Abstract
Substantial heterogeneity exists within cervical cancer that is generally infected by human papillomavirus (HPV). However, the most common histological subtype of cervical cancer, cervical squamous cell carcinoma (CSCC), is poorly characterized regarding the association between its heterogeneity and HPV oncoprotein expression. We filtered out 138 CSCC samples with infection of HPV16 only as the first step; then we compressed HPV16 E6/E7 expression as HPVpca and correlated HPVpca with the immunological profiling of CSCC based on supervised clustering to discover subtypes and to characterize the differences between subgroups in terms of the HPVpca level, pathway activity, epigenetic dysregulation, somatic mutation frequencies, and likelihood of responding to chemo/immunotherapies. Supervised clustering of immune signatures revealed two HPV16 subtypes (namely, HPV16-IMM and HPV16-KRT) that correlated with HPVpca and clinical outcomes. HPV16-KRT is characterized by elevated expression of genes in keratinization, biological oxidation, and Wnt signaling, whereas HPV16-IMM has a strong immune response and mesenchymal features. HPV16-IMM exhibited much more epigenetic silencing and significant mutation at FBXW7, while MUC4 and PIK3CA were mutated frequently for HPV16-KRT. We also imputed that HPV16-IMM is much more sensitive to chemo/immunotherapy than is HPV16-KRT. Our characterization tightly links the expression of HPV16 E6/E7 with biological and clinical outcomes of CSCC, providing valuable molecular-level information that points to decoding heterogeneity. Together, these results shed light on stratifications of CSCC infected by HPV16 and shall help to guide personalized management and treatment of patients.
Abbreviations: HPV, human papillomavirus; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CSCC, cervical squamous cell carcinoma; FDR, false discovery rate; FPKM, fragments per kilobase of nonoverlapped exon per million fragments mapped; GSEA, gene set enrichment analysis; GO, Gene Ontology; HR, hazard ratio; CI, confidence interval; ρ, Pearson correlation coefficient; TCGA, The Cancer Genome Atlas; CGI, CpG island; IC50, half-maximal inhibitory concentration
Introduction
Cervical cancer accounts for 528,000 new cases and 266,000 deaths worldwide annually, more than any other gynecological tumor [1]. In 2019, approximately 13,170 new cases and 4250 deaths of cervical cancer were estimated to occur in the United States [2]. Cervical cancer remains the second leading cause of cancer death in women between the ages of 20 and 39 years, with nine deaths per week in this age group [2]. Between 80% and 90% of cervical cancer cases involve squamous cells (squamous cell carcinoma). The remainder begin from glandular cells and are called adenocarcinomas. Ninety-five percent of all cases are caused by persistent infection with carcinogenic human papillomavirus (HPV) [3], which is one of the most common sexually transmitted diseases in both men and women worldwide [4]. Based on their association with cervical cancer and precursor lesions, HPVs can also be classified into high-risk and low-risk HPV types. Low-risk HPV types include types 6, 11, 42, 43, and 44. High-risk HPV types include types 16, 18, 31, 33, 34, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, 69, and 70. Similar to head and neck cancer, HPV type 16 (HPV16) is the most common high-risk type detected in tumors, accounting for 50% of cancers and their precursors, called high-grade squamous intraepithelial lesions [5], [6], [7]. Preliminary studies have suggested that variants of HPV16 may show varying degrees of association with cervical cancer [7].
HPV typically infects the basal layer of the epithelium and then exploits the epithelial-to-keratinocyte proliferation and differentiation pathways to accomplish the viral life cycle. HPV expresses two main viral oncoproteins, E6 and E7, which are necessary for malignant conversion and cooperatively inhibit apoptosis and enhance tumor cell growth and proliferation by inducing the degradation of tumor suppressor TP53 and disruption of function of the retinoblastoma protein (Rb1), respectively [8], [9]. Alteration of additional pathways, such as suppression of the immune response [10], cell adhesion [11], and oxidative stress [12], may also be essential for tumor transformation.
Routine HPV testing has revealed that most HPV infections resolve, suggesting that HPV infection is necessary but not sufficient to develop cervical neoplasia and that other events are required. The molecular features of cervical cancers are beginning to be described to decipher substantial heterogeneity [13], [14], [15], [16], [17]. Our previous work has minutely discussed the genomic differences of cervical cancer based on HPV status [18]. However, little is known about how the expression of HPV16 oncoproteins affects tumor development and their association with the heterogeneity of cervical cancer, especially for cervical squamous cell carcinoma (CSCC), which provided us a motivation. Therefore, we sought here to report a detailed analysis to determine the presence of potential subtypes of CSCC that were associated with the expression of HPV16 oncoproteins, and we further comprehensively characterized for the potential subtypes in a multiomics view including somatic mutation and DNA methylation. Similar to recent reports in head and neck cancers [19], [20], two subtypes of tumors were identified and designated as HPV16-IMM and HPV16-KRT. These two identified subtypes exhibited distinct molecular signaling pathway enrichment, DNA methylation profiling, and somatic mutation spectrum. Implications for CSCC with HPV16 progression and opportunities for personalized therapy are discussed.
Materials and Methods
Patients and Samples
Molecular data were obtained from The Cancer Genome Atlas Project (TCGA) patients diagnosed with cervical cancer. Transcriptome raw count data of the TCGA-CESC project were downloaded from the GDC data portal (https://portal.gdc.cancer.gov) with 307 samples including 304 tumor samples and 3 normal samples. The raw, paired-end reads in FASTQ were also obtained for virus detection. Methylation data assessed by TCGA using Infinium 450K arrays were downloaded from Xena Public Data Hubs (https://xena.ucsc.edu/public-hubs) with 312 tumor samples. Somatic mutation data were obtained from cBioPortal (http://www.cbioportal.org/datasets) with 281 tumor samples. Three hundred and four patients with sufficient clinical and pathologic information were available from Firehose (http://www.firehose.org/), and 252 patients with the histological type of CSCC were chosen for this study.
RNA Analysis
Data Preprocessing
-
a)
Raw counts for each gene of mRNAs from RNA-seq. Ensembl ID for genes (protein coding mRNAs) was annotated in GENCODE27 to generate Gene Symbol names. The gene type of protein coding was selected for mRNAs.
-
b)
Counts data normalization. Raw reads count data were normalized across samples using the R package “DESeq” [21]. Specifically, DESeq first estimates the effective library size, which is also called the size factor, by dividing each column by the geometric means of the rows given a matrix or data frame of raw count data. Next, the median of these ratios (skipping the genes with a geometric mean of zero) is used as the size factor for that column. With the estimation of size factors, DESeq then divides each column of the count table by the size factor for that column. Thus, the count values are brought to a common scale, making them comparable across samples. Furthermore, count data were transformed by the varianceStabilizingTransformation function provided in DESeq [21]. With this function, the standard deviation of each gene is roughly constant regardless of the gene expression magnitude.
-
c)
Low expression filtering. To reduce noise, we kept only mRNAs with normalized count equal to or above 1 in at least 10% of the samples for downstream analysis.
Virus Detection from RNA-Seq
The algorithm VirusSeq [22] was harnessed to computationally subtract human sequences and generate a set of nonhuman sequences (e.g., viruses) in RNA-Seq. The RNA-seq libraries were aligned to both human and HPV genomes to quantify the host and viral gene expression and determine the HPV status. Among all 304 tumor samples, we identified 168 HPV16, 38 HPV18, 1 HPV26, 1 HPV30, 7 HPV31, 8 HPV33, 2 HPV34, 6 HPV35, 22 HPV45, 1 HPV51, 8 HPV52, 1 HPV56, 7 HPV58, 3 HPV59, 7 HPV68, 1 HPV69, 2 HPV70, and 21 no virus. Seven HPV oncoproteins were quantified for expression: E1, E2, E5, E6, E7, L1, and L2. Viral gene expression was presented as fragments per kilobase of nonoverlapped exon per million fragments mapped (FPKM). A positive integration event, described by Zhang et al. [20], is a fusion candidate that has at least four discordant read pairs and at least one junction spanning read [22]. A tumor sample was called genic integration positive if it contained at least one identified integration event.
Definition of a Comprehensive HPV Variable
The joint action of HPV E6 and E7 oncoproteins is required for HPV-induced malignancy [23]. In the present study, we found that only oncoproteins E6 and E7 have an influence on the patient prognosis, and their expression levels were also confirmed to be highly associated, prompting us to focus on E6 and E7. We identified a comprehensive HPV variable to explain the original expression level of oncoprotein E6 and E7 that was calculated by principal component analysis (PCA). The new PCA-based variable HPVpca was derived from the first principal component that represented 98.9% of the variation in the original data. The coefficients (normalized loading) of E6 and E7 to the first principal component are shown below:
| HPV16 oncoprotein | E6 | E7 |
| Coefficient | 0.49 | 0.51 |
Mathematically, let Eij represent log2 (FPKM + 1) value of specific oncoprotein j in sample i and Cj denote the corresponding coefficient of HPV16 oncoprotein (HPVj, j ∈ {1, 2}). The HPVpca can be calculated as follows:
Molecular Characterization for Subtypes
Differential expression analysis was performed by the R package “DESeq2” using the standard comparison mode [24]. P values were adjusted for multiple testing using an embedded Benjamini-Hochberg procedure in the package. Gene set enrichment analysis (GSEA) and Gene Ontology (GO) annotation were performed using the R package “clusterProfiler” [25], [26] to characterize the subtype according to the mRNA expression profile. Redundant enriched GO terms were removed using the “simplify” function. To this end, Molecular Signature Database gene sets were tested. The enrichment scores of molecular pathways and gene expression signatures were evaluated using single-sample gene set enrichment analysis (R package “GSVA”) and NTP (Nearest Template Prediction, R package “CMScaller”) [27], [28], [29]. To computationally infer the infiltration level of specific immune cell types using RNA-seq data, we used 20 immune-related cell types from the literature that included three categories of adaptive immunity, innate immunity, and other component [30]. Supervised hierarchical clustering based on immune-related cell types was performed basically using the hclust() R function via Ward's clustering and 1-Pearson's correlation distance with k = 2 as the number of clusters. To calculate an E6 activity score, first, for each gene in the pathway, samples were ranked according to their expression levels. For each sample, the ranks of the genes were summed, and the resulting values were then centered by mean and scaled by the standard deviation across samples to yield the final scores. For E6 negatively regulated genes, the expression levels were ranked in descending order because the direction of regulation is known to be opposite. The Tumor Immune Dysfunction and Exclusion (TIDE) algorithm and subclass mapping were used to predict the clinical response to immune checkpoint blockade [31], [32].
Genetic and Epigenetic Analysis
We used the MutSigCV_v1.41 [33] (www.broadinstitute.org) to infer significant cancer mutated genes (q < 0.05) across the current identified two classes with default parameters. The tumor mutation burden was computed by summing all types of nonsilent mutations. The mutation landscape oncoprint was drawn by R package “ComplexHeatmap” [34]. The number of predicted neoantigens was extracted from previous TCGA research for cervical squamous cell carcinoma and endocervical adenocarcinoma [35]. Methylation analysis followed the chip analysis methylation pipeline (ChAMP) [36] with the required R package “IlluminaHumanMethylation450kanno.ilmn12.hg19” as annotation. Methylation probes were filtered by champ.filter() and champ.impute() with default parameters, and the β value was normalized by champ.norm() function. Differentially methylated probes were detected by champ.DMP(), and champ.GSEA was used to call GSEA results. Hypermethylated probes were identified by delta β greater than 0 and adjusted P < .05. Promoters located in a CpG island were determined by 450 k annotation file with the feature of TSS200 or TSS1500 and CpG island (CGI). For a gene with more than one probe mapping to its promoter, the median β value was considered. An epigenetically silenced gene was determined if a promoter CGI was hypermethylated and its corresponding gene's expression was upregulated. The DNA methylation-based immune infiltration score was extracted from TCGA previous research [37].
Chemotherapeutic Response Prediction
We predicted the chemotherapeutic response for each sample based on the largest publicly available pharmacogenomics database [the Genomics of Drug Sensitivity in Cancer (GDSC), https://www.cancerrxgene.org/]. Two commonly used chemo drugs, cisplatin and gemcitabine, were selected. The prediction process was implemented by R package “pRRophetic” where the samples' half-maximal inhibitory concentration (IC50) was estimated by ridge regression and the prediction accuracy was evaluated by 10-fold cross-validation based on the GDSC training set. All parameters were set by the default values with removal of the batch effect of “combat” and tissue type of “allSoldTumours,” and duplicate gene expression was summarized as mean value [38].
Statistical Analysis
All statistical tests were executed by R/3.5.2 using a χ2 or Fisher's exact test for categorical data when appropriate, a two-sample Wilcoxon test (Mann-Whitney test) for continuous data, a log-rank test Kaplan-Meier curve [39], and Cox regression [40] for the hazard ratio (HR). Survival analysis was performed using the R package “survival”. Fisher's exact test of independence was used to statistically test the association between categorical clinical information and defined subtypes. For all statistical analysis, a P value less than .05 was considered statistically significant.
Results
Overview of Sample Selection
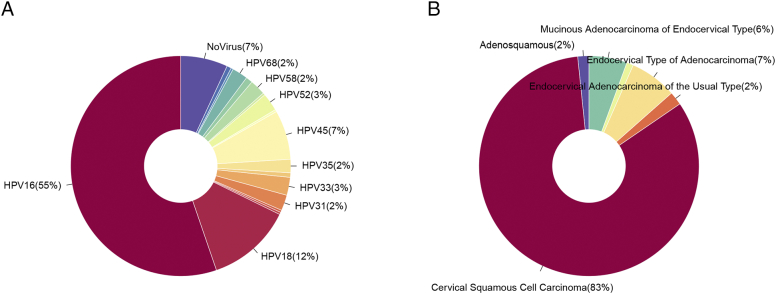
Among all 304 tumor samples, we identified 168 with HPV16, 115 with other types of HPV, and 21 samples with no HPV detected (Figure 1A). Five samples were diagnosed with a histological type of adenosquamous carcinoma, 252 with CSCC, 6 with endocervical adenocarcinoma of the usual type, 21 with endocervical type of adenocarcinoma, 3 with endometrioid adenocarcinoma of endocervix, and 17 with mucinous adenocarcinoma of endocervical type (Figure 1B). To avoid potential effects arising from histological type and virus type, we purified the samples used for analyzing and selected samples that were confirmed to be CSCC with infection of HPV16 virus only. By doing so, 138 HPV16 samples with full survival and clinicopathological information were selected for downstream analysis.
Figure 1.
Overview of sample selection. (A) Pie chart of the distribution of the detected HPV type and (B) histological type. The 138 selected samples were those diagnosed with CSCC and infected by HPV16.
Association Between Highly Correlated Oncoproteins E6/E7 and the Clinical Outcome
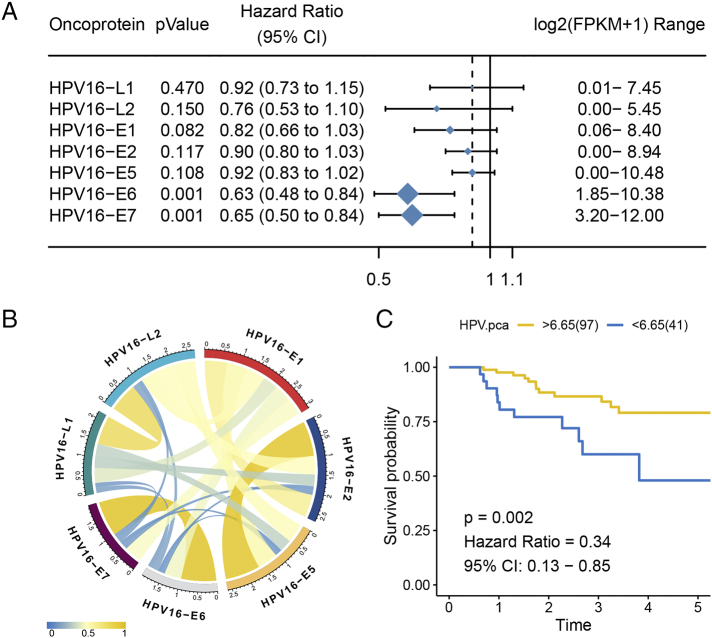
We performed univariate Cox regression to determine whether the seven detected HPV16 oncoproteins affected the patient outcome. As expected, oncoproteins E6 and E7 were significantly associated with overall survival, and their high expression could favor the prognosis of patients (E6: P = .001, HR = 0.63; E7: P = .0009, HR = 0.65) (Figure 2A). By calculating the Pearson correlation coefficient of the paired oncoproteins, we further found that the expression levels of HPV E6 and E7 were highly correlated in CSCC samples [ρ = 0.98 and false discovery rate (FDR) = 2.35e-94] (Figure 2B, Supplementary Table S1). Given the high correlation and significant impact on prognosis, a PCA-based variable, HPVpca, was calculated to compress the expression level of oncoproteins E6 and E7. Univariate Cox regression confirmed the correlation between HPVpca and overall survival [P = .0009, HR = 0.64, 95% confidence interval (CI) = 0.49-0.83]. Under the optimal cut point, we separated samples into two subsets: a 97-sample HPV16-H subset with relatively high expression of HPVpca and a 41-sample HPV16-L subset with relatively low expression. As expected, HPV16-H showed better survival than HPV16-L (P = .002, HR = 0.34, 95% CI = 0.13-0.85) (Figure 2C).
Figure 2.
Highly correlated oncoproteins E6 and E7 were associated with clinical outcome. (A) The forest plot shows only that HPV16 E6/E7 expression affects the patient overall survival. (B) Pearson correlation coefficient of pairwise HPV16 oncoproteins. (C) High level of HPVpca presented a favorable prognosis.
Differential expression analysis identified 360 differentially expressed genes with a threshold of P < .05, FDR < 0.25, and absolute log2(fold change) > log2(1.5), including 161 upregulated and 199 downregulated differentially expressed genes for HPV16-H compared with those for HPV16-L (Supplementary Table S2). Upregulated genes in HPV16-H were enriched for immune-related terms, such as positive regulation of T-cell activation (FDR = 1.03e-06), positive regulation of leukocyte activation (FDR = 1.89e-06), and MHC class II receptor activity (FDR = 7.64e-17); upregulated genes in HPV16-L demonstrated enrichment in several metabolic processes, such as glucan metabolic process (FDR = 0.034) and positive regulation of cAMP metabolic process (FDR = 0.015).
Association Between Two Supervised Clustering-Based HPV16 Subtypes and the Clinical Outcomes
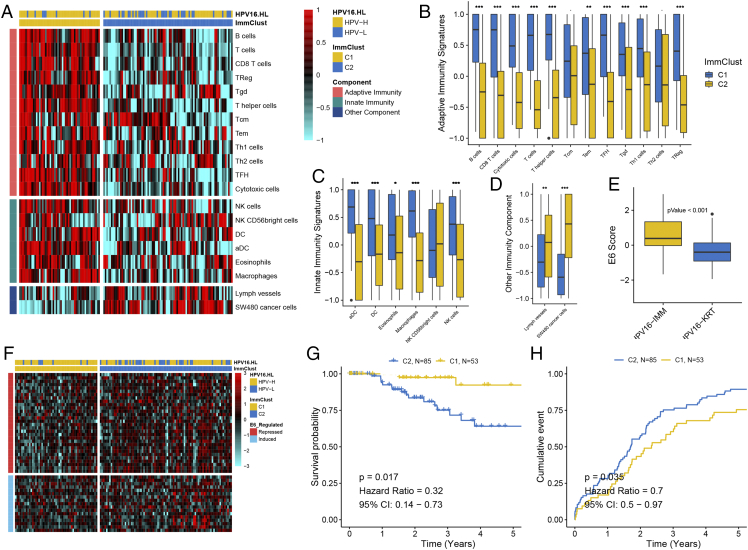
Now that a high level of HPVpca may correspond to a strong immune response from the above analysis, we wondered whether immunologic profiling could distinguish the heterogeneity of CSCC with HPV16. Thus, supervised clustering using 20 immune-related cell types was applied to all 138 HPV16 samples, and two distinct subtypes were revealed (Figure 3A). Specifically, a subtype, C1, comprising 53 samples was separated from another subtype, C2, containing 85 samples, exhibiting a high enrichment level for adaptive immunity signatures (Figure 3B) and innate immunity signatures (Figure 3C) and low enrichment for lymph vessels and SW480 cancer cells (Figure 3D).
Figure 3.
Immune signatures based on supervised clustering revealed two HPV16 subtypes that correlated with clinical outcomes. (A) Heatmap of 20 immune signatures across 138 CSCC samples distinguished two immunological patterns between the subtypes. The box plot shows different enrichment levels between the subtypes in (B) adaptive immunity signatures, (C) innate immunity signatures, and (D) other immunity components. E6 activity calculated by E6-repressed and -induced genes was compared and presented in (E) and (F). Kaplan-Meier curve showing the difference in (G) overall survival and (H) progression-free survival between the subtypes.
Subtype C1 with significantly higher HPVpca than C2 (P = .013) presented high enrichment for samples classified into the HPV16-H group, whereas subtype C2 was enriched for samples belonging to HPV16-L (P = .004). We next tested whether any other HPV16 oncoproteins (E1, E2, E5, L1, L2) were differentially expressed between C1 and C2, and no significance could be observed. Because the expression levels were quantified using RNA levels, they may not reflect the actual E6 or E7 protein activity levels in the cell. Fortunately, E6 and E7 were highly correlated in our study; thus, we used published literature [41] to calculate an E6 activity score for each sample to quantify E6 and E7 activity (Figure 3E). Overall, the E6 score was significantly higher in C1, indicating an elevated E6 activity (P = 1.36e-07) (Figure 3F). In particular, the genes downregulated by E6 in the literature were more repressed in C1 than in C2, whereas the upregulated genes were induced to a lesser degree. We next tried to determine whether a difference existed in HPV-integration events between the two subtypes, and no significance could be observed (P = .39).
Similar to HPV16-H, subtype C1 demonstrated a more favorable prognosis than subtype C2 (P = .017, HR = 0.32, 95% CI = 0.14-0.73) (Figure 3G). Regarding progression-free survival, subtype C1 showed a lower recurrence rate than subtype C2 (P = .035, HR = 0.7, 95% CI = 0.5-0.97) (Figure 3H).
Characterization of the HPV16 Subtypes Regarding Different Functional Pathways
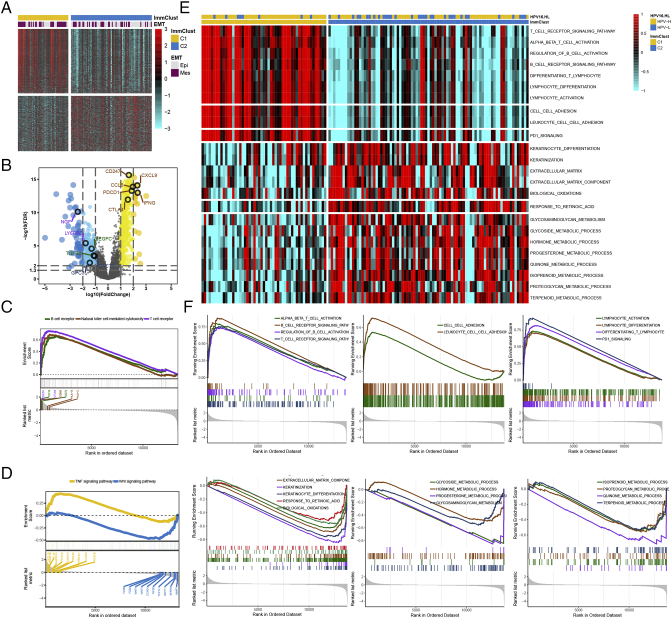
We next characterized the molecular differences between the two HPV16 subtypes. Differential expression analysis found 738 significantly differentially expressed genes, including 397 upregulated and 341 downregulated genes [absolute log2(fold change) > log2(2) and FDR < 0.05] (Figure 4A, Supplementary Table S3). GO enrichment analysis of biological processes for the upregulated genes in subtype C1 revealed global enrichment for “immune response” terms (Supplementary Table S4); upregulated genes in subtype C2 were most significantly enriched for “epithelial cell proliferation,” “Wnt signaling pathway,” and “metabolic process” such as retinol metabolism (Supplementary Table S5). The representative differentially expressed genes from each relevant GO term are shown in Figure 4B and include CD247, PDCD1, CTAL4, CCL5, CXCL9, and IFNG for “immune response”; TGFB2 and VEGFC for epithelial; DACT1 and GPC3 for Wnt signaling; and other genes for several metabolic processes. Kyoto Encyclopedia of Genes and Genomes enrichment analysis also showed remarkably elevated immune-related signaling pathways in C1 subtype, such as the T-cell and B-cell receptor signaling pathways and natural killer cell–mediated cytotoxicity (Figure 4C). We also found C1 enriched for the tumor necrosis factor (TNF) signaling pathway, whereas C2 showed high enrichment for the Wnt signaling pathway (Figure 4D, Supplementary Table S6). GSEA of the preranked gene list showed subtype C1 was enriched for immune-related cells (e.g., T cells, B cells, lymphocytes, the inflammatory response, and chemokines) or pathways (e.g., programmed cell death protein 1 signaling and cell adhesion), whereas subtype C2 was enriched for keratinocyte differentiation, biological oxidations, several metabolic processes (e.g., hormones and proteoglycan), and the retinoic acid pathway (Figure 4, E-F, Supplementary Table S7). Therefore, we designated C1 and C2 as HPV16-IMM and HPV16-KRT, respectively. Consistent with GO annotation, using a 315-gene epithelial-mesenchymal transition signature [42], more samples in the HPV16-IMM subtype were predicted to be in the mesenchymal class, whereas the HPV16-KRT subtype was enriched for more epithelial-like samples (P = .015).
Figure 4.
Differentially regulated genes and pathways between the HPV16 subtypes. (A) Heatmap of significant differentially expressed genes between the subtypes with different enrichment in the predicted epithelial-mesenchymal transition status. (B) The volcano plot shows representative genes within interested pathways. (C) Kyoto Encyclopedia of Genes and Genomes analysis demonstrated that the immune-related pathway and TNF signaling were enriched in the HPV16-IMM subtype, whereas the Wnt signaling was enriched in the HPV16-KRT subtype. (E) Heatmap of the enrichment level calculated by single-sample gene set enrichment analysis for interested pathways derived from GSEA and their corresponding GSEA plots in (F).
Significant Immune Infiltration for the HPV16-IMM Subtype Based on DNA Methylation
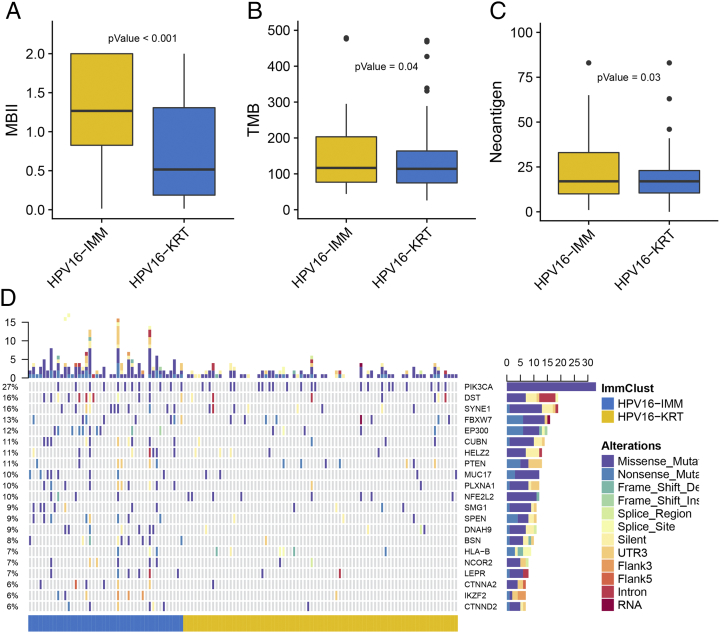
Considering the far-ranging differentially regulated genes and pathways between the HPV16 subtypes, we wondered if such dysregulation could mirror epigenetic alterations in CSCC with HPV16 because of the complicated biological correlation between gene expression and DNA methylation. After the probe-filtering process, 339,518 probes remained across 138 tumor samples. ChAMP identified 2512 hypermethylated probes in HPV16-IMM (Supplementary Table S8) that were enriched for immune processes such as T-cell, B-cell, and lymphocyte activation (Supplementary Table S9), a trend that was coincident with a significantly higher methylation-based immune infiltration score in HPV16-IMM than in HPV16-KRT (P = 8.23e-06) (Figure 5A). We further annotated 44,898 promoters located in CGIs, mapping to 10,269 genes. Integrative analysis by mRNA expression and promoter CGIs methylation identified 187 epigenetically silenced genes, including 183 for HPV16-IMM and only 4 for HPV16-KRT (Supplementary Table S10).
Figure 5.
Epigenetic and genetic alteration between the HPV16 subtypes in terms of (A) the methylation-based immune infiltration score, (B) tumor mutation burden, and (C) predicted number of neoantigens. (D) Oncoprint shows the somatic mutation landscape of MutSigCV-detected SMGs and other differentially mutated genes between the subtypes.
Differential Somatic Mutation Landscape Between the HPV16 Subtypes
To investigate whether differences exist in the somatic mutation frequencies between the HPV16 subtypes, we filtered genes with a nonsilent mutation rate greater than 5% and identified 16 genes differentially mutated between the HPV16 subtypes (P < .05, Supplementary Table S11). Under a stringent threshold of q < 0.05, MutSigCV detected four significantly mutated genes (SMGs) among all CSCC samples, including FBXW7 (q = 7.96e-09), PIK3CA (q = 2.37e-07), PTEN (q = 3.21e-05), and NFE2L2 (q = 3.15e-03) (Supplementary Table S12), all of which had been reported from previous TCGA research [17]. MutSigCV determined 215 and 232 significant mutations for HPV16-IMM and HPV16-KRT, respectively, under a loose threshold of P < .05, and only nine SMGs were shared. Additionally, only FBXW7 (q = 3.94e-03) was detected to be SMG for HPV16-IMM (Supplementary Table S13), whereas SMGs MUC4 (q = 5.33e-05) and PIK3CA (q = 0.035) could be targeted for HPV16-KRT (Figure 5D, Supplementary Table S14).
Additionally, recent analyses have described special links between the genomic landscape and antitumor immunity. Particularly, the tumor mutational burden (TMB) has emerged as a highly promising and clinically validated biomarker for immune checkpoint inhibitors, and presence of neoantigens could drive T-cell responses [35], [43], [44]. To verify whether the TMB and neoantigens may affect immunology in CSCC with HPV16, we analyzed the association between the HPV16 subtypes and count of somatic mutation or neoantigens. Basically, we found that HPV16-IMM presented a significantly higher count of TMB and neoantigens than HPV16-KRT (P = .04 for TMB and .03 for neoantigens) (Figure 5, B-C), suggesting that the HPV16-IMM subtype might be more sensitive to immune checkpoint inhibitor treatment.
More Sensitivity to Immuno/Chemotherapies for HPV16-IMM Subtype
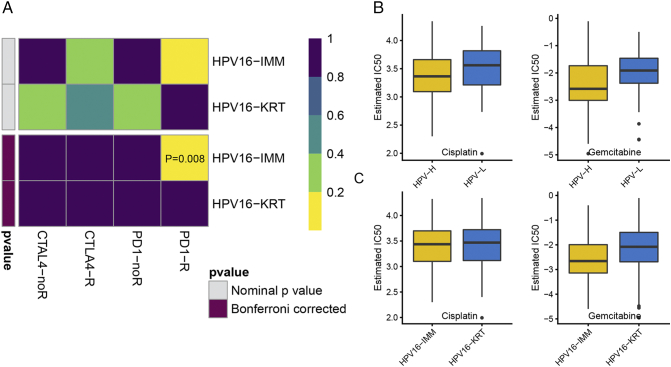
Examining TMB and neoantigens of the two HPV16 subtypes allowed us to further investigate the likelihood of responding to immunotherapy. Although immune checkpoint inhibitors have not yet been approved as routine drugs for cervical cancer, we therefore harnessed the TIDE algorithm to predict the likelihood of response to immunotherapy, and it demonstrated that HPV16-IMM (43%, 23/53) may be more likely to respond to immunotherapy than HPV16-KRT (26%, 22/85) (P = .04). In addition to the TIDE prediction, we also used subclass mapping to compare the expression profile of the two HPV16 subtypes we defined with another published dataset containing 47 patients with melanoma that responded to immunotherapies [45]. We were very delighted to see that HPV16-IMM is more promising to respond to anti–PD-1 therapy (Bonferroni corrected P = .008) (Figure 6A).
Figure 6.
Differential putative chemotherapeutic and immunotherapeutic response. The box plots of the estimated IC50 for cisplatin and gemcitabine are shown in (A) for HPVpca-based HPV-H and HPV-L and (B) for HPV16-IMM and HPV16-KRT. (C) Submap analysis manifested that HPV16-IMM could be more sensitive to the programmed cell death protein 1 inhibitor (Bonferroni-corrected P = .008).
Considering that chemotherapy is the common way to treat cervical cancer, we tried to assess the response of two HPV16 subtypes to two chemo drugs: cisplatin and gemcitabine. Thus, we trained the predictive model on the GDSC cell line data set by ridge regression with a satisfied predictive accuracy evaluated by 10-fold cross-validation. We estimated the IC50 for each sample in the TCGA dataset based on the predictive model of these two chemo drugs. We could observe a significant difference in the estimated IC50 between HPV16-H and HPV16-L for these two chemo drugs where HPV16-H could be more sensitive to commonly administered chemotherapies (P = .022 for cisplatin, P = .003 for gemcitabine) (Figure 6B). However, only gemcitabine could be observed to present a significant response sensitivity to HPV16-IMM compared with HPV16-KRT (P = .008, P = .516 for cisplatin) (Figure 6C).
Demographic Characteristics
The distributions of patient age, tumor grade, and clinical stage were not different between HPV16-IMM and HPV16-KRT. However, patients with HPV16-IMM tumors showed more tumor-free cases (P = .006), consistent with a favorable prognosis. The examined count of lymph node in HPV16-KRT was dramatically lower than that of HPV16I-IMM (P = 8.68e-08). We observed an elevated body mass index trend in HPV16-IMM (P = .087), but no significance could be drawn when considering four body mass index categories (Supplementary Table S15).
Discussion
Essentially all cervical cancers, including CSCC, are HPV positive by DNA, and HPV oncoprotein expression, which is critical for cancer development, has been widely accepted [4], [16]. It remains necessary to gain an in-depth understanding of the influence of HPV expression on the host and reveal the association between HPV and the inherent heterogeneity of this cancer, thus developing novel or improved personal treatment. This study, for the first time, is tightly linked to the expression of HPV16 E6/E7 with biological and clinical features using various types of data including TCGA public data and GDSC database resources, shedding new lights on targeted therapy in CSCC.
In the present study, we found that highly expression-related HPV16 E6 and E7 were significantly associated with the prognosis of CSCC patients. In general, high viral expression favored the patient clinical outcome. We compressed the expression of E6 and E7 into a new comprehensive PCA-based score, HPVpca. According to the optimal cutoff of prognosis, samples were divided into two groups with different HPVpca level. GO annotation indicated that the high-score group was enriched with “immune response” terms. Thus, we conducted supervised clustering on the 138 CSCC samples by specific immune cells and acquired two subtypes with distinct a pattern of immunologic profile. HPV16-IMM inhibited a significantly higher HPVpca level and more enriched TNF signaling, which is concordant with previous research that enhanced TNF pathway activity shall promote HPV E6/E7 expression [46]. The most significant difference in expression between these two subtypes is the upregulation of mesenchymal and immune-response genes in the HPV16-IMM subtype, which also presented higher E6 activity, keratinization, biological oxidation process, and metabolism dysregulation in HPV16-KRT. In fact, most of these pathways could be explained by the biology of HPV carcinogenesis where the HPV oncoprotein, especially E6, was reported to downregulate a mass of genes involved in keratinocyte differentiation and upregulate genes normally expressed in mesenchymal lineages, which is consistent with HPV16-IMM–enriched mesenchymal-like samples, whereas epithelial-like samples were enriched in another subtype, HPV16-KRT.
The higher immune response in HPV16-IMM was dramatically observed, featuring significant enrichment for immune signatures, such as T cells, B cells, lymphocytes, the inflammatory response, and chemokines. Together, these are hypothesized to promote a better response to the treatment of HPV16-infected patients, especially HPV16-IMM patients. In our study, we demonstrated that a stronger immune response could indeed predict a better overall survival and a lower recurrent rate. It was speculated that, when HPV shifts from the initial episomal form to an integrated transcribed form, the inflammatory/immune response towards HPV is simultaneously attenuated [20], but we could not observe a significant difference in HPV-integration events between the subtypes, which might be caused by the relatively small sample size. Anyway, the stronger inflammatory/immune response might be stimulated by highly expressed HPV16 in the HPV16-IMM subtype and may partially explain why HPV16-IMM may overall have a better prognosis. To decipher the poor prognosis of HPV16-KRT, we found enrichment for activated Wnt signaling. The Wnt pathway regulates cellular proliferation and differentiation processes and, thus, plays critical roles in pathologic conditions such as cancers. Activation of the Wnt pathway results in the accumulation of β-catenin, which in turn increases transcription of a broad range of genes to promote cell proliferation, achieving progression and malignant transformation [47]. Additionally, we found that HPV16-KRT is associated with retino-related pathways. Specifically, HPV16-KRT was enriched for the retinol metabolism pathway and retinoic acid, which might be of great value in the chemopreventive and therapeutic roles for cervical neoplasia [48] and should be investigated for the synthesis of new pharmacological agents [49]. Emerging evidence indicates that cancer is primarily a metabolic disease, and personalization of metabolic therapy as a broad-based cancer treatment strategy shall likely suggest more targets for exploration [50].
Analysis of DNA methylation suggested greater levels of immune cell infiltrates within HPV16-IMM, including T-cell, B-cell, and lymphocyte activation. Integrative analysis by mRNA expression and promoter CGI methylation manifested a significant broad spectrum of gene silencing in HPV16-IMM compared with that in HPV16-KRT (183 vs. 4). The differentially mutated gene SMG1, which we found to be frequently mutated in HPV16-IMM, was reported to correlate with improved survival when functional mutated [51]. EP300, which is also frequently mutated in HPV16-IMM, could affect growth-suppressive and proapoptotic functions driven by TGFβ and is only observed in 30% of squamous cell carcinomas based on the literature [17]. The two subtypes also presented different SMGs, of which PIK3CA being significantly mutated in HPV16-KRT is consistent with previous research of a co-occurrence condition for PIK3CA mutation and high keratin expression [17], and higher TMB in line with the submap result suggested that HPV16-IMM might have a high likelihood of responding to immunotherapy; however, no FDA-approved immune-based drugs for CSCC exists. Cervical cancers are usually treated with a combined regimen of platinum-based chemotherapy and radiation. Using the GDSC database, we imputed that HPV16-IMM could be more sensitive to commonly used chemotherapies than HPV16-KRT. The above discussion implicates that HPV16-IMM may benefit from the combination of chemotherapy and immunotherapy, and targeted therapy of FBXW7 mutation should also be considered, whereas HPV16-KRT could be more sensitive to targeted Wnt pathways, and MUC4 and PIK3CA mutation. Retino-related agents and metabolism-targeted therapies will also require fine-tuning to realize personalized treatment for patients in HPV16-KRT.
Briefly, we sought here to provide comprehensive understanding of how the expression of HPV16 oncoproteins affects tumor development and their association with the heterogeneity of CSCCs infected with HPV type 16. The molecular differences between the identified subtypes may favor the opportunity to be targeted under specific therapeutic approaches separately. However, this study has a few limitations. First, the inconspicuous sample size may make it difficult to observe the difference in clinicopathological features. Additionally, the TCGA data enrolled for analysis were mostly collected from patients with cervical cancer in developed countries but lacked data from developing countries.
Conclusions
Overall, our study tightly linked the expression of HPV16 E6/E7 with the biological and clinical outcomes of CSCC. Those patients with high expression of HPV16 E6/E7 could benefit from immunotherapy, which may accelerate the approval of immune checkpoint inhibitors for CSCC. We are poised for a further investigation and eagerly anticipate the verification of our findings in a larger cohort of patients.
Acknowledgments
Acknowledgements
The results published here are based upon data generated by TCGA, managed by the NCI and NHGRI. We are grateful to TCGA for this source of data. Information about TCGA can be found at http://cancergenome.nih.gov. We also thank Sergio Chavez of Department of Bioinformatics and Computational Biology (The University of Texas, MD Anderson Cancer Center) for editing the manuscript.
Data Accessibility
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations of Interest
None.
Footnotes
Funding sources: This work was supported by “Double First-Class” University project (CPU2018GY09).
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.04.003.
Contributor Information
Xiaoping Su, Email: xsu1@mdanderson.org.
Fangrong Yan, Email: f.r.yan@163.com.
Appendix A.
Supplementary Tables S1–S15
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Schiffman M, Wentzensen N, Wacholder S, Kinney W, Gage JC, Castle PE. Human papillomavirus testing in the prevention of cervical cancer. J Natl Cancer Inst. 2011;103:368–383. doi: 10.1093/jnci/djq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 5.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14:467–475. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 6.Walline HM, Komarck C, McHugh JB, Byrd SA, Spector ME, Hauff SJ, Graham MP, Bellile E, Moyer JS, Prince ME. High-risk human papillomavirus detection in oropharyngeal, nasopharyngeal, and oral cavity cancers: comparison of multiple methods. JAMA Otolaryngol Head Neck Surg. 2013;139:1320–1327. doi: 10.1001/jamaoto.2013.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hildesheim A, Schiffman M, Bromley C, Wacholder S, Herrero R, Rodriguez AC, Bratti MC, Sherman ME, Scarpidis U, Lin Q-Q. Human papillomavirus type 16 variants and risk of cervical cancer. J Natl Cancer Inst. 2001;93:315–318. doi: 10.1093/jnci/93.4.315. [DOI] [PubMed] [Google Scholar]
- 8.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 9.Yim E-K, Park J-S. The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res Treat. 2005;37:319–324. doi: 10.4143/crt.2005.37.6.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tindle RW. Immune evasion in human papillomavirus–associated cervical cancer. Nat Rev Cancer. 2002;2:59–65. doi: 10.1038/nrc700. [DOI] [PubMed] [Google Scholar]
- 11.Whiteside MA, Siegel EM, Unger ER. Human papillomavirus and molecular considerations for cancer risk. Cancer. 2008;113:2981–2994. doi: 10.1002/cncr.23750. [DOI] [PubMed] [Google Scholar]
- 12.Williams VM, Filippova M, Filippov V, Payne KJ, Duerksen-Hughes P. HPV16 E6* induces oxidative stress and DNA damage. J Virol. 2014;88:6751–6761. doi: 10.1128/JVI.03355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akagi K, Li J, Broutian TR, Padilla-Nash H, Xiao W, Jiang B, Rocco JW, Teknos TN, Kumar B, Wangsa D. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014;24:185–199. doi: 10.1101/gr.164806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio L, Cibulskis K, Bertelsen B. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang K-W, Alaei-Mahabadi B, Samuelsson T, Lindh M, Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat Commun. 2013;4:2513–2521. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rusan M, Li YY, Hammerman PS. Genomic landscape of human papillomavirus–associated cancers. Clin Cancer Res. 2015;21:2009–2019. doi: 10.1158/1078-0432.CCR-14-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Network CGAR. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Jiang Y, Lu X, Zhao H, Chen C, Wang Y, Hu W, Zhu Y, Yan H, Yan F. Genomic characterization of cervical cancer based on human papillomavirus status. Gynecol Oncol. 2019;152:629–637. doi: 10.1016/j.ygyno.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Tomar S, Graves CA, Altomare D, Kowli S, Kassler S, Sutkowski N, Gillespie MB, Creek KE, Pirisi L. Human papillomavirus status and gene expression profiles of oropharyngeal and oral cancers from European American and African American patients. Head Neck. 2016;38:E694–E704. doi: 10.1002/hed.24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Koneva LA, Virani S, Arthur AE, Virani A, Hall PB, Warden C, Carey TE, Chepeha DB, Prince M. Subtypes of HPV-positive head and neck cancers are associated with HPV characteristics, copy number alterations, PIK3CA mutation, and pathway signatures. Clin Cancer Res. 2016;22:4735–4745. doi: 10.1158/1078-0432.CCR-16-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S, Huber W. European Molecular Biology Laboratory (EMBL); Heidelberg, Germany: 2012. Differential expression of RNA-Seq data at the gene level–the DESeq package. [Google Scholar]
- 22.Chen Y, Yao H, Thompson EJ, Tannir NM, Weinstein JN, Su X. VirusSeq: software to identify viruses and their integration sites using next-generation sequencing of human cancer tissue. Bioinformatics. 2012;29:266–267. doi: 10.1093/bioinformatics/bts665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomaić V. Functional roles of E6 and E7 oncoproteins in HPV-induced malignancies at diverse anatomical sites. Cancer. 2016;8:95–116. doi: 10.3390/cancers8100095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550–570. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eide PW, Bruun J, Lothe RA, Sveen A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci Rep. 2017;7:16618–16626. doi: 10.1038/s41598-017-16747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7–21. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoshida Y. Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS One. 2010;5:15543–15551. doi: 10.1371/journal.pone.0015543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A. Spatiotemporal dynamics of intratumoural immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Hoshida Y, Brunet J-P, Tamayo P, Golub TR, Mesirov JP. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS One. 2007;2:1195–1203. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 35.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumours associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber A, Teschendorff AE. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017;33:3982–3984. doi: 10.1093/bioinformatics/btx513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Yang T-HO, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA. Vol. 48. 2018. The immune landscape of cancer Immunity; pp. 812–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geeleher P, Cox NJ, Huang RS. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014;15:R47. doi: 10.1186/gb-2014-15-3-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bland JM, Altman DG. Survival probabilities (the Kaplan-Meier method) BMJ. 1998;317:1572–1580. doi: 10.1136/bmj.317.7172.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fox J. 2002. Cox proportional-hazards regression for survival data: an R and S-PLUS companion to applied regression 2002. [Google Scholar]
- 41.Duffy CL, Phillips SL, Klingelhutz AJ. Microarray analysis identifies differentiation-associated genes regulated by human papillomavirus type 16 E6. Virology. 2003;314:196–205. doi: 10.1016/s0042-6822(03)00390-8. [DOI] [PubMed] [Google Scholar]
- 42.Tan TZ, Miow QH, Miki Y, Noda T, Mori S, Huang RYJ, Thiery JP. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med. 2014;6:1279–1293. doi: 10.15252/emmm.201404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steuer CE, Ramalingam SS. Tumour mutation burden: leading immunotherapy to the era of precision medicine? J Clin Oncol. 2018;36:631–632. doi: 10.1200/JCO.2017.76.8770. [DOI] [PubMed] [Google Scholar]
- 45.Roh W, Chen P-L, Reuben A, Spencer CN, Prieto PA, Miller JP, Gopalakrishnan V, Wang F, Cooper ZA, Reddy SM. Integrated molecular analysis of tumour biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aah3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaiotti D, Chung J, Iglesias M, Nees M, Baker PD, Evans CH, Woodworth CD. Tumour necrosis factor-α promotes human papillomavirus (HPV) E6/E7 RNA expression and cyclin-dependent kinase activity in HPV-immortalized keratinocytes by a ras-dependent pathway. Mol Carcinog. 2000;27:97–109. doi: 10.1002/(sici)1098-2744(200002)27:2<97::aid-mc5>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 47.Bahrami A, Hasanzadeh M, ShahidSales S, Yousefi Z, Kadkhodayan S, Farazestanian M, Joudi Mashhad M, Gharib M, Mahdi Hassanian S, Avan A. Clinical significance and prognosis value of Wnt signalling pathway in cervical cancer. J Cell Biochem. 2017;118:3028–3033. doi: 10.1002/jcb.25992. [DOI] [PubMed] [Google Scholar]
- 48.Abu J, Batuwangala M, Herbert K, Symonds P. Retinoic acid and retinoid receptors: potential chemopreventive and therapeutic role in cervical cancer. Lancet Oncol. 2005;6:712–720. doi: 10.1016/S1470-2045(05)70319-3. [DOI] [PubMed] [Google Scholar]
- 49.Nathan B, Yu-Jui Yvonne W. Retinoid pathway and cancer therapeutics. Adv Drug Deliv Rev. 2010;62:1285–1298. doi: 10.1016/j.addr.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seyfried TN, Flores RE, Poff AM, D'Agostino DP. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis. 2014;35:515–527. doi: 10.1093/carcin/bgt480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gubanova E, Brown BT, Ivanov SV, Helleday T, Mills GB, Yarbrough WG, Issaeva N. Downregulation of SMG-1 in HPV-positive head and neck squamous cell carcinoma due to promoter hypermethylation correlates with improved survival. Clin Cancer Res. 2012;18:1257–1267. doi: 10.1158/1078-0432.CCR-11-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1–S15
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.