Abstract
Objective
Medical word sense disambiguation (WSD) is challenging and often requires significant training with data labeled by domain experts. This work aims to develop an interactive learning algorithm that makes efficient use of expert’s domain knowledge in building high-quality medical WSD models with minimal human effort.
Methods
We developed an interactive learning algorithm with expert labeling instances and features. An expert can provide supervision in 3 ways: labeling instances, specifying indicative words of a sense, and highlighting supporting evidence in a labeled instance. The algorithm learns from these labels and iteratively selects the most informative instances to ask for future labels. Our evaluation used 3 WSD corpora: 198 ambiguous terms from Medical Subject Headings (MSH) as MEDLINE indexing terms, 74 ambiguous abbreviations in clinical notes from the University of Minnesota (UMN), and 24 ambiguous abbreviations in clinical notes from Vanderbilt University Hospital (VUH). For each ambiguous term and each learning algorithm, a learning curve that plots the accuracy on the test set against the number of labeled instances was generated. The area under the learning curve was used as the primary evaluation metric.
Results
Our interactive learning algorithm significantly outperformed active learning, the previous fastest learning algorithm for medical WSD. Compared to active learning, it achieved 90% accuracy for the MSH corpus with 42% less labeling effort, 35% less labeling effort for the UMN corpus, and 16% less labeling effort for the VUH corpus.
Conclusions
High-quality WSD models can be efficiently trained with minimal supervision by inviting experts to label informative instances and provide domain knowledge through labeling/highlighting contextual features.
Introduction
Medical documents contain many ambiguous terms, the meaning of which can only be determined from the context. For example, the word “ice” may refer to frozen water, methamphetamine (an addictive substance), or caspase-1 (a type of enzyme); and the acronym “PD” may stand for “peritoneal dialysis” (a treatment for kidney failure), “posterior descending” (a coronary artery), or “police department.” Assigning the appropriate meaning (a.k.a. “sense”) to an ambiguous word based on the context is referred to as the process of word sense disambiguation (WSD).1,2 WSD is a critical step for many medical natural language processing (NLP) applications, such as text indexing and categorization, named entity extraction, and computer-assisted chart review.
The research community has proposed and evaluated many WSD methods in the past, including supervised learning,3–5 semi-supervised learning,6–8 and knowledge-driven9,10 approaches. Collectively, these studies have shown that a substantial volume of high-quality training data annotated by human experts is required for existing WSD models to achieve desirable performance. However, annotating training data is a labor-intensive process, and the quality may deteriorate as the volume required to be annotated increases.11 This is particularly true for medical WSD, as assigning correct sense for ambiguous medical terms requires great attention and highly specialized domain knowledge.
To address this issue, the machine learning community has been exploring approaches that involve human experts just-in-time during a machine learning process, in contrast to conventional approaches wherein human experts are only involved in creating static annotated training or evaluation datasets. Such approaches are generally referred to as “active” learning. An active learning (AL) approach12 prioritizes instances to be labeled and presents to human experts the most informative ones that would help the algorithm achieve desirable performance with fewer iterations. This family of learning methods has shown far superior performance over that of random sampling in medical WSD tasks.13
In our previous work,14 we described ReQ-ReC (RR) expert, a step further by incorporating an information retrieval component in AL that allows human experts to identify and label typical instances using their domain knowledge through keyword search. It demonstrated better performance than AL in medical WSD tasks. However, even though experts are brought into the loop, existing interactive learning approaches still suffer from the “cold start” problem. That is, without any prior knowledge about a new WSD task, an algorithm based on artificial intelligence (i.e., a statistical WSD classifier) needs a large amount of training data to reach a reasonable accuracy. In contrast, well-trained human experts do not have the cold start problem because they come to a WSD task with established domain knowledge, which helps them directly determine the correct sense of an ambiguous word.
In this paper, we describe a novel interactive learning algorithm that is capable of directly acquiring domain knowledge from human experts by allowing them to articulate the evidence that leads to their sense tagging decisions (e.g., the presence of indicative words in the context that suggest the sense of the word). This knowledge is then applied in subsequent learning processes to help the algorithm achieve desirable performance with fewer iterations, thus solving the cold start problem. That is, besides labeling instances, the expert can provide domain knowledge by 2 means: (1) to specify informative words of a sense and (2) to highlight evidence words in labeled instances. These interaction modes enable experts to directly express their prior knowledge and thought process when they perform WSD, without adding much burden. The 2 channels complement each other: it is sometimes hard to specify strong informative words a priori, but easier to highlight these words in situ. The statistical classifier can learn from both labeled instances and informative words (i.e., labeled features), and query new labels using AL.
Simulated experiments on 3 WSD corpora show that expert’s domain knowledge gives the model a “warm start” at the beginning stage, significantly accelerating the learning process. On one biomedical literature corpus and two clinical notes corpora, the proposed algorithm makes better use of human experts in training WSD models than all existing approaches, achieving the state-of-the-art performance with least effort.
Methods
Instance Labeling vs Feature Determination
Below, we use an example to illustrate how the interactive learning algorithm works. Suppose the word “cold” (or its spelling variants, e.g., “COLD”) is mentioned across a set of medical documents. Depending on the context, it could mean “chronic obstructive lung disease,” “common colds,” or “low temperature.” The task of WSD is to determine the correct sense of each appearance of this word (i.e., each instance of the word).
A human expert performing this task may apply a number of rules based on her or his domain knowledge. For example, she or he may know that when all letters of the word are spelled in capital case, i.ee, “COLD,” it is more likely the acronym of “chronic obstructive lung disease” than any other possible senses. This judgment could be further strengthened when there are indicative words (or phrases) such as “chronic,” “obstructive,” or “lung” in the adjacent text. Likewise, if the word is not spelled in all capitals, and is accompanied by words such as “common,” “cough,” and “sneeze,” it likely means “common cold.” For certain senses, contextual cues may appear in other forms rather than indicative words. For example, a numeric value followed by a unit of temperature (e,g., “5 degrees C”) may give out that the word “cold” in the current context likely refers to “low temperature,” instead of a medical condition.
Unfortunately, such domain knowledge is not leveraged by conventional supervised learning approaches, which only ask human experts to label the sense of the instances of an ambiguous word, rather than capture how human experts make such judgments. In other words, conventional approaches only try to “infer” human wisdom from annotated results, instead of acquiring it directly—even if such wisdom is readily available and can be formalistically expressed. The interactive learning algorithm described in this paper addresses this limitation by allowing human experts to create labeled features in addition to labeling instances.
A labeled instance for an ambiguous word is a [context, sense] pair, following the conventional definition in supervised learning. For example, a labeled instance of the word “cold” can be:
[“Thepatientdevelopedcold and experienced cough and running nose.”, common cold].
A labeled feature for an ambiguous word is a [feature, sense] pair, where the feature is a textual pattern (a word, a phrase, a skip n-gram, or a regular expression in general). The pair encodes the (most likely) sense of the ambiguous word if the feature appears in its context. For example, human experts can express domain knowledge of the sense of “cold” by creating the following labeled features:
| [“COLD”: All cap, | chronic obstructive lung disease] |
| [“chronic”: Non all-cap, | chronic obstructive lung disease] |
| [“obstructive”: Non all-cap, | chronic obstructive lung disease] |
| [“lung”: Non all-cap, | chronic obstructive lung disease] |
| [“common”: Non all-cap, | common cold] |
| [“cough”: Non all-cap, | common cold] |
| [“sneeze”: Non all-cap, | common cold] |
| … |
Human experts can also express domain knowledge by highlighting a contextual cue after labeling an instance of “cold,” as in
[“Thetissuewas exposed to acold environment (5 degrees C).”, low temperature].
The highlighted text snippet essentially creates another labeled feature for “cold”:
[“<digit> degrees C”, low temperature].
A labeled feature encodes certain domain knowledge that human experts use to solve a WSD task, which can be directly applied to train machine-learning models. As a result, it improves WSD performance and, at the same time, reduces the amount of manual effort required to create a large quantity of labeled instances as training data.
Overall Workflow
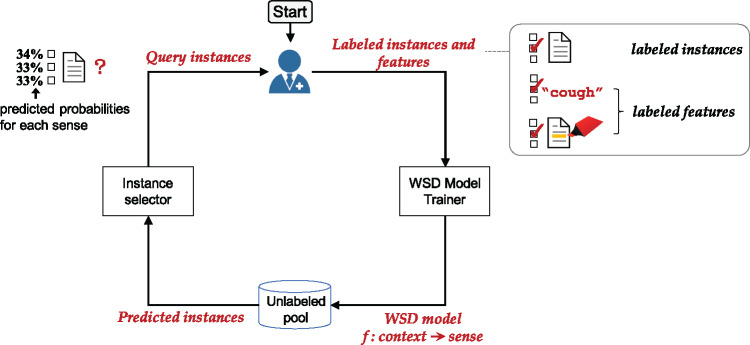
The interactive learning algorithm consists of several distinct components; illustrated in Figure 1.
Figure 1.
Interactive learning with labeled instances and features.
When the human expert can come up with good features for each sense of an ambiguous word, the algorithm can directly use them to train an initial WSD classifier. When such domain knowledge is not available, we assume that the human expert can identify at least one instance for each sense. She or he can then label the instance and highlight contextual cues in that instance. This kicks off the interactive learning process.
The algorithm contains an instance selector that determines how to best select instances from an unlabeled pool to present to the human expert. Then, the human expert labels the sense of the instance, followed by potentially suggesting features that were used as the “rationale” for the labeling decision (i.e., feature labeling). Next, the algorithm uses both labeled instances and labeled features to retrain the WSD classifier, then begins another iteration by selecting additional instances for manual labeling till a satisfactory WSD result is achieved. This process is described in more detail in the next few sections.
WSD Model Training
The algorithm of training and retraining a WSD model consists of 2 stages: feature representation and parameter estimation.
Dynamic feature representation
In conventional supervised learning, a model uses a fixed set of features throughout the training process. For text classification, this feature set is often all of the words in the corpus. In our interactive learning algorithm, labeled features may contain arbitrary textual patterns that are difficult to know ahead of time. Rather than trying to include all possible features from the beginning as conventional machine-learning methods do, we use a dynamic feature representation by starting with a set of base features and gradually expanding it as new features emerge. This method helps to prevent severe overfitting when the size of the feature set is large.
We use presence/absence of unigrams as the base features to represent an instance: , where V is the number of distinct unigrams. A labeled feature defines a real-valued function of an instance, such as “1 if the instance contains ‘COLD’ in all caps; 0 otherwise.” Suppose we have m labeled features at iteration t, then an instance is represented by a (V + m)-dimension vector .
Parameter estimation
We use logistic regression with linear kernel as the WSD classifier. If an ambiguous word has 2 senses, we build a binary classifier, otherwise there is a softmax multiclass classifier. Logistic regression classifiers output probability predictions in , which are then used by the AL algorithm.
Below, we describe the algorithm for training the logistic regression model. Suppose at a certain iteration, we have l labeled instances , and m labeled features . For an ambiguous word with k senses, or is a one-hot k-dimensional vector that encodes the assigned sense. We train a logistic regression model by minimizing the following loss function ( denotes the parameters of the model):
| (1) |
is the expectation for any instance containing feature to have sense c. Let Sj be the set of instances (both labeled and unlabeled) with non-zero feature values for , then
is the smooth version of feature label distribution, because unlike labeled instances, labeled features should be interpreted as preferences rather than as absolute assignments. and are trade-off weights for different loss terms. In this paper, we set .
In the loss function (1), the first term is the cross-entropy loss on labeled instances; the second term is the cross-entropy loss on labeled features; and the third term is a regularization term of parameter w. If the loss function only consists of the first and the third term, then it reduces to the loss function of a traditional softmax logistic regression classifier. The second term expresses a preference on the expected behavior of the WSD classifier, i.e., the presence of a feature strongly suggests a label (i.e., the most probable sense). This is a so-called generalized expectation criterion.15 Because of the second term, (1) is a nonconvex function. We use gradient descent to find a local minimum for the model parameter w. In practice, we find the local minimum yields a sufficiently performing classification model.
Instance Selection
The proposed algorithm kicks off the first iteration by a labeled feature for each sense. Once the WSD classifier is trained, AL can be applied to select a small set of unlabeled instances to present to human experts for labeling. Specifically, we use minimum margin-based AL as the instance selection algorithm which has shown superior performance in classification settings.12,14 It selects the unlabeled instance x that satisfies the smallest , where and are the most and second most probable senses. Intuitively, the classifier cannot determine whether or is the correct sense, therefore it needs to solicit input from human experts.
Evaluation Method
Evaluation corpora
In this study, we used three established medical corpora to evaluate the performance of the interactive learning algorithm.
The MSH corpus contains a set of MEDLINE abstracts automatically annotated using MSH indexing terms.16 Similar to how it was handled in previous work,13,14 for this corpus, we only included ambiguous words that have at least 100 instances, providing adequate data for training and evaluation. This gave us 198 ambiguous words, including 102 abbreviations, 86 nonabbreviated words, and 10 abbreviation-word combinations.
The University of Minnesota (UMN) corpus contains 74 ambiguous abbreviations from a total of 604 944 clinical notes created at the Fairview Health Services affiliated with the University of Minnesota; each abbreviation has 500 randomly sampled instances.17 Each instance is a paragraph in which the abbreviation appeared. Four abbreviations have a general English sense (FISH, IT, OR, US).
The Vanderbilt University Hospital (VUH) corpus contains ambiguous abbreviations from the admission notes created at the Vanderbilt University Hospital.18 Similar to the MSH corpus, we only retained 24 abbreviations that have more than 100 instances. Each instance is a sentence in which the abbreviation appeared. One abbreviation is a loanword in English (AD as in “ad lib”).
The summary statistics of these 3 evaluation corpora are shown in Table 1 (more details can be found in Supplementary Appendix Tables A1–A3). The MSH corpus has the richest context in an instance (i.e., highest average number of tokens per instance), and the least skewed distribution of senses (i.e., lowest proportion of dominating majority senses). Because the main objective of this study was to evaluate the performance of the interactive learning algorithm in comparison with other machine-learning algorithms, we did not further tune the context window size for each corpus. The 3 corpora share 3 abbreviations (SS, CA, RA). MSH and UMN share another 6 abbreviations. UMN and VUH share another 5 abbreviations. The same abbreviation may have different senses in different corpora.
Table 1.
Summary Statistics of Three Evaluation Corpora
| Corpus | Corpus size | Average number of instances per word | Average number of senses per word | Average number of tokens per instance | Average percentage of majority sense (%) |
|---|---|---|---|---|---|
| MSH | 198 | 190 | 2.1 | 202.84 | 54.2 |
| UMN | 74 | 500 | 5.5 | 60.59 | 73.4 |
| VUH | 24 | 194 | 4.3 | 18.73 | 78.3 |
Baseline Methods
To comparatively evaluate the performance of the interactive learning algorithm, we included 3 other machine-learning algorithms in the analysis. As shown in Table 2, these algorithms vary mainly based on how labeled instances or features are obtained from human experts.
Table 2.
Description of Baseline Methods
| Random sampling | Active learning | ReQ-ReC expert | Informed learning |
|---|---|---|---|
| The algorithm selects the next instance at random from the unlabeled pool | The algorithm selects the next instance using the minimum margin criterion.12,13 | The algorithm extends active learning by inviting human experts to search for typical instances for each sense using keywords14 | The proposed interactive learning algorithm |
| Start with one labeled instance for each sense | Start with one labeled instance for each sense | Start with one labeled feature for each sense | Start with one labeled feature (or one labeled instance with a highlighted feature) for each sense |
| Later iterations use random sampling to obtain instance labels | Later iterations use minimum margin to obtain instance labels | Later iterations use minimum margin to obtain instance labels | Later iterations use minimum margin to obtain instance labels |
Simulated human expert input
To derive evaluation metrics, we simulated human expert input using labeled data from each corpus, which is a method commonly used to evaluate AL algorithms.12 This method reduces potential influences that may be introduced due to performance variation by human experts. More specifically:
Labeling instances: We used the validated labels in these evaluation corpora as the oracle of instance labels.
Labeling features: To implement simulated human expert input (i.e., the “oracle”) that provides labeled features, we computed information gain for each unigram feature using the entire labeled corpus,19 and selected the most informative features as oracle features. A feature is associated with a sense when the feature co-occurs most frequently with the sense. To make it more realistic, we simulated the oracle that knows the q-th best feature among all unigram features, where q = 1, 5, 10. This oracle was also used in the “RR expert” algorithm when composing the first search query. The labeled features generated in this way were mostly the words in the definition of each sense.
Since, in reality, a human expert is unlikely able to come up with all features achieving the highest information gain, we also implemented a weaker, supplementary oracle that better resembles true human performance in realistic WSD tasks. It simulates the action of the expert highlighting a feature in a labeled instance while she or he is doing the annotation. In the first iteration, a random instance in each sense was given to the oracle. It identified the most informative n-gram (n = 1, 2, 3) feature in that instance. We used n-grams instead of unigrams to allow the oracle to highlight consecutive words in a sentence. To make the oracle more realistic, we simulated the oracle that knows the q-th best n-gram feature in that instance, where q = 1, 2, 3.
Evaluation metrics
We used learning curves to evaluate the cost-benefit performance of different learning algorithms. A learning curve plots the learning performance against the effort required in training the algorithm. In the context of this paper, learning performance is measured by classification accuracy on a test corpus; and effort is measured by the number of instances that need to be labeled by human experts. For each ambiguous word, we split its instances into an unlabeled set and a test set. When a learning algorithm is executed over the unlabeled set, a label is revealed only if the learning algorithm asks for it. With more and more labels becoming available, the WSD model is continuously updated and its accuracy continuously evaluated, producing a learning curve.
To reduce variation of the curve due to differences between the unlabeled set and the test set, we ran a 10-fold cross validation: 9 folds of the data are used as the unlabeled set and 1-fold used as the test set. The learning curve of the algorithm on a particular ambiguous word is produced by taking the average of the 10 curves. The overall aggregated learning curve of the algorithm is obtained by taking the average of all curves on all ambiguous words in an evaluation corpus.
In reality, human experts are unlikely to provide an inclusive set of features with the highest information gain prior to the annotation process. On the other hand, a well-trained human annotator should be able to identify the best (or one of the best) features after seeing and labeling an instance. Therefore, we hypothesize that the true performance of a human expert will be between the oracle that provides the best feature (best-case scenario) and the oracle that highlights the third best feature in a labeled instance (worst-case scenario). We average the learning curves of the best- and the worst-case scenarios to generate the learning curve of “informed learning.”
To summarize the performance of different learning algorithms using a composite score, we also generated a global Area under Learning Curve (ALC) for each algorithm on each corpus. This method was introduced in the 2010 Active Learning Challenge.20 The global ALC score was normalized by the area under the best achievable learning curve (constant 1.0 accuracy over all points).
To test the significance of performance difference between the algorithms in terms of average ALC scores, we used Wilcoxon signed rank test,21 a nonparametric test for paired examples. We set the type I error control at .
Results
The aggregated learning curves obtained by applying each of the learning algorithms on the evaluation corpora, including drill-down analyses of imperfect feature labeling and highlighting oracles, are exhibited in Figures 2–4.
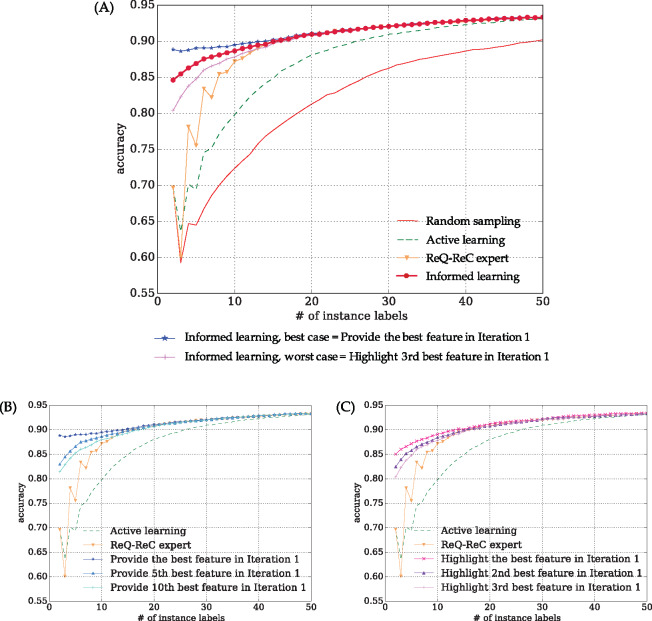
Figure 2.
Aggregated learning curves of 198 ambiguous words in the MSH corpus. (A) interactive learning algorithms in comparison, including the best- and worst-case scenarios of “informed learning”. To achieve 90% accuracy, “random sampling” required 49 instance labels, and “active learning” required 26 instance labels. “ReQ-ReC expert” used labeled features as instance search queries and required 17 instance labels to achieve 90% accuracy. “Informed learning” directly learned from feature labels and only required 15 instance labels to achieve 90% accuracy. (B and C) drill-down analysis of informed learning using imperfect feature labeling (highlighting) oracles, respectively. Even using imperfect feature labeling oracles, variants of “informed learning” still significantly outperformed both “active learning” and “ReQ-ReC expert,” according to Wilcoxon signed rank test (see Table 3).
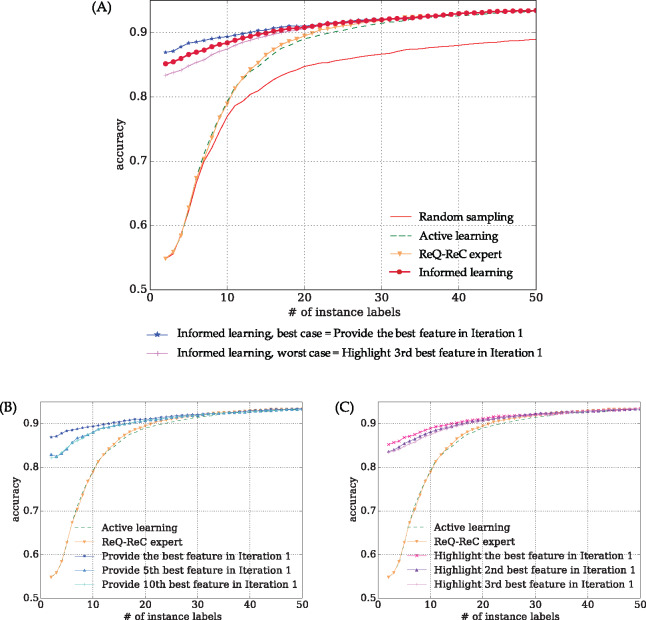
Figure 3.
Aggregated learning curves of 74 ambiguous words in the UMN corpus. (A) interactive learning algorithms in comparison, including the best- and worst-case scenarios of “informed learning”. To achieve 90% accuracy, “random sampling” required more than 50 instance labels, “active learning” required 23 instance labels, and “ReQ-ReC expert” required 21 instance labels. “Informed learning” required only 15 instance labels. (B and C) drill-down analysis of informed learning of imperfect feature labeling (highlighting) oracles, respectively. Even using imperfect feature oracles, variants of “informed learning” still significantly outperformed both “active learning” and “ReQ-ReC expert”, according to Wilcoxon signed rank test (see Table 3).
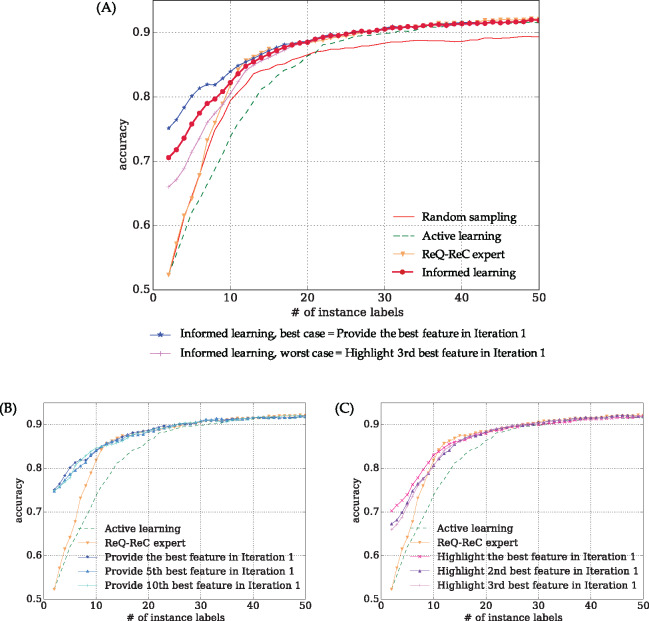
Figure 4.
Aggregated learning curves of 24 ambiguous words in the VUH corpus. (A) Interactive learning algorithms in comparison, including the best- and worst-case scenarios of “informed learning”. To achieve 90% accuracy, “random sampling” required more than 50 instance labels, “active learning” required 31 instance labels, “ReQ-ReC expert” and “Informed learning” required 26 labels. (B and C) drill-down analysis of learning curves of imperfect feature labeling (highlighting) oracles, respectively. Even using imperfect feature oracles, variants of “informed learning” still significantly outperformed “active learning”, according to Wilcoxon signed rank test (see Table 3).
The learning curves of the informed learning (IL) algorithm demonstrated a “warm start” substantially better than the other algorithms evaluated. This is as a result of applying directly acquired domain knowledge from human experts at the beginning of the learning process. The warm start not only helps to achieve desired performance faster with fewer instance labels, but also makes the proposed algorithm (potentially) less susceptible to highly skewed sense distribution. This is as shown by the curves on the 2 clinical WSD corpora, UMN, and VUH. To reach 90% accuracy, IL saved 42% instance labels compared to AL on the MSH corpus (15 vs 26), 35% instance labels on the UMN corpus (15 vs 23), and 16% instance labels on the VUH corpus (26 vs 31).
The ALC scores for each corpus and each learning algorithm, as well as the results of statistical significance tests, are reported in Table 3. On all 3 corpora, Wilcoxon signed rank test showed that the ALC scores of IL were statistically significantly better than margin-based AL. On 2 corpora (MSH and UMN), the ALC scores of IL were statistically significantly better than RR expert, the previous state of the art. These significance results hold even when the feature oracles were imperfect, demonstrating that the proposed algorithm was applicable in a broad range of conditions.
Table 3.
Area under learning curve (ALC) scores of evaluated interactive learning algorithms
| Learning algorithm | MSH | UMN | VUH |
|---|---|---|---|
| Random sampling | 0.8159 | 0.8146 | 0.8311 |
| Active learning | 0.8676 | 0.8522 | 0.8309 |
| ReQ-ReC expert | 0.8928 | 0.8550 | 0.8524 |
| Informed learning | 0.9094*,† | 0.9074*,† | 0.8706* |
| Provide the best feature in Iteration 1 | 0.9141*,† | 0.9122*,† | 0.8792* |
| Provide fifth best feature in Iteration 1 | 0.9087*,† | 0.9038*,† | 0.8773* |
| Provide 10th best feature in Iteration 1 | 0.9052*,† | 0.9029*,† | 0.8777* |
| Highlight the best feature in Iteration 1 | 0.9119*,† | 0.9091*,† | 0.8675* |
| Highlight second best feature in Iteration 1 | 0.9072*,† | 0.9035*,† | 0.8639* |
| Highlight third best feature in Iteration 1 | 0.9047*,† | 0.9004*,† | 0.8620* |
The bottom 2 sections are variants of “Informed learning” with different feature labeling (highlighting) oracles. “*” means the score is significant compared to “Active learning” at level . “†” means the score is significant compared to “ReC-ReQ expert” at level .
Discussion
Warm-start Effect
The IL algorithm is perfectly positioned to address the “cold start” problem. AL works best when the model has a reasonably good “understanding” of the problem space so that the selected instances are the most informative. At the beginning, the model trained on very few labeled instances can perform poorly and waste data selection. In IL, human experts can start the learning process by specifying an informative keyword of a sense, which essentially provides weak labels for many instances containing that keyword, resulting in a “warm start.” It significantly reduces the total number of instance labels to reach high accuracy.
Error Analysis
In Table 4, we break down the performance of each algorithm on different subsets of words in three corpora. In the MSH corpus, as abbreviations often co-occur with its full forms, they were easier to disambiguate than nonabbreviated words. The abbreviations in UMN and VUH were harder to disambiguate than those in MSH, because the unbalanced sense distribution presented a challenge to machine learning models.
Table 4.
Average ALC Scores of Evaluated Interactive Learning Algorithms Across Different Subsets of Ambiguous Words
| Subsets of ambiguous words in each corpus | Average ALC score |
ALC advantage (%) |
||||
|---|---|---|---|---|---|---|
| Random sampling | Active learning | ReQ-ReC expert | Informed learning | Informed over Active (%) | Informed over ReQ-ReC (%) | |
| MSH | ||||||
| 102 abbreviations | 0.8617 | 0.9189 | 0.9349 | 0.9548 | 101/102 (99) | 98/102 (96) |
| 10 abbreviation-word combinations | 0.8265 | 0.8623 | 0.8922 | 0.9150 | 10/10 (100) | 10/10 (100) |
| 86 nonabbreviated words | 0.7603 | 0.8074 | 0.8430 | 0.8549 | 86/86 (100) | 66/86 (77) |
| UMN | ||||||
| 70 abbreviations | 0.8145 | 0.8520 | 0.8545 | 0.9076 | 70/70 (100) | 70/70 (100) |
| 4 abbreviation-word combinations | 0.8176 | 0.8540 | 0.8635 | 0.9048 | 4/4 (100 | 4/4 (100) |
| VUH | ||||||
| 23 abbreviations | 0.8332 | 0.8343 | 0.8552 | 0.8710 | 21/23 (91) | 18/23 (78) |
| 1 abbreviation-word combination | 0.7820 | 0.7535 | 0.7877 | 0.8490 | 1/1 (100) | 1/1 (100) |
We studied the cases where IL underperformed AL or RR expert. The main reason was that the simulated feature oracle sometimes provided low-quality labeled features. In fact, words with high information gain could be rare words, not generalizing to many examples; they could also be common words (e.g., “that,” “of”), which happened to appear more frequently in one sense than others but were too noisy to be useful in classification. IL works well when a labeled feature is representative of and specific to a sense. We hypothesize that real human experts are more capable of providing such high-quality features than simulated experts.
AL and RR start learning with an equal number of instances in each sense, i.e., assuming a uniform prior distribution over senses. As for IL, initial labeled features induce a sense distribution through feature popularity (a frequent feature indicates a major sense), naturally giving rise to a skewed sense distribution. When the true sense distribution is indeed uniform (MSH), AL, and RR may have an advantage over IL. However, when the true sense distribution is skewed (UMN and VUH), AL and RR may suffer as they need more instance labels to correct their uniform prior assumption.
In this study, we set 90% accuracy as the target and measured the number of instances required for achieving that performance. In secondary analysis of Electronic Health Records (EHRs) data for clinical research, NLP systems with over 90% accuracy are often viewed as reasonable22–24 and have been widely used. However, for NLP systems that will be used for clinical practice (e.g., clinical decision support systems), higher performance would be required. Therefore, the target performance is dependent on specific tasks. In the future, we will further investigate our approaches when required performance changes.
Conclusion
This paper introduces a novel interactive machine learning algorithm that can learn from domain knowledge to rapidly build statistical classifiers for medical WSD. Human experts can express domain knowledge by either prescribing informative words for a sense, or highlighting evidence words when labeling an instance. In addition, active learning technique is employed to query instance labels. Experiments using three biomedical WSD corpora showed that the algorithm delivered significantly better performance than strong baseline methods. In the future, we will conduct evaluation studies to assess the performance of the algorithm using real-world scenarios with real human experts.
Funding
This work was supported by the National Science Foundation under grant numbers 1054199 and 1633370; and the National Library of Medicine under grant number 2R01LM010681-05.
Competing interests
None.
Contributors
YW preprocessed the data, designed and implemented the interactive learning algorithms, conducted experiments and statistical significance tests, and drafted and revised the manuscript. KZ revised the experimental design, interpreted the results, and extensively revised the manuscript. HX conceived the research project, provided the data, and extensively revised the manuscript. QM conceived the research project, designed the algorithmic framework and evaluation methodology, and extensively revised the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
References
- 1. Ide N, Véronis J. Introduction to the special issue on word sense disambiguation: the state of the art. Comput Linguist 1998; 241: 2–40. [Google Scholar]
- 2. Schuemie MJ, Kors JA, Mons B. Word sense disambiguation in the biomedical domain: an overview. J Comput Biol 2005; 125: 554–565. [DOI] [PubMed] [Google Scholar]
- 3. Liu H, Teller V, Friedman C. A multi-aspect comparison study of supervised word sense disambiguation. J Am Med Inform Assoc 2004; 114: 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu H, Markatou M, Dimova R et al. Machine learning and word sense disambiguation in the biomedical domain: design and evaluation issues. BMC Bioinformatics 2006; 71: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu Y, Xu J, Zhang Y et al. Clinical abbreviation disambiguation using neural word embeddings. In: proceedings of the 2015 Workshop on Biomedical Natural Language Processing (BioNLP), Beijing, China: July, 2015: 171–176. [Google Scholar]
- 6. Liu H, Lussier YA, Friedman C. Disambiguating ambiguous biomedical terms in biomedical narrative text: an unsupervised method. J Biomed Inform 2001; 344: 249–261. [DOI] [PubMed] [Google Scholar]
- 7. Xu H, Stetson PD, Friedman C. Combining corpus-derived sense profiles with estimated frequency information to disambiguate clinical abbreviations. In: AMIA Ann Symp Proc, Vol. 2012 AMIA; 2012: 1004–1013. [PMC free article] [PubMed] [Google Scholar]
- 8. Finley GP, Pakhomov SV, McEwan R et al. Towards comprehensive clinical abbreviation disambiguation using machine-labeled training data. In: AMIA Ann Symp Proc, Vol. 2016 AMIA; 2016: 560–569. [PMC free article] [PubMed] [Google Scholar]
- 9. Liu H, Johnson SB, Friedman C. Automatic resolution of ambiguous terms based on machine learning and conceptual relations in the UMLS. J Am Med Inform Assoc 2002; 96: 621–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu H, Kim W, Hatzivassiloglou V et al. Using MEDLINE as a knowledge source for disambiguating abbreviations and acronyms in full-text biomedical journal articles. J Biomed Inform 2007; 402: 150–159. [DOI] [PubMed] [Google Scholar]
- 11. Pustejovsky J, Stubbs A. Natural Language Annotation for Machine Learning: A Guide to Corpus-building for Applications. Sebastopol, CA, USA: O'Reilly Media, Inc.; 2012. [Google Scholar]
- 12. Settles B. Active Learning Literature Survey. University of Wisconsin-Madison, 2009, Computer Sciences Technical Report 1648. [Google Scholar]
- 13. Chen Y, Cao H, Mei Q et al. Applying active learning to supervised word sense disambiguation in MEDLINE. J Am Med Inform Assoc 2013; 205: 1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Y, Zheng K, Xu H et al. Clinical word sense disambiguation with interactive search and classification. In: AMIA Ann Symp Proc, Vol. 2016 AMIA; 2016: 2062–2071. [PMC free article] [PubMed] [Google Scholar]
- 15. Druck G, Mann G, McCallum A. Learning from labeled features using generalized expectation criteria. In: proceedings of the 31st Annual International ACM SIGIR Conference on Research and Development in Information Retrieval . Singapore: ACM; July, 2008: 595–602. [Google Scholar]
- 16. Jimeno-Yepes AJ, McInnes BT, Aronson AR. Exploiting MeSH indexing in MEDLINE to generate a data set for word sense disambiguation. BMC Bioinformatics 2011; 12: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moon S, Pakhomov S, Liu N et al. A sense inventory for clinical abbreviations and acronyms created using clinical notes and medical dictionary resources. J Am Med Inform Assoc 2014; 212: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu Y, Denny J, Rosenbloom ST et al. A prototype application for real-time recognition and disambiguation of clinical abbreviations. In: Proceedings of the 7th International Workshop on Data and Text Mining in Biomedical Informatics. New York, NY, USA: ACM; 2013: 7–8. [Google Scholar]
- 19. Yang Y, Pedersen JO. A comparative study on feature selection in text categorization. In: Proceedings of the 14th International Conference on Machine Learning. Nashville, TN, USA: ACM; July, 1997: 412–420. [Google Scholar]
- 20. Guyon I, Cawley G, Dror G et al. Results of the Active Learning Challenge. JMLR: Workshop Conf Proc 2011; 16: 19–45. [Google Scholar]
- 21. Wilcoxon F. Individual comparisons by ranking methods. Biometrics Bull 1945; 16: 80–83. [Google Scholar]
- 22. Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010; 171: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 185: 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc 2013; 205: 806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.