The mechanisms by which MGMT-deficient glioblastomas acquire resistance to temozolomide are unclear. Yi et al. show that retrograde nuclear transportation of the DNA damage repair proteins XPC and CBX5 confers temozolomide resistance in MGMT-deficient glioblastoma cells. Development of DHC2 inhibitors could be a promising strategy for overcoming acquired temozolomide resistance.
Keywords: DHC2, XPC, CBX5, acquired TMZ resistance, glioblastoma
Abstract
The acquisition of temozolomide resistance is a major clinical challenge for glioblastoma treatment. Chemoresistance in glioblastoma is largely attributed to repair of temozolomide-induced DNA lesions by O6-methylguanine-DNA methyltransferase (MGMT). However, some MGMT-deficient glioblastomas are still resistant to temozolomide, and the underlying molecular mechanisms remain unclear. We found that DYNC2H1 (DHC2) was expressed more in MGMT-deficient recurrent glioblastoma specimens and its expression strongly correlated to poor progression-free survival in MGMT promotor methylated glioblastoma patients. Furthermore, silencing DHC2, both in vitro and in vivo, enhanced temozolomide-induced DNA damage and significantly improved the efficiency of temozolomide treatment in MGMT-deficient glioblastoma. Using a combination of subcellular proteomics and in vitro analyses, we showed that DHC2 was involved in nuclear localization of the DNA repair proteins, namely XPC and CBX5, and knockdown of either XPC or CBX5 resulted in increased temozolomide-induced DNA damage. In summary, we identified the nuclear transportation of DNA repair proteins by DHC2 as a critical regulator of acquired temozolomide resistance in MGMT-deficient glioblastoma. Our study offers novel insights for improving therapeutic management of MGMT-deficient glioblastoma.
Introduction
Glioblastoma multiforme (GBM) is the most common form of primary malignant brain tumour in adults and is also the most lethal cancer of the CNS (Wen and Kesari, 2008; Louis et al., 2016). Despite advances in understanding the molecular mechanisms underlying tumorigenesis, most current treatments are still ineffective, including targeted treatments such as EGFR- and VEGF-targeted agents (Gilbert et al., 2014; Reifenberger et al., 2017; Weller et al., 2017). Temozolomide (TMZ) is the only chemotherapeutic drug that has been confirmed to significantly prolong the overall survival of patients with GBM, but the median survival is still only 12–15 months after receiving a standard treatment course (Stupp et al., 2005, 2017). Resistance to TMZ therapy is an important issue and also a major cause of treatment failure, indicating that overcoming TMZ resistance is critical to improve treatment outcomes.
TMZ is an alkylating agent that methylates DNA at the N7 position of guanine, the O3 position of adenine, and the O6 position of guanine. The cytotoxicity of this drug is mostly due to O6-methylguanine-induced DNA damage (Esteller et al., 2000; Friedman et al., 2000). The lack of response to TMZ treatment is typically due to intrinsic and acquired resistance of tumours to the drug. Expression of the demethylating enzyme O6-methylguanine-DNA methyltransferase (MGMT) is a key pathway that has been implicated in intrinsic TMZ resistance. MGMT induces resistance to TMZ by removing alkyl groups from the O6 position of guanine directly (Pegg and Byers, 1992; Weller et al., 2010). The current consensus is that MGMT expression is inhibited by the methylation of MGMT promoter, which results in the TMZ-resistance. Recent studies reported some new mechanisms for MGMT regulation, such as K-M enhancer activation (Chen et al., 2018), highlighting the complexity of TMZ-resistance. Nevertheless, MGMT protein is the key factor for TMZ-resistance and the expression remains relatively stable during the course of glioblastoma (Brandes et al., 2017). Our previous study also showed that both MGMT promoter methylation status and MGMT protein expression were not changed in MGMT-negative U87 TMZ-resistant cells (Yi et al., 2016). In addition, analysis of MGMT promoter methylation revealed that only approximately half of GBMs expressed MGMT (Hegi et al., 2005; Brennan et al., 2013). Many studies have shown that low levels of MGMT in GBMs were sufficient to confer TMZ resistance (Sarkaria et al., 2008; Agnihotri et al., 2014). These findings suggest the existence of a MGMT-independent mechanism for TMZ resistance that has yet to be reported.
Our previous studies found that GBM cells with acquired TMZ resistance showed activated autophagy process (Lu et al., 2015), significant morphological changes (Yi et al., 2016) and cytoskeletal rearrangements that were associated with increased expression of DYNC2H1 (dynein, cytoplasmic 2, heavy chain 1, also known as DHC2) (Wang et al., 2016). DHC2 belongs to the cytoplasmic dynein protein family and encodes the heavy chain of the cytoplasmic dynein-2 motor required for trafficking cargo from the tip to the base of the cilium (Ichikawa et al., 2011; Ocbina et al., 2011). Mutations in human DHC2 are associated with the ciliopathic disorders, short rib-polydactyly syndrome type III and Jeune asphyxiating thoracic dystrophy (JATD) (Dagoneau et al., 2009). An association between DHC2 and TMZ-sensitivity of GBM was confirmed in our previous study (Wang et al., 2016), but the mechanism remains elusive. In this study, we used a combination of subcellular proteomics and in vitro analyses to verify DHC2 as a critical regulator of acquired TMZ resistance in MGMT-deficient GBM cells. We also showed that the underlying mechanism involved nuclear transportation of XPC and CBX5, which augmented DNA damage repair activity.
Materials and methods
Cell lines, human tumour tissues and study approval
The human GBM cell line U87 (ATCC lot number: 63710285) and T98G (ATCC lot number: 70000188) cells were purchased from the American Type Culture Collection (ATCC). GBM1 and GBM2 are primary GBM cells isolated from fresh tumour tissues. Both GBM1 and U87 are MGMT-deficient GBM cells, and GBM2 and T98G are MGMT-positive GBM cells, which was verified by western blotting and pyrosequencing methylation assays. All GBM patient samples and matching clinical data were obtained from Nanfang Hospital, Southern Medical University (Supplementary Tables 1 and 2). This study was approved by the ethics committee of Southern Medical University.
Gene expression profiling and survival analysis
Gene expression profiling of DYNC2H1 in gliomas, Kaplan-Meier analysis and survival curves were carried out using the R2 microarray analysis and visualization platform (http://r2.amc.nl). All of the Kaplan-Meier survival curves were generated using the Kaplan scan algorithm of the R2 database for the most optimal mRNA cut-off expression level to discriminate between a good and bad prognosis, and the listed P-values were calculated without using a Bonferroni correction. The cBioportal tool (http://www.cbioportal.org/) was used for the analysis of co-expression with DHC2 in the TCGA (The Cancer Genome Atlas) GBM database.
Cell viability and colony formation assays
Cells with target gene knockout or knockdown and control cells (105 cells) were plated into six-well dishes in 2 ml of Dulbecco’s modified Eagle medium (DMEM) with foetal bovine serum (FBS) in the presence or absence of TMZ for 7 days. Cells were collected and cell count and cell viability were analysed using Trypan blue (Solarbio Inc). Additional viability assays were performed in 96-well assays at 103 cells per well culture using the CCK-8 cell viability kit (Dojindo Laboratories) according to the manufacturer’s instructions.
For the colony formation assay, DHC2 knockout or knockdown cells and the control cells were cultured in six-well plates with 200 cells per well in the presence or absence of TMZ treatment for 24 h. After 2 weeks, visible colonies were fixed with 100% methanol and stained with 0.1% crystal violet in 20% methanol for 15 min. The area of colonies was calculated using ImageJ software (1.48v, National Institutes of Health, USA).
In vivo analyses
The details on subcutaneous, orthotopic xenograft studies and therapeutic regimens in mice are provided in the Supplementary material. Investigators were blinded to the treatment groups. All animal experiments were conducted with the approval from the Southern Medical University Institutional Committee for Animal Research and conformed with the national guidelines for the care and use of laboratory animals.
MRI of orthotopic mouse tumours
Intracranial tumour growth was monitored in vivo in isoflurane-anaesthetized mice by MRI after inoculation using a Bruker 7.0 T scanner (Bruker BioSpin GmbH) with a 16 cm bore. T2-weighted images were acquired by a rapid acquisition relaxation-enhanced sequence.
Immunohistochemistry
Immunohistochemistry assays were carried out on GBM samples or nude mouse xenograft tumour tissues to detect and score DHC2, MGMT, γH2AX, XPC and CBX5 expression. Paraffin-embedded blocks were cut into 4-μm sections and deparaffinized and rehydrated. Antigen retrieval was performed by pressure cooking for 5 min in citrate buffer (pH 6.0), followed by blocking of endogenous peroxidase in 0.3% H2O2. After blocking with 5% bovine serum albumin (BSA) for 1 h, sections were incubated sequentially with primary antibodies and horseradish peroxidase-linked secondary antibody. Sections were covered with diaminobenzidine for visualizing the staining and then counterstained with haematoxylin before being examined by microscope.
Immunofluorescence
For immunofluorescence, 104 cells were grown on 15 mm confocal petri dishes and treated with TMZ for the indicated time. Cells were fixed for 10 min in 4% paraformaldehyde, permeabilized with 0.5% Triton™ X for 10 min and blocked with 5% BSA for 1 h. After removal of BSA, cells were incubated with the indicated primary antibody (γH2AX, DHC2, XPC and CBX5) and detected using Alexa Fluor® 488- or 546-labelled secondary antibodies (Invitrogen). Cells were also stained with DAPI. Images were captured on a Carl-Zeiss confocal microscope.
Ubiquitination assay
For the in vitro ubiquitination assay, U87 DHC2(−/−) cells were transfected with FLAG-tagged ubiquitin plasmids and treated with TMZ. At 72 h after treatment, cells were treated with 10 mM of the proteasome inhibitor MG132 for 6 h. Cells were then lysed in Cell Lysis Buffer (Cell Signaling Technology) for western blotting and immunoprecipitation.
Statistical analysis
Statistical evaluations were carried out using SPSS statistical software, version 19.0 (SPSS Inc., Chicago, IL, USA). All experiments were performed in triplicate with the mean and standard error of the mean (SEM) reported. Where appropriate, ANOVA was conducted for multigroup comparisons, and direct comparisons were conducted using an unpaired two-tailed Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001; ns = no significance).
Data availability
The data supporting the findings of this study are available within the article and its Supplementary material.
Results
DYNC2H1 expression correlates with response to TMZ and prognosis of MGMT-deficient GBM patients
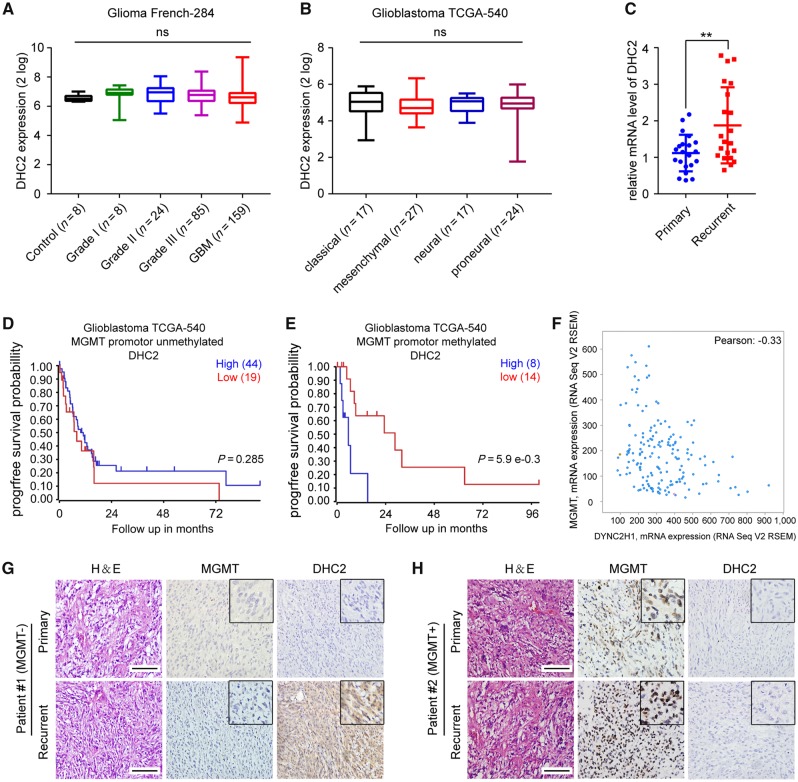
We sought to determine whether DYNC2H1 expression was associated with prognosis in GBM patients. To this end, we analysed data from the Glioblastoma TCGA-540 database and found that high DYNC2H1 expression was associated with poor overall survival of GBM patients and was also associated with poor prognosis of other tumours, such as adrenocortical carcinoma, pancreatic adenocarcinoma and colon tumours (Supplementary Fig. 1), suggesting that DHC2 may play an oncogenic role. Next, to assess whether DYNC2H1 expression was also related to glioma progression, we analysed the expression data from the Glioma French-284 database and Glioblastoma TCGA-540 database, which are available from the online R2 genomic analysis and visualization platform. No differences were observed in DYNC2H1 expression across different grades of malignant glioma tissues, in comparison to adjacent normal brain tissues (Fig. 1A). There were also no observed differences in DYNC2H1 expression among various GBM subtypes (Fig. 1B). These results indicate a lack of association of DHC expression with glioma progression.
Figure 1.
Analysis of the correlation of DHC2 expression to MGMT expression and TMZ treatment responses in GBM patients. (A) DYNC2H1/DHC2 expression across glioma tissues of different grades and healthy tissues from the tumour Glioma French-284 database. ns = no significance. (B) DYNC2H1/DHC2 expression across different GBM subtypes in the Glioblastoma TCGA-540 database. (C) qRT-PCR analysis of DYNC2H1 mRNA expression levels in 21 primary GBMs without TMZ treatment and 21 recurrent GBMs with standard TMZ treatment. GAPDH expression was used for normalization. **P < 0.01. (D) Progression-free (progrfree) survival analysis of GBM patients without MGMT promoter methylation using the Glioblastoma TCGA-540 database. (E) Progression-free survival analysis of GBM patients with MGMT promoter methylation using the Glioblastoma TCGA-540 database. (F) Co-expression analysis of DYNC2H1 and MGMT mRNA expression using the Glioblastoma TCGA-provisional database. (G and H) Representative images of immunohistochemical analysis of DYNC2H1 and MGMT in six paired primary and recurrent GBM tissues. Scale bars = 100 μm. H&E = haematoxylin and eosin.
We previously demonstrated using immunohistochemistry that DHC2 expression was higher in GBM tissues from TMZ-treated patients with recurrent GBM than in primary GBM tissues from patients that did not receive TMZ treatment (Wang et al., 2016). To confirm these findings, we performed the same comparison by evaluating mRNA expression of DYNC2H1 and observed that it was expressed at higher levels in TMZ-treated recurrent GBM in comparison to primary GBM without TMZ treatment (Fig. 1C). The data on the patient cohort can be seen in Supplementary Table 1. These findings suggest that DHC2 may be involved in TMZ-treatment response of GBM patients.
Intriguingly, we further observed that high DYNC2H1 expression was significantly related to poor progression-free survival (PFS) in GBM patients with MGMT promoter methylation, and no significant relationship to PFS was observed in GBM patients with no MGMT promoter methylation (Fig. 1D and E). Furthermore, we found that mRNA expression of DHC2 was negatively related to MGMT expression after analysing the Glioblastoma TCGA-Provisional database (Fig. 1F). Next, immunohistochemical analysis of MGMT and DHC2 was performed in paired primary and recurrent GBM tumour tissues collected from the same patient receiving standard TMZ therapy. We observed that recurrent tumours with high levels of MGMT tended to express low levels of DHC2, whereas tumours with low levels of MGMT tended to express high levels of DHC2 (Fig. 1G and H). Taken together, these findings demonstrate a negative correlation between DHC2 expression and MGMT expression, and indicate a strong correlation of DHC2 with TMZ response and survival probability in MGMT-deficient GBM patients.
Sensitivity of MGMT-negative GBM cells to TMZ is associated with regulation of DNA damage by DHC2
To investigate how the negative correlation between DHC2 and MGMT impacts response of GBM cells to TMZ, we knocked down DHC2 expression in two MGMT-negative cell lines (U87, GBM1) and two MGMT-positive cell lines (T98G, GBM2). Both GBM1 and GBM2 are primary tumour cell lines isolated from GBM tumours, and GFAP, GSC markers (SOX2, SOX9, CD133, CD56) and pericyte/MSC markers (CD248, CD105) were checked to confirm these cultures were GBM (Supplementary Fig. 2A). MGMT promoter methylation status and protein expression of these cell lines are showed in Supplementary Fig. 2B, C and Supplementary Table 2. The quantity of MGMT during the TMZ treatment course in our experimental models was also detected, the MGMT expression was quite stable before and after TMZ treatment (Supplementary Fig. 2D). Furthermore, to strengthen our findings, CRISPR/Cas9-mediated genome editing was used to knockout DYNC2H1 in MGMT-deficient GBM cell lines, including U87 and GBM1 cells. However, the DYNC2H1 gene was successfully knocked out only in U87 cells (Supplementary Fig. 2E–G), but failed in GBM1 cells. As the U87 cell line is a commonly used GBM cell line tool, DYNC2H1 knockout U87 cells served as a key experimental model to further clarify the mechanism of DHC2 in TMZ resistance. Efficiency of short hairpin RNA (shRNA)-mediated knockdown of DYNC2H1 was evaluated by qRT-PCR (Supplementary Fig. 3A).
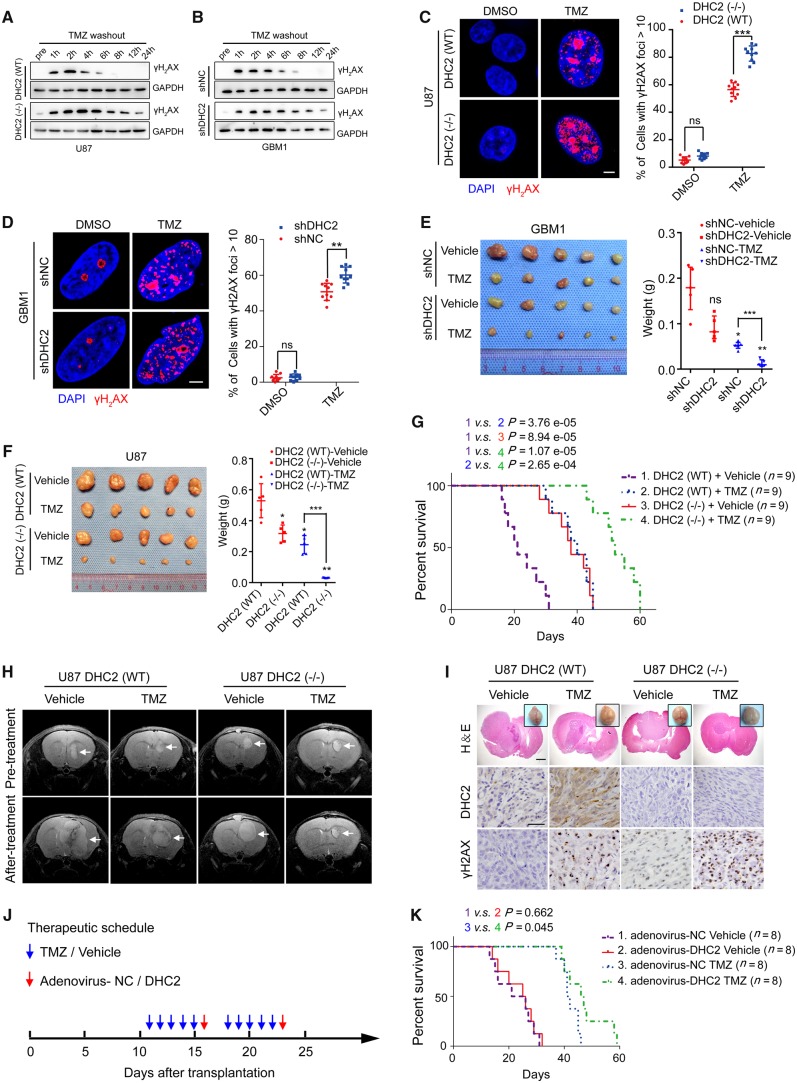
While treatment of all four cell lines with TMZ at the IC50 concentration (Supplementary Fig. 3B; 246.77 μM for U87, 457.89 μM for GBM1; 756.82 μM for T98G; 2298.56 μM for GBM2) led to a significant decrease in the cell viability, knockdown or knockout of DHC2 in the MGMT-negative (U87, GBM1), but not in the MGMT-positive (T98G, GBM2) cell lines treated with TMZ led to significant reduction in cell survival (Supplementary Fig. 3C). Next, we assessed the DNA damage status of U87 and GBM1 cells by analysing the phosphorylation of H2AX (γH2AX). Western blotting analysis showed that depletion of DHC2 in both U87 and GBM1 cells resulted in persistently high levels of γH2AX up to 24 h after TMZ treatment, in comparison to the control cells where γH2AX could not be detected beyond 8 h post-TMZ (Fig. 2A and B). Consistent with these results, immunofluorescence analysis revealed that the active foci of γH2AX were significantly increased upon TMZ treatment in DHC2-depleted U87 and GBM1 cells (Fig. 2C and D).
Figure 2.
DHC2 mediates TMZ-induced DNA damage repair of MGMT-deficient GBM cells. (A) Wild-type (WT) or CRISPR/Cas9-mediated DYNC2H1/DHC2 knockout (−/−) U87 cells were treated with 200 μM TMZ for 24 h, then the TMZ-contained culture media were replaced with common culture media and the TMZ-treated GBM cells were allowed to repair for indicated time points, and followed by western blotting analysis of cellular lysates for the γH2AX expression, using GAPDH as loading control. pre = pretreatment, indicates U87 cells without TMZ treatment. (B) The same experimental processes were conducted in shNC (normal control shRNA-transduced) and shDHC2 (DHC2 knockdown shRNA-transduced) GBM1 cells (400 μM TMZ for treatment), western blotting analysis of γH2AX expression and GAPDH served as the internal loading control. (C and D) DYNC2H1DHC2(WT) or DYNC2H1/DHC2(−/−) U87 cells (C) and shNC or shDHC2-transduced GBM1 cells (D) were treated with the indicated concentration of TMZ (200 μM for U87, 400 μM for GBM1) for 24 h and recover for 6 h with TMZ washing out. Immunofluorescence staining of γH2AX foci and the percentage of cells containing >10 γH2AX foci in 10 random microscopic fields was calculated. Scale bar = 5 μm. **P < 0.01, ***P < 0.001, ns = no significance. (E and F) Representative images of subcutaneous xenografts in nude mice derived from GBM1 shNC and shDHC2 cells (E), or U87 DHC2(WT) and DHC2(−/−) cells (F) with or without TMZ treatment (intraperitoneal injection of 20 mg/kg TMZ in saline) on the 42nd day, shNC-vehicle or DHC2(WT)-vehicle group served as the control and the comparison symbol above the bar represented the statistical results compared with the control group. *P < 0.05; **P < 0.01; ***P < 0.001; ns = no significance. (G) T2-weighted MRI of intracranial xenografts (arrows) in mice bearing DHC2(WT) or DHC2(−/−) U87 cells before and after treatment with vehicle alone or TMZ. (H) Brain images, haematoxylin and eosin staining and immunohistochemical analyses for DHC2 and γH2AX expression in sections of representative intracranial tumour-bearing mice from each treatment arm. Scale bar = 1 mm in haematoxylin and eosin, 25 μm in immunohistochemistry images. (I) Survival curve of mice with DHC2(WT) and DHC2(−/−) U87 cell-derived intracranial xenografts treated with vehicle or TMZ. (J) Schematic representation of the therapeutic schedule followed. In each treatment course, 20 mg/kg TMZ/vehicle was injected intraperitoneally into mice, and 109 units of shDHC2/shNC adenovirus were injected intratumourally. (K) Survival curve of mice harbouring the U87 cell-derived intracranial xenografts and subjected to the therapeutic schedule in J.
We next analysed the tumorigenic potential of DHC2 knockdown cells upon TMZ treatment by colony formation assays and xenograft mouse models. Depletion of DHC2 in both U87 and GBM1 cells resulted in significantly reduced colony formation upon TMZ treatment, in comparison to DHC2-expressing control cells with TMZ treatment (Supplementary Fig. 3D and E). Our in vivo results in Fig. 2E–G indicate that knockdown or knockout of DYNC2H1 had significant effects on tumorigenic potential and survival without TMZ treatment. Furthermore, DHC2 also had an impact on TMZ sensitivity in GBM cells in vivo. Nude mice subcutaneously transplanted with DHC2-depleted U87 and GBM1 cells developed tumours with significantly reduced volumes and weights upon treatment with TMZ, indicating that loss of DHC2 sensitized U87 and GBM1 cells to TMZ treatment in vivo (Fig. 2E and F). Additionally, mice bearing intracranial xenografts derived from DHC2 knockout U87 cells showed a drastic decrease in tumour volume upon treatment with TMZ, as revealed by T2-weighted MRI (Fig. 2H). We next determined whether DYNC2H1 silencing could mediate TMZ sensitivity in MGMT-positive T98G and GBM2 cells. In contrast to our findings with MGMT-deficient cells (U87, GBM1), we found that DYNC2H1 knockdown in MGMT-positive cells (T98G, GBM2) did not affect cell viability (Supplementary Fig. 3C), CFU formation (Supplementary Fig. 3F and G) or DNA damage status (Supplementary Fig. 3H and I) after TMZ treatment. In addition, we also found that overexpression of the MGMT protein in DYNC2H1 knockout MGMT-deficient U87 cells prevented TMZ-induced CFU reduction and DNA damage (Supplementary Fig. 3J and K).
Histological analysis showed that all intracranial tumours originating from DHC2-depleted U87 cells (Fig. 2I) or subcutaneous tumours originating from DHC2-depleted GBM1 cells (Supplementary Fig. 3L) exhibited significant upregulation of γH2AX upon TMZ treatment. Reduction in tumour progression upon TMZ treatment correlated with survival. The TMZ-treated, DHC2-depleted xenograft group exhibited significantly increased survival compared with the TMZ-treated control xenograft group (Fig. 2G).
In summary, our results showed that loss of DHC2 in MGMT-negative cells results in increased DNA damage, increased sensitivity to TMZ and impaired tumorigenesis.
DHC2-targeted therapy sensitizes mice with GBM to TMZ
To evaluate the therapeutic potential of DHC2, we used a high titre adenovirus (1.7 × 1011 pfu/ml) for DYNC2H1 knockdown in vivo. U87 cells were injected intracranially into mice, and these tumour-bearing mice were treated with control shRNA (shNC) or shDHC2 adenovirus in the presence or absence of TMZ. The therapeutic schedule is shown in Fig. 2J. Consistent with the results in vitro, mice treated with TMZ and shDHC2 adenovirus exhibited a significant increase in overall survival compared to mice treated with TMZ alone (Fig. 2K). These findings shed light on the clinical applicability of DHC2-targeted therapy in improving TMZ sensitivity.
Subcellular proteomic analysis identifies XPC and CBX5 as potential regulatory partners of DHC2
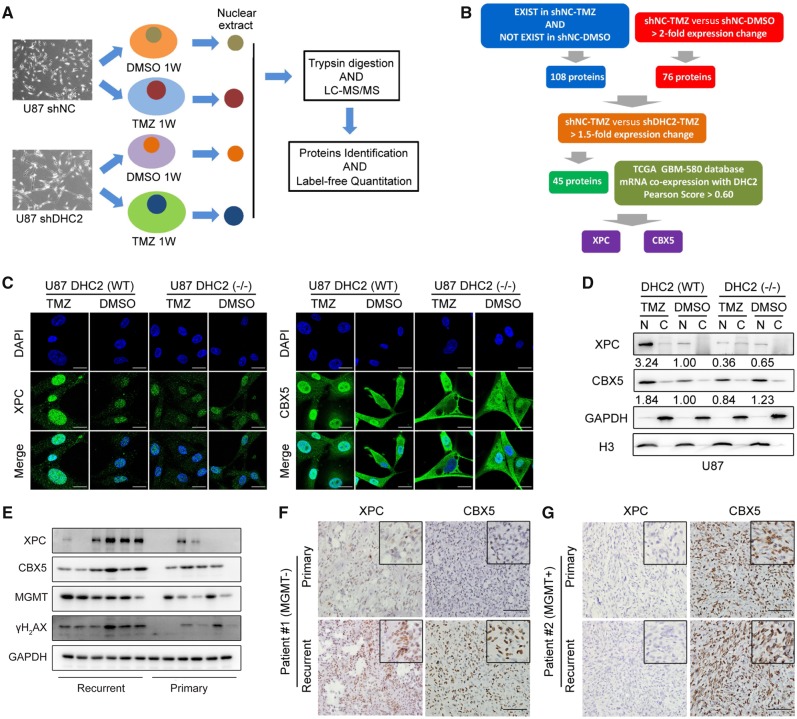
To explore the mechanism of DHC2-mediated TMZ-induced DNA damage response in MGMT-deficient GBM cells, a proteomic analysis was performed on DHC2-depleted TMZ-treated cells to identify components involved in DHC2-mediated responses to TMZ. We treated DHC2-knockdown and control U87 cells with TMZ or dimethyl sulphoxide (DMSO) for 1 week and then harvested the nuclear fractions for proteomic analysis (Fig. 3A). Over 2000 proteins were identified, and we set a criterion to select the potential partners of DHC2 among the large amount of proteins. A total of 45 differentially expressed proteins (DEPs) were identified and the selection criteria are described in the Supplementary material and the ‘Materials and methods’ section. A protein–protein interaction (PPI) network of the 45 DEPs was constructed (Supplementary Fig. 4A). Gene Ontology Biological Process indicated that the 45 DEPs were enriched in chromosome organization process, cellular response to DNA damage stimulus process, DNA repair process and others (Supplementary Fig. 4B). To identify the DEPs that most strongly correlated with DHC2 expression, we analysed the TCGA GBM-540 DYNC2H1 mRNA co-expression dataset and set the cut-off value to a Pearson score > 0.6, which predicted strong correlation with DHC2. Ultimately, both XPC (xeroderma pigmentosum group C-complementing protein, DNA repair protein complementing XP-C cells) and CBX5 (chromobox protein homologue 5, heterochromatin protein 1 homologue alpha) were identified as the most correlated potential regulatory partners of DHC2 with Pearson scores >0.6 (Supplementary Fig. 4C). XPC plays an important role in nucleotide excision repair and high expression of XPC is correlated with poor overall survival of GBM patients according to our analysis of the TCGA GBM-540 database (Supplementary Fig. 4D). CBX5 is a component of heterochromatin. Both XPC and CBX5 have been reported to be involved in DNA damage repair in previous studies (Min and Pavletich, 2007; Dinant and Luijsterburg, 2009; Zarebski et al., 2009; Nemzow et al., 2015). On this basis, we identified XPC and CBX5 as being strongly correlated with DHC2 and to be potential interacting/regulatory partners of DHC2. A schematic representation of this analysis is shown in Fig. 3B.
Figure 3.
Proteomic analysis to identify potential regulatory partners of DHC2 and subsequent validation of targets. (A) Schematic representation of protocol followed for proteomic analysis. shNC and shDHC2 U87 cells were treated with 200 μM TMZ or dimethyl sulphoxide (DMSO) for 1 week, the nuclear fractions were harvested for proteomic analysis, experimental procedures of mass spectrometry proteomic were described in ‘Materials and methods’ section. (B) Schematic of DHC2 downstream analysis. First, to select the related proteins with TMZ-treatment response in GBM cells, DEPs were identified by meeting either of the following criteria: (i) with a significant fold change of >2 in the U87-TMZ group compared with the U87-DMSO group (U87 shNC-TMZ/U87 shNC-DMSO > 2); or (ii) only present in the U87-TMZ group but not in the U87-DMSO group (indicated proteins related to TMZ treatment). A total of 184 DEPs were identified in this study. Next, we sorted proteins correlated with DHC2 expression among these 184 DEPs. We filtered the 184 DEPs with >1.5-fold changes in the U87-TMZ group compared with the DHC2 knockdown U87-TMZ group (U87 shNC-TMZ/U87 shDHC2-TMZ >1.5; also indicated DEPs downregulated in DHC2 knockdown U87-TMZ group compared with U87-TMZ group), and a total of 45 DEPs were identified as candidate regulatory proteins of DHC2. Finally, we analysed the TCGA GBM-540 DYNC2H1/DHC2 mRNA co-expression dataset and set the cut-off value to a Pearson score >0.6 to identify the DEPs that most strongly correlated with DHC2 expression. Both XPC and CBX5 were then identified. (C) Immunofluorescence staining to detect the localization of XPC and CBX5 in DHC2(WT) and DHC2(−/−) U87 cells after 200 μM TMZ treatment or DMSO treatment for 1 week. Scale bar = 20 μm. (D) Western blotting analysis of XPC and CBX5 expression in cytoplasmic (C) and nuclear (N) fractions of DHC2(WT) and DHC2(−/−) U87 cells after TMZ or DMSO treatment for 1 week. GAPDH and Histone H3 served as the internal loading controls. (E) Western blotting analyses of XPC, CBX5, MGMT and γH2AX expression in six primary and recurrent GBM tissues. GAPDH served as the internal loading control. (F and G) Representative images of immunohistochemical staining of XPC and CBX5 in six paired primary and recurrent GBM tumours. Patient 1: MGMT-deficient (F), Patient 2: MGMT-positive (G). Scale bar = 100 μm.
DHC2 interacts with XPC and CBX5 and regulates their nuclear localization
To verify if XPC and CBX5 are interacting partners of DHC2, we examined their intracellular distribution in U87 and GBM1 cells treated with different concentrations of TMZ according to in vivo concentrations in humans (Hammond et al., 1999; Rosso et al., 2009) and in vitro IC50 concentrations. Immunofluorescence analysis revealed that both XPC and CBX5 accumulated in the nucleus in U87 (50, 100, 200 μM TMZ or DMSO treatment) and GBM1 cells (50, 100, 400 μM TMZ or DMSO treatment), while nuclear translocation was not observed in DHC2-knockout U87 or DHC2-knockdown GBM1 cells after TMZ treatment (Fig. 3C and Supplementary Fig. 5A and B). Western blotting analysis of cellular fractions gave the same results (Fig. 3D and Supplementary Fig. 5C). Because of the limited transfection efficacy of lentivirus, we observed shDHC2-transfected (GFP expression and DHC2 knockdown) and shDHC2-non-transfected (no GFP expression and normal expression of DHC2) U87 cells in the same microscopic field. XPC was translocated into the nucleus in control shRNA (shNC)-transfected U87 cells after TMZ treatment, but the translocation was not observed in shDHC2-transfected U87 cells after TMZ treatment (Supplementary Fig. 5D). In MGMT-positive T98G and GBM2 cells, there were no changes in the intracellular distribution of XPC and CBX5, regardless of DHC2 knockdown and TMZ treatment (Supplementary Fig. 6A and B). Establishing cell lines from tumours that retain cancer-initiating stem cell properties would provide a valuable and accurate model of the human GBM. These cancer-initiating stem cells would provide valuable insights into the origin of GBM heterogeneity and enable more refined analysis of molecular mechanisms that regulate transformation, self-renewal, commitment, and differentiation (Pollard et al., 2009; Behnan et al., 2017). To strengthen our findings, another four MGMT-deficient primary GBMs were established in serum-free culture conditions. Both GBM1 and GBM2 cells were also cultured in serum-free conditions and formed spheres. The expression of GSC markers (SOX2, SOX9, CD133, CD56) and pericyte/MSC markers (CD248, CD105) were checked to confirm the origin of GBM cells and GSC formation of these spheres (Supplementary Fig. 7). The effects of TMZ treatment on the expression of XPC and CBX5 were detected. The results also demonstrated that XPC and CBX5 expression were upregulated, and both XPC and CBX5 were translocated into nucleus upon TMZ treatment compared with the control group in MGMT-deficient GSCs, but not observed in MGMT-positive GSCs (Supplementary Fig. 8).
We also examined the expression of XPC, CBX5, MGMT and γH2AX in six primary GBM tissues without TMZ treatment and six recurrent GBM tissues with standard TMZ treatment. Western blotting analyses revealed that XPC, CBX5 and γH2AX were highly expressed in recurrent GBM tissues and their expression negatively correlated with MGMT expression (Fig. 3E). Consistent with the findings of XPC and CBX5, nuclear translocation in TMZ-treated MGMT-deficient cells, immunohistochemistry analysis in six paired primary and recurrent GBM tumour tissues revealed higher expression of both XPC and CBX5 in the nucleus of MGMT-deficient recurrent GBM cells than in the paired primary GBM cells, whereas no difference in XPC and CBX5 expression was observed between paired MGMT-positive expressed primary and recurrent GBM cells (Fig. 3F and G).
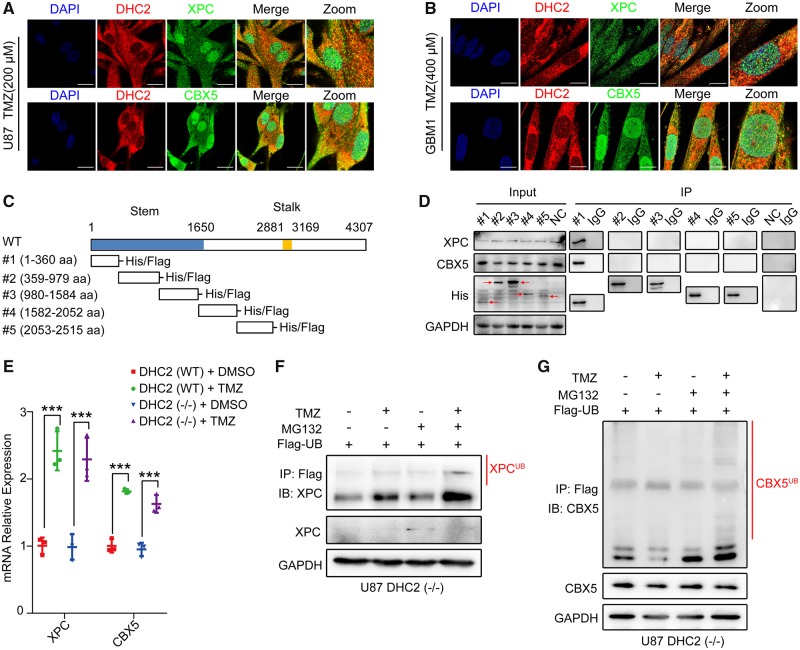
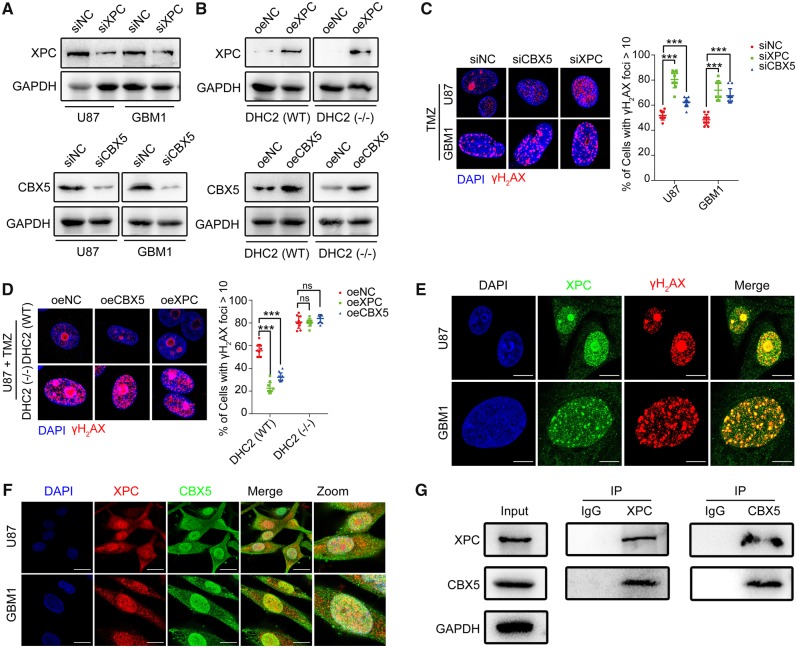
To examine if DHC2 interacts with XPC and CBX5, we analysed the localization of DHC2, XPC and CBX5 by confocal microscopy in TMZ-treated U87 and GBM1 cells. Both XPC and CBX5 co-localized with DHC2, and this co-localization was prominently observed in the cytoplasm. A small part was also observed in the nucleus (Fig. 4A and B). Next, we analysed which region of DHC2 interacts with XPC and CBX5. Previous studies reported that each heavy chain of dynein consists of a C-terminal globular head together with two elongated flexible structures, called the stalk (the microtubule-binding domain) and the N-terminal tail, called the stem (the cargo-binding domain) (Habura et al., 1999; King, 2000; Oiwa and Sakakibara, 2005). DHC2 belongs to dynein heavy chain 1; the predicted stem region comprises amino acids (aa) 1–1650 and the predicted stalk region comprises aa 2881–3169 (Uniprot database: www.uniprot.org) (Fig. 4C). To map the specific region of DHC2 required for its interaction with XPC and CBX5 and considering that DHC2 consisted of 4307 amino acids with a large molecular weight of 492 kDa, we constructed five fragments of DHC2 harbouring amino acids from the stem or stalk region with His tags (#1, aa 1–360; #2, aa 359–979; #3, aa 980–1584; #4, aa 1582–2052; and #5, aa 2053–2515) (Fig. 4C). These fragments covered the first 2515 amino acids of DHC2 that comprised the stem region. U87 cells over-expressing each fragment of DHC2 were treated with TMZ, followed by pull-down with anti-His antibody to co-immunoprecipitate XPC and CBX5. Interaction was seen in the aa 1–360 region (N-terminal tail, stem domain), indicating that the aa 1–360 region interacted with XPC and CBX5 (Fig. 4D).
Figure 4.
DHC2 mediates nuclear transportation of XPC and CBX5 into the nucleus. (A and B) Immunofluorescence staining to examine co-localization of XPC, CBX5 and DHC2 in U87 cells (A) and GBM1 cells (B) after indicated TMZ treatment (200 μM for U87, 400 μM for GBM1) for 48 h. Scale bar = 20 μm. (C) Schematic representation of the domains in full-length DHC2 protein with amino acid (aa) residue numbering; the stem (aa 1–1650) and stalk (aa 2881–3169) domains are as indicated. Five fragments of DHC2 were constructed: #1 (aa 1–360), #2 (aa 359–979), #3 (aa 980–1584), #4 (aa 1582–2052) and #5 (aa 2053–2515), with His and Flag tags. (D) The five fragments of DHC2 were each transfected into U87 cells followed by treatment with 200 μM TMZ for 48 h. Empty plasmid transfected U87 cells with the same treatment served as the normal control (NC). Cell lysates were immunoprecipitated (IP) with anti-His antibody-conjugated magnetic beads and immunoblotted with anti-His, anti-XPC and anti-CBX5 antibodies. (E) qRT-PCR for analysis of XPC and CBX5 mRNA expression in DHC2(WT) and DHC2(−/−) U87 cells after 200 μM TMZ treatment for 1 week. GAPDH expression was used for normalization. ***P < 0.001. (F and G) FLAG-tagged ubiquitin was transfected into U87 cells, in the presence or absence of 200 µM TMZ treatment for 48 h, followed by treatment with 10 mM MG132 or vehicle for 6 h. Cellular lysates were harvested and immunoprecipitated (IP) with anti-FLAG antibody, and the ubiquitin-conjugated XPC (F) and CBX5 (G) were subsequently detected by immunoblotting (IB) using the indicated antibodies.
Intriguingly, an absence of nuclear transport of XPC and CBX5 in TMZ-treated cells upon DHC2 silencing was not associated with increased cytoplasmic retention of XPC and CBX5 (Fig. 3C and D and Supplementary Fig. 5). Despite no elevated expression of XPC and CBX5 proteins, the mRNA levels of XPC and CBX5 were still increased in TMZ-treated DHC2 knockout U87 cells compared to the control cells (Fig. 4E). As previous studies reported that both XPC and CBX5 undergo ubiquitylation for proteasomal degradation (Sugasawa et al., 2005; Wang et al., 2005; Chaturvedi and Parnaik, 2010; He et al., 2014), we decided to examine if XPC and CBX5 underwent cytoplasmic degradation in DHC2 knockout U87 cells after TMZ treatment. Ubiquitinated XPC and CBX5 could be detected in DHC2 knockout U87 cells overexpressing FLAG-ubiquitin, and increased ubiquitination was observed with the proteasome inhibitor MG132 in the presence of TMZ (Fig. 4F and G). These results indicated that XPC and CBX5 were degraded in the cytoplasm of DHC2 knockout U87 cells after TMZ treatment via a proteasome-dependent pathway.
XPC and CBX5 mediate repair of TMZ-induced DNA damage
As previous studies reported that both XPC and CBX5 were involved in DNA damage repair (Min and Pavletich, 2007; Dinant and Luijsterburg, 2009; Zarebski et al., 2009; Nemzow et al., 2015), we decided to examine if XPC and CBX5 were also involved in repair of TMZ-mediated DNA damage in GBM. We knocked down or overexpressed XPC and CBX5 in U87 and GBM1 cells (Fig. 5A and B), followed by TMZ treatment and immunofluorescence staining of γH2AX. Both U87 and GBM1 cells with XPC or CBX5 knockdown had increased number of γH2AX foci (Fig. 5C) and decreased cell survival after TMZ treatment (Supplementary Fig. 9A). Overexpression of XPC or CBX5 in TMZ-treated U87 and GBM1 cells resulted in reduction of γH2AX foci (Fig. 5D and Supplementary Fig. 9B) and increased cell survival (Supplementary Fig. 9C and D), indicating that both XPC and CBX5 are involved in repair of TMZ-induced DNA damage. However, overexpression of XPC or CBX5 did not protect DHC2 knockout U87 cells from TMZ-induced DNA damage (Fig. 5D) or cell death (Supplementary Fig. 9C), which indicated that DHC2 is required for XPC- and CBX5-mediated regulation of DNA damage repair. Immunofluorescence staining of γH2AX and XPC in U87 and GBM1 cells after TMZ treatment showed that XPC co-localized to the DNA damage sites where γH2AX foci were observed (Fig. 5E). Immunofluorescence staining of CBX5 and XPC showed that CBX5 co-localized with XPC (Fig. 5F), and co-immunoprecipitation assay revealed that XPC and CBX5 formed a complex (Fig. 5G). Taken together, our findings suggest that XPC and CBX5 are part of a complex that can mediate repair of TMZ-induced DNA damage.
Figure 5.
Both XPC and CBX5 were involved in repair of TMZ-mediated DNA damage. (A) U87 and GBM1 cells were transfected with siRNA using Lipofectamine™ 2000. Cellular lysates were harvested at 72 h after transfection, followed by immunoblotting analysis for the efficiency of XPC and CBX5 knockdown by RNA interference. siNC = normal control siRNA. (B) Immunoblotting for XPC and CBX5 in U87 DHC2(WT) and DHC2(−/−) cells demonstrating effective overexpression (oe). NC = normal control; empty plasmid transfected. (C) GBM1 and U87 cells with XPC or CBX5 knockdown were treated with TMZ (200 μM for U87, 400 μM for GBM1) for 24 h and allowed to recover for 6 h. Immunofluorescence staining of γH2AX foci, and the percentage of cells containing >10 γH2AX foci in 10 random microscopic fields was calculated. Scale bar = 5 μm. ***P < 0.001. (D) U87 DHC2(WT) and DHC2(−/−) cells with XPC or CBX5 overexpression were treated with 200 μM TMZ for 24 h and allowed to recover for 6 h. Immunofluorescence staining of γH2AX foci and percentage of cells containing >10 γH2AX foci in 10 random microscopic fields was calculated. Scale bar = 5 μm. ***P < 0.001, ns = no significance. (E) GBM1 and U87 cells were treated with the indicated concentration of TMZ (200 μM for U87, 400 μM for GBM1) for 24 h. Immunofluorescence staining of γH2AX foci and XPC shows co-localization. Scale bar = 10 μm. (F) Immunofluorescence staining to examine co-localization of XPC and CBX5 in U87 and GBM1 cells after TMZ treatment (200 μM for U87, 400 μM for GBM1) for 48 h. Scale bar = 20 μm. (G) U87 cells were exposed to 200 µM TMZ or DMSO for 48 h, and cell lysates were immunoprecipitated (IP) with anti-XPC or anti-CBX5 antibody and immunoblotted with the indicated antibodies.
Figure 6.
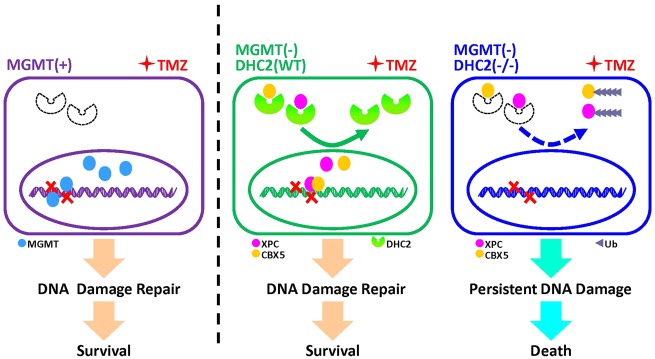
A mechanistic model for DHC2-mediated acquired TMZ resistance in MGMT-negative GBM cells. In MGMT-positive GBM cells, DHC2 is low or not expressed. MGMT mediates TMZ-induced DNA damage repair and results in TMZ-resistance. In MGMT-deficient GBM cells upon TMZ treatment, DHC2 is upregulated and mediated nuclear transportation of DNA repair proteins XPC and CBX5, further contributes to DNA damage repair and TMZ-resistance. Once DHC2 is depleted in MGMT-deficient GBM cells, XPC and CBX5 are degraded via a proteasome-dependent pathway, and result in persistent DNA damage and improvement of TMZ sensitivity.
Discussion
Currently, TMZ is the only chemotherapeutic drug that is confirmed to significantly prolong the overall survival of GBM patients. GBMs with MGMT expression are innately resistant to TMZ therapy. Although a small nucleoside inhibitor, O6-benzylguanine (O6BG), has been shown to effectively deplete MGMT activity and restore GBM cell sensitivity to TMZ in clinical studies (Quinn et al., 2005, 2009), its clinical applicability is limited as only approximately half of GBMs express MGMT on the basis of MGMT promoter methylation status. Most MGMT-negative GBMs that are initially responsive to TMZ develop therapeutic resistance, leading to treatment failure. The molecular mechanisms of acquired resistance remain poorly understood and there are no related therapeutic strategies to overcome such acquired TMZ-resistance.
Our previous studies showed that persistent TMZ treatment can induce cytoskeletal rearrangement of GBM cells and upregulate the expression of DHC2 (Wang et al., 2016; Yi et al., 2016). In this study, we identified DHC2 as a key regulator of acquired TMZ resistance in MGMT-deficient GBM cells. We found that DHC2 was highly upregulated in recurrent GBM tissues from patients that received TMZ treatment. After further analysing the TCGA-540 GBM dataset, we found that DHC2 expression significantly influenced the progression-free survival of MGMT-deficient GBM patients but had no effect on MGMT-expressing GBM patients. The mRNA co-expression analysis showed that DHC2 expression was negatively correlated with MGMT expression. Immunohistochemical assays of DHC2 and MGMT in six paired primary and recurrent GBM tissues also confirmed that DHC2 expression negatively correlated with MGMT expression and was substantially enhanced in recurrent MGMT-deficient TMZ-refractory GBMs when compared with paired primary GBMs. In vivo and in vitro experiments demonstrated that silencing DHC2 expression can enhance TMZ-induced DNA damage and increase TMZ sensitivity in MGMT-deficient GBM cells but not MGMT-positive GBM cells.
DHC2 plays a role in intracellular cargo transportation and DNA damage repair. We performed subcellular proteomic and bioinformatic analyses to identify the potential products that DHC2 transport and also to explore the underlying molecular mechanisms of DHC2-mediated DNA damage repair. We found that XPC and CBX5 could be cargo transported into the nucleus by DHC2. Bioinformatic analysis showed that XPC and CBX5 co-expressed with DHC2. Is a common transcription factor motif in the promoters of these three genes? After searching the JASPAR and QIAGEN databases, we found that TBP (TATA-binding proteins), NKX3–2 (NK3 homeobox 2) and NFATC3 (nuclear factor of activated T cells 3) may be the common transcription factors of these three genes. We also speculate on two other potential underlying mechanisms. First, as DHC2 belongs to the cytoplasmic dynein protein family and consists of an N-terminal tail called the stem (the cargo-binding domain), DHC2 may mediate the nuclear trans-localization of some transcription factors or transcription-related proteins, which regulate the transcription and expression of XPC and CBX5 genes. Second, upregulation of XPC or CBX5 expression induced DHC2 by some feedback mechanism. However, the exact mechanism underlying the coordinated gene expression of DYNC2H1 with XPC and DYNC2H1 with CBX5 remains elusive. Immunohistochemical assays of clinical GBM specimens confirmed that XPC and CBX5 are the interacting partners of DHC2. Co-immunoprecipitation and co-localization analysis showed that both XPC and CBX5 can bind to DHC2 directly. However, the intracellular distribution of XPC and CBX5 showed no changes in MGMT-positive GBM cells regardless of TMZ exposure, indicating MGMT-independent regulation.
XPC plays an important role in nucleotide excision repair (NER) by acting as a damage sensing and DNA-binding factor component of the XPC complex (Batty et al., 2000; Sugasawa et al., 2009). Mutations in the XPC gene or other NER components can result in xeroderma pigmentosum (Li et al., 1993; Chavanne et al., 2000). Many studies suggested that the NER pathway is an important mechanism of TMZ-induced O6-methylguanine repair, in addition to MGMT (Fu et al., 2012; Hombach-Klonisch et al., 2018), and contributes to the acquired TMZ resistance in GBM (Nagel et al., 2017). Researchers reported that XPC silencing can sensitize U87 cells to arsenic trioxide by increasing oxidative damage (Liu et al., 2010). XPC mRNA was upregulated in U87 cells exposed to alkylating agents (Batista et al., 2007). However, to our knowledge, no other studies focused on the mechanism of the XPC and NER pathways on the TMZ resistance in GBM.
CBX5 is a component of heterochromatin that recognizes and binds histone H3 tails methylated at Lys-9 (H3K9me), leading to epigenetic repression (Dawson et al., 2009). Several studies have reported that CBX5 is also linked to DNA damage response. CBX5 is recruited to DNA damage sites to accelerate DNA damage response (Dinant and Luijsterburg, 2009; Zarebski et al., 2009; Baldeyron et al., 2011; Soria and Almouzni, 2013; Gilmore et al., 2016). CBX5 also promotes the function of DNA repair proteins and enhances the DNA damage repair pathway (Lee et al., 2013). Roberto Papait et al. (2009) reported that TMZ treatment increased the amount of CBX5 bound to chromatin, which was consistent with our findings. However, the effect of CBX5 on repair of TMZ-induced DNA damage in GBM cells remains unclear.
Our findings suggested that knockdown of either XPC or CBX5 can aggravate TMZ-induced DNA damage in MGMT-deficient GBM cells and that enhancing XPC or CBX5 expression could accelerate TMZ-induced DNA damage repair. However, the protective effects of XPC and CBX5 overexpression were not observed in DHC2 knockout U87 cells, indicating a requirement for DHC in mediating these effects. We also observed proteasome-dependent degradation of XPC and CBX5 in the cytoplasm of DHC2 knockout U87 cells after TMZ treatment. These findings suggested that both XPC and CBX5 must be transported into the nucleus by DHC2 to be functional, as in the absence of DHC2 they remain non-functional and are subject to cytoplasmic degradation. However, the mechanism underlying proteasome recruitment and degradation of XPC and CBX5 in DHC2 knockout GBM cells is unknown. As XPC plays a key role in the NER pathway (Batty et al., 2000; Sugasawa et al., 2009), we performed immunofluorescence staining of XPC and γH2AX, and showed that XPC can co-localize with TMZ-induced DNA damage sites directly, suggesting that the XPC and NER pathways may be involved in TMZ-induced DNA damage repair. Given that CBX5 can promote the function of DNA damage repair proteins (Lee et al., 2013), we also examined co-localization and interaction between XPC and CBX5, and found that CBX5 co-localized with XPC, indicating that CBX5 may enhance the activity of the XPC-mediated NER pathway.
As DHC2 can interact with XPC and CBX5, we also identified the interaction domains between DHC2 and its partners (XPC and CBX5) by performing a co-immunoprecipitation assay on five fragments of DHC2. We found that the aa 1–360 region of DHC2 interacted with XPC and CBX5. To evaluate the therapeutic potential of DHC2, we injected a high titre shDHC2 adenovirus into tumour-bearing mice intracranially, followed by TMZ or vehicle treatment. We found that DHC2-targeted therapy improved TMZ sensitivity dramatically and showed a significant increase in overall survival, indicating DHC2 could be a promising therapeutic target for improving TMZ sensitivity. The results need to be further verified in more strictly designed preclinical therapeutic assay due to the limitation of animal models, experimental sample size and some potential bias.
In conclusion, we identified a key mechanism that contributes to the acquired TMZ resistance in MGMT-deficient GBM cells. Upregulation of DHC2 induces nuclear transportation of XPC and CBX5 into the nucleus, which enhances the repair of TMZ-induced DNA damage and subsequently results in TMZ resistance. In addition, we also identified the interaction domain between DHC2 and its partners (XPC and CBX5), which provided a promising therapeutic target for overcoming acquired TMZ-resistance in MGMT-deficient GBM.
Funding
This study was supported by the National Natural Science Foundation of China (Grant nos. 81773290, 81472315, and 81802505), Guangdong Science and Technology Department (2017A030313497, 2016A040403053).
Competing interests
The authors report no competing interests.
Supplementary Material
Glossary
Abbreviations
- DEPs
differentially expressed proteins
- DHC2
dynein cytoplasmic 2 heavy chain 1
- GBM
glioblastoma multiforme
- MGMT
O6-methylguanine-DNA methyltransferase
- TCGA
The Cancer Genome Atlas
- TMZ
temozolomide
References
- Agnihotri S, Burrell K, Buczkowicz P, Remke M, Golbourn B, Chornenkyy Y et al. ATM regulates 3-methylpurine-DNA glycosylase and promotes therapeutic resistance to alkylating agents. Cancer Discov 2014; 4: 1198–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol 2011; 193: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista LF, Roos WP, Christmann M, Menck CF, Kaina B. Differential sensitivity of malignant glioma cells to methylating and chloroethylating anticancer drugs: p53 determines the switch by regulating xpc, ddb2, and DNA double-strand breaks. Cancer Res 2007; 67: 11886–95. [DOI] [PubMed] [Google Scholar]
- Batty D, Rapic’-Otrin V, Levine AS, Wood RD. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J Mol Biol 2000; 300: 275–90. [DOI] [PubMed] [Google Scholar]
- Behnan J, Stangeland B, Hosainey SA, Joel M, Olsen TK, Micci F et al. Differential propagation of stroma and cancer stem cells dictates tumorigenesis and multipotency. Oncogene 2017; 36: 570–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes AA, Franceschi E, Paccapelo A, Tallini G, De Biase D, Ghimenton C et al. Role of MGMT methylation status at time of diagnosis and recurrence for patients with glioblastoma: clinical implications. Oncologist 2017; 22: 432–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR et al. The somatic genomic landscape of glioblastoma. Cell 2013; 155: 462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P, Parnaik VK. Lamin A rod domain mutants target heterochromatin protein 1alpha and beta for proteasomal degradation by activation of F-box protein, FBXW10. PLoS One 2010; 5: e10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavanne F, Broughton BC, Pietra D, Nardo T, Browitt A, Lehmann AR et al. Mutations in the XPC gene in families with xeroderma pigmentosum and consequences at the cell, protein, and transcript levels. Cancer Res 2000; 60: 1974–82. [PubMed] [Google Scholar]
- Chen X, Zhang M, Gan H, Wang H, Lee JH, Fang D et al. A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nat Commun 2018; 9: 2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagoneau N, Goulet M, Genevieve D, Sznajer Y, Martinovic J, Smithson S et al. DYNC2H1 mutations cause asphyxiating thoracic dystrophy and short rib-polydactyly syndrome, type III. Am J Hum Genet 2009; 84: 706–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature 2009; 461: 819–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant C, Luijsterburg MS. The emerging role of HP1 in the DNA damage response. Mol Cell Biol 2009; 29: 6335–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 2000; 343: 1350–4. [DOI] [PubMed] [Google Scholar]
- Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clin Cancer Res 2000; 6: 2585–97. [PubMed] [Google Scholar]
- Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer 2012; 12: 104–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med 2014; 370: 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore JM, Sardiu ME, Groppe BD, Thornton JL, Liu X, Dayebgadoh G et al. WDR76 Co-Localizes with Heterochromatin Related Proteins and Rapidly Responds to DNA Damage. PLoS One 2016; 11: e0155492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habura A, Tikhonenko I, Chisholm RL, Koonce MP. Interaction mapping of a dynein heavy chain. Identification of dimerization and intermediate-chain binding domains. J Biol Chem 1999; 274: 15447–53. [DOI] [PubMed] [Google Scholar]
- Hammond LA, Eckardt JR, Baker SD, Eckhardt SG, Dugan M, Forral K et al. Phase I and pharmacokinetic study of temozolomide on a daily-for-5-days schedule in patients with advanced solid malignancies. J Clin Oncol 1999; 17: 2604–13. [DOI] [PubMed] [Google Scholar]
- He J, Zhu Q, Wani G, Sharma N, Han C, Qian J et al. Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquitinating XPC protein and preventing XPC protein from undergoing ultraviolet light-induced and VCP/p97 protein-regulated proteolysis. J Biol Chem 2014; 289: 27278–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005; 352: 997–1003. [DOI] [PubMed] [Google Scholar]
- Hombach-Klonisch S, Mehrpour M, Shojaei S, Harlos C, Pitz M, Hamai A et al. Glioblastoma and chemoresistance to alkylating agents: Involvement of apoptosis, autophagy, and unfolded protein response. Pharmacol Ther 2018; 184: 13–41. [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Watanabe Y, Murayama T, Toyoshima YY. Recombinant human cytoplasmic dynein heavy chain 1 and 2: observation of dynein-2 motor activity in vitro. FEBS Lett 2011; 585: 2419–23. [DOI] [PubMed] [Google Scholar]
- King SM. AAA domains and organization of the dynein motor unit. J Cell Sci 2000; 113 (Pt 14): 2521–6. [DOI] [PubMed] [Google Scholar]
- Lee YH, Kuo CY, Stark JM, Shih HM, Ann DK. HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response. Nucleic Acids Res 2013; 41: 5784–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Bales ES, Peterson CA, Legerski RJ. Characterization of molecular defects in xeroderma pigmentosum group C. Nat Genet 1993; 5: 413–7. [DOI] [PubMed] [Google Scholar]
- Liu SY, Wen CY, Lee YJ, Lee TC. XPC silencing sensitizes glioma cells to arsenic trioxide via increased oxidative damage. Toxicol Sci 2010; 116: 183–93. [DOI] [PubMed] [Google Scholar]
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 2016; 131: 803–20. [DOI] [PubMed] [Google Scholar]
- Lu Y, Xiao L, Liu Y, Wang H, Li H, Zhou Q et al. MIR517C inhibits autophagy and the epithelial-to-mesenchymal (-like) transition phenotype in human glioblastoma through KPNA2-dependent disruption of TP53 nuclear translocation. Autophagy 2015; 11: 2213–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature 2007; 449: 570–5. [DOI] [PubMed] [Google Scholar]
- Nagel ZD, Kitange GJ, Gupta SK, Joughin BA, Chaim IA, Mazzucato P et al. DNA repair capacity in multiple pathways predicts chemoresistance in glioblastoma multiforme. Cancer Res 2017; 77: 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemzow L, Lubin A, Zhang L, Gong F. XPC: Going where no DNA damage sensor has gone before. DNA Repair (Amst) 2015; 36: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocbina PJ, Eggenschwiler JT, Moskowitz I, Anderson KV. Complex interactions between genes controlling trafficking in primary cilia. Nat Genet 2011; 43: 547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oiwa K, Sakakibara H. Recent progress in dynein structure and mechanism. Curr Opin Cell Biol 2005; 17: 98–103. [DOI] [PubMed] [Google Scholar]
- Papait R, Magrassi L, Rigamonti D, Cattaneo E. Temozolomide and carmustine cause large-scale heterochromatin reorganization in glioma cells. Biochem Biophys Res Commun 2009; 379: 434–9. [DOI] [PubMed] [Google Scholar]
- Pegg AE, Byers TL. Repair of DNA containing O6-alkylguanine. FASEB J 1992; 6: 2302–10. [DOI] [PubMed] [Google Scholar]
- Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 2009; 4: 568–80. [DOI] [PubMed] [Google Scholar]
- Quinn JA, Desjardins A, Weingart J, Brem H, Dolan ME, Delaney SM et al. Phase I trial of temozolomide plus O6-benzylguanine for patients with recurrent or progressive malignant glioma. J Clin Oncol 2005; 23: 7178–87. [DOI] [PubMed] [Google Scholar]
- Quinn JA, Jiang SX, Reardon DA, Desjardins A, Vredenburgh JJ, Rich JN et al. Phase II trial of temozolomide plus o6-benzylguanine in adults with recurrent, temozolomide-resistant malignant glioma. J Clin Oncol 2009; 27: 1262–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat Rev Clin Oncol 2017; 14: 434–52. [DOI] [PubMed] [Google Scholar]
- Rosso L, Brock CS, Gallo JM, Saleem A, Price PM, Turkheimer FE et al. A new model for prediction of drug distribution in tumor and normal tissues: pharmacokinetics of temozolomide in glioma patients. Cancer Res 2009; 69: 120–7. [DOI] [PubMed] [Google Scholar]
- Sarkaria JN, Kitange GJ, James CD, Plummer R, Calvert H, Weller M et al. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res 2008; 14: 2900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Almouzni G. Differential contribution of HP1 proteins to DNA end resection and homology-directed repair. Cell Cycle 2013; 12: 422–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005; 352: 987–96. [DOI] [PubMed] [Google Scholar]
- Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA 2017; 318: 2306–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: directional binding of the XPC complex and DNA strand scanning. Mol Cell 2009; 36: 642–53. [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 2005; 121: 387–400. [DOI] [PubMed] [Google Scholar]
- Wang H, Feng W, Lu Y, Li H, Xiang W, Chen Z et al. Expression of dynein, cytoplasmic 2, heavy chain 1 (DHC2) associated with glioblastoma cell resistance to temozolomide. Sci Rep 2016; 6: 28948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QE, Zhu Q, Wani G, El-Mahdy MA, Li J, Wani AA. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res 2005; 33: 4023–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller M, Butowski N, Tran DD, Recht LD, Lim M, Hirte H et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial. Lancet Oncol 2017; 18: 1373–85. [DOI] [PubMed] [Google Scholar]
- Weller M, Stupp R, Reifenberger G, Brandes AA, van den Bent MJ, Wick W et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol 2010; 6: 39–51. [DOI] [PubMed] [Google Scholar]
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med 2008; 359: 492–507. [DOI] [PubMed] [Google Scholar]
- Yi GZ, Liu YW, Xiang W, Wang H, Chen ZY, Xie SD et al. Akt and beta-catenin contribute to TMZ resistance and EMT of MGMT negative malignant glioma cell line. J Neurol Sci 2016; 367: 101–6. [DOI] [PubMed] [Google Scholar]
- Zarebski M, Wiernasz E, Dobrucki JW. Recruitment of heterochromatin protein 1 to DNA repair sites. Cytometry A 2009; 75: 619–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the article and its Supplementary material.