In the title compound, C—H⋯O hydrogen bonds and weak C—H⋯π interactions link adjacent molecules into a three-dimensional supramolecular network.
Keywords: chalcone, crystal structure, hydrogen bond, Hirshfeld surface analysis
Abstract
A novel chalcone, C20H20O, derived from benzylidenetetralone, was synthesized via Claissen–Schmidt condensation between tetralone and 2,4,6-trimethylbenzaldehyde. In the crystal, molecules are linked by C—H⋯O hydrogen bonds, producing R 2 2(20) and R 2 4(12) ring motifs. In addition, weak C—H⋯π and π-stacking interactions are observed. The intermolecular interactions were investigated using Hirshfeld surface analysis and two-dimensional fingerprint plots, revealing that the most important contributions for the crystal packing are from H⋯H (66.0%), H⋯C/ C⋯H (22.3%), H⋯O/O⋯H (9.3%), and C⋯C (2.4%) interactions. Shape-index plots show π–π stacking interactions and the curvedness plots show flat surface patches characteristic of planar stacking.
Chemical context
Chalcone (systematic name 1,3-diphenyl-2-propene-1-one) is an aromatic ketone that represents the central core for various derivatives with interesting properties, known as chalcones (Kostanecki & Tambor, 1899 ▸). For example, chalcones are found in fruits, vegetables, spices, tea or soy, and find applications as pharmaceuticals (Di Carlo et al., 1999 ▸). Chalcones are also major intermediates in the synthesis of natural products and are widely used in synthetic and pharmaceutical chemistry (Dhar, 1981 ▸; Ansari et al., 2005 ▸) because they have antitumor (Modzelewska et al., 2006 ▸), antifungal (López et al., 2001 ▸), anti-inflammatory (Lee et al., 2006 ▸), anti-bacterial (Batovska et al., 2009 ▸) or antitubercular properties (Lin et al., 2002 ▸). In general, chalcones consist of two aromatic rings that are linked by a three-carbon α,β-unsaturated carbonyl system, leading to a completely delocalized π-electron system. Recently, chalcones have also been used in the field of materials science as non-linear optical devices (Raghavendra et al., 2017 ▸). As part of our studies in this area, we report herein the synthesis, crystal structure and Hirshfeld surface analysis of a new chalcone.
Structural commentary
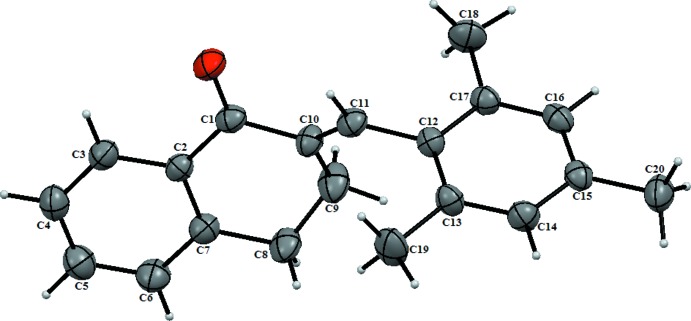
In the title molecule (Fig. 1 ▸), the cyclohexanone ring (C1/C2,C7/C8,C9/C10) has an envelope conformation with the flap atom C9 deviating by 0.280 (3) Å from the least-squares plane through the ring. The cyclohexanone ring is nearly co-planar with the benzene ring (C2–C7) being fused at a dihedral angle of 4.70 (18)°, but is inclined to the other benzene ring (C12–C17) by 74.95 (13)°. Torsion angles involving the methylene group C10=C11 are 83.3 (5)° (C17—C12—C11—C10), 129.8 (4)° (C11—C10—C9—C8) and 27.7 (6)° (O1—C1—C10—C11).
Figure 1.
The molecular structure of the title compound, with the atom labelling. Displacement ellipsoids are drawn at the 50% probability level.
Supramolecular features
The main intermolecular interactions in the crystal structure of the title compound are of type C—H⋯O, C—H⋯π (Table 1 ▸) and π–π. Interactions between a methyl group and the carbonyl O atom (C20—H20C⋯O1ii) as well as between an aromatic H atom and the carbonyl atom (C16—H16⋯O1i) lead to  (20) and
(20) and  (12) motifs (Fig. 2 ▸), linking adjacent molecules parallel to (001) (Table 2 ▸, Fig. 2 ▸). A weak C9—H9A⋯Cg2iii (Cg2 is the centroid of the C2–C7 benzene ring) interaction is also present (Fig. 2 ▸), along with weak aromatic π-stacking interactions [Cg2⋯Cg2(−2 − x, −y, −1 − z) = 3.887 (3) Å] that consolidate the three-dimensional packing.
(12) motifs (Fig. 2 ▸), linking adjacent molecules parallel to (001) (Table 2 ▸, Fig. 2 ▸). A weak C9—H9A⋯Cg2iii (Cg2 is the centroid of the C2–C7 benzene ring) interaction is also present (Fig. 2 ▸), along with weak aromatic π-stacking interactions [Cg2⋯Cg2(−2 − x, −y, −1 − z) = 3.887 (3) Å] that consolidate the three-dimensional packing.
Table 1. Hydrogen-bond geometry (Å, °).
Cg2 is the centroid of the C2–C7 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C16—H16⋯O1i | 0.93 | 2.69 | 3.493 (5) | 145 |
| C20—H20C⋯O1ii | 0.96 | 2.60 | 3.535 (5) | 165 |
| C9—H9A⋯Cg2iii | 0.97 | 2.90 | 3.865 (6) | 175 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Figure 2.
A view along the a axis of the title structure. Blue dashed lines denote the C—H⋯O hydrogen bonds which form  (20) and
(20) and  (12) ring motifs. C—H⋯π interactions are shown as green dashes lines.
(12) ring motifs. C—H⋯π interactions are shown as green dashes lines.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C20H20O |
| M r | 276.36 |
| Crystal system, space group | Triclinic, P

|
| Temperature (K) | 293 |
| a, b, c (Å) | 8.728 (2), 8.757 (2), 12.094 (3) |
| α, β, γ (°) | 77.768 (19), 80.822 (19), 61.929 (18) |
| V (Å3) | 795.2 (4) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.07 |
| Crystal size (mm) | 0.64 × 0.51 × 0.33 |
| Data collection | |
| Diffractometer | Stoe IPDS 2 |
| Absorption correction | Integration (X-RED32; Stoe & Cie, 2002 ▸) |
| T min, T max | 0.956, 0.982 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 8143, 2726, 1102 |
| R int | 0.088 |
| (sin θ/λ)max (Å−1) | 0.595 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.061, 0.155, 0.91 |
| No. of reflections | 2726 |
| No. of parameters | 194 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.25, −0.14 |
Database survey
A search of the Cambridge Structural Database (CSD, version 5.40, update November 2018; Groom et al., 2016 ▸) using (E)-2-(4-methylbenzylidene)-3,4-dihydronaphthalen-1(2H)-one as the main skeleton revealed the presence of four structures containing the chalcone moiety with different substituents that are similar to the title compound: (E)-4-[(1-oxo-3,4-dihydronaphthalen-2(1H)-ylidene)methyl]benzonitrile (QEVMAI; Baddeley et al., 2017 ▸); (E)-4-[(5-methoxy-1-oxo-3,4-dihydronaphthalen-2(1H)-ylidene)methyl]benzonitrile (QEVMEM; Baddeley et al., 2017 ▸); (E)-4-[(6-methoxy-1-oxo-3,4-dihydronaphthalen-2(1H)-ylidene)methyl]benzonitrile (QEVMIQ; Baddeley et al., 2017 ▸); 1′-(4-bromophenyl)-4′-{4-[(1-oxo-3,4-dihydronaphthalen-2(1H)-ylidene) methyl]phenyl}-3′′,4′′-dihydro-1′′H,2H-dispiro(acenaphthylene-1,2′-pyrrolidine-3′,2′′-naphthalene)-1′′,2-dione (VUZXOE; Saravanan et al., 2010 ▸). QEVMAI and VUZXOE both crystallize in space group P
 , while QEVMEM and QEVMIQ crystallize in space group P21/c. In the structures of QEVMAI, QEVMEM and QEVMIQ, the dihedral angles between the phenyl groups are 45.66 (5), 55.06 (7) and 69.78 (5)°, respectively. In the structure of VUZXOE, the central benzene ring makes a dihedral angle of 42.71 (7)° with the bromophenyl ring.
, while QEVMEM and QEVMIQ crystallize in space group P21/c. In the structures of QEVMAI, QEVMEM and QEVMIQ, the dihedral angles between the phenyl groups are 45.66 (5), 55.06 (7) and 69.78 (5)°, respectively. In the structure of VUZXOE, the central benzene ring makes a dihedral angle of 42.71 (7)° with the bromophenyl ring.
Hirshfeld surface analysis
A Hirshfeld surface analysis (Spackman & Jayatilaka, 2009 ▸) and the associated two-dimensional fingerprint plots (McKinnon et al., 2007 ▸) were performed with CrystalExplorer17 (Turner et al., 2017 ▸), using standard surface resolution with the three-dimensional d norm surfaces plotted over a fixed colour scale of −0.0870 (red) to 1.2944 (blue) a.u.. The three-dimensional d norm surface of the title molecule is illustrated in Fig. 3 ▸ a and 4 ▸. The pale-red spots symbolize short contacts and negative d norm values on the surface correspond to the C—H⋯O interactions described above (Table 1 ▸). The overall two-dimensional fingerprint plot is illustrated in Fig. 5 ▸ a. The Hirshfeld surfaces mapped over d norm are shown for the H⋯H, H⋯C/ C⋯H, H⋯O/O⋯H, C⋯C contacts (McKinnon et al., 2007 ▸), and the two-dimensional fingerprint plots are shown in Fig. 5 ▸ b and 5c, respectively, associated with their relative contributions to the Hirshfeld surface. The largest contribution to the overall crystal packing is from H⋯H interactions (66.0%); H⋯H contacts are shown in the middle region 1.10 Å < (d i + d e) < 1.18 Å. H⋯C/C⋯H contacts contribute 22.3% to the Hirshfeld surface, resulting in two pairs of characteristic wings in the fingerprint plot. The pair of tips appears at 1.10 Å < (d i + d e) < 1.65 Å. H⋯O/O⋯H contacts make a 9.3% contribution to the Hirshfeld surface. The contacts are represented by a pair of sharp spikes in the region 1.05 Å < (d i + d e) < 1.4 Å in the fingerprint plot. The C⋯C contacts are a measure of π–π stacking interactions and contribute 2.4% to the Hirshfeld surface. They appear as an arrow-shaped distribution at 1.80 Å < (d i + d e) < 2.0 Å.
Figure 3.
(a) dnorm mapped on Hirshfeld surfaces for visualizing the intermolecular interactions; (b) shape-index map and (c) curvedness map of the title compound.
Figure 4.
dnorm mapped on Hirshfeld surfaces for visualizing the intermolecular interactions.
Figure 5.

(a) The overall two-dimensional fingerprint plot and (b) Hirshfeld surface representations with the function d norm plotted onto the surface for (i) H⋯H, (ii) H⋯C/C⋯H, (iii) H⋯O/O⋯H and (iv) C⋯C interactions. (c) The two-dimensional fingerprint plots for the title compound, delineated into (i) H⋯H, (ii) H⋯C/ C⋯H, (iii) H⋯O/O⋯H, (iv) C⋯C interactions.
The shape-index map of the title molecule (Fig. 3 ▸ b) was generated in the ranges −1 to 1 Å. The convex blue regions symbolize hydrogen-donor groups and concave red regions symbolize hydrogen-acceptor groups. π–π interactions on the shape-index map are indicated by adjacent red and blue triangles. As can be seen in Fig. 3 ▸ b, there are π–π interactions present between adjacent molecules in the title complex.
The curvedness map of the title compound (Fig. 3 ▸ c) was generated in the range −4 to 0.4 Å. The large green regions represent a relatively flat (i.e. planar) surface area, while the blue regions demonstrate areas of curvature. The presence of π–π stacking interactions is also evident as flat regions around the rings on the Hirshfeld surface plotted over curvedness.
Synthesis and crystallization
2,4,6-Trimethylbenzylidenetetralone was prepared according to a literature protocol (Kumar et al., 2017 ▸). 10 ml of a NaOH solution (40%wt) was slowly added to a mixture of tetralone (1 mmol) and 2,4,6-trimethylbenzaldehyde (1 mmol) in ethanol (10 ml) at room temperature and stirred overnight. Then ice-cold water was added to the reaction mixture. The resulting precipitate was filtered off and dried in vacuo. The compound was purified by crystallization from ethanol, resulting in colourless prismatic crystals.
Yield 85%, m.p. 358 K; IR (ν, cm−1): 3060 (C—H, aromatic), 2920 (C—H, aliphatic), 1670 (C=O), 1620 (C=C, aromatic); 1H NMR (300 MHz, DMSO-d 6, δ, ppm): 7.9 (1H, d, =C—H), 7.58 (1H, s, =C—H), 7.50 (1H, t, =C—H), 7.38 (1H,t, =C—H), 7.30 (1H, d, =C—H), 6.82 (2H, s, =C—H), 2.8 (2H, t, —CH2), 2.4 (2H, t, —CH2), 2.2 (3H, s,—CH3), 2.02 (6H, s, 2 CH3); 13C NMR (75 MHz, DMSO-d 6, δ, ppm): 186.9, 144.5, 138.0, 137.2, 135.9, 135.6, 134.2, 133.5, 132.4, 129.3, 128.6, 128.0, 127.6, 28.9, 27.4, 21.3, 20.5. Analysis calculated for C20H20O: C, 86.92%; H, 7.29%; O, 5.79%. Found: C, 86.99%; H, 7.35%; O, 5.90%.
Refinement details
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. Hydrogen atoms were fixed geometrically and treated as riding, with C—H = 0.97 Å for methyl groups, 0.96 Å for methylene groups, 0.93 Å for aromatic hydrogen atoms and 0.98 Å for methine groups, with U iso(H) = 1.2U eq(C) or 1.5U eq(C-methyl).
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989019006182/wm5495sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019006182/wm5495Isup3.hkl
Supporting information file. DOI: 10.1107/S2056989019006182/wm5495Isup3.cml
CCDC reference: 1913649
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C20H20O | Z = 2 |
| Mr = 276.36 | F(000) = 296 |
| Triclinic, P1 | Dx = 1.154 Mg m−3 |
| a = 8.728 (2) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 8.757 (2) Å | Cell parameters from 12610 reflections |
| c = 12.094 (3) Å | θ = 2.7–30.2° |
| α = 77.768 (19)° | µ = 0.07 mm−1 |
| β = 80.822 (19)° | T = 293 K |
| γ = 61.929 (18)° | Prism, colorless |
| V = 795.2 (4) Å3 | 0.64 × 0.51 × 0.33 mm |
Data collection
| Stoe IPDS 2 diffractometer | 1102 reflections with I > 2σ(I) |
| Detector resolution: 6.67 pixels mm-1 | Rint = 0.088 |
| rotation method scans | θmax = 25.0°, θmin = 2.7° |
| Absorption correction: integration (X-RED32; Stoe & Cie, 2002) | h = −10→10 |
| Tmin = 0.956, Tmax = 0.982 | k = −10→10 |
| 8143 measured reflections | l = −14→14 |
| 2726 independent reflections |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.061 | w = 1/[σ2(Fo2) + (0.0601P)2] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.155 | (Δ/σ)max < 0.001 |
| S = 0.91 | Δρmax = 0.25 e Å−3 |
| 2726 reflections | Δρmin = −0.14 e Å−3 |
| 194 parameters | Extinction correction: SHELXL2018 (Sheldrick, 2015b), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.016 (4) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | −1.0524 (3) | −0.2385 (4) | −0.7862 (2) | 0.0977 (9) | |
| C12 | −0.4930 (4) | −0.5186 (5) | −0.8354 (3) | 0.0635 (9) | |
| C2 | −1.0387 (4) | −0.2214 (4) | −0.5971 (3) | 0.0639 (9) | |
| C17 | −0.4278 (4) | −0.6907 (5) | −0.8516 (3) | 0.0705 (10) | |
| C1 | −0.9638 (4) | −0.2867 (5) | −0.7056 (3) | 0.0715 (10) | |
| C10 | −0.7746 (4) | −0.4083 (4) | −0.7133 (3) | 0.0695 (10) | |
| C13 | −0.3805 (5) | −0.4437 (5) | −0.8456 (3) | 0.0744 (10) | |
| C11 | −0.6837 (4) | −0.4120 (4) | −0.8119 (3) | 0.0736 (11) | |
| H11 | −0.745928 | −0.339613 | −0.873942 | 0.088* | |
| C15 | −0.1364 (4) | −0.7151 (5) | −0.8870 (3) | 0.0711 (10) | |
| C7 | −0.9379 (4) | −0.2800 (5) | −0.5040 (3) | 0.0743 (10) | |
| C16 | −0.2502 (5) | −0.7860 (5) | −0.8766 (3) | 0.0762 (11) | |
| H16 | −0.206551 | −0.902035 | −0.886559 | 0.091* | |
| C14 | −0.2038 (5) | −0.5443 (5) | −0.8704 (3) | 0.0798 (11) | |
| H14 | −0.128920 | −0.494447 | −0.875919 | 0.096* | |
| C3 | −1.2113 (4) | −0.0905 (5) | −0.5883 (3) | 0.0792 (11) | |
| H3 | −1.280000 | −0.051417 | −0.649452 | 0.095* | |
| C9 | −0.7027 (5) | −0.5193 (6) | −0.6044 (3) | 0.1051 (15) | |
| H9A | −0.745452 | −0.606334 | −0.583241 | 0.126* | |
| H9B | −0.576855 | −0.580756 | −0.615353 | 0.126* | |
| C8 | −0.7503 (4) | −0.4167 (5) | −0.5111 (3) | 0.0951 (13) | |
| H8A | −0.676588 | −0.358974 | −0.520287 | 0.114* | |
| H8B | −0.726257 | −0.496759 | −0.439895 | 0.114* | |
| C4 | −1.2801 (5) | −0.0192 (5) | −0.4895 (4) | 0.0909 (13) | |
| H4 | −1.394381 | 0.067973 | −0.484398 | 0.109* | |
| C6 | −1.0115 (5) | −0.2062 (6) | −0.4058 (3) | 0.0942 (13) | |
| H6 | −0.945571 | −0.244732 | −0.343260 | 0.113* | |
| C5 | −1.1799 (6) | −0.0771 (6) | −0.4000 (4) | 0.0988 (14) | |
| H5 | −1.226312 | −0.028553 | −0.333787 | 0.119* | |
| C20 | 0.0576 (4) | −0.8225 (6) | −0.9157 (3) | 0.1039 (15) | |
| H20A | 0.094884 | −0.940932 | −0.877619 | 0.156* | |
| H20B | 0.121224 | −0.772547 | −0.891430 | 0.156* | |
| H20C | 0.078722 | −0.821651 | −0.996162 | 0.156* | |
| C18 | −0.5449 (5) | −0.7765 (5) | −0.8436 (4) | 0.1077 (15) | |
| H18A | −0.589023 | −0.792507 | −0.766309 | 0.162* | |
| H18B | −0.479794 | −0.888378 | −0.868695 | 0.162* | |
| H18C | −0.640231 | −0.703345 | −0.890605 | 0.162* | |
| C19 | −0.4477 (5) | −0.2554 (5) | −0.8281 (4) | 0.1150 (16) | |
| H19A | −0.531484 | −0.178397 | −0.882365 | 0.173* | |
| H19B | −0.352349 | −0.226633 | −0.838182 | 0.173* | |
| H19C | −0.501571 | −0.241994 | −0.752796 | 0.173* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0782 (16) | 0.112 (2) | 0.088 (2) | −0.0233 (15) | −0.0269 (14) | −0.0175 (16) |
| C12 | 0.069 (2) | 0.057 (2) | 0.060 (2) | −0.0271 (19) | −0.0034 (16) | −0.0044 (18) |
| C2 | 0.061 (2) | 0.066 (2) | 0.066 (2) | −0.0311 (19) | −0.0055 (18) | −0.0045 (19) |
| C17 | 0.079 (2) | 0.061 (3) | 0.073 (3) | −0.032 (2) | −0.0094 (18) | −0.0086 (19) |
| C1 | 0.073 (2) | 0.071 (3) | 0.069 (3) | −0.031 (2) | −0.019 (2) | −0.001 (2) |
| C10 | 0.062 (2) | 0.070 (3) | 0.062 (2) | −0.0218 (19) | −0.0091 (18) | 0.0017 (19) |
| C13 | 0.082 (3) | 0.058 (3) | 0.081 (3) | −0.032 (2) | 0.0010 (19) | −0.014 (2) |
| C11 | 0.072 (2) | 0.072 (3) | 0.072 (3) | −0.029 (2) | −0.0174 (19) | −0.001 (2) |
| C15 | 0.077 (2) | 0.070 (3) | 0.058 (2) | −0.026 (2) | −0.0030 (18) | −0.012 (2) |
| C7 | 0.071 (2) | 0.084 (3) | 0.067 (2) | −0.037 (2) | −0.005 (2) | −0.006 (2) |
| C16 | 0.095 (3) | 0.059 (3) | 0.069 (2) | −0.028 (2) | −0.010 (2) | −0.0122 (19) |
| C14 | 0.082 (3) | 0.081 (3) | 0.085 (3) | −0.045 (2) | 0.0018 (19) | −0.015 (2) |
| C3 | 0.069 (2) | 0.077 (3) | 0.091 (3) | −0.032 (2) | −0.013 (2) | −0.008 (2) |
| C9 | 0.092 (3) | 0.101 (3) | 0.079 (3) | −0.013 (2) | −0.015 (2) | 0.003 (3) |
| C8 | 0.080 (3) | 0.099 (3) | 0.081 (3) | −0.018 (2) | −0.024 (2) | −0.003 (3) |
| C4 | 0.076 (3) | 0.085 (3) | 0.102 (3) | −0.030 (2) | 0.008 (3) | −0.022 (3) |
| C6 | 0.095 (3) | 0.109 (3) | 0.072 (3) | −0.040 (3) | −0.010 (2) | −0.012 (3) |
| C5 | 0.101 (3) | 0.107 (4) | 0.085 (3) | −0.045 (3) | 0.008 (3) | −0.025 (3) |
| C20 | 0.076 (3) | 0.113 (4) | 0.098 (3) | −0.020 (2) | 0.003 (2) | −0.030 (3) |
| C18 | 0.114 (3) | 0.093 (3) | 0.141 (4) | −0.065 (3) | −0.010 (3) | −0.023 (3) |
| C19 | 0.107 (3) | 0.073 (3) | 0.172 (5) | −0.044 (2) | 0.007 (3) | −0.038 (3) |
Geometric parameters (Å, º)
| O1—C1 | 1.218 (4) | C3—C4 | 1.383 (5) |
| C12—C17 | 1.384 (4) | C3—H3 | 0.9300 |
| C12—C13 | 1.393 (4) | C9—C8 | 1.477 (5) |
| C12—C11 | 1.491 (4) | C9—H9A | 0.9700 |
| C2—C7 | 1.396 (5) | C9—H9B | 0.9700 |
| C2—C3 | 1.404 (4) | C8—H8A | 0.9700 |
| C2—C1 | 1.473 (4) | C8—H8B | 0.9700 |
| C17—C16 | 1.390 (4) | C4—C5 | 1.359 (5) |
| C17—C18 | 1.510 (5) | C4—H4 | 0.9300 |
| C1—C10 | 1.486 (4) | C6—C5 | 1.371 (5) |
| C10—C11 | 1.319 (4) | C6—H6 | 0.9300 |
| C10—C9 | 1.490 (5) | C5—H5 | 0.9300 |
| C13—C14 | 1.389 (4) | C20—H20A | 0.9600 |
| C13—C19 | 1.519 (5) | C20—H20B | 0.9600 |
| C11—H11 | 0.9300 | C20—H20C | 0.9600 |
| C15—C14 | 1.373 (5) | C18—H18A | 0.9600 |
| C15—C16 | 1.377 (5) | C18—H18B | 0.9600 |
| C15—C20 | 1.524 (4) | C18—H18C | 0.9600 |
| C7—C6 | 1.390 (5) | C19—H19A | 0.9600 |
| C7—C8 | 1.508 (5) | C19—H19B | 0.9600 |
| C16—H16 | 0.9300 | C19—H19C | 0.9600 |
| C14—H14 | 0.9300 | ||
| C17—C12—C13 | 119.7 (3) | C10—C9—H9A | 109.0 |
| C17—C12—C11 | 120.0 (3) | C8—C9—H9B | 109.0 |
| C13—C12—C11 | 120.3 (3) | C10—C9—H9B | 109.0 |
| C7—C2—C3 | 119.0 (3) | H9A—C9—H9B | 107.8 |
| C7—C2—C1 | 121.2 (3) | C9—C8—C7 | 114.6 (4) |
| C3—C2—C1 | 119.8 (4) | C9—C8—H8A | 108.6 |
| C12—C17—C16 | 119.0 (3) | C7—C8—H8A | 108.6 |
| C12—C17—C18 | 121.7 (3) | C9—C8—H8B | 108.6 |
| C16—C17—C18 | 119.3 (4) | C7—C8—H8B | 108.6 |
| O1—C1—C2 | 121.3 (3) | H8A—C8—H8B | 107.6 |
| O1—C1—C10 | 121.8 (3) | C5—C4—C3 | 119.6 (4) |
| C2—C1—C10 | 116.9 (4) | C5—C4—H4 | 120.2 |
| C11—C10—C1 | 119.9 (3) | C3—C4—H4 | 120.2 |
| C11—C10—C9 | 125.0 (3) | C5—C6—C7 | 120.9 (4) |
| C1—C10—C9 | 115.1 (3) | C5—C6—H6 | 119.6 |
| C14—C13—C12 | 119.3 (3) | C7—C6—H6 | 119.6 |
| C14—C13—C19 | 119.7 (4) | C4—C5—C6 | 121.0 (4) |
| C12—C13—C19 | 121.1 (3) | C4—C5—H5 | 119.5 |
| C10—C11—C12 | 127.7 (3) | C6—C5—H5 | 119.5 |
| C10—C11—H11 | 116.2 | C15—C20—H20A | 109.5 |
| C12—C11—H11 | 116.2 | C15—C20—H20B | 109.5 |
| C14—C15—C16 | 117.6 (3) | H20A—C20—H20B | 109.5 |
| C14—C15—C20 | 121.2 (4) | C15—C20—H20C | 109.5 |
| C16—C15—C20 | 121.2 (4) | H20A—C20—H20C | 109.5 |
| C6—C7—C2 | 118.9 (3) | H20B—C20—H20C | 109.5 |
| C6—C7—C8 | 120.4 (4) | C17—C18—H18A | 109.5 |
| C2—C7—C8 | 120.6 (3) | C17—C18—H18B | 109.5 |
| C15—C16—C17 | 122.4 (4) | H18A—C18—H18B | 109.5 |
| C15—C16—H16 | 118.8 | C17—C18—H18C | 109.5 |
| C17—C16—H16 | 118.8 | H18A—C18—H18C | 109.5 |
| C15—C14—C13 | 122.0 (4) | H18B—C18—H18C | 109.5 |
| C15—C14—H14 | 119.0 | C13—C19—H19A | 109.5 |
| C13—C14—H14 | 119.0 | C13—C19—H19B | 109.5 |
| C4—C3—C2 | 120.6 (4) | H19A—C19—H19B | 109.5 |
| C4—C3—H3 | 119.7 | C13—C19—H19C | 109.5 |
| C2—C3—H3 | 119.7 | H19A—C19—H19C | 109.5 |
| C8—C9—C10 | 112.8 (4) | H19B—C19—H19C | 109.5 |
| C8—C9—H9A | 109.0 | ||
| C13—C12—C17—C16 | 0.7 (5) | C3—C2—C7—C8 | −178.2 (3) |
| C11—C12—C17—C16 | 178.2 (3) | C1—C2—C7—C8 | −2.0 (5) |
| C13—C12—C17—C18 | −178.9 (3) | C14—C15—C16—C17 | 0.8 (5) |
| C11—C12—C17—C18 | −1.3 (5) | C20—C15—C16—C17 | −179.4 (3) |
| C7—C2—C1—O1 | 178.4 (4) | C12—C17—C16—C15 | −0.7 (5) |
| C3—C2—C1—O1 | −5.3 (5) | C18—C17—C16—C15 | 178.9 (3) |
| C7—C2—C1—C10 | −3.8 (5) | C16—C15—C14—C13 | −1.0 (5) |
| C3—C2—C1—C10 | 172.5 (3) | C20—C15—C14—C13 | 179.2 (3) |
| O1—C1—C10—C11 | 27.7 (6) | C12—C13—C14—C15 | 1.0 (5) |
| C2—C1—C10—C11 | −150.1 (3) | C19—C13—C14—C15 | −179.9 (4) |
| O1—C1—C10—C9 | −152.2 (4) | C7—C2—C3—C4 | 0.7 (5) |
| C2—C1—C10—C9 | 30.0 (5) | C1—C2—C3—C4 | −175.6 (3) |
| C17—C12—C13—C14 | −0.8 (5) | C11—C10—C9—C8 | 129.8 (4) |
| C11—C12—C13—C14 | −178.4 (3) | C1—C10—C9—C8 | −50.3 (5) |
| C17—C12—C13—C19 | −179.9 (4) | C10—C9—C8—C7 | 43.9 (5) |
| C11—C12—C13—C19 | 2.6 (5) | C6—C7—C8—C9 | 163.5 (4) |
| C1—C10—C11—C12 | 177.2 (3) | C2—C7—C8—C9 | −18.7 (6) |
| C9—C10—C11—C12 | −2.9 (7) | C2—C3—C4—C5 | −0.4 (6) |
| C17—C12—C11—C10 | 83.3 (5) | C2—C7—C6—C5 | −0.3 (6) |
| C13—C12—C11—C10 | −99.2 (5) | C8—C7—C6—C5 | 177.5 (4) |
| C3—C2—C7—C6 | −0.4 (5) | C3—C4—C5—C6 | −0.3 (7) |
| C1—C2—C7—C6 | 175.9 (3) | C7—C6—C5—C4 | 0.7 (7) |
Hydrogen-bond geometry (Å, º)
Cg2 is the centroid of the C2–C7 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C16—H16···O1i | 0.93 | 2.69 | 3.493 (5) | 145 |
| C20—H20C···O1ii | 0.96 | 2.60 | 3.535 (5) | 165 |
| C9—H9A···Cg2iii | 0.97 | 2.90 | 3.865 (6) | 175 |
Symmetry codes: (i) x+1, y−1, z; (ii) −x−1, −y−1, −z−2; (iii) −x+1, −y, −z+1.
Funding Statement
This work was funded by Ondokuz Mayis Üniversitesi grant PYO.FEN.1906.19.001.
References
- Ansari, F. L., Nazir, S., Noureen, H. & Mirza, B. (2005). Chem. Biodivers. 2, 1656–1664. [DOI] [PubMed]
- Baddeley, T. C., Gomes, L. R., Low, J. N., Turner, A. B., Wardell, J. L. & Watson, G. J. R. (2017). Z. Kristallogr. 232, 317–333.
- Batovska, D., Parushev, S., Stamboliyska, B., Tsvetkova, I., Ninova, M. & Najdenski, H. (2009). Eur. J. Med. Chem. 44, 2211–2218. [DOI] [PubMed]
- Dhar, D. N. (1981). The Chemistry of Chalcones and Related Compounds. New York: Wiley.
- Di Carlo, G., Mascolo, N., Izzo, A. A. & Capasso, F. (1999). Life Sci. 65, 337–353. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Kostanecki, S. V. & Tambor, J. (1899). Ber. Dtsch. Chem. Ges. 32, 1921–1926.
- Kumar, B., Smita, K. & Flores, L. C. (2017). Arabian J. Chem. 10, S2335–S2342.
- Lee, S. H., Seo, G. S., Kim, Y., Jin, X. Y., Kim, H. D. & Sohn, D. H. (2006). Eur. J. Pharmacol. 532, 178–186. [DOI] [PubMed]
- Lin, Y. M., Zhou, Y., Flavin, M. T., Zhou, L. M., Nie, W. & Chen, F. C. (2002). Bioorg. Med. Chem. 10, 2795–2802. [DOI] [PubMed]
- López, S. N., Castelli, M. V., Zacchino, S. A., Dom\?ínguez, J. N., Lobo, G., Charris-Charris, J., Cortés, J. C. G., Ribas, J. C., Devia, C., Rodr\?íguez, A. M. & Enriz, R. D. (2001). Bioorg. Med. Chem. 9, 1999–2013. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. pp. 3814–3816. [DOI] [PubMed]
- Modzelewska, A., Pettit, C., Achanta, G., Davidson, N. E., Huang, P. & Khan, S. R. (2006). Bioorg. Med. Chem. 14, 3491–3495. [DOI] [PubMed]
- Raghavendra, S., Chidan Kumar, C. S., Shetty, T. C. S., Lakshminarayana, B. N., Quah, C. K., Chandraju, S., Ananthnag, G. S., Gonsalves, R. A. & Dharmaprakash, S. M. (2017). Results Phys. 7, 2550–2556.
- Saravanan, B., Rajesh, R., Raghunathan, R., Chakkaravarthi, G. & Manivannan, V. (2010). Acta Cryst. E66, o2801. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (2002). X-AREA and X-RED32. Stoe & Cie GmbH, Darmstadt, Germany.
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. University of Western Australia. http://hirshfeldsurface.net
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989019006182/wm5495sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019006182/wm5495Isup3.hkl
Supporting information file. DOI: 10.1107/S2056989019006182/wm5495Isup3.cml
CCDC reference: 1913649
Additional supporting information: crystallographic information; 3D view; checkCIF report