Summary
This review describes how direct visualization of the dynamic interactions of cells with different extracellular matrix microenvironments can provide novel insights into complex biological processes. Recent studies have moved characterization of cell migration and invasion from classical 2D culture systems into 1D and 3D model systems, revealing multiple differences in mechanisms of cell adhesion, migration and signalling—even though cells in 3D can still display prominent focal adhesions. Myosin II restrains cell migration speed in 2D culture but is often essential for effective 3D migration. 3D cell migration modes can switch between lamellipodial, lobopodial and/or amoeboid depending on the local matrix environment. For example, “nuclear piston” migration can be switched off by local proteolysis, and proteolytic invadopodia can be induced by a high density of fibrillar matrix. Particularly, complex remodelling of both extracellular matrix and tissues occurs during morphogenesis. Extracellular matrix supports self‐assembly of embryonic tissues, but it must also be locally actively remodelled. For example, surprisingly focal remodelling of the basement membrane occurs during branching morphogenesis—numerous tiny perforations generated by proteolysis and actomyosin contractility produce a microscopically porous, flexible basement membrane meshwork for tissue expansion. Cells extend highly active blebs or protrusions towards the surrounding mesenchyme through these perforations. Concurrently, the entire basement membrane undergoes translocation in a direction opposite to bud expansion. Underlying this slowly moving 2D basement membrane translocation are highly dynamic individual cell movements. We conclude this review by describing a variety of exciting research opportunities for discovering novel insights into cell‐matrix interactions.
Keywords: 3D culture, basement membrane remodelling, branching morphogenesis, cell migration, extracellular matrix, invasion
1. INTRODUCTION
The extracellular matrix is now acknowledged to be a key regulator of a wide range of cell biological processes, including signalling and tissue remodelling. Although numerous, elegant biochemical and cell biological studies have documented key roles played by extracellular matrix molecules in embryonic development, tissue remodelling and disease, there have been fewer studies involving direct real‐time visualization of the dynamic interactions of cells and tissues with the extracellular matrix. Others and we have recently applied increasingly powerful live‐cell and live‐tissue fluorescence microscopy technologies to try to identify novel and unexpected cell‐matrix interactions in development and malignancy. This short review will highlight approaches that have yielded altered or new understanding of complex tissue processes. Its focus will be on examples chosen from research from our own laboratory, but we emphasize that there are a number of other outstanding studies in this research area (eg see the following Ref.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14).
2. MOVING INTO THE THIRD DIMENSION
Many important insights have been gained using regular cell culture on flat two‐dimensional surfaces of glass or plastic, which can be termed “2D culture.” However, work during the past couple of decades has moved the study of cell‐matrix interactions—and even the development of early organ primordia—into 3D tissue culture to attempt to understand the complex events that occur in vivo at the molecular and microscopic physical level. Such 3D tissue models provide insights into the mechanisms of 3D cell migration, embryonic development and pathology. This approach permits the application of many of the same powerful technologies used in regular 2D cell culture, including transfection to increase or decrease the expression of a specific protein, antibody inhibition studies and analyses of the biological effects of interactions of cells with different types of extracellular matrix molecules in various topologies (reviews include the following Reference.15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25
3. 1D MIGRATION
Besides 2D and 3D cell culture, however, the use of “1D” systems has proven to be surprisingly relevant to studies of functional cell interactions with the fibrillar structures characteristic of interstitial matrix. As depicted in Figure 1, it has been known for many decades that cells readily migrate along matrix fibrils using a process known as “contact guidance”.26 When viewed in cross section, the migration of a cell along a fibril can be modelled as 1D migration, though such in vitro “1D” lines usually have a width of 1‐2 μm. On such 1D substrates, cells form long, continuous cell‐matrix adhesions and exhibit hyperactive lamellipodial activity27, 28 that can be accompanied by unusual fin‐like membrane protrusive waves.29 Not surprisingly, many of the cell biological characteristics of cell migration along a 1D substrate can closely mimic the behaviour of cells in 3D matrix, but not on flat 2D substrates (Figure 2). These similarities in morphology and cell dynamics may well be due to the capacity of simple 1D model systems to mimic at least partially the fibrillar nature of the interstitial matrix. Similarities between 1D and 3D systems include individual cell morphology, mode of cell migration, orientation of the centrosome and dependence on non‐muscle myosin II.28 Importantly, these studies also identify cell‐matrix adhesions that closely resemble classical focal adhesions characterized in 2D cultures that are used by migrating cells on both 1D and 3D matrices. In fact, the number of focal adhesions and the level of activated β1 integrins can be even higher in cells migrating in 3D collagen matrix than on a 2D substrate.27
Figure 1.
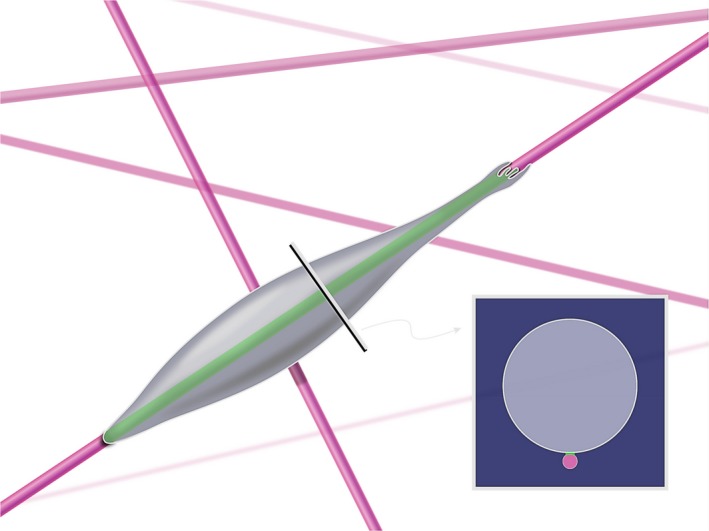
Cell migration on matrix fibres resembles one‐dimensional (1D) migration. A cell translocating along a single fibre or fibril has a very small region of contact (see the inset), which can be mimicked by engineering very narrow 1D lines on a cell culture surface and coating them with a matrix protein confined to that line [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 2.
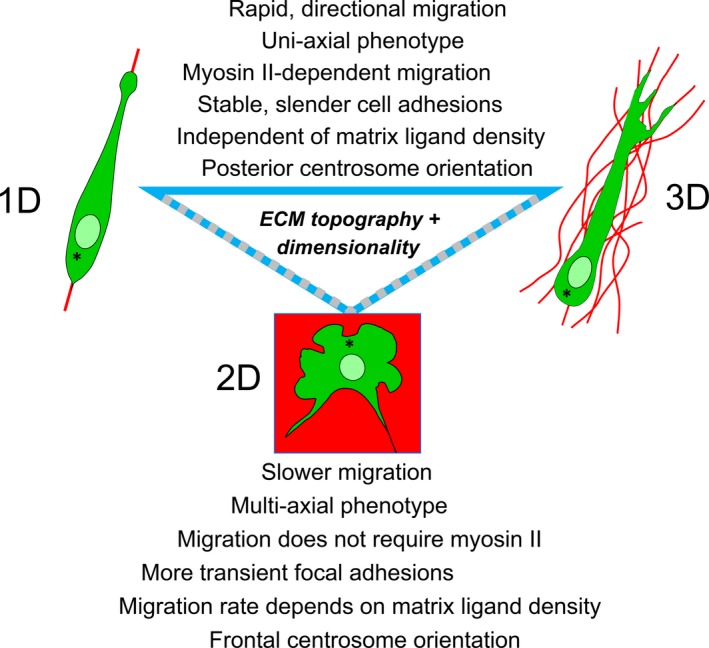
Direct comparisons of cells migrating in 1D, 2D and 3D models in vitro. Many specific cell biological behavioural features of fibroblasts migrating within a fibronectin‐rich 3D cell‐derived matrix are mimicked by migration of these cells on fibronectin‐coated 1D lines, but not migration on 2D glass substrates coated with fibronectin or with serum proteins. Figure re‐drawn from Doyle et al28 [Colour figure can be viewed at wileyonlinelibrary.com]
4. MODES OF 3D CELL MIGRATION
There are multiple modes of migration by cells in various different 3D extracellular matrix environments. For example, primary human fibroblasts, which are flat and spread out on 2D substrates, often use lamellipodial migration characterized by large, flat lamellipodia driven by actin polymerization. These same cells in 3D collagen gels use tiny, multiple lamellipodia with filopodia. In contrast, when these cells migrate in a 3D cell‐derived matrix, they switch to 3D “lobopodial” migration30 (Figure 3). A number of normal human immune cells can migrate by a mode of migration that resembles the locomotion and morphology of an amoeba, a process termed amoeboid migration; some tumour cells can also use this mode of migration (reviewed by Ref.2, 31, 32, 33
Figure 3.
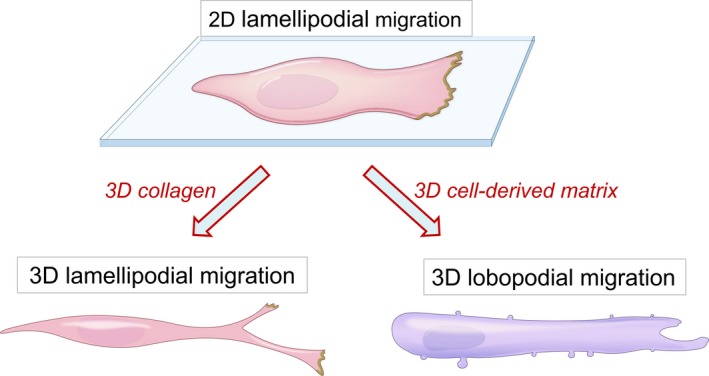
Cell morphologies in 2D vs 3D collagen or 3D cell‐derived matrix environments. Cells such as human fibroblasts that migrate on flat substrates are flattened in morphology and display lamellipodia at their leading edge, which promote migration by actin polymerization and cell protrusion. The same cell type in 3D collagen gels become spindle‐shaped and display multiple tiny lamellipodia at the tip of extending cell processes at the leading edge. In 3D cell‐derived matrix, however, these cells have a more tubular shape with lateral blebs and a leading edge that lacks lamellipodia as they migrate using lobopodial migration. Cells can be switched from lobopodial to lamellipodial migration by mild proteolysis of the cell‐derived matrix. Figure re‐drawn from Petrie et al30 [Colour figure can be viewed at wileyonlinelibrary.com]
A recently described mode of migration occurs in a confining or constricting 3D matrix that induces lobopodial migration. This migration mode is characterized by protrusion of a blunt leading edge of the cell devoid of classical actin‐driven lamellipodia, and it depends on differential intracellular pressure driven by a “nuclear piston”.30, 34 In the lobopodial mode of migration (Figure 4), a cell is embedded within an extracellular matrix that produces an elongated cell with its plasma membrane pressed close to the nucleus to seal off the anterior from the posterior regions of the cell. This diffusion barrier/hydraulic pressure seal appears to require either very close nucleus‐plasma membrane apposition or an extensive membranous vesicular barrier. Although not characterized in detail, the mechanism by which the nucleus is pulled forward to pressurize the anterior portion of the cell appears to be via myosin II linked to vimentin intermediate filaments, which are in turn anchored to the nucleus by nesprin‐3.34
Figure 4.
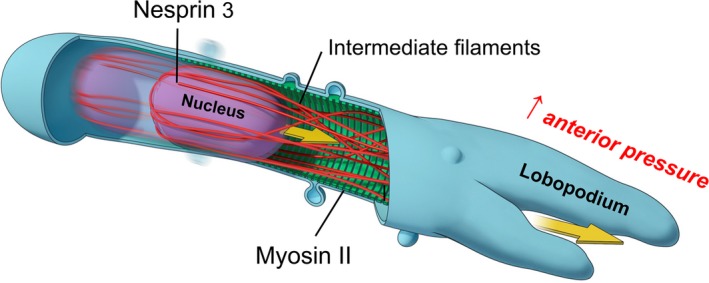
Nuclear piston 3D cell migration. Human fibroblasts migrating within a confining 3D cell‐derived matrix switch to lobopodial migration, a migration mode in which the nucleus can serve as a piston. The nucleus is pulled forward by myosin II contractility via vimentin intermediate filaments that link to nesprin‐3 on the nucleus. This pulling forward of the nucleus pressurizes the anterior end of the cell to protrude a lobopodial process. New cell‐matrix adhesions then form at the cell anterior to anchor the cells, and the cycle can repeat with another round of nuclear piston movement [Colour figure can be viewed at wileyonlinelibrary.com]
5. CANCER
The malignant counterpart of normal human fibroblasts, fibrosarcoma cells, fails to undergo nuclear piston migration in a cell‐derived matrix until their protease activity is inhibited for stabilization of the adjacent extracellular matrix.35 In fact, treatment of cell‐derived matrix with proteases can abolish lobopodial migration by normal human fibroblasts.30 Experimentally enhancing expression of the protease MT1‐MMP to promote local proteolysis at only 3‐4 times normal levels will cause loss of lobopodial migration by these normal human cells. Inhibition of MMPs restores normal lobopodial migration to these protease‐overexpressing cells.35 Thus, cells can switch their modes of migration depending on the level of proteolytic activity affecting cell interactions with locally adjacent extracellular matrix.
Invasion of human tumour cells into extracellular matrix has been studied extensively by many laboratories. Local invasion of these cells can be facilitated by microscopic cellular extensions termed invadopodia (Figure 5). These filopodia‐like cell processes often contain the MT1‐MMP protease for digestion of local extracellular matrix.36, 37 Real‐time imaging reveals that these microscopic processes are highly dynamic,38 which may account for their ability to degrade zones of matrix in their vicinity. Interestingly, besides being regulated by oncogenes and Rho GTPase activity, invadopodia can be induced dramatically by merely elevating the concentration of local fibrillar collagen matrix to a local concentration of 15 mg/mL or higher.39 This induction can surprisingly also occur with normal human fibroblasts in serum‐free medium in vitro. Although the complex regulation of invadopodia remains to be characterized further, this phenomenon of invadopodia induction by high‐density collagen involves a complex series of changes in protein phosphorylation, including phosphorylation of the integrin‐associated regulator kindlin‐2.39
Figure 5.
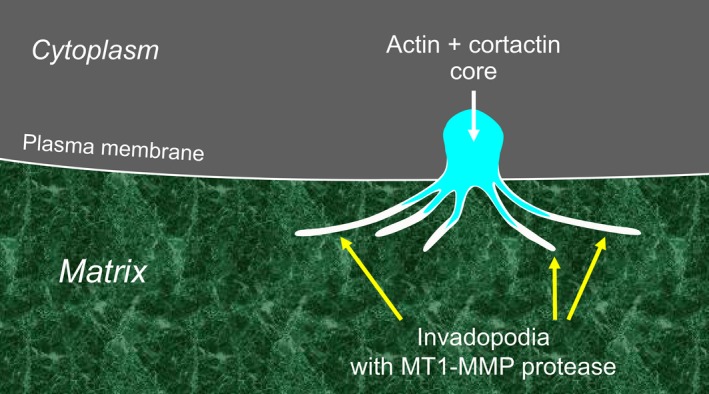
Invadopodia: dynamic micro‐invasive structures. Invadopodia are generated from an actin‐cortactin core at the plasma membrane and are used by cancer cells to degrade the extracellular matrix locally to promote invasion. The protease MT1‐MMP is expressed on the thin, filopodia‐like processes, which can degrade the matrix proteins and structures that they touch [Colour figure can be viewed at wileyonlinelibrary.com]
It has been known for many years that normal cells will often migrate towards regions of increasing stiffness in a process termed “durotaxis”.40 A recent study reveals that a variety of human tumour cells can also undergo durotaxis as efficiently as normal human fibroblasts and that the efficiency of durotaxis can be greatest at regions of low matrix stiffness.41 Another interesting feature of durotaxis is that it appears to be most efficient as a collective cell process, perhaps because a gradient of stiffness can be detected most efficiently by combining weak stiffness signals over a larger distance spanning multiple cells.42
6. BRANCHING MORPHOGENESIS
A particularly complex series of cell‐matrix interactions occurs during embryonic development of multiple organs in the highly dynamic process termed branching morphogenesis.43, 44, 45, 46, 47, 48, 49 This process converts a simple single epithelial bud to highly branched structures that greatly enhance epithelial surface area to provide sufficient exchange of gases in lungs, produce copious saliva by salivary glands and excrete litres of urine by kidneys. Branching morphogenesis involves both subdivision of an initial single epithelial bud by the formation of clefts or additional buds and the outward expansion of the newly formed branches of the organ (Figure 6). One striking feature of this type of development of early embryonic organs is the transient acquisition of cell motility by initially quiescent epithelial bud cells.50, 51 This high level of cell migratory activity during the process of branching morphogenesis may be important to permit tissue plasticity in analogy to the physical process of jamming and unjamming observed during expansion of the early embryonic axis.52 In addition, however, this high level of cell motility combined with cell‐cell adhesive interactions can contribute to tissue self‐organization. For example, completely dissociated and separated epithelial cells (Figure 7A,B) can self‐aggregate if provided with a 3D Matrigel matrix and growth factors, eventually forming organoids with bud‐like protrusions (Figure 7C).53 These buds display nearly identical patterns of cell adhesion molecules and F‐actin as never‐dissociated epithelial buds. This process of self‐assembly has been extended to salivary gland tissue engineering, as well as numerous organoid models, including mini‐brains and mini‐guts.54, 55, 56, 57, 58 These approaches may permit tissue engineering and regenerative therapy. From the point of view of matrix biology, however, much remains to be learned about the specific contributions of matrix to organoid formation and how it can provide an appropriate niche in vivo.
Figure 6.
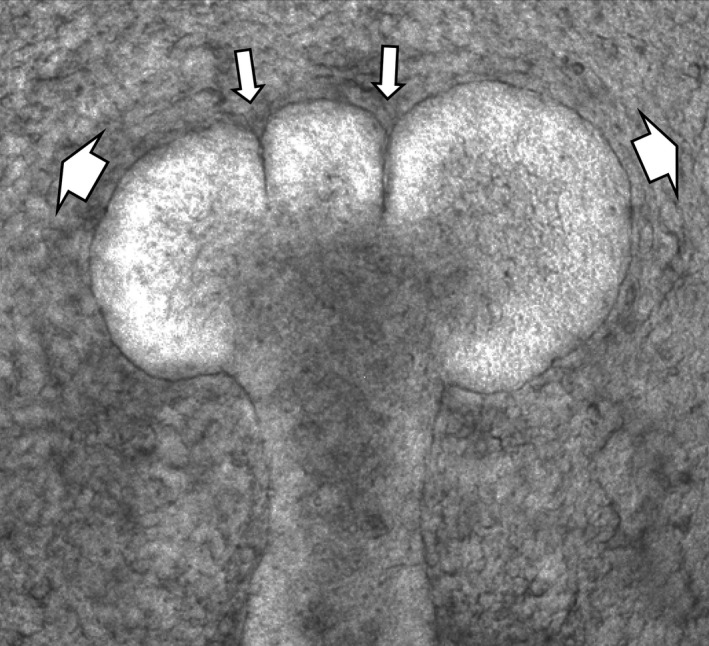
Early step of mammalian branching morphogenesis. This example shows a mouse salivary gland with two forming clefts (narrow arrows) and forming buds expanding outward (wide arrows) into the surrounding mesenchyme
Figure 7.
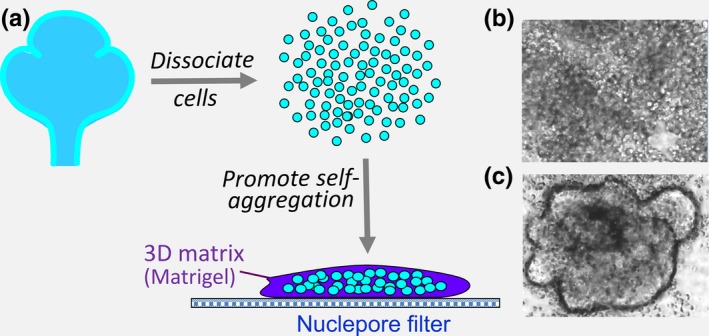
Self‐assembly of dissociated epithelial cells into bud‐like organoid structures. A, Isolated embryonic salivary gland epithelia were dissociated into single cells and then cultured within a small 3D Matrigel microenvironment on a nuclepore membrane filter. Phase‐contrast time‐lapse microscopy shows rapid self‐aggregation of the initially dissociated cells (B) into clusters that merge into large aggregates, from which bud‐like structures protrude (C) during a process of self‐organization [Colour figure can be viewed at wileyonlinelibrary.com]
7. BASEMENT MEMBRANE DYNAMICS
Basement membranes are well‐known structural features of many tissues, providing a substrate for epithelial cell attachment and organization, as well as separating epithelial and mesenchymal tissues.59, 60, 61, 62 The basement membrane provides a well‐known barrier to epithelial (carcinoma) cell invasion, and a classical feature of malignancy is tumour cell invasion across the basement membrane. Although this process is thought to involve proteases, physical force by a cellular process against the basement membrane can also contribute to cellular invasion across the basement membrane.63, 64, 65
One puzzle in developmental biology has been how tough, sheet‐like basement membrane barriers can be transiently transformed into sufficiently flexible sheets during embryonic development in order to permit rapid local tissue expansion without tissue mixing, for example, during the outward expansion of buds during branching morphogenesis (Figure 8A,B). Degradation of basement membrane proteins and proteoglycans by hydrolytic enzymes has been known for many decades to produce thinning of the basement membrane to allow local tissue expansion,66, 67 but exactly how this process is mediated was not clear. Direct examination of basement membrane structure and dynamics during active branching morphogenesis reveals a dramatic process of proteolytic and actomyosin‐dependent generation of numerous microscopic perforations or holes in the region of the basement membrane located at the tip of an expanding bud. These perforations generate a lace‐like meshwork (Figures 8C and 9) that permits extensive local distensibility of the basement membrane as visualized directly by time‐lapse confocal microscopy.68 These basement membrane perforations vary considerably in size but often average only 1‐2 μm2 in area, which is considerably less than the 25 μm2 average area of the adjacent epithelial cells. Because of the small size of these numerous perforations, the epithelial cells are restrained behind a highly flexible basement membrane.
Figure 8.
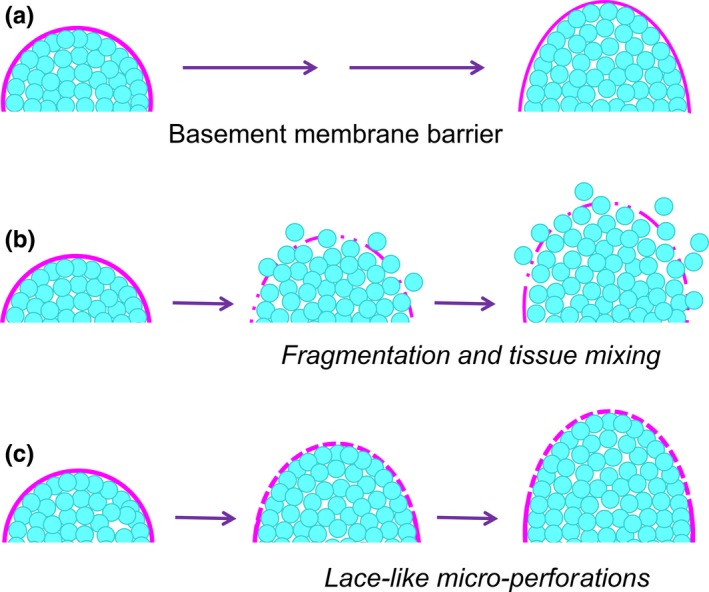
The challenge of embryonic tissue expansion within a confining basement membrane. A, The basement membrane barrier between epithelial and mesenchymal tissues must be able to expand along with bud expansion or extension during branching morphogenesis. B, Although degradation and thinning of the basement membrane are known to occur, absence of a controlled process would lead to tissue fragmentation and mixing of the highly motile epithelial cells into the surrounding mesenchyme—but this mixing does not occur in vivo. C, Basement membranes can become flexible by the formation of numerous microscopic holes or perforations that produce a lace‐like meshwork of basement membrane that can expand while still confining the epithelial cells [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 9.
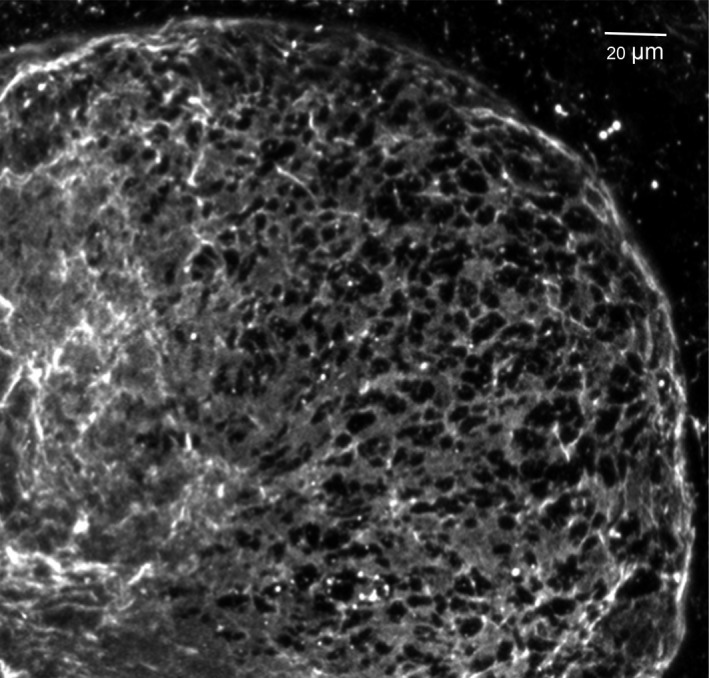
Perforated basement membrane meshwork at a bud tip. This image shows an embryonic salivary gland bud that was expanding towards the right with its basement membrane stained for collagen IV (light grey). Note the intact basement membrane on the left that becomes perforated by numerous microscopic holes (black) towards the righthand tip of an expanding bud
Intriguingly, however, the epithelial cells very frequently extend cellular blebs or elongated processes up to 5 μm in length through the perforations towards the surrounding mesenchyme cells. These highly active, extending and retracting cell processes may contribute to formation or maintenance of the perforations,68 but they may also correspond to the previously described direct contacts between epithelial and mesenchymal cells through the basement membrane (basal lamina).69, 70 In fact, a classical analysis of epithelial‐mesenchymal interaction reported that such close cell‐cell interactions may be required for successful development, in addition to the currently extensively studied growth factor interactions known to be involved in branching morphogenesis.71
The embryonic basement membrane, however, undergoes bulk translocation. In embryonic salivary glands, the basement membrane moves as a seemingly intact structure replete with the perforations in a direction opposite to that of bud expansion. Buds expand outward at approximately 5 μm/h, whereas the basement membrane translocates in the opposite direction towards the secondary duct at a rate of approximately 7‐8 μm/h (Figure 10). Both the formation of basement membrane perforations and the rearward translocation of the entire basement membrane require general MMP‐associated proteolytic activity and myosin II‐dependent actomyosin contractility.68 A major puzzle involves how this global translocation of basement membrane is driven, since local cell motility is not obviously directional so as to provide motive forces to help move the basement membrane.
Figure 10.
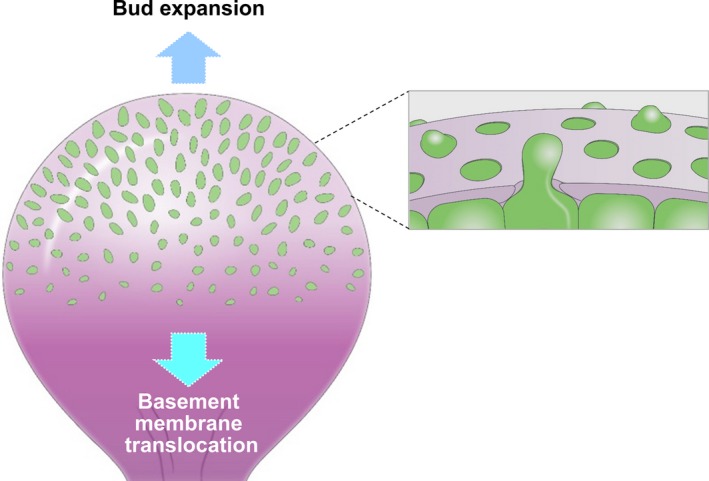
Basement membrane dynamics during embryonic branching morphogenesis. As buds expand outward, the basement membrane is perforated by numerous microscopic holes through which epithelial cell blebs and elongated processes protrude towards the mesenchyme. Concurrently, the entire basement membrane translocates backward towards the secondary duct [Colour figure can be viewed at wileyonlinelibrary.com]
8. FUTURE CHALLENGES AND OPPORTUNITIES
Direct visualization of cell‐extracellular matrix interactions involving single cells or tissues has provided novel insights into otherwise puzzlingly complex processes. The combination of direct real‐time visualization and a variety of biochemical, biophysical and genetic approaches promises to open new avenues of research for understanding multiple complicated biological processes. In the following section, we suggest a series of intriguing future avenues of research into cell‐matrix interactions. These and many other exciting opportunities should provide novel insights for many decades into the future.
8.1. Migration, invasion and matrix assembly
Recent investigations have characterized differing modes of 3D migration, for example, amoeboid, mesenchymal and lobopodial. However, the field will need to examine in‐depth the spatiotemporal dynamics of cells as they interact with their surrounding matrix microenvironment, in order to characterize the biophysics and biomechanics associated with these modes of migration. In addition, because contractility is known to be required for rapid 3D cell migration by numerous cell types, it will be important to understand how differences in cell interactions with different types of extracellular matrix affect cellular mechanosensing and mechanotransduction during 3D cell migration.
The diversity of protein composition and architecture in different types of 3D matrix can alter cell adhesion, mechanosensing/mechanotransduction and cell migration or invasion. Comparing in vivo matrix molecular components and architecture of different tissues and then developing increasingly complex and realistic in vitro 3D models will be important for understanding the roles of different types of matrix and their biophysical properties in biological processes. One example involves the known effects of matrix density on increased risk for breast cancer.72, 73, 74, 75, 76 Because local tissue invasion initiates tumour progression and metastasis, further elucidation of cell interactions with the basement membrane will be informative. For example, how are local dynamics of basement membrane and associated extracellular matrix involved in the transformation of carcinoma in situ into invasive carcinoma?
8.2. Organ morphogenesis
Work from our own and other laboratories has demonstrated extensive basement membrane remodelling during normal branching morphogenesis of mammalian organs during embryonic development. However, it remains unclear how cells regulate the production and assembly of new basement membrane for the coordinated surface expansion of epithelium during organ branching. An interesting future direction will be to examine how basement membrane components are produced and secreted using live imaging approaches. CRISPR/Cas‐mediated genetic perturbations will likely help to clarify regulatory mechanisms of this process. A related question for both developing and adult tissues is how basement membranes accumulate such substantial levels of fibronectin immediately at the surface facing mesenchymal tissues. Live imaging of cell interactions with basement membranes should clarify this and other questions about cell‐basement membrane interactions.
Additional approaches to understanding the mechanisms of branching morphogenesis will include increasingly in‐depth characterizations of RNA regulatory biology. For example, transcriptomic approaches will be valuable, including single‐cell sequencing to characterize the diverse cells of different early embryonic organs and their capacity to secrete specific matrix proteins as they self‐organize during development into complex, functionally distinct tissues and organs. MicroRNAs have also emerged as important regulators during branching morphogenesis,44, 77 but also in cancer initiation/progression78; some miRNAs may be implicated in both processes. Their roles in extracellular matrix remodelling during branching morphogenesis and cancer progression are poorly understood. Direct comparisons of these biological processes should provide new mechanistic insights and potential therapeutic approaches to targeting matrix remodelling in different diseases.
We previously implicated the protein BTBD7 in branching morphogenesis,79 but the extent of its functions and their molecular mechanisms remains unclear. BTBD7 is part of a large group of >200 BTB/POZ domain‐containing proteins encoded by the human genome that serve central biological roles ranging from organogenesis, gastrulation and stem cell differentiation to ribonucleotide damage responses and cell cycle regulation.80, 81, 82, 83 Determining the function of BTBD7 and the mechanisms through which it regulates cellular phenotypes, such as partial epithelial‐mesenchymal‐transition (EMT), should open new insight into its role in cell‐matrix interactions and organ development.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
Research in the authors’ laboratory is supported by the Intramural Research Program of the NIH, NIDCR.
Yamada KM, Collins JW, Cruz Walma DA, et al. Extracellular matrix dynamics in cell migration, invasion and tissue morphogenesis. Int. J. Exp. Path. 2019;100:144–152. 10.1111/iep.12329
REFERENCES
- 1. Bachir AI, Horwitz AR, Nelson WJ, Bianchini JM. Actin‐based adhesion modules mediate cell interactions with the extracellular matrix and neighboring cells. Cold Spring Harb Perspect Biol. 2017;9 10.1101/cshperspect.a023234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Charras G, Sahai E. Physical influences of the extracellular environment on cell migration. Nat Rev Mol Cell Biol. 2014;15:813‐824. [DOI] [PubMed] [Google Scholar]
- 3. Dzamba BJ, DeSimone DW. Extracellular matrix (ECM) and the sculpting of embryonic tissues. Curr Top Dev Biol. 2018;130:245‐274. [DOI] [PubMed] [Google Scholar]
- 4. Gauthier NC, Roca‐Cusachs P. Mechanosensing at integrin‐mediated cell‐matrix adhesions: from molecular to integrated mechanisms. Curr Opin Cell Biol. 2018;50:20‐26. [DOI] [PubMed] [Google Scholar]
- 5. Haak AJ, Tan Q, Tschumperlin DJ. Matrix biomechanics and dynamics in pulmonary fibrosis. Matrix Biol. 2018;73:64‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Humphries JD, Chastney MR, Askari JA, Humphries MJ. Signal transduction via integrin adhesion complexes. Curr Opin Cell Biol. 2018;56:14‐21. [DOI] [PubMed] [Google Scholar]
- 7. Hynes RO, Yamada KM. Extracellular matrix biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2012:397. [Google Scholar]
- 8. Loganathan R, Rongish BJ, Smith CM, et al. Extracellular matrix motion and early morphogenesis. Development. 2016;143:2056‐2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu Y, Kamel‐El Sayed SA, Wang K, et al. Live imaging of type I Collagen assembly dynamics in osteoblasts stably expressing GFP and mCherry‐tagged Collagen constructs. J Bone Miner Res. 2018;33:1166‐1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsubayashi Y, Louani A, Dragu A, et al. A moving source of matrix components is essential for de novo basement membrane formation. Curr Biol. 2017;27:3526‐3534.e3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nerger BA, Nelson CM. 3D culture models for studying branching morphogenesis in the mammary gland and mammalian lung. Biomaterials. 2019;198:135‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwarzbauer JE, DeSimone DW. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb Perspect Biol. 2011;3 10.1101/cshperspect.a005041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Helvert S, Storm C, Friedl P. Mechanoreciprocity in cell migration. Nat Cell Biol. 2018;20:8‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walraven M, Hinz B. Therapeutic approaches to control tissue repair and fibrosis: extracellular matrix as a game changer. Matrix Biol. 2018;71‐72:205‐224. [DOI] [PubMed] [Google Scholar]
- 15. Bardsley K, Deegan AJ, El Haj A, Yang Y. Current state‐of‐the‐art 3D tissue models and their compatibility with live cell imaging. Adv Exp Med Biol. 2017;1035:3‐18. [DOI] [PubMed] [Google Scholar]
- 16. Barrila J, Crabbe A, Yang J, et al. Modeling host‐pathogen interactions in the context of the microenvironment: three‐dimensional cell culture comes of age. Infect Immun. 2018;86 10.1128/IAI.00282-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bertrand P, Girard N, Delpech B, Duval C, Danjou J, Dauce JP. Hyaluronan (Hyaluronic‐Acid) and Hyaluronectin in the Extracellular‐Matrix of Human Breast Carcinomas ‐ Comparison between Invasive and Noninvasive Areas. Int J Cancer. 1992;52:1‐6. [DOI] [PubMed] [Google Scholar]
- 18. Bissell MJ. Goodbye flat biology ‐ time for the 3rd and the 4th dimensions. J Cell Sci. 2017;130:3‐5. [DOI] [PubMed] [Google Scholar]
- 19. Bissell MJ, Barcellos‐Hoff MH. The influence of extracellular matrix on gene expression: is structure the message? J Cell Sci Suppl. 1987;8:327‐343. [DOI] [PubMed] [Google Scholar]
- 20. Duval K, Grover H, Han LH, et al. Modeling physiological events in 2D vs. 3D cell culture. Physiology (Bethesda). 2017;32:266‐277. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 21. Eyckmans J, Chen CS. 3D culture models of tissues under tension. J Cell Sci. 2017;130:63‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lintz M, Munoz A, Reinhart‐King CA. The mechanics of single cell and collective migration of tumor cells. J Biomech Eng. 2017;139 10.1115/1.4035121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat Rev Genet. 2018;19:671‐687. [DOI] [PubMed] [Google Scholar]
- 24. Saglam‐Metiner P, Gulce‐Iz S, Biray‐Avci C. Bioengineering‐inspired three‐dimensional culture systems: Organoids to create tumor microenvironment. Gene. 2019;686:203‐212. [DOI] [PubMed] [Google Scholar]
- 25. Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601‐610. [DOI] [PubMed] [Google Scholar]
- 26. Weiss P. Experiments on cell and axon orientation invitro ‐ the role of colloidal exudates in tissue organization. J Exp Zool. 1945;100:353‐386. [DOI] [PubMed] [Google Scholar]
- 27. Doyle AD, Carvajal N, Jin A, Matsumoto K, Yamada KM. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility‐dependent adhesions. Nat Commun. 2015;6:8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doyle AD, Wang FW, Matsumoto K, Yamada KM. One‐dimensional topography underlies three‐dimensional fibrillar cell migration. J Cell Biol. 2009;184:481‐490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guetta‐Terrier C, Monzo P, Zhu J, et al. Protrusive waves guide 3D cell migration along nanofibers. J Cell Biol. 2015;211:683‐701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Petrie RJ, Gavara N, Chadwick RS, Yamada KM. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J Cell Biol. 2012;197:439‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paluch EK, Aspalter IM, Sixt M. Focal adhesion‐independent cell migration. Annu Rev Cell Dev Biol. 2016;32:469‐490. [DOI] [PubMed] [Google Scholar]
- 32. Ruprecht V, Wieser S, Callan‐Jones A, et al. Cortical contractility triggers a stochastic switch to fast amoeboid cell motility. Cell. 2015;160:673‐685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Te Boekhorst V, Preziosi L, Friedl P. Plasticity of cell migration in vivo and in silico. Annu Rev Cell Dev Biol. 2016;32:491‐526. [DOI] [PubMed] [Google Scholar]
- 34. Petrie RJ, Koo H, Yamada KM. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 2014;345:1062‐1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Petrie RJ, Harlin HM, Korsak LI, Yamada KM. Activating the nuclear piston mechanism of 3D migration in tumor cells. J Cell Biol. 2017;216:93‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Artym VV, Zhang Y, Seillier‐Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 2006;66:3034‐3043. [DOI] [PubMed] [Google Scholar]
- 37. Chen WT, Wang JY. Specialized surface protrusions of invasive cells, invadopodia and lamellipodia, have differential MT1‐MMP, MMP‐2, and TIMP‐2 localization. Ann N Y Acad Sci. 1999;878:361‐371. [DOI] [PubMed] [Google Scholar]
- 38. Artym VV, Matsumoto K, Mueller SC, Yamada KM. Dynamic membrane remodeling at invadopodia differentiates invadopodia from podosomes. Eur J Cell Biol. 2011;90:172‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Artym VV, Swatkoski S, Matsumoto K, et al. Dense fibrillar collagen is a potent inducer of invadopodia via a specific signaling network. J Cell Biol. 2015;208:331‐350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J . 2000;79:144‐152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. DuChez BJ, Doyle AD, Dimitriadis EK, Yamada KM. Durotaxis by human cancer cells. Biophys J . 2019;116(4):670‐683. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sunyer R, Conte V, Escribano J, et al. Collective cell durotaxis emerges from long‐range intercellular force transmission. Science. 2016;353:1157‐1161. [DOI] [PubMed] [Google Scholar]
- 43. Ghabrial A, Luschnig S, Metzstein MM, Krasnow MA. Branching morphogenesis of the Drosophila tracheal system. Annu Rev Cell Dev Biol. 2003;19:623‐647. [DOI] [PubMed] [Google Scholar]
- 44. Hauser BR, Hoffman MP. Regulatory Mechanisms Driving Salivary Gland Organogenesis. Curr Top Dev Biol. 2015;115:111‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ochoa‐Espinosa A, Affolter M. Branching morphogenesis: from cells to organs and back. Cold Spring Harb Perspect Biol. 2012;4 10.1101/cshperspect.a008243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pozzi A, Zent R. Extracellular matrix receptors in branched organs. Curr Opin Cell Biol. 2011;23:547‐553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Varner VD, Nelson CM. Cellular and physical mechanisms of branching morphogenesis. Development. 2014;141:2750‐2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang S, Sekiguchi R, Daley WP, Yamada KM. Patterned cell and matrix dynamics in branching morphogenesis. J Cell Biol. 2017;216:559‐570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Warburton D, El‐Hashash A, Carraro G, et al. Lung organogenesis. Curr Top Dev Biol. 2010;90:73‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14:570‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. J Cell Sci. 2006;119:3376‐3384. [DOI] [PubMed] [Google Scholar]
- 52. Mongera A, Rowghanian P, Gustafson HJ, et al. A fluid‐to‐solid jamming transition underlies vertebrate body axis elongation. Nature. 2018;561:401‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wei C, Larsen M, Hoffman MP, Yamada KM. Self‐organization and branching morphogenesis of primary salivary epithelial cells. Tissue Eng. 2007;13:721‐735. [DOI] [PubMed] [Google Scholar]
- 54. Amin ND, Pasca SP. Building models of brain disorders with three‐dimensional organoids. Neuron. 2018;100:389‐405. [DOI] [PubMed] [Google Scholar]
- 55. Artegiani B, Clevers H. Use and application of 3D‐organoid technology. Hum Mol Genet. 2018;27:R99‐R107. [DOI] [PubMed] [Google Scholar]
- 56. Barkauskas CE, Chung MI, Fioret B, Gao X, Katsura H, Hogan BL. Lung organoids: current uses and future promise. Development. 2017;144:986‐997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li M, Izpisua Belmonte JC. Organoids ‐ preclinical models of human disease. N Engl J Med. 2019;380:569‐579. [DOI] [PubMed] [Google Scholar]
- 58. Tanaka J, Ogawa M, Hojo H, et al. Generation of orthotopically functional salivary gland from embryonic stem cells. Nat Commun. 2018;9:4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brown KL, Cummings CF, Vanacore RM, Hudson BG. Building collagen IV smart scaffolds on the outside of cells. Protein Sci. 2017;26:2151‐2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jayadev R, Sherwood DR. Basement membranes. Curr Biol. 2017;27:R207‐R211. [DOI] [PubMed] [Google Scholar]
- 61. Pozzi A, Yurchenco PD, Iozzo RV. The nature and biology of basement membranes. Matrix Biol. 2017;57‐58:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sekiguchi R, Yamada KM. Basement membranes in development and disease. Curr Top Dev Biol. 2018;130:143‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Caceres R, Bojanala N, Kelley LC, et al. Forces drive basement membrane invasion in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2018;115:11537‐11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chang TT, Thakar D, Weaver VM. Force‐dependent breaching of the basement membrane. Matrix Biol. 2017;57‐58:178‐189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kelley LC, Chi Q, Caceres R, et al. Adaptive F‐actin polymerization and localized ATP production drive basement membrane invasion in the absence of MMPs. Dev Cell. 2019;48:313‐328.e318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bernfield M, Banerjee SD. The turnover of basal lamina glycosaminoglycan correlates with epithelial morphogenesis. Dev Biol. 1982;90:291‐305. [DOI] [PubMed] [Google Scholar]
- 67. Moore KA, Polte T, Huang S, et al. Control of basement membrane remodeling and epithelial branching morphogenesis in embryonic lung by Rho and cytoskeletal tension. Dev Dyn. 2005;232:268‐281. [DOI] [PubMed] [Google Scholar]
- 68. Harunaga JS, Doyle AD, Yamada KM. Local and global dynamics of the basement membrane during branching morphogenesis require protease activity and actomyosin contractility. Dev Biol. 2014;394:197‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bluemink JG, Van Maurik P, Lawson KA. Intimate cell contacts at the epithelial/mesenchymal interface in embryonic mouse lung. J Ultrastruct Res. 1976;55:257‐270. [DOI] [PubMed] [Google Scholar]
- 70. Cutler LS, Chaudhry AP. Intercellular contacts at the epithelial‐mesenchymal interface during the prenatal development of the rat submandibular gland. Dev Biol. 1973;33:229‐240. [DOI] [PubMed] [Google Scholar]
- 71. Wartiovaara J, Nordling S, Lehtonen E, Saxen L. Transfilter induction of kidney tubules: correlation with cytoplasmic penetration into nucleopore filters. J Embryol Exp Morphol. 1974;31:667‐682. [PubMed] [Google Scholar]
- 72. Barcus CE, Keely PJ, Eliceiri KW, Schuler LA. Stiff collagen matrices increase tumorigenic prolactin signaling in breast cancer cells. J Biol Chem. 2013;288:12722‐12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Carey SP, Martin KE, Reinhart‐King CA. Three‐dimensional collagen matrix induces a mechanosensitive invasive epithelial phenotype. Sci Rep. 2017;7:42088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Levental KR, Yu H, Kass L, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891‐906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Morris BA, Burkel B, Ponik SM, et al. Collagen matrix density drives the metabolic shift in breast cancer cells. EBioMedicine. 2016;13:146‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Provenzano PP, Inman DR, Eliceiri KW, et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6 10.1186/1741-7015-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Carraro G, El‐Hashash A, Guidolin D, et al. miR‐17 family of microRNAs controls FGF10‐mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E‐Cadherin distribution. Dev Biol. 2009;333:238‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369‐5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Onodera T, Sakai T, Hsu JC, Matsumoto K, Chiorini JA, Yamada KM. Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science. 2010;329:562‐565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Canning P, Sorrell FJ, Bullock AN. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med. 2015;88:101‐107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Choi J, Lee K, Ingvarsdottir K, et al. Loss of KLHL6 promotes diffuse large B‐cell lymphoma growth and survival by stabilizing the mRNA decay factor roquin2. Nat Cell Biol. 2018;20:586‐596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Werner A, Iwasaki S, McGourty CA, et al. Cell‐fate determination by ubiquitin‐dependent regulation of translation. Nature. 2015;525:523‐527. [DOI] [PMC free article] [PubMed] [Google Scholar]


