Abstract
Canonical epigenetic modifications, including DNA methylation, histone modification and chromatin remodeling, play a role in numerous life processes, particularly neurodevelopment. Epigenetics explains the development of cells in an organism with the same DNA sequence into different cell types with various functions. However, previous studies on epigenetics have only focused on the chromatin level. Recently, epigenetic modifications of RNA, which mainly include 6-methyladenosine (m6A), pseudouridine, 5-methylcytidine (m5C), inosine (I), 2′-O-ribosemethylation, and 1-methyladenosine (m1A), have gained increasing attention. Circular RNAs (circRNAs), which are a type of non-coding RNA without a 5′ cap or 3′ poly (A) tail, are abundantly found in the brain and might respond to and regulate synaptic function. Also, circRNAs have various functions, such as microRNA sponge, regulation of gene transcription and interaction with RNA binding protein. In addition, circRNAs are methylated by N6-methyladenosine (m6A). In this review, we discuss the crucial roles of epigenetic modifications of circRNAs, such as m6A, in the genesis and development of neurons and in synaptic function and plasticity. Thus, this type of changes in circRNAs might be a therapeutic target in central nervous system (CNS) disorders and could aid the diagnosis and treatment of these disorders.
Keywords: circular RNA, epigenomics, neurodevelopment, m6A, non-coding RNA
Introduction
Epigenomics, which refers to all the molecular pathways that modulate the expression of a genotype into a particular phenotype without any changes to the genome, plays an important role in the growth and development of mammals (Dupont et al., 2009). Research on canonical epigenetics has concentrated on DNA modifications and chromatin variations, and RNA epigenetic modifications, particularly those in non-coding RNAs, have recently garnered increasing attention. With the development of RNA deep sequencing technology and bioinformatics approaches, circRNAs have become increasing important among non-coding RNAs. Unlike linear RNAs, circRNAs have covalently closed loop structures without 5′ caps or 3′ poly-A tails due to back-splicing (Bolisetty and Graveley, 2013). Because of their stability (Suzuki and Tsukahara, 2014), evolutionary conservatism (Jeck et al., 2013) and high abundance (Gruner et al., 2016), circRNAs act as miRNA sponges (Hansen et al., 2013; Memczak et al., 2013), factors of RNA splicing (Ashwal-Fluss et al., 2014), and modulators of the expression of parental genes (Li Z. et al., 2015). circRNAs can also serve as biomarkers for numerous diseases (Meng et al., 2017). The latest studies have demonstrated that circRNAs can be methylated by m6A (Yang et al., 2017; Zhou et al., 2017), and its translation is enhanced by METTL3 and METTL14, and inhibited by FTO (Yang et al., 2017). Both circRNAs and m6A are involved in RNA processing and are related to neurodevelopment (Dominissini et al., 2012; Meyer et al., 2012; Rybak-Wolf et al., 2015). So, this review describes the effect of canonical epigenetics in neurodevelopment, summarizes the progress on RNA epigenetics and circRNAs, and suggests the relationship between neurodevelopment and circRNA epigenetics.
Canonical Epigenetics and Neurodevelopment
Epigenetics is involved in many vital biological processes and plays an important role in the growth and development of organisms. Epigenetics explains how cells that carry the same genetic information differentiate into different cell types with various functions (Gapp et al., 2014). It is difficult to succeed in the treatment of neurological diseases, such as Parkinson’s disease, Alzheimer’s disease (AD), gliomas, and epilepsy. Thus, a study of the relationship between epigenetics and neurodevelopment contributes to our understanding of the occurrence and development of these diseases. The traditional epigenetics processes include DNA methylation, histone modification and chromatin remodeling. In this section, we demonstrate that DNA methylation, histone modification and chromatin remodeling play a role in neurodevelopment.
DNA Methylation and Neurodevelopment
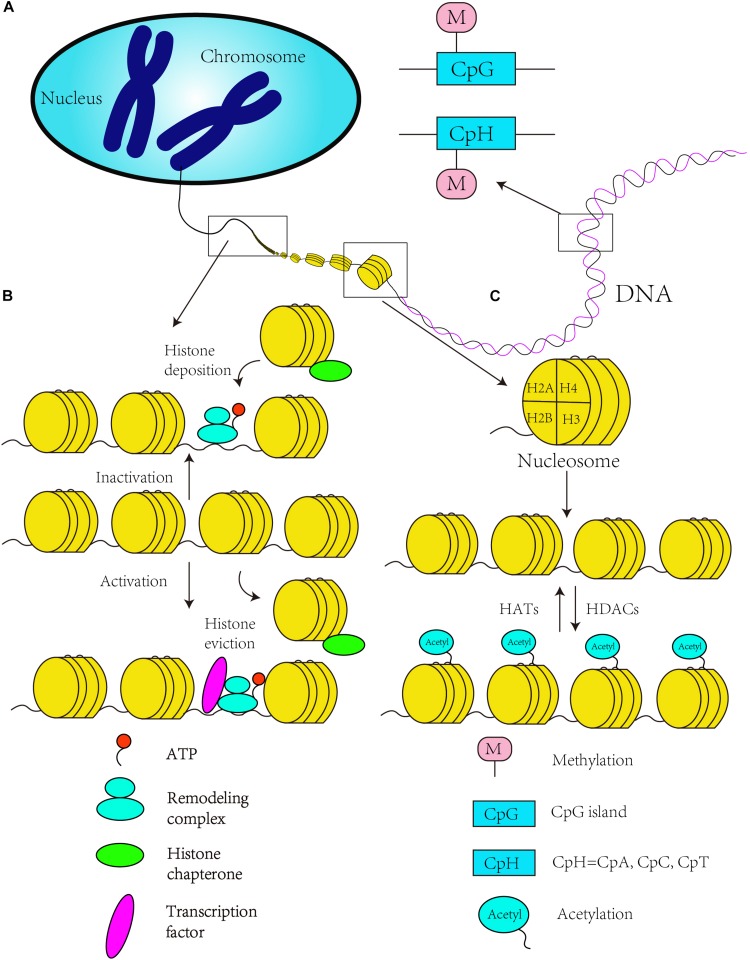
DNA methylation, as a covalent modification of genomic DNA, modifies gene expression and provides a mechanism for transmitting and perpetuating epigenetic information through DNA replication and cell division (Harman and Martin, 2019). Early development includes two stages of epigenetic programming: the first stage involves DNA demethylation or remethylation and the reprograming of histone PTMs in somatic cells, and the second stage guarantees and rebuilds parental imprints during germ cell development through DNA methylation (van Montfoort et al., 2012; Gapp et al., 2014). In addition, the neurodevelopment process also highly depends on DNA methylation (Gapp et al., 2014). Specifically, DNA methylation is observed at CpG islands in the mammalian genome, where it can modulate gene expression (Jones, 2012), through the addition of a methyl group to m5C molecules (Pulido Fontes et al., 2015). Brain DNA has one of the highest levels of m5Cs in human organs (Kriaucionis and Heintz, 2009; LaSalle et al., 2013), and DNA methylation is required for neuronal differentiation in mammals (Takizawa et al., 2001; Mohn et al., 2008). In addition, researchers have found that DNA methylation changes with LTP and m5C methyltransferase regulate synaptic plasticity in the hippocampus (Levenson et al., 2006). Some recent studies confirmed that in the presence of LTP, the methylation status of LTP genes undergoes widespread changes in the adult brain (Maag et al., 2017). These studies also showed that the methylation of Bdnf CpG islands is related to isoform switching from transcripts (Maag et al., 2017). Coincidentally, some researchers have identified that CpG island enriched for genes related to development and neurodifferentiation in schizophrenia patients, and widespread DNA methylation changes in schizophrenia-associated CpGs, were related to the transition from fetal brain cortex to postnatal development (Jaffe et al., 2016) (Figure 1A). Sleep deprivation can alter the cortical genome-wide distribution of DNA methylation, and these differences are enriched in gene pathways involving in the synapse formation and synaptic plasticity (Massart et al., 2014). In addition, some studies have found that gene-specific DNA methylation occurs in response to folic acid supplementation during pregnancy and is related to brain development and function (Caffrey et al., 2018).
FIGURE 1.
Canonical epigenetics mechanism. (A) DNA methylation in the CpG and CpH context. (B) Chromatin remodeling is ATP dependent. ATP-dependent remodeling complexes regulate DNA accessibility, and histone chaperones move and transfer histones on and off a locus. An open chromatin structure enables RNA polymerase II to catalyze the transcription when the histone is evicted, while chromatin state change to inactivity to inhibit gene transcription with the histone deposition of chromosome. (C) The acetylation of histone tails by histone acetyltransferases (HATs) establishes a more relaxed state in the chromatin that results in transcriptional activation (activated state). Histone deacetylation by HDACs reverses the activating chromatin state to a “inactive” chromatin state to inhibit gene transcription.
Histone Modification and Neurodevelopment
The basic structural unit of chromatin is the nucleosome, which is composed of one H3/H4 tetramer, two H2A and H2B dimers, and one H1 molecule. Histone modification refers to the set of covalent PTMs of histone proteins, and these modifications, which mainly include methylation, acetylation, phosphorylation, ubiquitination, sumoylation, and ADP-ribosylation (Berger, 2007), have been proven to be important in stem cell differentiation and neurodevelopment (Podobinska et al., 2017). For instance, class I and II HDACs, which contain two catalytic domains, act as the regulators of histone acetylation in mammals (Zhang et al., 2006; Podobinska et al., 2017). Many protein complexes that contain HDACs, such as SIN3/HDAC (Silverstein and Ekwall, 2005), the NuRD (Xue et al., 1998), co-repressor for element-1-silencing transcription factor (CoREST) (Shi et al., 2004) and the nuclear receptor co-repressor (N-CoR) (Jepsen et al., 2007), play important roles in neurodevelopment. These complexes not only catalyze the deacetylation of histones but are also associated with the activation of gene expression (Shimbo et al., 2013) or involved in the neural commitment and differentiation of stem cells (Podobinska et al., 2017). Learning and memory are vitally important processes in the growth and development of individual, and are necessary in brain development. Some animal experiments have shown that lacking HDAC2 or HDAC3 could improve learning (Guan et al., 2009; McQuown et al., 2011; Narayan and Dragunow, 2017), while loss of HDAC4 and HDAC5 has been shown to damage memory function (Kim et al., 2012; Sando, et al., 2012; Agis-Balboa et al., 2013), especially, HDAC4, which also participates in experience-dependent plasticity of synaptic (Agis-Balboa et al., 2013). Conversely, in the aged brain, HDACs and HATs, as transcription repressors to catalyze histone deacetylation, have been reported to be altered, and could be linked to age-related altered gene transcription (Barter and Foster, 2018). Some scientists have proven that the same isoforms of HDAC3 and HDAC4 were undetectable in the human AD prefrontal cortex compared to mouse models of AD, which had relatively high concentrations (Anderson et al., 2015). And comparison to the control cases, HDAC1 and HDAC2 were decreased but HDAC5 and HDAC6 were significantly increased in AD patient (Anderson et al., 2015), which implied that isoform selectivity of HDACs could be a target of therapy (Narayan and Dragunow, 2017). Although HDAC4 is undetectable in normal human brain, the expression of brain tumor tissue was increased (de Ruijter et al., 2003). Interestingly, deletion or mutation of HDAC4 results in reduced expression of RAI1 can cause mental retardation, such as Smith–Magenis syndrome (Williams et al., 2010) (Figure 1C).
Chromatin Remodeling and Neurodevelopment
Chromatin remodeling refers to dynamic modifications of the chromatin architecture that regulate transcription through displacement and rearrangement of the nucleosome. The process of chromatin remodeling is driven by ATP (Zaghlool et al., 2016), and chromatin remodeling complexes can be classified into four main classes: SWI/SNF, ISWI, INO80, and Mi2/CHD (Olave et al., 2002; Choi et al., 2015) (Figure 1B). A few recent studies have focused on chromatin remodeling and neurodevelopment. The Wnt signaling pathway is one of the most important pathways in embryonic development and axis patterning (Salinas, 2012), and some researchers have found that this signaling pathway can be repressed by one of the BRG1-associated factors in the ARID1B chromatin remodeling complex (also known as the SWI/SNF-A complex) (Vasileiou et al., 2015). This signaling pathway can be affected by a mutation in bromodomain adjacent to zinc finger domain protein 1A (BAZ1A), which encodes ATP-utilizing chromatin assembly and remodeling factor 1 (ACF1) (Zaghlool et al., 2016), and the mutation in BAZ1A also affects the development of proper synaptic functions (Zaghlool et al., 2016). Furthermore, chromatin remodeling might be influenced by persistent exposure to 6OH-BDE-47 (brominated diphenyl ether, BDE) and thereby affects downstream processes, such as synapse development and the overall functional maturity of neurons (Poston et al., 2018). Also, chromatin remodeling can be regulated by non-coding RNAs. For example, microRNA-9 and microRNA-124a could inhibit the expression of gene BAF53a (also known as ACTL6a), which is a component of SWI/SNF chromatin remodeling complexes, by corresponding to the recognition sites of 3′ untranslated region. Ultimately, neural progenitor proliferation was repressed (Yoo et al., 2009). As for the aging brain, chromatin remodeling driven by histone modifications is tightly related to the enzymes which can regulate the process of modifications added or removed (Harman and Martin, 2019). However, regulation and function of these enzymes is altered during brain aging leading to changes in the epigenome (Pal and Tyler, 2016). To some extent, these studies have demonstrated that chromatin remodeling plays a role in neurodevelopment and can affect embryonic development.
Rna Modification
Epigenomics refers to stable and heritable changes in gene expression that do not alter the DNA sequence (Berger et al., 2009). However, epigenetic modifications occur not only in DNA but also in RNA, called the epitranscriptome, but the heritability of RNA modifications needs further study. Epitranscriptome includes more than 100 types of RNA modifications (Sun et al., 2016), and researchers have found that RNA modifications are abundant in tRNAs, rRNAs, and snRNAs but relatively rare in mRNAs (Lee et al., 2014; Sun et al., 2016). But in the last several years, technological advances improving our ability to identify mRNA modifications and recent studies of the cellular transcriptome have focused attention on epitranscription (Flamand and Meyer, 2019). Many of these modified transcripts in the brain are associated with autism and other neurodevelopmental disorders, and have implied that the epitranscriptome may impact the development and maturation of synapses (Washbourne, 2015; Flamand and Meyer, 2019). To a great extent, these modifications enrich the functions of RNA and genetic diversity (Maden, 1990; Wang X. et al., 2014; Zhang and Jia, 2016), and the common RNA modifications include pseudouridine (Ψ), m6A, 5-methylcytosine (m5C), m7G, N1-methyladenosine, and Nm (Sun et al., 2016; Zhang and Jia, 2016). This section summarizes the most common types of epigenetic modifications of RNA.
N6-Methyladenosine (m6A)
N6-Methyladenosine, which refers to the methylation of position N6 of adenosine, is one of the most abundant modifications of mRNAs found in all eukaryotes. Early studies used mass spectrometry to detect this modification and revealed that the relative m6A content ranged from 0.1 to 0.4% (Wei et al., 1975), which corresponds to the modification of approximately three to five sites in each mRNA (Wei et al., 1975; Lee et al., 2014). The m6A modification, which is post-transcriptionally decoded by m6A methyltransferase, is a prevalent internal modification in eukaryotic mRNA (Wang X. et al., 2014), and always occurs in the consensus sequence RRACH (R = G or A; H = A, C or U) (Niu et al., 2013). To detect and analyze the location of m6A, researchers have developed a m6A-specific MeRIP-Seq approach and found that m6A is mainly concentrated in the 3′ UTRs of mRNAs, long internal exons and the stop codons (Meyer et al., 2012). The distribution of m6A in tissue-specific sites has also been investigated, and the results revealed that this modification is most abundantly found in the heart, brain and kidney (Meyer et al., 2012). Furthermore, the distribution of m6A is richer in the adult brain than in the fetal brain (Meyer et al., 2012). Coincidentally, Dominissini D et al. used an m6A-seq approach and found that the sites modified by m6A are highly conserved in humans and mice (Dominissini et al., 2012). Antibody-based crosslinking strategies have been developed in recent years to increase the resolution of m6A (Chen K. et al., 2015; Linder et al., 2015; Roundtree and He, 2016).
To more accurately describe the process of m6A, researchers have used the terms “writer,” “eraser,” and “readers,” and these terms are extensively used for many types of modifications, not just m6A methyltransferase. This modification is considered a “writer,” which uses the SAM cofactor as the methyl donor, and this cofactor is post-transcriptionally methylated at the N6 position of adenosine. m6A methyltransferase consists of METTL3, METTL14 and the regulatory subunit WTAP (Bokar et al., 1994; Liu et al., 2014; Ping et al., 2014). METTL14 has enzymatic activity (Liu et al., 2014), interacts with METTL3 and preferentially methylates the conserved GGACU and GGAUU sequences (Liu et al., 2014). Even though it does not have the activity of methyltransferase due to the lack of a catalytic center, WTAP can locate the methyltransferase complex to nuclear speckles by interacting with METTL3 and METTL14 (Ping et al., 2014). The knockdown of METTL3 causes changes in the splicing patterns and alternative polyadenylation, and influences RNA stability, transcriptional silencing, and translation (Dominissini et al., 2012; Schwartz et al., 2014; Ke et al., 2015; Meyer et al., 2015; Zhou et al., 2015; Lin et al., 2016; Patil et al., 2016; Wang X. et al., 2016; Pendleton et al., 2017). A recent study revealed a new mechanism of m6A: METTL16, a long unknown U6 snRNA methyltransferase able to control the SAM levels, which influence the level of m6A in most cells by regulating the expression of human MAT2A (Pendleton et al., 2017).
The discovery of m6A demethylating enzymes, named “erasers,” focused on FTO (Dina et al., 2007) and ALKBH5, which are proteins that belong to the Fe (II) and 2-oxoglutarate-dependent oxygenase superfamily (Jia et al., 2011; Zheng et al., 2013) and oxidize m6A through N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (f6A) intermediates (Fu et al., 2013). Recent studies have shown that FTO participates in many vital life processes, such as the regulation of dopaminergic signaling in the brain (Hess et al., 2013), the mRNA splicing of adipogenetic regulatory factors (Ben-Haim et al., 2015), adipogenesis (Zhao et al., 2014), and the enhancement of leukemic oncogene-mediated cell transformation and leukemogenesis (Li Z. et al., 2017). Both FTO and ALKBH5 are important in cells, and in HeLa cells, these demethylating enzymes also affect the processing, nuclear export and metabolism of mRNA (Zheng et al., 2013).
The effector proteins of m6A, which are called “readers,” include the YT521-B homology (YTH) family, which encodes five proteins, namely the YTH domain family (YTHDF) proteins 1, 2 and 3 and the YTH domain-containing (YTHDC) proteins 1 and 2 in mammals (Zhang et al., 2010; Li Z. et al., 2017). To date, four of these proteins have been shown to exhibit m6A selectivity in vitro and in vivo (Wang X. et al., 2014; Xu et al., 2014; Roundtree and He, 2016). YTHDF2 and YTHDC1 have a conserved hydrophobic binding pocket specific for m6A and participate in the process regulating the methylation and transcript fate of mRNA (Luo and Tong, 2014; Wang X. et al., 2014; Xu et al., 2014). In addition, the high-resolution mapping of transcription-binding sites has revealed that YTHDF1 and YTHDF2 prefer to bind to the GGACU conserved sequence motif in mRNA, which shows substantial overlap with sites of m6A methylation (Zhu et al., 2014; Wang et al., 2015; Roundtree and He, 2016).
N6-Methyladenosine plays a critical role in the development of an organism, and changes in the levels of m6A have an impact on many life processes, including tissue development, stem cell self-renewal (Wang Y. et al., 2014; Zhao et al., 2014) and differentiation (Geula et al., 2015). m6A can also control the heat shock response (Zhou et al., 2015), circadian clocks (Fustin et al., 2013), and processes associated with the fate and function of RNAs, such as the stability, splicing, transport, localization and translation of RNAs (Zheng et al., 2013; Wang X. et al., 2014; Wang Y. et al., 2014; Wang et al., 2015; Zhao et al., 2014; Meyer et al., 2015; Zhou et al., 2015), primary microRNA processing (Alarcon et al., 2015; Chen T. et al., 2015), and RNA-protein interactions (Dominissini et al., 2012; Meyer et al., 2012; Liu et al., 2015). Similarly, in neurodevelopment, m6A still has a critical role in reducing brain volume (Ho et al., 2010). In the developing cortex, m6A is abundant and controls the ample transcripts involved in neurogenesis and neuronal differentiation (Yoon et al., 2017; Flamand and Meyer, 2019). With the age growing, the m6A levels is increasing, especially in adulthood via controlling synaptic plasticity in the mature brain (Meyer et al., 2012). However, a considerable body of evidence indicates a relationship between m6A and diseases. In fact, it has been demonstrated that m6A is related to obesity, diabetes and cancer (Klungland and Dahl, 2014; Zelinski et al., 2014). Meanwhile, FTO also participate in regulation of learning and memory. For instance, decreasing the expression of Fto in the hippocampus causes the enhanced contextual fear memory and impaired LTP (Walters et al., 2017; Engel et al., 2018). Recent studies have shown that ALKBH5 and the depletion of m6A drive the formation of cancer stem cells (Jaffrey and Kharas, 2017). Coincidentally, a study conducted in 2017 revealed that m6A is relevant to the self-renewal and tumorigenesis of glioblastoma stem cells (Cui et al., 2017).
Other RNA Modifications
Most studies on m5C have focused on DNA, and m5C is rare in RNA (Roundtree and He, 2016). However, researchers have discovered that m5C is enriched in the 3′-UTRs (Chhabra, 2015). 3-Methylcytidine (m3C) was first discovered in total RNA from Saccharomyces cerevisiae (Hall, 1963). The studies discovered that METTL2 and METTL6 have m3C modifications in specific tRNAs and that METTL8 only induces m3C modifications in mRNA in humans and mice (Xu et al., 2017). Some researchers successfully characterized RNA methylation in mixtures of either isomers of RNA or non-isomeric RNA forms and identified the RNA methylation modifications, including m6A, m5C, m3U, and m5U, by top-down mass spectrometry (Glasner et al., 2017).
Pseudouridine is also a relatively abundant type of RNA modification, and the relative amount of pseudouridine in RNA is in the range of 0.2–0.6% (Li X. et al., 2015). Pseudouridine formation involves two mechanisms: one is dependent on tRNA-pseudouridine synthase I and the other relies on a type of H/ACA box snoRNA (Charette and Gray, 2000; Ofengand, 2002). In rRNA, pseudouridine mainly appears in PTCs, decoding centers and the A-site finger region (ASF) (Jack et al., 2011). Thus, this modification might participate in the processing of rRNA, the assembly of ribosomes and the maintenance of advanced structures (Kiss et al., 2010). Studies have shown that in snRNA (U1, U2, U3, U4, U5, and U6), pseudouridine is highly conserved in different types of species (Yu et al., 2011). In 2011, some researchers showed that stop codons could be transformed into sense codons by pseudouridylation (Karijolich and Yu, 2011).
Inosine is a normal and essential post-transcriptional RNA modification introduced by specific deaminases (Alseth et al., 2014). In tRNA, this process is catalyzed by ADAT, whereas in mRNA and non-coding RNA, ADAR catalyze the process (Bass et al., 1997). In fact, A-to-I RNA editing plays a significant physiological role in neuronal function (Behm and Ohman, 2016). RNA encoding glioma-associated oncogene 1 (GLI1) is edited such that an arginine is changed to a glycine (R/G) in the protein. The GLI1 mRNA is highly edited, which induces an increase in the capacity of GLI1 to activate transcription by adenosine deamination in the normal cerebellum, but the process is obviously decreased in cell lines originating from cerebellar tumors (Shimokawa et al., 2013). Researchers have found that ADAR2 auto-editing is increased during mouse brain development and in rat primary cortical neuronal cultures, which suggests that ADAR2 activity is globally elevated (Hang et al., 2008; Behm and Ohman, 2016). In addition, modulated GluA2-4 R/G editing and alternative splicing generates AMPA receptors, which can adapt to differential rapid fast-synaptic transmission during development (Grosskreutz et al., 2003).
Epigenetic Modifications of Circular RNAs
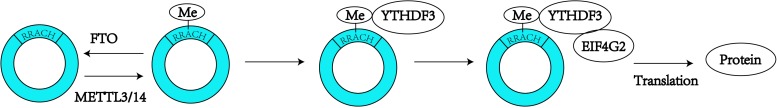
In recent years, circRNAs have been one of the most frequently studied types of non-coding RNA. Due to their unique features, which are described above, circRNAs are known as miRNA sponges (Hansen et al., 2013; Memczak et al., 2013) and might also serve as potential biomarkers for a number of diseases, particularly cancers (Meng et al., 2017). Although numerous biological functions of circRNAs remain unknown, this field of research is being continuously explored. Some of the endogenous circRNAs identified to date have the capability of being translated into proteins through a process driven by the IRESs (Legnini et al., 2017). In early 2017, a group of researchers found that circRNAs can be widely methylated by m6A, as determined through the m6A immunoprecipitation of RNA samples treated with the RNase R exoribonuclease, and are efficiently translated through short sequences consisting of the m6A site as IRESs in human cells (Yang et al., 2017). The initiation of this m6A-mediated translation requires the eIF4G2 initiation factor and the YTHDF3 m6A reader, and the translation process is enhanced by METTL3/14 and inhibited by FTO (Yang et al., 2017). These researchers also inferred that proteins translated by circRNAs can be correlated with environmental stress (Yang et al., 2017). Coincidentally, other researchers designed a computational pipeline named AutoCirc to analyze the results from RNA and m6A immunoprecipitation and further demonstrated that m6A modifications are extensively observed in circRNAs. These researchers also showed that m6A circRNAs have highly cell-specific expression (Zhou et al., 2017), and revealed that circRNAs with m6A modifications also have long single exons. In addition, the researchers compared m6A circRNAs and m6A mRNAs and validated that the methylated exons in mRNAs are different from the exons that form m6A circRNAs (Zhou et al., 2017). Additionally, m6A circRNAs are related to mRNA stability through interaction with YTHDF1/YTHDF2 (Figure 2; Zhou et al., 2017). These studies expand the yield of RNA modifications and circRNAs, and more questions regarding circRNA modifications need to be expounded.
FIGURE 2.
N6-Methyladenosine modification is always found to occur in the consensus sequence identified as: RRACH (R = G or A; H = A, C or U). M6A modification can promote the translation of the circRNAs.
CircRNAs and Neurodevelopment
In 2015, researchers used high-resolution in situ hybridization to verify that circRNAs are most abundantly found in the human brain (You et al., 2015), and some researchers have attempted to determine the reason for the enrichment of circRNAs in the brain (Chen and Schuman, 2016). It is well known that there are long introns (Jeck et al., 2013; Liang and Wilusz, 2014; Zhang et al., 2014) that flank circularized exons; thus, researchers have inferred that brain-specific genes might carry additional sequence features that can promote circRNA formation (Chen and Schuman, 2016). Although many functions of the circRNAs in brain remain unclear, it is undisputed that the circRNA levels in neurons are dynamically modulated. These studies illustrate that circRNAs play a vital role in neurodevelopment through these mechanism.
MicroRNA Sponge and Interaction With RNA Binding Proteins
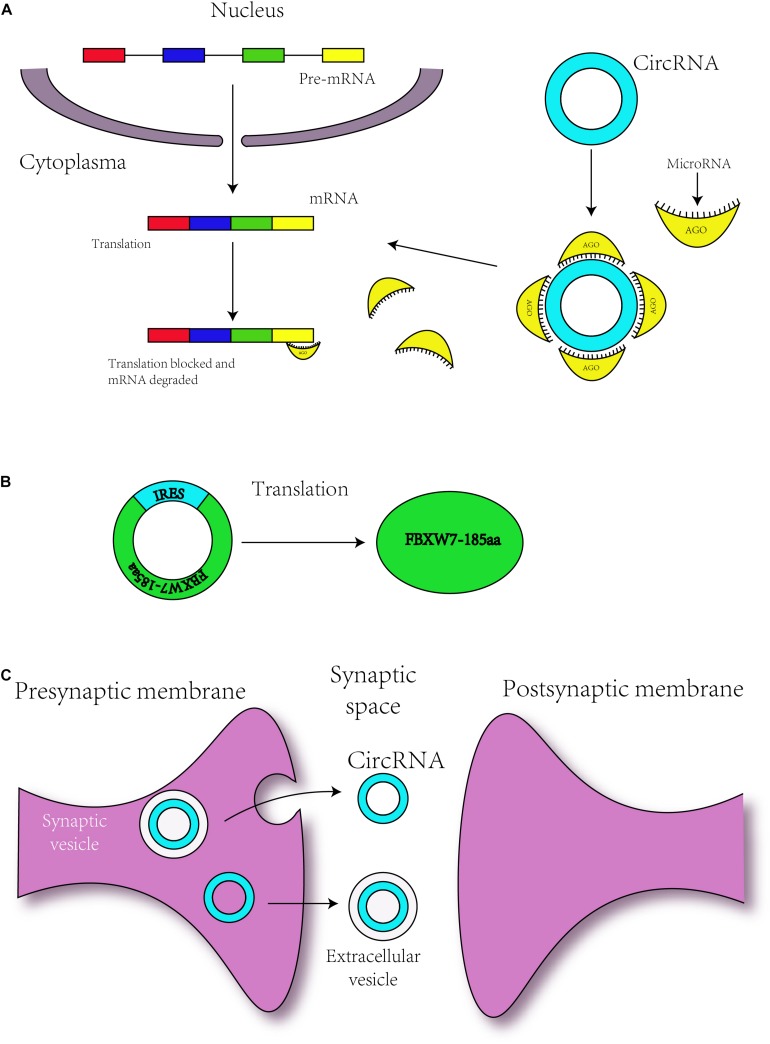
Serving as a microRNA sponge was the first discovered function of circRNAs in 2013. The ciRS-7 contains more than 70 conserved binding sites for miRNA-7, and ciRS-7 can bind with Argonaute (AGO) protein (Figure 3A; Hansen et al., 2013). Additionally, the co-expression of ciRS-7 and miR-7 is distinctly high in neocortical and hippocampal neurons, which implies a high degree of endogenous interaction (Hansen et al., 2013). In 2017, researchers established the Cdr1as-loss mouse model by CRISPR/Cas9 (Piwecka and Glazar, 2017). Circ-Cdr1as reportedly binds to miR-7 and miR-671 (Hansen et al., 2011, 2013), and researchers have found that miR-7 and miR-671 are deregulated post-transcriptionally in the Cdr1as-knock-off brain. Furthermore, the expression of immediate early genes, such as Fos, which is the direct target of miR-7, is increased in the Cdr1as-loss mouse brain (Piwecka and Glazar, 2017). Cdr1as-knockout mice show defects in neuropsychiatric behaviors, which suggests that Cdr1as might be crucial for neuron-controlled behavior (Piwecka and Glazar, 2017). To ensure its interaction with RNA-binding proteins, Circ-Foxo3 is able to bind with many types of proteins, such as cell cycle-related proteins (Du et al., 2016), and thus, might participate in neuronal cell division in neurodevelopment.
FIGURE 3.
Circular RNAs are related to neurodevelopment through different mechanisms. (A) circRNAs regulate the translation of mRNAs by serving as miRNA sponges. (B) Some circRNAs with an IRES sequence can translate proteins, and this translation is driven by IRES. (C) circRNAs may transmit the information to environment by synaptosomes or extracellular vesicle such as exosomes.
Regulation of Gene Transcription
Circular RNAs can also regulate gene transcription, but these effects are achieved via varied mechanisms. These circRNAs always exist in the nucleus. For example, in HeLa and HEK293 cells, circ-EIF3J and circ-PAIP2 are exon-intron circRNAs or EIciRNAs and interact with U1 snRNP to promote transcription of their parental genes (Li Z. et al., 2015). In 2013, researchers found a circular intronic RNA denoted ci-ankrd52 and found that this circRNA might affect the rate or efficiency of transcription as a positive regulator of Pol II transcription (Zhang et al., 2013). Therefore, these circRNAs might be related to transcription in neurons (Li Z. et al., 2015; van Rossum et al., 2016).
Translation
As mentioned above, m6A can mediate the translation of circRNAs, but circRNAs can also be translated via other mechanisms. Some circRNAs contain the IRES responsible for driving translation, for instance, circ-ZNF609 and circMbl3 were found to translate proteins (Legnini et al., 2017; Pamudurti et al., 2017). In addition, in 2017, some researchers have found that circ-FBXW7 is abundantly expressed in the normal human brain and can encode a novel 21-kDa protein, the translation of which is driven by IRES (Figure 3B; Yang et al., 2018). The upregulation of this new protein can inhibit the proliferation and cell cycle acceleration of glioma cells (Yang et al., 2018). As a consequence, circRNAs might translate the proteins to regulate the process of neurodevelopment.
Neurodevelopment and Synaptic Function
In 2015, some researchers have confirmed that circRNAs with biological functions are correlated with synaptic function and are significantly enriched in synapses, parts of the synapse, presynaptic active zones, presynaptic membranes and postsynaptic density (Figure 3C; You et al., 2015). circDscam, circKlhl2, circElavl3, circNlgn1, circGigyf2, circNbea, and circRmst are derived from synapse-related genes (You et al., 2015), indicating a relationship between circRNAs and synaptic function. In addition, highly expressed circRNAs are derived from synaptic genes, such as Dscam and Homer1, and might participate in Wnt signaling, axon guidance and TGF-β signaling (Veno et al., 2015; You et al., 2015). During hippocampal and Drosophila brain development, the expression of circRNAs is developmentally upregulated (Westholm et al., 2014; You et al., 2015) and can be regulated by neural plasticity (You et al., 2015). These findings revealed the role of circRNAs in brain development. Other researchers have found that the expression level of circRNAs shows differences among various brain areas (Rybak-Wolf et al., 2015).
The Potential Roles of circRNA Epigenetic Modifications in Neurodevelopment
As mentioned above, m6A is one of the most abundant methylation patterns in mRNA and is also present in circRNAs (Yang et al., 2017; Zhou et al., 2017). In addition, FTO, as m6A demethylating enzymes, were found to display dynamic expression in postnatal neurodevelopment (Li L. et al., 2017). FTO deficiency not only results in a decreased brain size and a reduced body weight but also leads to impairments in learning and memory (Li L. et al., 2017). Further studies have illustrated that m6A is indispensable for the regulation of RNA fate and function, which are central to differentiation and growth (Geula et al., 2015). In addition, the majority of circRNAs are upregulated during the development of the Drosophila brain, but some circRNAs are downregulated (You et al., 2015; Zhou et al., 2017). By serving as miRNA sponges, circRNAs are involved in the regulation of RNA processing, such as alternative splicing, pre-RNA splicing and RNA editing (Hansen et al., 2013; Starke et al., 2015; van Rossum et al., 2016). Their expression level is regulated by synaptic plasticity during neurodevelopment. As a result, we hypothesized various mechanisms through which epigenetic circRNAs affect neurodevelopment. First, the epigenetic modification of circRNAs might occupy miRNA sites, which can prevent miRNA-mRNA binding. Second, during neurodevelopment, epigenetic circRNAs might transmit information to the microenvironment by exosomes. Exosomes were first found in 1983 as a type of 50-nm vesicles (Harding et al., 2013), and play a significant role in intracellular and extracellular communication. Some studies have demonstrated that pre-miRNAs with Dicer, AGO2, and trans-activation response RNA binding protein (TRBP) are present in exosomes of breast cancer cells (Melo et al., 2014). Therefore, circRNAs with epigenomic changes might regulate the biogenesis and contents of exosomes to participate in the early formation and plasticity of synapses. Third, some circRNAs can translate the protein, and these proteins might play a role in RNA processing. Thus, we can infer that epigenetic modifications of circRNAs, such as m6A, might play a vital role in genesis and neurodevelopment by impacting the alternative splicing of RNAs and in synaptic function and plasticity by influencing RNA processing. In addition, circRNAs participate in many CNS diseases, such as GBM (Zhu et al., 2017), CNS lymphoma (Baraniskin et al., 2016), cerebral ischemia (Ouyang et al., 2013), stroke (Ouyang et al., 2013), Alzheimer’s disease (Wu et al., 2013), Huntington’s disease (Wu et al., 2013), and Parkinson’s disease (Wu et al., 2013). This finding indicates that circRNAs might serve as biomarkers of CNS disorders (Qu et al., 2015; Lu and Xu, 2016). Therefore, in view of these data, changes in the epigenetic modifications of circRNAs might influence RNA stability and lead to neuronal disorders. circRNAs might be a therapeutic target of CNS disorders and can potentially aid the diagnosis of various diseases.
Conclusion
In conclusion, although changes in the epigenetic modifications of circRNAs could exert an effect on neurodevelopment and CNS diseases, considerable studies are needed to confirm this finding. It is thus important to identify the changes in circRNA epigenetic modifications in neurodevelopment and to find the mechanisms of these modifications, which could reveal the roles of circRNAs in CNS diseases. These studies might aid the diagnosis and treatment of CNS diseases.
Author Contributions
ZF, ZX, and YT collected the related manuscript. SM drafted and revised the manuscript. HZ drew the figures. MW participated in the design of the review and helped to draft and revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- ADAR
adenosine deaminases acting on RNA
- ADAT
adenosine deaminases acting on tRNA
- ALKBH5
AlkB homolog 5
- ARID1B
AT-rich interactive domain-containing protein 1B
- Bdnf
brain-derived neurotrophic factor
- CHD
chromodomain helicase DNA-binding protein
- circRNA
circular RNA
- ciRS-7
circular RNA sponge for miR-7
- CNS
central nervous system
- CoREST
corepressor for element-1-silencing transcription factor
- DSCAM
Down Syndrome cell adhesion molecule
- FTO
fat mass-and-obesity-associated protein
- GBM
glioblastoma multiforme
- HATs
histone acetyl-transferases
- HDACs
histone deacetylases
- IRESs
internal ribosome entry sites
- ISWI
Imitation SWItch
- LTP
long-term potentiation
- m1A
1-methyladenosine
- m5C
5-methylcytidine
- m5C
carbon 5 of cytosine
- m6A
N6-methyladenosine
- m7G
N7-methylguanosine
- MeRIP-Seq
methylated RNA immunoprecipitation with next-generation sequencing
- METTL14
methyltransferase-like 14
- METTL3
methyltransferase-like 3
- miRNA
microRNA
- N-CoR
nuclear receptor corepressor
- Nm
2 ′ -O-methylation
- NuRD
nucleosome remodeling and deacetylase complex of histone acetylation
- PTC
peptidyl-transferase center
- PTMs
post-translational modifications
- SAM
S-adenosyl methionine
- SWI/SNF
SWItch/Sucrose Non-Fermentable
- TRBP
transactivation response RNA-binding protein
- UTRs
untranslated regions
- WTAP
Wilms’tumor 1-associating protein
- YTHDC
YTH domain-containing protein
- YTHDF
YTH domain-containing family.
Footnotes
Funding. This work was supported by the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (2014BAI04B02) to MW.
References
- Agis-Balboa R. C., Pavelka Z., Kerimoglu C., Fischer A. (2013). Loss of HDAC5 impairs memory function: implications for Alzheimer’s disease. J. Alzheimers. Dis. 33 35–44. 10.3233/jad-2012-121009 [DOI] [PubMed] [Google Scholar]
- Alarcon C. R., Lee H., Goodarzi H., Halberg N., Tavazoie S. F. (2015). N6-methyladenosine marks primary microRNAs for processing. Nature 519 482–485. 10.1038/nature14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseth I., Dalhus B., Bjoras M. (2014). Inosine in DNA and RNA. Curr. Opin. Genet. Dev. 26 116–123. 10.1016/j.gde.2014.07.008 [DOI] [PubMed] [Google Scholar]
- Anderson K. W., Chen J., Wang M., Mast N., Pikuleva I. A., Turko I. V. (2015). Quantification of histone deacetylase isoforms in human frontal cortex, human retina, and mouse brain. PLoS One 10:e0126592. 10.1371/journal.pone.0126592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R., Meyer M., Pamudurti N. R., Ivanov A., Bartok O., Hanan M., et al. (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56 55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Baraniskin A., Zaslavska E., Nopel-Dunnebacke S., Ahle G., Seidel S., Schlegel U., et al. (2016). Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for primary central nervous system lymphoma. Neuro. Oncol. 18 361–367. 10.1093/neuonc/nov144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barter J. D., Foster T. C. (2018). Aging in the brain: new roles of epigenetics in cognitive decline. Neuroscientist 24 516–525. 10.1177/1073858418780971 [DOI] [PubMed] [Google Scholar]
- Bass B. L., Nishikura K., Keller W., Seeburg P. H., Emeson R. B., O’Connell M. A., et al. (1997). A standardized nomenclature for adenosine deaminases that act on RNA. RNA 3 947–949. [PMC free article] [PubMed] [Google Scholar]
- Behm M., Ohman M. (2016). RNA editing: a contributor to neuronal dynamics in the mammalian brain. Trends Genet. 32 165–175. 10.1016/j.tig.2015.12.005 [DOI] [PubMed] [Google Scholar]
- Ben-Haim M. S., Moshitch-Moshkovitz S., Rechavi G. (2015). FTO: linking m6A demethylation to adipogenesis. Cell Res. 25 3–4. 10.1038/cr.2014.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447 407–412. 10.1038/nature05915 [DOI] [PubMed] [Google Scholar]
- Berger S. L., Kouzarides T., Shiekhattar R., Shilatifard A. (2009). An operational definition of epigenetics. Genes Dev. 23 781–783. 10.1101/gad.1787609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar J. A., Rath-Shambaugh M. E., Ludwiczak R., Narayan P., Rottman F. (1994). Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 269 17697–17704. [PubMed] [Google Scholar]
- Bolisetty M. T., Graveley B. R. (2013). Circuitous route to transcription regulation. Mol. Cell 51 705–706. 10.1016/j.molcel.2013.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey A., Irwin R. E., McNulty H., Strain J. J., Lees-Murdock D. J., McNulty B. A., et al. (2018). Gene-specific DNA methylation in newborns in response to folic acid supplementation during the second and third trimesters of pregnancy: epigenetic analysis from a randomized controlled trial. Am. J. Clin. Nutr. 107 566–575. 10.1093/ajcn/nqx069 [DOI] [PubMed] [Google Scholar]
- Charette M., Gray M. W. (2000). Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49 341–351. 10.1080/152165400410182 [DOI] [PubMed] [Google Scholar]
- Chen K., Lu Z., Wang X., Fu Y., Luo G. Z., Liu N., et al. (2015). High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew. Chem. 54 1587–1590. 10.1002/anie.201410647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Hao Y. J., Zhang Y., Li M. M., Wang M., Han W., et al. (2015). m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem. Cell 1 289–301. 10.1016/j.stem.2015.01.016 [DOI] [PubMed] [Google Scholar]
- Chen W., Schuman E. (2016). Circular RNAs in brain and other tissues: a functional enigma. Trends Neurosci. 39 597–604. 10.1016/j.tins.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Chhabra R. (2015). miRNA and methylation: a multifaceted liaison. Chembiochem 16 195–203. 10.1002/cbic.201402449 [DOI] [PubMed] [Google Scholar]
- Choi K. Y., Yoo M., Han J. H. (2015). Toward understanding the role of the neuron-specific BAF chromatin remodeling complex in memory formation. Exp. Mol. Med. 47:e155. 10.1038/emm.2014.129 [DOI] [PubMed] [Google Scholar]
- Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., et al. (2017). m6A RNA Methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18 2622–2634. 10.1016/j.celrep.2017.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruijter A. J., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburg A. B. (2003). Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370(Pt 3), 737–749. 10.1042/bj20021321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina C., Meyre D., Gallina S., Durand E., Korner A., Jacobson P., et al. (2007). Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 39 724–726. 10.1038/ng2048 [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Du W. W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B. B. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic. Acids Res. 44 2846–2858. 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C., Armant D. R., Brenner C. A. (2009). Epigenetics: definition, mechanisms and clinical perspective. Semin. Reprod. Med. 27 351–357. 10.1055/s-0029-1237423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel M., Eggert C., Kaplick P. M., Eder M., Roh S., Tietze L., et al. (2018). The role of m(6)A/m-RNA methylation in stress response regulation. Neuron 99 389.e9–403.e9. 10.1016/j.neuron.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamand M. N., Meyer K. D. (2019). The epitranscriptome and synaptic plasticity. Curr. Opin. Neurobiol. 59 41–48. 10.1016/j.conb.2019.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Jia G., Pang X., Wang R. N., Wang X., Li C. J., et al. (2013). FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 4:1798. 10.1038/ncomms2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin J. M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., et al. (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155 793–806. 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- Gapp K., Woldemichael B. T., Bohacek J., Mansuy I. M. (2014). Epigenetic regulation in neurodevelopment and neurodegenerative diseases. Neuroscience 264 99–111. 10.1016/j.neuroscience.2012.11.040 [DOI] [PubMed] [Google Scholar]
- Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A. A., Kol N., Salmon-Divon M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347 1002–1006. 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- Glasner H., Riml C., Micura R., Breuker K. (2017). Label-free, direct localization and relative quantitation of the RNA nucleobase methylations m6A, m5C, m3U, and m5U by top-down mass spectrometry. Nucleic Acids Res. 45 8014–8025. 10.1093/nar/gkx470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskreutz J., Zoerner A., Schlesinger F., Krampfl K., Dengler R., Bufler J. (2003). Kinetic properties of human AMPA-type glutamate receptors expressed in HEK293 cells. Eur. J. Neurosci. 17 1173–1178. 10.1046/j.1460-9568.2003.02531.x [DOI] [PubMed] [Google Scholar]
- Gruner H., Cortes-Lopez M., Cooper D. A., Bauer M., Miura P. (2016). CircRNA accumulation in the aging mouse brain. Sci. Rep. 6:38907. 10.1038/srep38907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J. S., Haggarty S. J., Giacometti E., Dannenberg J. H., Joseph N., Gao J., et al. (2009). HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459 55–60. 10.1038/nature07925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. H. (1963). Isolation Of 3-Methyluridine And 3-Methylcytidine from solubleribonucleic acid. Biochem. Biophys. Res. Commun. 12 361–364. 10.1016/0006-291x(63)90105-0 [DOI] [PubMed] [Google Scholar]
- Hang P. N., Tohda M., Matsumoto K. (2008). Developmental changes in expression and self-editing of adenosine deaminase type 2 pre-mRNA and mRNA in rat brain and cultured cortical neurons. Neurosci. Res. 61 398–403. 10.1016/j.neures.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Hansen T. B., Wiklund E. D., Bramsen J. B., Villadsen S. B., Statham A. L., Clark S. J., et al. (2011). miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30 4414–4422. 10.1038/emboj.2011.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C. V., Heuser J. E., Stahl P. D. (2013). Exosomes: looking back three decades and into the future. J. Cell Biol. 200 367–371. 10.1083/jcb.201212113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman M. F., Martin M. G. (2019). Epigenetic mechanisms related to cognitive decline during aging. J. Neurosci. Res. 10.1002/jnr.24436 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Hess M. E., Hess S., Meyer K. D., Verhagen L. A., Koch L., Bronneke H. S., et al. (2013). The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16 1042–1048. 10.1038/nn.3449 [DOI] [PubMed] [Google Scholar]
- Ho A. J., Stein J. L., Hua X., Lee S., Hibar D. P., Leow A. D., et al. (2010). A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc. Natl. Acad. Sci. U.S.A. 107 8404–8409. 10.1073/pnas.0910878107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack K., Bellodi C., Landry D. M., Niederer R. O., Meskauskas A., Musalgaonkar S., et al. (2011). rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 44 660–666. 10.1016/j.molcel.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A. E., Gao Y., Deep-Soboslay A., Tao R., Hyde T. M., Weinberger D. R., et al. (2016). Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci. 19 40–47. 10.1038/nn.4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey S. R., Kharas M. G. (2017). Emerging links between m6A and misregulated mRNA methylation in cancer. Genome Med. 9:2 10.1186/s13073-016-0395-398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd C. E., Liu J., et al. (2013). Circular NAs are abundant, conserved, and associated with ALU repeats. RNA 19 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K., Solum D., Zhou T., McEvilly R. J., Kim H. J., Glass C. K., et al. (2007). SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450 415–419. 10.1038/nature06270 [DOI] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A. (2012). Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13 484–492. 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- Karijolich J., Yu Y. T. (2011). Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474 395–398. 10.1038/nature10165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S., Alemu E. A., Mertens C., Gantman E. C., Fak J. J., Mele A., et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 29 2037–2053. 10.1101/gad.269415.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. S., Akhtar M. W., Adachi M., Mahgoub M., Bassel-Duby R., Kavalali E. T., et al. (2012). An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J. Neurosci. 32 10879–10886. 10.1523/jneurosci.2089-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Fayet-Lebaron E., Jady B. E. (2010). Box H/ACA small ribonucleoproteins. Mol. Cell 37 597–606. 10.1016/j.molcel.2010.01.032 [DOI] [PubMed] [Google Scholar]
- Klungland A., Dahl J. A. (2014). Dynamic RNA modifications in disease. Curr. Opin. Genet. Dev. 26 47–52. 10.1016/j.gde.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Kriaucionis S., Heintz N. (2009). The nuclear DNA base 5-hydroxymethylcytosine is present in purkinje neurons and the brain. Science 324 929–930. 10.1126/science.1169786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle J. M., Powell W. T., Yasui D. H. (2013). Epigenetic layers and players underlying neurodevelopment. Trends Neurosci. 36 460–470. 10.1016/j.tins.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Kim B., Kim V. N. (2014). Emerging roles of RNA modification: m(6)A and U-tail. Cell 158 980–987. 10.1016/j.cell.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., et al. (2017). Circ-ZNF609 Is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66 22.e9–37.e9. 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson J. M., Roth T. L., Lubin F. D., Miller C. A., Huang I. C., Desai P., et al. (2006). Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 281 15763–15773. 10.1074/jbc.M511767200 [DOI] [PubMed] [Google Scholar]
- Li L., Zang L., Zhang F., Chen J., Shen H., Shu L., et al. (2017). Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 26 2398–2411. 10.1093/hmg/ddx128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhu P., Ma S., Song J., Bai J., Sun F., et al. (2015). Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11 592–597. 10.1038/nchembio.1836 [DOI] [PubMed] [Google Scholar]
- Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22 256–264. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- Li Z., Weng H., Su R., Weng X., Zuo Z., Li C., et al. (2017). FTO plays an oncogenic role in acute myeloid leukemia as a n6-methyladenosine rna demethylase. Cancer Cell 31 127–141. 10.1016/j.ccell.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., Wilusz J. E. (2014). Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 28 2233–2247. 10.1101/gad.251926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Choe J., Du P., Triboulet R., Gregory R. I. (2016). The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62 335–345. 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B., Grozhik A. V., Olarerin-George A. O., Meydan C., Mason C. E., Jaffrey S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12 767–772. 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518 560–564. 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Xu A. D. (2016). Mini review: circular rnas as potential clinical biomarkers for disorders in the central nervous system. Front. Genet. 7:53 10.3389/fgene.2016.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Tong L. (2014). Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. U.S.A. 111 13834–13839. 10.1073/pnas.1412742111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag J. L., Kaczorowski D. C., Panja D., Peters T. J., Bramham C. R., Wibrand K., et al. (2017). Widespread promoter methylation of synaptic plasticity genes in long-term potentiation in the adult brain in vivo. BMC Genomics 18:250. 10.1186/s12864-017-3621-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E. (1990). The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 39 241–303. 10.1016/s0079-6603(08)60629-7 [DOI] [PubMed] [Google Scholar]
- Massart R., Freyburger M., Suderman M., Paquet J., El Helou J., Belanger-Nelson E., et al. (2014). The genome-wide landscape of DNA methylation and hydroxymethylation in response to sleep deprivation impacts on synaptic plasticity genes. Transl. Psychiatry 4:e347. 10.1038/tp.2013.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuown S. C., Barrett R. M., Matheos D. P., Post R. J., Rogge G. A., Alenghat T., et al. (2011). HDAC3 is a critical negative regulator of long-term memory formation. J. Neurosci. 31 764–774. 10.1523/jneurosci.5052-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo S. A., Sugimoto H., O’Connell J. T., Kato N., Villanueva A., Vidal A., et al. (2014). Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26 707–721. 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Li P., et al. (2017). CircRNA: functions and properties f a novel potential biomarker for cancer. Mol. Cancer 16:94 10.1186/s12943-017-0663-662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Patil D. P., Zhou J., Zinoviev A., Skabkin M. A., Elemento O., et al. (2015). 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 163 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., Jaffrey S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149 1635–1646. 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F., Weber M., Rebhan M., Roloff T. C., Richter J., Stadler M. B., et al. (2008). Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 30 755–766. 10.1016/j.molcel.2008.05.007 [DOI] [PubMed] [Google Scholar]
- Narayan P., Dragunow M. (2017). Alzheimer’s disease and histone code alterations. Adv. Exp. Med. Biol. 978 321–336. 10.1007/978-3-319-53889-1_17 [DOI] [PubMed] [Google Scholar]
- Niu Y., Zhao X., Wu Y. S., Li M. M., Wang X. J., Yang Y. G. (2013). N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteom. Bioinform. 11 8–17. 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengand J. (2002). Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 514 17–25. 10.1016/s0014-5793(02)02305-0 [DOI] [PubMed] [Google Scholar]
- Olave I. A., Reck-Peterson S. L., Crabtree G. R. (2002). Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 71 755–781. 10.1146/annurev.biochem.71.110601.135507 [DOI] [PubMed] [Google Scholar]
- Ouyang Y. B., Stary C. M., Yang G. Y., Giffard R. (2013). MicroRNAs: innovative targets for cerebral ischemia and stroke. Curr. Drug Targets 14 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Tyler J. K. (2016). Epigenetics and aging. Sci. Adv. 2:e1600584. 10.1126/sciadv.1600584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti N. R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., et al. (2017). Translation of CircRNAs. Mol. Cell 66 9.e7–21.e7. 10.1016/j.molcel.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil D. P., Chen C. K., Pickering B. F., Chow A., Jackson C., Guttman M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 369–373. 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton K. E., Chen B., Liu K., Hunter O. V., Xie Y., Tu B. P., et al. (2017). The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169 824.e14–835.e14. 10.1016/j.cell.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping X. L., Sun B. F., Wang L., Xiao W., Yang X., Wang W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 177–189. 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka M., Glazar P. (2017). Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357:eaam8526. 10.1126/science.aam8526 [DOI] [PubMed] [Google Scholar]
- Podobinska M., Szablowska-Gadomska I., Augustyniak J., Sandvig I., Sandvig A., Buzanska L. (2017). Epigenetic modulation of stem cells in neurodevelopment: the role of methylation and acetylation. Front. Cell Neurosci. 11:23 10.3389/fncel.2017.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poston R. G., Dunn C. J., Sarkar P., Saha R. N. (2018). Persistent 6-OH-BDE-47 exposure impairs functional neuronal maturation and alters expression of neurodevelopmentally-relevant chromatin remodelers. Environ. Epigenet. 4:dvx020. 10.1093/eep/dvx020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido Fontes L., Quesada Jimenez P., Mendioroz Iriarte M. (2015). Epigenetics and epilepsy. Neurologia 30 111–118. 10.1016/j.nrl.2014.03.012. [DOI] [PubMed] [Google Scholar]
- Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., et al. (2015). Circular RNA: a new star of noncoding RNAs. Cancer Lett. 365 141–148. 10.1016/j.canlet.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Roundtree I. A., He C. (2016). RNA epigenetics–chemical messages for posttranscriptional gene regulation. Curr. Opin. Chem. Biol. 30 46–51. 10.1016/j.cbpa.2015.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., et al. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58 870–885. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- Salinas P. C. (2012). Wnt signaling in the vertebrate central nervous system: from axon guidance to synaptic function. Cold Spring Harb. Perspect. Biol. 4:a008003. 10.1101/cshperspect.a008003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando R., Gounko N., Pieraut S., Liao L., Yates J., Maximov A. (2012). HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell 151 821–834. 10.1016/j.cell.2012.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Mumbach M. R., Jovanovic M., Wang T., Maciag K., Bushkin G. G., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 8 284–296. 10.1016/j.celrep.2014.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., et al. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119 941–953. 10.1016/j.cell.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Shimbo T., Du Y., Grimm S. A., Dhasarathy A., Mav D., Shah R. R., et al. (2013). MBD3 localizes at promoters, gene bodies and enhancers of active genes. PLoS Genet. 9:e1004028. 10.1371/journal.pgen.1004028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa T., Rahman M. F., Tostar U., Sonkoly E., Stahle M., Pivarcsi A., et al. (2013). RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA Biol. 10 321–333. 10.4161/rna.23343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein R. A., Ekwall K. (2005). Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 47 1–17. 10.1007/s00294-004-0541-545 [DOI] [PubMed] [Google Scholar]
- Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L. H., et al. (2015). Exon circularization requires canonical splice signals. Cell Rep. 10 103–111. 10.1016/j.celrep.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Sun W. J., Li J. H., Liu S., Wu J., Zhou H., Qu L. H., et al. (2016). RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 44 D259–D265. 10.1093/nar/gkv1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Tsukahara T. (2014). A view of pre-mRNA splicing from RNase R resistant RNAs. Int. J. Mol. Sci. 15 9331–9342. 10.3390/ijms15069331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T., Nakashima K., Namihira M., Ochiai W., Uemura A., Yanagisawa M., et al. (2001). DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell 1 749–758. 10.1016/s1534-5807(01)00101-0 [DOI] [PubMed] [Google Scholar]
- van Montfoort A. P., Hanssen L. L., de Sutter P., Viville S., Geraedts J. P., de Boer P. (2012). Assisted reproduction treatment and epigenetic inheritance. Hum. Reprod. Update 18 171–197. 10.1093/humupd/dmr047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum D., Verheijen B. M., Pasterkamp R. J. (2016). Circular RNAs: novel regulators of neuronal development. Front. Mol. Neurosci. 9:74 10.3389/fnmol.2016.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileiou G., Ekici A. B., Uebe S., Zweier C., Hoyer J., Engels H., et al. (2015). Chromatin-remodeling-factor arid1b represses wnt/beta-catenin signaling. Am. J. Hum. Genet. 97 445–456. 10.1016/j.ajhg.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veno M. T., Hansen T. B., Veno S. T., Clausen B. H., Grebing M., Finsen B., et al. (2015). Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 16:245 10.1186/s13059-015-0801-803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters B. J., Mercaldo V., Gillon C. J., Yip M., Neve R. L., Boyce F. M., et al. (2017). The role of the RNA demethylase FTO (Fat Mass and Obesity-Associated) and mRNA Methylation in hippocampal memory formation. Neuropsychopharmacology 42 1502–1510. 10.1038/npp.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534 575–578. 10.1038/nature18298 [DOI] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., Hon G. C., Yue Y., Han D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 117–120. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B. S., Roundtree I. A., Lu Z., Han D., Ma H., et al. (2015). N(6)-methyladenosine modulates messenger rna translation efficiency. Cell 161 1388–1399. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Y., Toth J. I., Petroski M. D., Zhang Z., Zhao J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16 191–198. 10.1038/ncb2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washbourne P. (2015). Synapse assembly and neurodevelopmental disorders. Neuropsychopharmacology 40 4–15. 10.1038/npp.2014.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C. M., Gershowitz A., Moss B. (1975). Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell 4 379–386. 10.1016/0092-8674(75)90158-0 [DOI] [PubMed] [Google Scholar]
- Westholm J. O., Miura P., Olson S., Shenker S., Joseph B., Sanfilippo P., et al. (2014). Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 9 1966–1980. 10.1016/j.celrep.2014.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. R., Aldred M. A., Der Kaloustian V. M., Halal F., Gowans G., McLeod D. R., et al. (2010). Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am. J. Hum. Genet. 87 219–228. 10.1016/j.ajhg.2010.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Zuo X., Deng H., Liu X., Liu L., Ji A. (2013). Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain. Res. Bull. 97 69–80. 10.1016/j.brainresbull.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Xu C., Wang X., Liu K., Roundtree I. A., Tempel W., Li Y., et al. (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10 927–929. 10.1038/nchembio.1654 [DOI] [PubMed] [Google Scholar]
- Xu L., Liu X., Sheng N., Oo K. S., Liang J., Chionh Y. H., et al. (2017). Three distinct 3-methylcytidine (m3C) methyltransferases modify tRNA and mRNA in mice and humans. J. Biol. Chem. 292 14695–14703. 10.1074/jbc.M117.798298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Wong J., Moreno G. T., Young M. K., Cote J., Wang W. (1998). NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2 851–861. 10.1016/s1097-2765(00)80299-3 [DOI] [PubMed] [Google Scholar]
- Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., et al. (2017). Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27 626–641. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., et al. (2018). Novel role of FBXW7 Circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 110. 304–315. 10.1093/jnci/djx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A. S., Staahl B. T., Chen L., Crabtree G. R. (2009). MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460 642–646. 10.1038/nature08139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K. J., Ringeling F. R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., et al. (2017). Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell 171 877.e17–889.e17. 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., et al. (2015). Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 18 603–610. 10.1038/nn.3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A. T., Ge J., Yu Y. T. (2011). Pseudouridines in spliceosomal snRNAs. Protein Cell 2 712–725. 10.1007/s13238-011-1087-1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghlool A., Halvardson J., Zhao J. J., Etemadikhah M., Kalushkova A., Konska K., et al. (2016). A Role for the chromatin-remodeling factor BAZ1A in neurodevelopment. Hum. Mutat. 37 964–975. 10.1002/humu.23034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelinski E. L., Deibel S. H., McDonald R. J. (2014). The trouble with circadian clock dysfunction: multiple deleterious effects on the brain and body. Neurosci. Biobehav. Rev. 40 80–101. 10.1016/j.neubiorev.2014.01.007 [DOI] [PubMed] [Google Scholar]
- Zhang X. O., Wang H. B., Zhang Y., Lu X., Chen L. L., Yang L. (2014). Complementary sequence-mediated exon circularization. Cell 159 134–147. 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Zhang X., Jia G. F. (2016). RNA epigenetic modification: N6-methyladenosine. Yi Chuan 38. 275–288. 10.16288/j.yczz.16-049 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Gilquin B., Khochbin S., Matthias P. (2006). Two catalytic domains are required for protein deacetylation. J. Biol. Chem. 281 2401–2404. 10.1074/jbc.C500241200 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang X. O., Chen T., Xiang J. F., Yin Q. F., Xing Y. H., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51 792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Theler D., Kaminska K. H., Hiller M., de la Grange P., Pudimat R., et al. (2010). The YTH domain is a novel RNA binding domain. J. Biol. Chem. 285 14701–14710. 10.1074/jbc.M110.104711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Yang Y., Sun B. F., Shi Y., Yang X., Xiao W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24 1403–1419. 10.1038/cr.2014.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Molinie B., Daneshvar K., Pondick J. V., Wang J., Van Wittenberghe N., et al. (2017). Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 20 2262–2276. 10.1016/j.celrep.2017.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wan J., Gao X., Zhang X., Jaffrey S. R., Qian S. B. (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526 591–594. 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Ye J., Zhang L., Xia L., Hu H., Jiang H., et al. (2017). Differential expression of circular RNAs in glioblastoma multiforme and its correlation with prognosis. Transl. Oncol. 10 271–279. 10.1016/j.tranon.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Roundtree I. A., Wang P., Wang X., Wang L., Sun C., et al. (2014). Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 24 1493–1496. 10.1038/cr.2014.152 [DOI] [PMC free article] [PubMed] [Google Scholar]