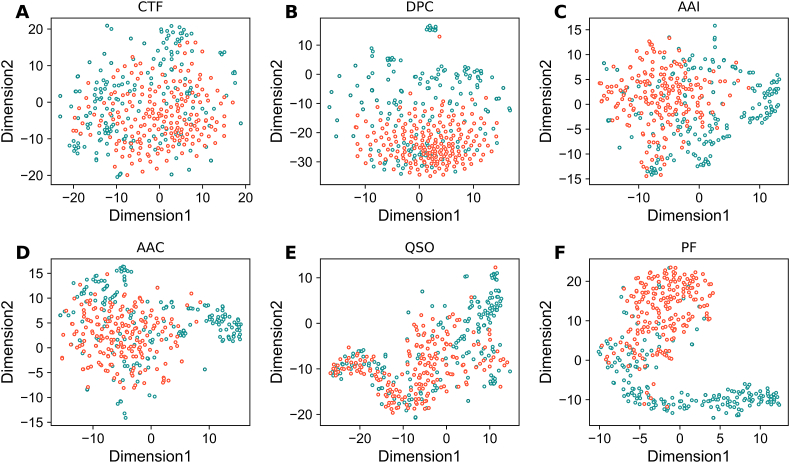
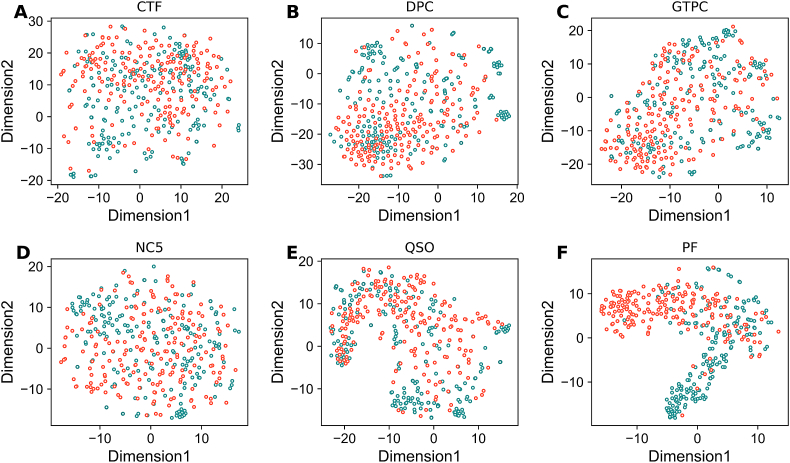
To explain the improved performance after feature optimization, we compared the spatial distribution between the optimal and original features. For an intuitive comparison, T-distributed stochastic neighbor embedding (t-SNE) implemented in Scikit with default parameters (n_components = 2, perplexity = 30, and learning_rate = 100) was employed for each encoding to reduce the multi-dimensional space into a two-dimensional one. Here, we compared nine probabilistic features that were obtained from the first layer with the top five feature encodings (CTD, DPC, AAI, AAC, and QSO) on AntiTB_RD dataset. Fig. 6 shows t-SNE distribution of the original and optimal features in the two-dimensional space. As shown in Fig. 6A–E, the positive (AtbPs) and negative (non-AtbPs) samples in the original feature space overlapped, indicating that the original feature space cannot effectively separate AtbPs from non-AtbPs. Conversely, probabilistic features (Fig. 6E) showed that most of the positives and negative samples in the feature space could be easily differentiated when compared to the original feature space, thus improving the performance. Furthermore, we computed t-SNE distribution for AntiTB_MD (Fig. 7) and observed similar trends with the AntiTB_RD results.
Fig. 6.
t-SNE visualization of the AntiTb_RD in a two-dimensional feature space. The dark cyan circles and salmon circles represent AtbPs and non-AtbPs, respectively. (A) CTF, (B) DPC, (C) AAI, (D) AAC, (E) QSO, and (F) nine-dimensional probabilistic features (PF).
Fig. 7.
t-SNE visualization of the AntiTb_MD in a two-dimensional feature space. The dark cyan and salmon circles respectively represent AtbPs and non-AtbPs. (A) CTF, (B) DPC, (C) GTPC, (D) NC5, (E) QSO, and (F) the nine-dimensional probabilistic features (PF).
15. Implementation of a Webserver
As mentioned in [57] and suggested in a series of publications [[58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68]] highlighting the importance in the development of webservers, we established a user-friendly webserver, AtbPpred (http:/thegleelab.org/AtbPpred), which is aimed at reaching a wide research community. To validate our findings, all data sets utilized in this study can be freely downloaded from our web server. Below, we provide a simple three-step guideline in the utility of our webserver to obtain final predicted outcomes. In the first step, users can select any one of the two prediction models. In the second step, submit the query sequences in the input query box. Note that the input sequences should be in FASTA format. Examples of FASTA-formatted sequences can be seen below the input box. In the final step, the ‘Submit’ button is clicked to provide the prediction results as the output. For each run, users can submit a maximum of 3000 peptides for a single run. Moreover, we scanned the entire APD database [69] and AMPfun dataset [70] and built a list of potential anti-tubercular peptides, which is available in our webserver (http://thegleelab.org/AtbPpred/AtbPData.html).
16. Conclusion
In this study, we developed a novel sequence-based two-layer predictor called AtbPpred for the identification of AtbPs from the provided peptide sequences. In this predictor, the optimal feature set was identified individually from nine different feature encodings and developed their corresponding prediction models in the first layer. Subsequently, all these models predicted scores were further considered as features and developed the final prediction model in the second layer. Unlike the previous method [7], AtbPpred integrates different aspects of sequence information through nine prediction models, thereby overcoming each model limitations and generating more stable predictions. Hence, AtbPpred showed consistent performance using both training and independent datasets, demonstrating the practicability and benefits of our proposed method. Two explanations could shed light on the robustness of our method. Firstly, we utilized ERT classifier for training. When compared to other ML algorithms, ERT showed better performance. Secondly, the probabilistic features used in the second layer can more effectively distinguish AtbPs and non-AtbPs in feature space, when compared to the feature encodings used in the first layer. Besides AtbP prediction, our proposed framework could be further extended to other peptide sequence-based predictors and applied to diverse computational biology fields [[71], [72], [73]]. Furthermore, our proposed method along with the increasing availability of experimentally verified data and novel features will greatly improve the prediction of AtbPs. To enable its wide use in the research community, we made AtbPpred available as a user-friendly public web server. AtbPpred is expected to be a valuable tool in the identification of hypothetical AtbPs in a high-throughput and cost-effective manner, further enabling the characterization of their functional mechanisms.
Funding
This work was supported by the National Natural Science Foundation of China (No. 61701340), the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education, Science, and Technology [2018R1D1A1B07049572 and 2018R1D1A1B07049494] and ICT & Future Planning [2016M3C7A1904392].
Declarations of Competing Interests
None declared.
Acknowledgements
The authors would like to thank Da Yeon Lee (Ajou University) and Chanyoung Ahn (UC Berkeley) for assistance with the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.06.024.
Contributor Information
Leyi Wei, Email: weileyi@tju.edu.cn.
Gwang Lee, Email: glee@ajou.ac.kr.
Appendix A. Supplementary data
Supplementary material
References
- 1.Banuls A.L., Sanou A., Anh N.T., Godreuil S. Mycobacterium tuberculosis: ecology and evolution of a human bacterium. J Med Microbiol. 2015;64:1261–1269. doi: 10.1099/jmm.0.000171. [DOI] [PubMed] [Google Scholar]
- 2.Mandal N., Anand P.K., Gautam S., Das S., Hussain T. Diagnosis and treatment of paediatric tuberculosis: an insight review. Crit Rev Microbiol. 2017;43:466–480. doi: 10.1080/1040841X.2016.1262813. [DOI] [PubMed] [Google Scholar]
- 3.Khusro A., Aarti C., Agastian P. Anti-tubercular peptides: a quest of future therapeutic weapon to combat tuberculosis. Asian Pac J Trop Med. 2016;9:1023–1034. doi: 10.1016/j.apjtm.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Pinto L., Menzies D. Treatment of drug-resistant tuberculosis. Infect Drug Resist. 2011;4:129–135. doi: 10.2147/IDR.S10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khusro A., Aarti C., Barbabosa-Pliego A., Salem A.Z.M. Neoteric advancement in TB drugs and an overview on the anti-tubercular role of peptides through computational approaches. Microb Pathog. 2018;114:80–89. doi: 10.1016/j.micpath.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 6.Teng T., Liu J., Wei H. Anti-mycobacterial peptides: from human to phage. Cell Physiol Biochem. 2015;35:452–466. doi: 10.1159/000369711. [DOI] [PubMed] [Google Scholar]
- 7.Usmani S.S., Bhalla S., Raghava G.P.S. Prediction of Antitubercular peptides from sequence information using ensemble classifier and hybrid features. Front Pharmacol. 2018;9:954. doi: 10.3389/fphar.2018.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Usmani S.S., Kumar R., Kumar V., Singh S., Raghava G.P.S. Database (Oxford). 2018. 2018. AntiTbPdb: a knowledgebase of anti-tubercular peptides. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar V., Agrawal P., Kumar R., Bhalla S., Usmani S.S., Varshney G.C. Prediction of cell-penetrating potential of modified peptides containing natural and chemically modified residues. Front Microbiol. 2018;9:725. doi: 10.3389/fmicb.2018.00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagpal G., Chaudhary K., Agrawal P., Raghava G.P.S. Computer-aided prediction of antigen presenting cell modulators for designing peptide-based vaccine adjuvants. J Transl Med. 2018;16:181. doi: 10.1186/s12967-018-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagpal G., Usmani S.S., Raghava G.P.S. A web resource for designing subunit vaccine against major pathogenic species of bacteria. Front Immunol. 2018;9:2280. doi: 10.3389/fimmu.2018.02280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Usmani S.S., Kumar R., Bhalla S., Kumar V., Raghava G.P.S. In silico tools and databases for designing peptide-based vaccine and drugs. Adv Protein Chem Struct Biol. 2018;112:221–263. doi: 10.1016/bs.apcsb.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Yang B., Leier A., Marquez-Lago T.T., Hayashida M., Rocker A. Bastion6: a bioinformatics approach for accurate prediction of type VI secreted effectors. Bioinformatics. 2018;34:2546–2555. doi: 10.1093/bioinformatics/bty155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J., Li J., Yang B., Xie R., Marquez-Lago T.T., Leier A. Bastion3: a two-layer ensemble predictor of type III secreted effectors. Bioinformatics. 2019;35(12):2017–2028. doi: 10.1093/bioinformatics/bty914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen J., Zhang J., Luo X., Zhu W., Yu K., Chen K. Predicting protein-protein interactions based only on sequences information. Proc Natl Acad Sci U S A. 2007;104:4337–4341. doi: 10.1073/pnas.0607879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawashima S., Kanehisa M. AAindex: amino acid index database. Nucleic Acids Res. 2000;28:374. doi: 10.1093/nar/28.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawashima S., Pokarowski P., Pokarowska M., Kolinski A., Katayama T., Kanehisa M. AAindex: amino acid index database, progress report 2008. Nucleic Acids Res. 2008;36:D202–D205. doi: 10.1093/nar/gkm998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saha I., Maulik U., Bandyopadhyay S., Plewczynski D. Fuzzy clustering of physicochemical and biochemical properties of amino acids. Amino Acids. 2012;43:583–594. doi: 10.1007/s00726-011-1106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manavalan B., Govindaraj R.G., Shin T.H., Kim M.O., Lee G. iBCE-EL: a new ensemble learning framework for improved linear B-cell epitope prediction. Front Immunol. 2018;9:1695. doi: 10.3389/fimmu.2018.01695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manavalan B., Basith S., Shin T.H., Wei L., Lee G. Meta-4mCpred: a sequence-based meta-predictor for accurate DNA 4mC site prediction using effective feature representation. Mol Ther Nucleic Acids. 2019;16:733–744. doi: 10.1016/j.omtn.2019.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan J.-X., Dao F.-Y., Lv H., Feng P.-M., Ding H. Identifying phage Virion proteins by using two-step feature selection methods. Molecules. 2018;23:2000. doi: 10.3390/molecules23082000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boopathi V., Subramaniyam S., Malik A., Lee G., Manavalan B., Yang D.C. mACPpred: a support vector machine-based meta-predictor for identification of anticancer peptides. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20081964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei L., Luan S., Nagai L.A.E., Su R., Zou Q. Exploring sequence-based features for the improved prediction of DNA N4-methylcytosine sites in multiple species. Bioinformatics. 2019;35:1326–1333. doi: 10.1093/bioinformatics/bty824. [DOI] [PubMed] [Google Scholar]
- 24.Manavalan B., Shin T.H., Kim M.O., Lee G. PIP-EL: a new ensemble learning method for improved proinflammatory peptide predictions. Front Immunol. 2018;9:1783. doi: 10.3389/fimmu.2018.01783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manavalan B., Shin T.H., Kim M.O., Lee G. AIPpred: sequence-based prediction of anti-inflammatory peptides using random Forest. Front Pharmacol. 2018;9:276. doi: 10.3389/fphar.2018.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manavalan B., Shin T.H., Lee G. PVP-SVM: sequence-based prediction of phage Virion proteins using a support vector machine. Front Microbiol. 2018;9:476. doi: 10.3389/fmicb.2018.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manavalan B., Basith S., Shin T.H., Wei L., Lee G. mAHTPred: a sequence-based meta-predictor for improving the prediction of anti-hypertensive peptides using effective feature representation. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty1047. (in press) [DOI] [PubMed] [Google Scholar]
- 28.Qiang X., Zhou C., Ye X., Du P.F., Su R., Wei L. CPPred-FL: a sequence-based predictor for large-scale identification of cell-penetrating peptides by feature representation learning. Brief Bioinform. 2018 doi: 10.1093/bib/bby091. (in press) [DOI] [PubMed] [Google Scholar]
- 29.Wei L., Chen H., Su R. M6APred-EL: a sequence-based predictor for identifying N6-methyladenosine sites using ensemble learning. Mol Ther Nucleic Acids. 2018;12:635–644. doi: 10.1016/j.omtn.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei L., Zhou C., Chen H., Song J., Su R. ACPred-FL: a sequence-based predictor based on effective feature representation to improve the prediction of anti-cancer peptides. Bioinformatics. 2018;34(23):4007–4016. doi: 10.1093/bioinformatics/bty451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geurts P., Ernst D., Wehenkel L. Extremely randomized trees. Mach Learn. 2006;63:3–42. [Google Scholar]
- 32.Basith S., Manavalan B., Shin T.H., Lee G. iGHBP: computational identification of growth hormone binding proteins from sequences using extremely randomised tree. Comput Struct Biotechnol J. 2018;16:412–420. doi: 10.1016/j.csbj.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abraham A., Pedregosa F., Eickenberg M., Gervais P., Mueller A., Kossaifi J. Machine learning for neuroimaging with scikit-learn. Front Neuroinform. 2014;8:14. doi: 10.3389/fninf.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dao F.Y., Lv H., Wang F., Feng C.Q., Ding H., Chen W. Identify origin of replication in Saccharomyces cerevisiae using two-step feature selection technique. Bioinformatics. 2019;35:2075–2083. doi: 10.1093/bioinformatics/bty943. [DOI] [PubMed] [Google Scholar]
- 35.Feng C.Q., Zhang Z.Y., Zhu X.J., Lin Y., Chen W., Tang H. iTerm-PseKNC: a sequence-based tool for predicting bacterial transcriptional terminators. Bioinformatics. 2019;35:1469–1477. doi: 10.1093/bioinformatics/bty827. [DOI] [PubMed] [Google Scholar]
- 36.Chou K.-C. Some remarks on protein attribute prediction and pseudo amino acid composition. J Theor Biol. 2011;273:236–247. doi: 10.1016/j.jtbi.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen W., Feng P., Yang H., Ding H., Lin H., Chou K.C. iRNA-3typeA: identifying three types of modification at RNA's adenosine sites. Mol Ther Nucleic Acids. 2018;11:468–474. doi: 10.1016/j.omtn.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W., Tang H., Ye J., Lin H., Chou K.C. iRNA-PseU: identifying RNA pseudouridine sites. Mol Ther Nucleic Acids. 2016;5 doi: 10.1038/mtna.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng P.M., Chen W., Lin H., Chou K.C. iHSP-PseRAAAC: identifying the heat shock protein families using pseudo reduced amino acid alphabet composition. Anal Biochem. 2013;442:118–125. doi: 10.1016/j.ab.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 40.Chen W., Yang H., Feng P., Ding H., Lin H. iDNA4mC: identifying DNA N4-methylcytosine sites based on nucleotide chemical properties. Bioinformatics. 2017;33:3518–3523. doi: 10.1093/bioinformatics/btx479. [DOI] [PubMed] [Google Scholar]
- 41.Lin H., Ding C., Song Q., Yang P., Ding H., Deng K.J. The prediction of protein structural class using averaged chemical shifts. J Biomol Struct Dyn. 2012;29:643–649. doi: 10.1080/07391102.2011.672628. [DOI] [PubMed] [Google Scholar]
- 42.Yang H., Tang H., Chen X.X., Zhang C.J., Zhu P.P., Ding H. Identification of secretory proteins in Mycobacterium tuberculosis using pseudo amino acid composition. Biomed Res Int. 2016;2016:5413903. doi: 10.1155/2016/5413903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y.W., Su Z.D., Yang W., Lin H., Chen W., Tang H. IonchanPred 2.0: a tool to predict ion channels and their types. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18091838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manavalan B., Basith S., Shin T.H., Choi S., Kim M.O., Lee G. MLACP: machine-learning-based prediction of anticancer peptides. Oncotarget. 2017;8:77121–77136. doi: 10.18632/oncotarget.20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei L., Ding Y., Su R., Tang J., Zou Q. Prediction of human protein subcellular localization using deep learning. J Parallel Distrib Comput. 2018;117:212–217. [Google Scholar]
- 46.Wei L., Tang J., Zou Q. Local-DPP: an improved DNA-binding protein prediction method by exploring local evolutionary information. Inform Sci. 2017;384:135–144. [Google Scholar]
- 47.Wei L., Wan S., Guo J., Wong K.K. A novel hierarchical selective ensemble classifier with bioinformatics application. Artif Intell Med. 2017;83:82–90. doi: 10.1016/j.artmed.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Su R., Hu J., Zou Q., Manavalan B., Wei L. Empirical comparison and analysis of web-based cell-penetrating peptide prediction tools. Brief Bioinform. 2019 doi: 10.1093/bib/bby124. (in press) [DOI] [PubMed] [Google Scholar]
- 49.Xu Z.C., Feng P.M., Yang H., Qiu W.R., Chen W., Lin H. iRNAD: a computational tool for identifying D modification sites in RNA sequence. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz358. (in press) [DOI] [PubMed] [Google Scholar]
- 50.Zhao X., Chen L., Lu J. A similarity-based method for prediction of drug side effects with heterogeneous information. Math Biosci. 2018;306:136–144. doi: 10.1016/j.mbs.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 51.Chen L., Pan X., Zhang Y.H., Kong X., Huang T., Cai Y.D. Tissue differences revealed by gene expression profiles of various cell lines. J Cell Biochem. 2018 doi: 10.1002/jcb.27977. (in press) [DOI] [PubMed] [Google Scholar]
- 52.Chen L., Pan X., Hu X., Zhang Y.H., Wang S., Huang T. Gene expression differences among different MSI statuses in colorectal cancer. Int J Cancer. 2018 doi: 10.1002/ijc.31554. (in press) [DOI] [PubMed] [Google Scholar]
- 53.Chen W., Lv H., Nie F., Lin H. i6mA-Pred: identifying DNA N6-methyladenine sites in the rice genome. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz015. (in press) [DOI] [PubMed] [Google Scholar]
- 54.Wei L., Su R., Luan S., Liao Z., Manavalan B., Zou Q. Iterative feature representations improve N4-methylcytosine site prediction. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz408. (in press) [DOI] [PubMed] [Google Scholar]
- 55.Wei L., Zhou C., Chen H., Song J., Su R. ACPred-FL: a sequence-based predictor using effective feature representation to improve the prediction of anti-cancer peptides. Bioinformatics. 2018;34:4007–4016. doi: 10.1093/bioinformatics/bty451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei L., Hu J., Li F., Song J., Su R., Zou Q. Comparative analysis and prediction of quorum-sensing peptides using feature representation learning and machine learning algorithms. Brief Bioinform. 2018 doi: 10.1093/bib/bby107. (in press) [DOI] [PubMed] [Google Scholar]
- 57.Chou K.-C., Shen H.-B. Recent advances in developing web-servers for predicting protein attributes. Nat Sci. 2009;1:63. [Google Scholar]
- 58.Cao R., Adhikari B., Bhattacharya D., Sun M., Hou J., Cheng J. QAcon: single model quality assessment using protein structural and contact information with machine learning techniques. Bioinformatics. 2017;33:586–588. doi: 10.1093/bioinformatics/btw694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao R., Bhattacharya D., Hou J., Cheng J. DeepQA: improving the estimation of single protein model quality with deep belief networks. BMC Bioinformatics. 2016;17:495. doi: 10.1186/s12859-016-1405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao R., Freitas C., Chan L., Sun M., Jiang H., Chen Z. ProLanGO: protein function prediction using neural machine translation based on a recurrent neural network. Molecules. 2017;22 doi: 10.3390/molecules22101732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zou Q., Sr., Xing P., Wei L., Liu B. Gene2vec: gene subsequence embedding for prediction of mammalian N6-Methyladenosine sites from mRNA. RNA. 2019;25:205–218. doi: 10.1261/rna.069112.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shoombuatong W., Schaduangrat N., Pratiwi R., Nantasenamat C. THPep: a machine learning-based approach for predicting tumor homing peptides. Comput Biol Chem. 2019;80:441–451. doi: 10.1016/j.compbiolchem.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 63.Schaduangrat N., Nantasenamat C., Prachayasittikul V., Shoombuatong W. ACPred: a computational tool for the prediction and analysis of anticancer peptides. Molecules. 2019;24 doi: 10.3390/molecules24101973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khatun M.S., Hasan M.M., Kurata H. PreAIP: computational prediction of anti-inflammatory peptides by integrating multiple complementary features. Front Genet. 2019;10:129. doi: 10.3389/fgene.2019.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hasan M.M., Rashid M.M., Khatun M.S., Kurata H. Computational identification of microbial phosphorylation sites by the enhanced characteristics of sequence information. Sci Rep. 2019;9:8258. doi: 10.1038/s41598-019-44548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Win T.S., Schaduangrat N., Prachayasittikul V., Nantasenamat C., Shoombuatong W. PAAP: a web server for predicting antihypertensive activity of peptides. Future Med Chem. 2018;10:1749–1767. doi: 10.4155/fmc-2017-0300. [DOI] [PubMed] [Google Scholar]
- 67.Hasan M.M., Kurata H. GPSuc: global prediction of generic and species-specific Succinylation sites by aggregating multiple sequence features. PLoS One. 2018;13 doi: 10.1371/journal.pone.0200283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hasan M.M., Khatun M.S., Mollah M.N.H., Yong C., Dianjing G. NTyroSite: computational identification of protein Nitrotyrosine sites using sequence evolutionary features. Molecules. 2018;23 doi: 10.3390/molecules23071667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang G., Li X., Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chung C.R., Kuo T.R., Wu L.C., Lee T.Y., Horng J.T. Characterization and identification of antimicrobial peptides with different functional activities. Brief Bioinform. 2019 doi: 10.1093/bib/bbz043. (in press) [DOI] [PubMed] [Google Scholar]
- 71.Chen X., Chou W.C., Ma Q., Xu Y. SeqTU: a web server for identification of bacterial transcription units. Sci Rep. 2017;7:43925. doi: 10.1038/srep43925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X., Yu B., Ma A., Chen C., Liu B., Ma Q. Protein-protein interaction sites prediction by ensemble random forests with synthetic minority oversampling technique. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty995. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang J., Chen X., McDermaid A., Ma Q. DMINDA 2.0: integrated and systematic views of regulatory DNA motif identification and analyses. Bioinformatics. 2017;33:2586–2588. doi: 10.1093/bioinformatics/btx223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material