In the crystal, the molecules are linked by pairs of O—H⋯O hydrogen bonds, forming dimers with an  (18) ring motif. The dimers are linked by pairs of C—H⋯O contacts with an
(18) ring motif. The dimers are linked by pairs of C—H⋯O contacts with an  (10) ring motif, forming ribbons extended along the [2
(10) ring motif, forming ribbons extended along the [2 0] direction.
0] direction.
Keywords: crystal structure, Schiff base, hydrogen bonding, Hirshfeld surface analysis
Abstract
The title compound, C14H12N2O4, is a Schiff base that exists in the keto–enamine tautomeric form and adopts a Z configuration. The molecule is almost planar, the rings making a dihedral angle of 4.99 (7)°. The molecular structure is stabilized by an intramolecular N—H⋯O hydrogen bond forming an S(6) ring motif. In the crystal, inversion-related molecules are linked by pairs of O—H⋯O hydrogen bonds, forming dimers with an R
2
2(18) ring motif. The dimers are linked by pairs of C—H⋯O contacts with an R
2
2(10) ring motif, forming ribbons extended along the [2 0] direction. Hirshfeld surface analysis, two-dimensional fingerprint plots and the molecular electrostatic potential surfaces were used to analyse the intermolecular interactions present in the crystal, indicating that the most important contributions for the crystal packing are from H⋯H (33.9%), O⋯H/H⋯O (29.8%) and C⋯H/H⋯C (17.3%) interactions.
0] direction. Hirshfeld surface analysis, two-dimensional fingerprint plots and the molecular electrostatic potential surfaces were used to analyse the intermolecular interactions present in the crystal, indicating that the most important contributions for the crystal packing are from H⋯H (33.9%), O⋯H/H⋯O (29.8%) and C⋯H/H⋯C (17.3%) interactions.
Chemical context
Compounds containing the RHC=NR fragment, obtained by the condensation reaction of primary amines with aldehydes or ketones under proper conditions, are named Schiff bases after Hugo Schiff (Schiff, 1864 ▸). Schiff bases have a wide variety of applications in many areas such as analytical, biological, and inorganic chemistry (Jain et al., 2008 ▸; Lozier et al., 1975 ▸; Calligaris & Randaccio, 1987 ▸). Many Schiff bases are biologically active and some bases show phototochromism which can be used for radiation intensity measurements, display systems or optical devices (Hadjoudis et al., 1987 ▸). In the present study, a new Schiff base, (Z)-6-[(2-hydroxy-5-nitroanilino)methylidene]-4-methylcyclohexa-2,4-dien-1-one, was obtained in crystalline form from the reaction of 2-amino-4-nitrophenol with 2-hydroxy-5-methylbenzaldehyde. We report here the synthesis and the crystal and molecular structures of the title compound along with the results of a Hirshfeld surface analysis.
Structural commentary
Fig. 1 ▸ illustrates the molecular structure of the title compound. Its asymmetric unit contains one independent molecule, which adopts the keto–enamine tautomeric form. The molecule is almost planar, the C1–C6 and C8–C13 rings making a dihedral angle of 4.99 (7)°. The O4=C9, C9—C8, C8=C7, C7—N2 and N2—C5 bond lengths are typical of double and single bonds, respectively (Table 1 ▸), thus indicating that the title molecule exists as a keto–enamine tautomer (Kansiz et al., 2018 ▸). The bond lengths at the N1 atom are typical of aromatic nitro groups. The molecular structure is stabilized by the intramolecular N—H⋯O hydrogen bond involving the keto O4 and amine N2 atoms (Fig. 1 ▸, Table 2 ▸).
Figure 1.
The molecular structure of the title compound, with the atom-labelling scheme. Displacement ellipsoids are drawn at the 40% probability level. Dashed lines denote the intramolecular N—H⋯O hydrogen bond forming an S(6) ring motif.
Table 1. Selected bond lengths (Å).
| O3—C4 | 1.3329 (14) | O2—N1 | 1.2213 (15) |
| O4—C9 | 1.2887 (15) | N1—C1 | 1.4530 (15) |
| N2—C7 | 1.3070 (15) | C7—C8 | 1.4054 (16) |
| N2—C5 | 1.4035 (15) | C8—C9 | 1.4340 (15) |
| O1—N1 | 1.2202 (15) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2⋯O4 | 0.86 | 1.86 | 2.5661 (13) | 139 |
| O3—H3⋯O4i | 0.82 | 1.77 | 2.5465 (12) | 156 |
| C6—H6⋯O1ii | 0.93 | 2.51 | 3.4383 (16) | 172 |
| C7—H7⋯O1ii | 0.93 | 2.40 | 3.3182 (14) | 172 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Supramolecular features
The most important intermolecular interactions in the title structure are the medium–strong O3—H3⋯O4i hydrogen bonds, which link inversion-related molecules into dimers with an  (18) ring motif (Table 2 ▸). These dimers are further connected by pairs of weak C—H⋯O hydrogen bonds with an
(18) ring motif (Table 2 ▸). These dimers are further connected by pairs of weak C—H⋯O hydrogen bonds with an  (10) ring motif to form ribbons extended along the [2
(10) ring motif to form ribbons extended along the [2 0] direction (Fig. 2 ▸).
0] direction (Fig. 2 ▸).
Figure 2.
A view of the crystal packing of the title compound. Dashed lines denote the intermolecular C—H⋯O and O—H⋯O hydrogen bonds forming dimers with  (10) and
(10) and  (18) ring motifs (Table 1 ▸).
(18) ring motifs (Table 1 ▸).
Database survey
A search of the Cambridge Structural Database (CSD, version 5.40, update November 2018; Groom et al., 2016 ▸) for the 2-[(2-hydroxyphenyliminio)methyl]phenolate fragment revealed 25 hits where this fragment adopts the keto–enamine tautomeric form. The enamine (N2—C7) and keto (C9—O4) bond lengths in the title compound are the same within standard uncertainties as the corresponding bond lengths in the structures of 2-{[(2-hydroxyphenyl)iminio]methyl}-4-methoxyphenolate (BALGUR02; Makal et al., 2011 ▸), 4-bromo-2-{[(2-hydroxy-5-methylphenyl)iminio]methyl}phenolate (EYUSIC; Takjoo et al., 2017 ▸), 2-hydroxy-6-{[(2-hydroxyphenyl)iminio]methyl}phenolate methanol solvate (HEKSIC; Ezeorah et al., 2018 ▸) and 2-{(E)-[(5-bromo-2-hydroxyphenyl)methylidene]amino}-4-chlorophenol (SEFKUL; Ebrahimipour et al., 2012 ▸). In the structures of these typical keto–enamine tautomers, the bonds corresponding to C7—C8 in the title structure are distinctly longer, being in the range of 1.416–1.423 Å. As for the C9—O4 bond, its length compares well with 1.286 (2) Å for HEKSIC and 1.291 (2) Å for SEFKUL, but this bond is shorter than 1.298 (2) Å for EYUSIC and 1.310 (2) Å for BALGUR02. It is likely that the intermolecular O—H⋯O hydrogen bond, where the keto O atom acts as a hydrogen-bond acceptor, is an important prerequisite for the tautomeric shift toward the keto–enamine form. In fact, in all 25 structures of the keto–enamine tautomers, hydrogen bonds of this type are observed.
Hirshfeld surface analysis
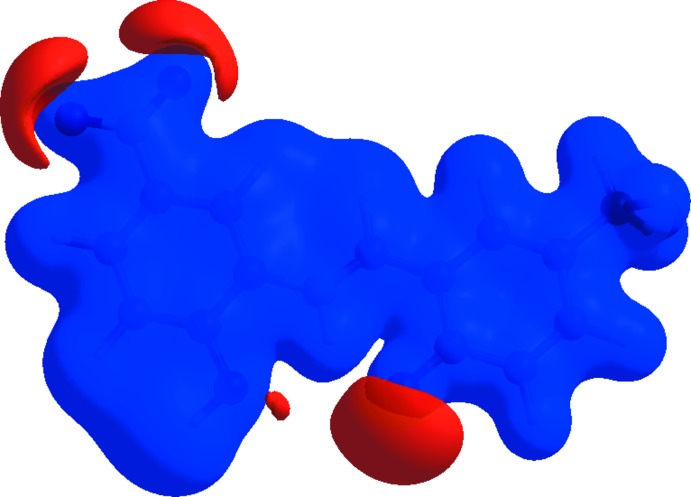
The Hirshfeld surface analysis (Spackman & Jayatilaka, 2009 ▸) and the associated two-dimensional fingerprint plots (McKinnon et al., 2007 ▸) were performed with CrystalExplorer17 (Turner et al., 2017 ▸). The Hirshfeld surfaces were mapped with different properties namely, d norm, electrostatic potential, d i and d e (Fig. 3 ▸). The Hirshfeld surfaces mapped over dnorm utilize the function of normalized distances d e and d i, where d e and d i are the distances from a given point on the surface to the nearest atom outside and inside, respectively. The blue, white and red colour conventions used for the d norm-mapped Hirshfeld surfaces recognize long intermolecular contacts, the contacts at the van der Waals separations, and short intermolecular contacts, respectively. The red region is apparent around the keto oxygen atom (O4) participating in the O—H⋯O and N—H⋯O contacts mentioned above (Fig. 3 ▸, Table 2 ▸). Fig. 4 ▸ illustrates the Hirshfeld surface of the molecule in the crystal, with the evident hydrogen-bonding interactions indicated as intense red spots.
Figure 3.
The Hirshfeld surfaces of the title compound mapped over (a) d norm, (b) electrostatic potential, (c) d i and (d) d e.
Figure 4.
d norm mapped on the Hirshfeld surfaces to visualize the intermolecular interactions for the title compound.
The two-dimensional fingerprint plot derived from a Hirshfeld surface provides a convenient visual summary of the frequency of each combination of d e and d i across the surface of a molecule. A fingerprint plot delineated into specific interatomic contacts contains information related to specific intermolecular interactions (Tan et al., 2019 ▸). The blue colour refers to the frequency of occurrence of the (d i, d e) pairs with the full fingerprint outlined in grey. Fig. 5 ▸ a shows the two-dimensional fingerprint of the sum of the contacts contributing to the Hirshfeld surface represented in normal mode. Individual fingerprint plots (Fig. 5 ▸ b) reveal that the H⋯H contacts clearly make the most significant contribution to the Hirshfeld surface (33.9%). It is usually the case that the main contribution to the overall surface arises from H⋯H contacts. In addition, O⋯H/H⋯O and C⋯H/H⋯C contacts contribute 29.8% and 17.3%, respectively, to the Hirshfeld surface. In particular, the O⋯H/H⋯O and C⋯H/H⋯C contacts indicate the presence of intermolecular O—H⋯O and C—H⋯O interactions, respectively. Much weaker C⋯O/O⋯C (6.8%) and C⋯C (4.8%) contacts also occur.
Figure 5.
Two-dimensional fingerprint plots for the title compound, with a d norm view and the relative contributions of the atom pairs to the Hirshfeld surface.
The view of the electrostatic potential obtained using CrystalExplorer17 enables the visualization of the donors and acceptors of intermolecular interactions through blue and red regions around the participating atoms corresponding to positive and negative electrostatic potential, respectively, on the surface. The view of the electrostatic potential in the range −0.0500 to 0.0500 a.u., calculated for the title compound at the HF/STO-3G level, is shown in Fig. 6 ▸. The acceptors for N—H⋯O and O—H⋯O hydrogen bonds are shown as red areas around the O4 atom related with negative electrostatic potentials (Fig. 6 ▸).
Figure 6.
A view of the molecular electrostatic potential of the title compound in the range −0.0500 to 0.0500 a.u. calculated at the HF/STO-3 G level.
Synthesis and crystallization
The title compound was prepared by mixing the solutions of 2-hydroxy-5-methylbenzaldehyde (34.0 mg, 0.25 mmol) in ethanol (15 ml) and 2-amino-4-nitrophenol (38.5 mg, 0.25 mmol) in ethanol (15 ml) with subsequent stirring for 5 h under reflux. Single crystals of the title compound suitable for X-ray analysis were obtained by slow evaporation of an ethanol solution (yield 65%, m.p. 523–525 K).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The hydroxy H atom was located in a difference-Fourier map, and the OH group was allowed to rotate during the refinement procedure (AFIX 147). The C-bound H atoms were positioned geometrically and refined using a riding model: C—H = 0.93 Å with U iso(H) = 1.2U eq(C) for aromatic H atoms and C—H = 0.96 Å with U iso(H) = 1.5U eq(C) for methyl H atoms. The amine H atom was also refined using a riding model: N—H = 0.86 Å with U iso(H) = 1.2U eq(N).
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | C14H12N2O4 |
| M r | 272.26 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 296 |
| a, b, c (Å) | 6.0052 (4), 7.8206 (5), 26.2985 (19) |
| β (°) | 90.303 (5) |
| V (Å3) | 1235.07 (14) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.11 |
| Crystal size (mm) | 0.57 × 0.43 × 0.19 |
| Data collection | |
| Diffractometer | Stoe IPDS 2 |
| Absorption correction | Integration (X-RED32; Stoe & Cie, 2002 ▸) |
| T min, T max | 0.946, 0.981 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 8536, 3300, 2436 |
| R int | 0.028 |
| (sin θ/λ)max (Å−1) | 0.686 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.041, 0.121, 1.08 |
| No. of reflections | 3300 |
| No. of parameters | 183 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.15, −0.19 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S205698901900673X/yk2122sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901900673X/yk2122Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901900673X/yk2122Isup3.cml
CCDC reference: 1915380
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C14H12N2O4 | F(000) = 568 |
| Mr = 272.26 | Dx = 1.464 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.0052 (4) Å | Cell parameters from 9358 reflections |
| b = 7.8206 (5) Å | θ = 2.6–29.6° |
| c = 26.2985 (19) Å | µ = 0.11 mm−1 |
| β = 90.303 (5)° | T = 296 K |
| V = 1235.07 (14) Å3 | Plate, orange |
| Z = 4 | 0.57 × 0.43 × 0.19 mm |
Data collection
| STOE IPDS 2 diffractometer | 3300 independent reflections |
| Radiation source: sealed X-ray tube, 12 x 0.4 mm long-fine focus | 2436 reflections with I > 2σ(I) |
| Detector resolution: 6.67 pixels mm-1 | Rint = 0.028 |
| rotation method scans | θmax = 29.2°, θmin = 2.7° |
| Absorption correction: integration (X-RED32; Stoe & Cie, 2002) | h = −7→8 |
| Tmin = 0.946, Tmax = 0.981 | k = −10→10 |
| 8536 measured reflections | l = −27→36 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.041 | H-atom parameters constrained |
| wR(F2) = 0.121 | w = 1/[σ2(Fo2) + (0.0685P)2 + 0.0139P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.08 | (Δ/σ)max < 0.001 |
| 3300 reflections | Δρmax = 0.15 e Å−3 |
| 183 parameters | Δρmin = −0.19 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O3 | 0.85290 (16) | 0.10060 (14) | 0.53689 (4) | 0.0593 (3) | |
| H3 | 0.926262 | 0.063519 | 0.560850 | 0.089* | |
| O4 | 0.85793 (15) | 0.05497 (13) | 0.40914 (4) | 0.0573 (3) | |
| N2 | 0.57371 (16) | 0.20069 (12) | 0.46738 (4) | 0.0413 (2) | |
| H2 | 0.697995 | 0.148587 | 0.462636 | 0.050* | |
| O1 | 0.00525 (18) | 0.53706 (15) | 0.56524 (4) | 0.0687 (3) | |
| O2 | 0.0910 (2) | 0.50110 (16) | 0.64372 (4) | 0.0716 (3) | |
| N1 | 0.12631 (18) | 0.47944 (14) | 0.59848 (4) | 0.0482 (3) | |
| C5 | 0.52750 (19) | 0.24767 (14) | 0.51773 (4) | 0.0377 (2) | |
| C1 | 0.32008 (19) | 0.38097 (14) | 0.58331 (5) | 0.0400 (3) | |
| C6 | 0.34505 (19) | 0.34263 (14) | 0.53221 (4) | 0.0394 (2) | |
| H6 | 0.241371 | 0.379997 | 0.508283 | 0.047* | |
| C7 | 0.45170 (19) | 0.22613 (14) | 0.42678 (4) | 0.0405 (3) | |
| H7 | 0.315808 | 0.281942 | 0.430109 | 0.049* | |
| C4 | 0.68303 (19) | 0.19257 (15) | 0.55447 (5) | 0.0423 (3) | |
| C8 | 0.51880 (18) | 0.17203 (15) | 0.37826 (4) | 0.0397 (2) | |
| C13 | 0.3761 (2) | 0.20126 (16) | 0.33610 (5) | 0.0440 (3) | |
| H13 | 0.242313 | 0.258453 | 0.341190 | 0.053* | |
| C9 | 0.72607 (19) | 0.08425 (16) | 0.37131 (5) | 0.0432 (3) | |
| C12 | 0.4290 (2) | 0.14798 (17) | 0.28810 (5) | 0.0476 (3) | |
| C2 | 0.4709 (2) | 0.32843 (16) | 0.61998 (5) | 0.0457 (3) | |
| H2A | 0.450177 | 0.356630 | 0.653984 | 0.055* | |
| C3 | 0.6520 (2) | 0.23381 (16) | 0.60524 (5) | 0.0480 (3) | |
| H3A | 0.754588 | 0.197052 | 0.629487 | 0.058* | |
| C11 | 0.6332 (2) | 0.06220 (19) | 0.28190 (5) | 0.0541 (3) | |
| H11 | 0.672078 | 0.024806 | 0.249571 | 0.065* | |
| C10 | 0.7763 (2) | 0.03164 (19) | 0.32125 (5) | 0.0540 (3) | |
| H10 | 0.909625 | −0.024955 | 0.315049 | 0.065* | |
| C14 | 0.2767 (3) | 0.1771 (2) | 0.24318 (5) | 0.0638 (4) | |
| H14A | 0.141128 | 0.229334 | 0.254508 | 0.096* | |
| H14B | 0.243485 | 0.069530 | 0.227267 | 0.096* | |
| H14C | 0.348636 | 0.250904 | 0.219161 | 0.096* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O3 | 0.0503 (5) | 0.0818 (6) | 0.0458 (5) | 0.0315 (5) | −0.0095 (4) | −0.0046 (5) |
| O4 | 0.0477 (5) | 0.0811 (6) | 0.0430 (5) | 0.0271 (5) | −0.0075 (4) | −0.0024 (4) |
| N2 | 0.0384 (5) | 0.0503 (5) | 0.0352 (5) | 0.0132 (4) | −0.0015 (4) | −0.0015 (4) |
| O1 | 0.0591 (6) | 0.0891 (7) | 0.0578 (6) | 0.0363 (5) | −0.0010 (5) | −0.0009 (5) |
| O2 | 0.0699 (7) | 0.0984 (8) | 0.0467 (6) | 0.0202 (6) | 0.0138 (5) | −0.0104 (5) |
| N1 | 0.0454 (6) | 0.0538 (6) | 0.0455 (6) | 0.0084 (5) | 0.0058 (5) | −0.0033 (5) |
| C5 | 0.0383 (5) | 0.0422 (5) | 0.0327 (5) | 0.0055 (4) | −0.0029 (4) | −0.0001 (4) |
| C1 | 0.0389 (6) | 0.0429 (6) | 0.0382 (6) | 0.0042 (4) | 0.0009 (4) | −0.0006 (4) |
| C6 | 0.0375 (5) | 0.0456 (6) | 0.0351 (6) | 0.0068 (4) | −0.0033 (4) | 0.0022 (4) |
| C7 | 0.0386 (5) | 0.0459 (6) | 0.0369 (6) | 0.0103 (4) | −0.0031 (4) | −0.0002 (4) |
| C4 | 0.0379 (5) | 0.0479 (6) | 0.0409 (6) | 0.0088 (5) | −0.0056 (5) | −0.0004 (5) |
| C8 | 0.0392 (6) | 0.0443 (6) | 0.0356 (5) | 0.0068 (4) | −0.0014 (4) | 0.0011 (4) |
| C13 | 0.0416 (6) | 0.0527 (6) | 0.0378 (6) | 0.0091 (5) | −0.0042 (5) | 0.0025 (5) |
| C9 | 0.0390 (6) | 0.0521 (6) | 0.0384 (6) | 0.0080 (5) | −0.0023 (5) | 0.0011 (5) |
| C12 | 0.0492 (7) | 0.0590 (7) | 0.0344 (6) | 0.0014 (5) | −0.0037 (5) | 0.0033 (5) |
| C2 | 0.0513 (7) | 0.0517 (6) | 0.0339 (6) | 0.0043 (5) | −0.0029 (5) | −0.0021 (5) |
| C3 | 0.0477 (6) | 0.0579 (7) | 0.0382 (6) | 0.0091 (5) | −0.0125 (5) | 0.0007 (5) |
| C11 | 0.0537 (7) | 0.0733 (9) | 0.0354 (6) | 0.0060 (6) | 0.0043 (5) | −0.0040 (6) |
| C10 | 0.0453 (7) | 0.0722 (8) | 0.0445 (7) | 0.0148 (6) | 0.0044 (5) | −0.0040 (6) |
| C14 | 0.0669 (9) | 0.0863 (10) | 0.0381 (7) | 0.0072 (8) | −0.0102 (6) | 0.0039 (7) |
Geometric parameters (Å, º)
| O3—C4 | 1.3329 (14) | C8—C13 | 1.4162 (16) |
| O3—H3 | 0.8200 | C8—C9 | 1.4340 (15) |
| O4—C9 | 1.2887 (15) | C13—C12 | 1.3683 (18) |
| N2—C7 | 1.3070 (15) | C13—H13 | 0.9300 |
| N2—C5 | 1.4035 (15) | C9—C10 | 1.4134 (18) |
| N2—H2 | 0.8600 | C12—C11 | 1.4077 (19) |
| O1—N1 | 1.2202 (15) | C12—C14 | 1.5074 (19) |
| O2—N1 | 1.2213 (15) | C2—C3 | 1.3726 (17) |
| N1—C1 | 1.4530 (15) | C2—H2A | 0.9300 |
| C5—C6 | 1.3789 (15) | C3—H3A | 0.9300 |
| C5—C4 | 1.4077 (15) | C11—C10 | 1.3629 (19) |
| C1—C2 | 1.3823 (17) | C11—H11 | 0.9300 |
| C1—C6 | 1.3857 (16) | C10—H10 | 0.9300 |
| C6—H6 | 0.9300 | C14—H14A | 0.9600 |
| C7—C8 | 1.4054 (16) | C14—H14B | 0.9600 |
| C7—H7 | 0.9300 | C14—H14C | 0.9600 |
| C4—C3 | 1.3873 (18) | ||
| C4—O3—H3 | 109.5 | C12—C13—H13 | 119.0 |
| C7—N2—C5 | 128.19 (10) | C8—C13—H13 | 119.0 |
| C7—N2—H2 | 115.9 | O4—C9—C10 | 122.28 (11) |
| C5—N2—H2 | 115.9 | O4—C9—C8 | 121.13 (11) |
| O1—N1—O2 | 122.72 (11) | C10—C9—C8 | 116.59 (11) |
| O1—N1—C1 | 118.29 (10) | C13—C12—C11 | 117.30 (12) |
| O2—N1—C1 | 118.99 (11) | C13—C12—C14 | 122.28 (12) |
| C6—C5—N2 | 124.25 (10) | C11—C12—C14 | 120.42 (12) |
| C6—C5—C4 | 120.05 (10) | C3—C2—C1 | 118.70 (11) |
| N2—C5—C4 | 115.70 (10) | C3—C2—H2A | 120.6 |
| C2—C1—C6 | 122.58 (11) | C1—C2—H2A | 120.6 |
| C2—C1—N1 | 119.25 (11) | C2—C3—C4 | 120.53 (11) |
| C6—C1—N1 | 118.17 (10) | C2—C3—H3A | 119.7 |
| C5—C6—C1 | 118.33 (10) | C4—C3—H3A | 119.7 |
| C5—C6—H6 | 120.8 | C10—C11—C12 | 122.80 (12) |
| C1—C6—H6 | 120.8 | C10—C11—H11 | 118.6 |
| N2—C7—C8 | 122.26 (10) | C12—C11—H11 | 118.6 |
| N2—C7—H7 | 118.9 | C11—C10—C9 | 121.30 (12) |
| C8—C7—H7 | 118.9 | C11—C10—H10 | 119.4 |
| O3—C4—C3 | 124.50 (10) | C9—C10—H10 | 119.4 |
| O3—C4—C5 | 115.68 (10) | C12—C14—H14A | 109.5 |
| C3—C4—C5 | 119.81 (10) | C12—C14—H14B | 109.5 |
| C7—C8—C13 | 119.15 (10) | H14A—C14—H14B | 109.5 |
| C7—C8—C9 | 120.86 (10) | C12—C14—H14C | 109.5 |
| C13—C8—C9 | 119.97 (10) | H14A—C14—H14C | 109.5 |
| C12—C13—C8 | 122.04 (11) | H14B—C14—H14C | 109.5 |
| C7—N2—C5—C6 | −4.9 (2) | C9—C8—C13—C12 | 0.05 (19) |
| C7—N2—C5—C4 | 175.61 (12) | C7—C8—C9—O4 | −1.11 (19) |
| O1—N1—C1—C2 | 173.86 (12) | C13—C8—C9—O4 | −179.47 (12) |
| O2—N1—C1—C2 | −6.20 (19) | C7—C8—C9—C10 | 178.60 (12) |
| O1—N1—C1—C6 | −6.51 (18) | C13—C8—C9—C10 | 0.25 (18) |
| O2—N1—C1—C6 | 173.42 (12) | C8—C13—C12—C11 | −0.1 (2) |
| N2—C5—C6—C1 | −179.62 (11) | C8—C13—C12—C14 | 179.34 (13) |
| C4—C5—C6—C1 | −0.16 (17) | C6—C1—C2—C3 | −0.3 (2) |
| C2—C1—C6—C5 | 0.28 (18) | N1—C1—C2—C3 | 179.26 (11) |
| N1—C1—C6—C5 | −179.33 (10) | C1—C2—C3—C4 | 0.3 (2) |
| C5—N2—C7—C8 | 179.95 (11) | O3—C4—C3—C2 | −179.58 (13) |
| C6—C5—C4—O3 | 179.56 (11) | C5—C4—C3—C2 | −0.2 (2) |
| N2—C5—C4—O3 | −0.93 (17) | C13—C12—C11—C10 | −0.1 (2) |
| C6—C5—C4—C3 | 0.12 (18) | C14—C12—C11—C10 | −179.58 (15) |
| N2—C5—C4—C3 | 179.62 (11) | C12—C11—C10—C9 | 0.4 (2) |
| N2—C7—C8—C13 | 178.88 (11) | O4—C9—C10—C11 | 179.23 (13) |
| N2—C7—C8—C9 | 0.51 (19) | C8—C9—C10—C11 | −0.5 (2) |
| C7—C8—C13—C12 | −178.33 (12) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···O4 | 0.86 | 1.86 | 2.5661 (13) | 139 |
| O3—H3···O4i | 0.82 | 1.77 | 2.5465 (12) | 156 |
| C6—H6···O1ii | 0.93 | 2.51 | 3.4383 (16) | 172 |
| C7—H7···O1ii | 0.93 | 2.40 | 3.3182 (14) | 172 |
Symmetry codes: (i) −x+2, −y, −z+1; (ii) −x, −y+1, −z+1.
Funding Statement
This work was funded by Ondokuz Mayis Üniversitesi grant PYO.FEN.1906.19.001.
References
- Calligaris, M. & Randaccio, L. (1987). Comprehensive Coordination Chemistry, Vol. 2, edited by G. Wilkinson, pp. 715–738. London: Pergamon.
- Ebrahimipour, S. Y., Mague, J. T., Akbari, A. & Takjoo, R. (2012). J. Mol. Struct. 1028, 148–155.
- Ezeorah, J. C., Ossai, V., Obasi, L. N., Elzagheid, M. I., Rhyman, L., Lutter, M., Jurkschat, K., Dege, N. & Ramasami, P. (2018). J. Mol. Struct. 1152, 21–28.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hadjoudis, E., Vittorakis, M. & Moustakali-Mavridis, I. (1987). Tetrahedron, 43, 1345–1360.
- Jain, J., Masand, N., Sinha, R., Garg, V. & Patil, V. (2008). J. Cell Tissue Res. 8, 1431–1431.
- Kansiz, S., Çakmak, Ş., Dege, N., Meral, G. & Kütük, H. (2018). X-ray Struct. Anal. Online, 34, 17–18.
- Lozier, R. H., Bogomolni, R. A. & Stoeckenius, W. (1975). Biophys. J. 15, 955–962. [DOI] [PMC free article] [PubMed]
- Makal, A., Schilf, W., Kamieński, B., Szady-Chelmieniecka, A., Grech, E. & Woźniak, K. (2011). Dalton Trans. 40, 421–430. [DOI] [PubMed]
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. pp. 3814–3816. [DOI] [PubMed]
- Schiff, H. (1864). Ann. Chem. Pharm. 131, 118–119.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (2002). X-AREA and X-RED32. Stoe & Cie GmbH, Darmstadt, Germany.
- Takjoo, R., Akbari, A., Ebrahimipour, S. Y., Kubicki, M., Mohamadi, M. & Mollania, N. (2017). Inorg. Chim. Acta, 455, 173–182.
- Tan, S. L., Jotani, M. M. & Tiekink, E. R. T. (2019). Acta Cryst. E75, 308–318. [DOI] [PMC free article] [PubMed]
- Turner, M. J., MacKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17.5. University of Western Australia. http://hirshfeldsurface.net.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S205698901900673X/yk2122sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901900673X/yk2122Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901900673X/yk2122Isup3.cml
CCDC reference: 1915380
Additional supporting information: crystallographic information; 3D view; checkCIF report