Abstract
As histone deacetylase inhibitors such as romidepsin (depsipeptide, FK228) complete successful Phase I clinical trials in pediatric solid tumors, it is important that their mechanisms of action are delineated in order to inform the development of subsequent clinical trials as single agents or in combination therapies. In this study, we evaluate the effect of romidepsin as a single agent on a number of different neuroblastoma (NB) cell lines. We find that the growth of 6/6 human NB tumor cell lines but not an immortalized fibroblast cell line (NIH3T3) is inhibited by romidepsin (IC50 = 1–6.5 ng/ml) after 72 h of treatment. Romidepsin shows selective dose-dependent cytotoxicity in both single copy and N-myc amplified NB cell lines, in cell lines with wild-type or mutant p53 and those containing Alk mutations. The decrease in cell proliferation is accompanied by caspase-dependent apoptosis as shown by PARP cleavage, an accumulation of cells in the sub-G1 phase of the cell cycle and the ability of a pan-caspase inhibitor to reduce cell death. Romidepsin inhibits the growth of subcutaneous NB xenografts in a dose dependent manner in immunocompromised mice. Furthermore, romidepsin induces expression of genes such as p21 and expression of p75 and NTRK (TrkA) which are more highly expressed in the tumors from NB patients that have a good prognosis. These studies support continued investigations into the therapeutic activity of romidepsin in NB.
Keywords: Romidepsin, FK228, Depsipeptide, neuroblastoma, apoptosis, HDAC inhibitor
Introduction
Neuroblastoma (NB) is the third most common pediatric cancer and is the most common extracranial solid tumor of childhood. This tumor is responsible for approximately 15% of all childhood cancer deaths.1,2 The prognosis of NB, especially patients in the high-risk group, remains poor despite intensive chemotherapy and bone marrow transplantation. In NB, amplification of the N-myc gene is seen in 20% of tumors, and is associated with more aggressive tumors and poor prognosis.2 More recently, genetic alterations in Alk, including mutations or gene amplification that lead to increased expression are associated with poor prognosis.3 Yet, despite these genetic alterations, agents which regulate gene expression programs, such as HDAC inhibitors or retinoids, arrest NB tumor growth and induce differentiation and/or apoptosis.
Several lines of evidence suggest that histone acetylation via alterations in chromatin structure plays a role in transcriptional regulation4, 5 Histone acetylation has been associated with transcriptional activation, whereas deacetylation of histones is associated with gene silencing and transcriptional repression. The e-amino groups of specific lysine residues within histones are modified by histone acetyltransferase (HAT) and histone deacetylase (HDAC) enzymes. The overall level of histone acetylation depends on the balance of activity of these enzymes.4 Because the acetylation state of histones is associated with transcriptional activity, the role of histone acetylation in regulating re-expression of silenced tumor suppressor genes has been studied widely.4–6 Recent studies indicate that acetylation of other proteins may affect their activities and this includes transcription factors in which acetylation may alter DNA binding and transcriptional activity.7, 8
HDAC inhibitors9 include short-chain fatty acids (sodium butyrate and valproic acid), the organic hydroxamic acids (trichostatin A (TSA) and suberanilohydroxamic acid (SAHA)), benzamides (CI-994 and MS-27–275), cyclic tetrapeptides (trapoxin A) and the bicyclic peptides, such as romidepsin (FK228 or FR901228).10 Romidepsin, originally isolated from Chromobacterium violaceum, was the first histone deacetylase inhibitor to demonstrate clinical anti-tumor activity in patients.11 Although TSA and romidepsin target the same pathway, the anti-proliferative effect of romidepsin is 10-fold greater than that of TSA, and the IC50 of romidepsin on histone acetylation is much lower than that of TSA.12 Similar to other HDAC inhibitors, romidepsin has been shown to induce cell cycle arrest, cellular differentiation, apoptosis and alter gene expression in a variety of adult malignancies.10, 12, 13 A pediatric phase I investigation of romidepsin has determined the maximally tolerated dose14 and a preliminary assessment indicated inhibition of tumor cell growth in 3 of 4 NB cell lines.15
We have shown that HDAC inhibitors such as MS-27–275 can mediate potent in vitro and in vivo antitumor activity against a broad panel of pediatric solid tumors including NB.16 Previous studies focused on regulation of NB tumor cell growth;15 in this study, we detail mechanisms of cell cycle regulation and induction of apoptosis and gene regulation induced by romidepsin in NB tumor cells.
Results
Romidepsin inhibits NB cell growth in a dose-dependent manner
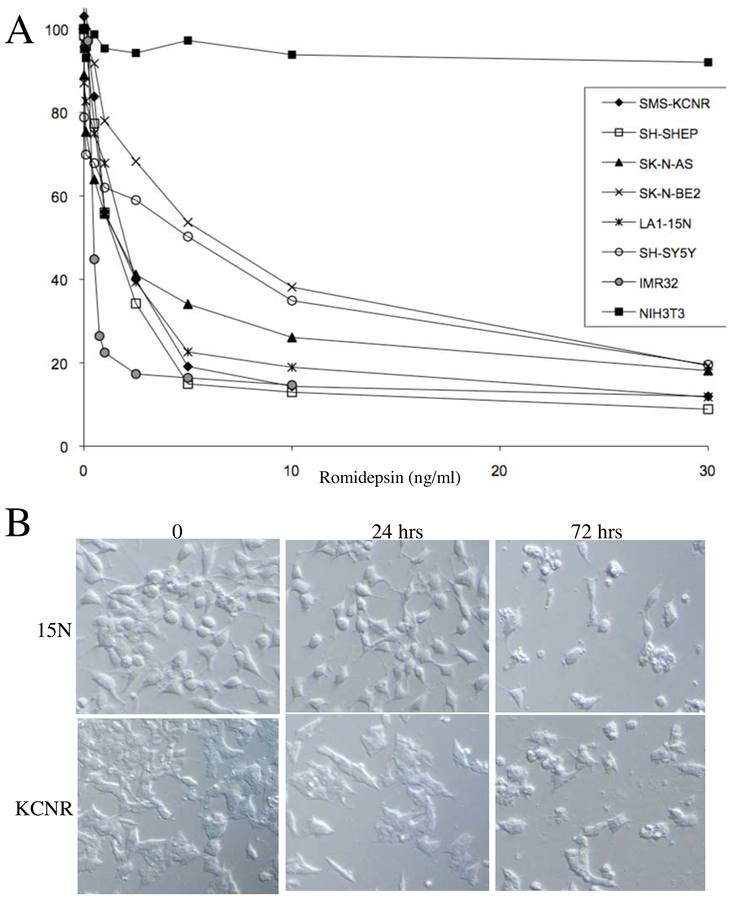
The characteristics of the NB cell lines used in this study are detailed in Table 1. We initially investigated whether romidepsin could inhibit cell proliferation. Cells cultured with various concentrations of romidepsin for 72 h. Romidepsin (0.5–30 ng/mL) resulted in a dose-dependent decrease in cell viability of all NB cell lines as measured by the MTT or MTS assay (Fig 1A). Both MYCN amplified and non-amplified cell lines showed similar dose-dependent inhibition of growth in vitro, including cell lines with p53 mutations (SK-N-BE2, AS, LA1–15N). The romidepsin IC50 ranged from 1 – 6.5 ng/ml in different NB cell lines (Table 1) and all were more sensitive to romidepsin than the non-transformed fibroblast cell line NIH3T3 (Fig 1A).
Table 1.
Neuroblastoma Cell Lines
| Cell Line | Chromosomal Alteration | NMA* | p53 | Alk | IC50 (ng/ml) | IC80 (ng/ml) | Ref |
|---|---|---|---|---|---|---|---|
| IMR32 | 1p LOH | + | wt | wt | 1 | 2 | 22 |
| SK-N-BE(2) | 1p LOH, t(3,17)(p21,q21) | + | mt | wt | 6.3 | 30 | 18 |
| SMS-KCNR | 1p LOH, t(17,20)(p21,q13) | + | wt | mt, R1275Q | 2 | 5 | 17 |
| SK-N-AS | 1p del, 1p(36.33) | − | wt | wt | 1.8 | 30 | 20 |
| SH-SY5Y | 1p LOH | − | wt | mt, F1174L | 5 | 30 | 19 |
| SH-SHEP | 1p LOH | − | wt | mt, F1174L | 1 | 4 | 22 |
| LA1–15N | 1p LOH | + | mt | mt, F1174L | 1.2 | 10 | 21 |
NMA-MYCN amplified; LOH= loss of hterozygosity; wt=wild-type; mt=mutant
Figure 1-.
Romidepsin treatment of NB cell lines. Panel A. Dose response analysis of 8 NB and NIH3T3 cell lines in the presence of 0.5–30 ng/ml concentration of romidepsin by MTT/MTS assay at 72 h. Each point represents the mean of 2–4 experiments for each time point. Panel B. Morphologic effects of romidepsin in 2 representative NB cell lines-15N and KCNR at 24 h and 72 h after treatment with IC80 concentration of romidepsin.
Morphological examination by light microscopy revealed that all of the NB cell lines treated with romidepsin had a dose-dependent decrease in cell number and extensive change in morphology to rounded, denser, and non-adherent cells (Fig 1B). This change in morphology was visible as early as 24 h after treatment. By 72 h, there were numerous round floating cells with pyknotic nuclei in each of the cell lines and these cells did not exclude trypan blue. There was no morphologic evidence of differentiation (neuritic extension) during the entire time course.
Romidepsin increases acetylation of histone H3
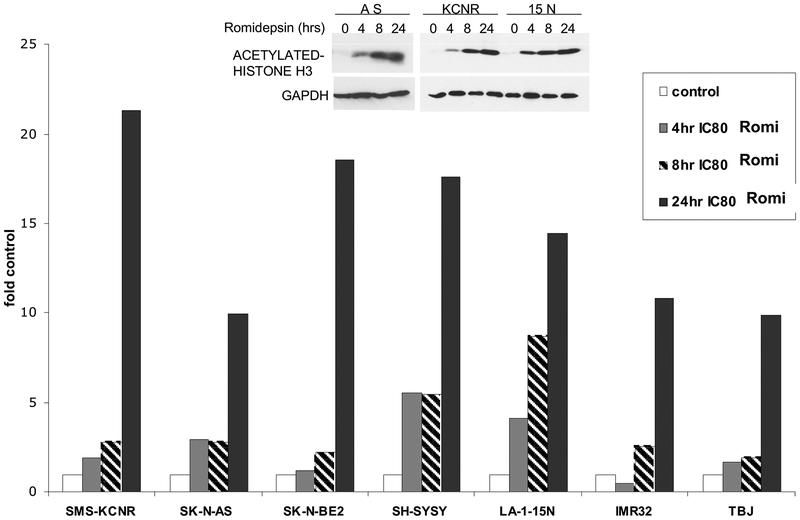
To assess the effects of romidepsin on the acetylation of histones in NB cells, seven cell lines were treated in vitro with the IC80 concentration of romidepsin for 4, 8 and 24 h. Protein lysates were evaluated for acetylation by monitoring the acetylation of lysines on histone H3 (Ac-H3) by a quantitative-immunoblot analysis (Fig 2) with selected examples of Western analysis detailed in Fig. 2 inset. Accumulation of acetylated histones was seen as early as 4 h after romidepsin treatment in all cell lines (except IMR32) and increased further at 24 h.
Figure 2-.
Acetylation of histones after romidepsin treatment. NB cell lines were treated with IC80 concentration of romidepsin for 4, 8 and 24 h, protein was extracted and analyzed for Ac-H3 analysis by immunoblot assay. Blots were reprobed for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels as loading controls. Normalized values are plotted as ratio of control. Inset- Representative western analyses of acetylated Histone H3.
Romidepsin induces apoptosis
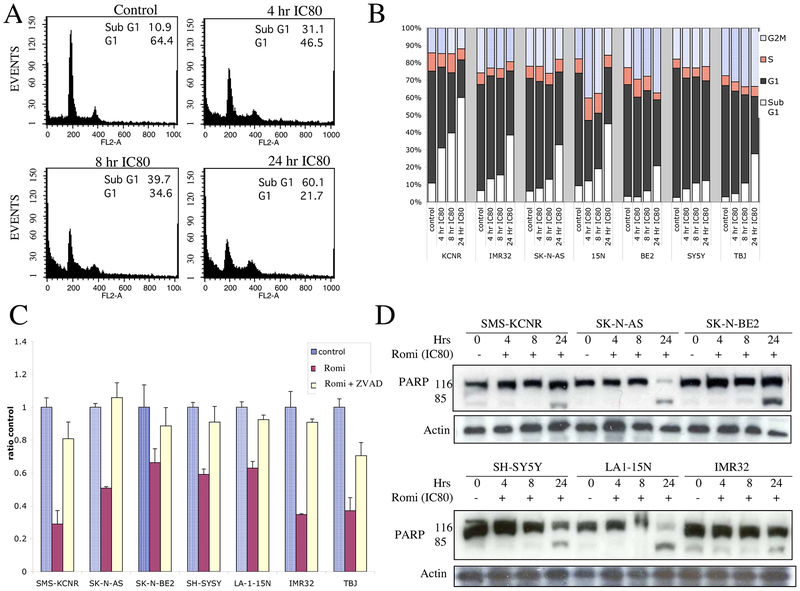
Because inhibition of cell growth may be due to cell cycle arrest or induction of apoptosis, NB cell lines were exposed to romidepsin (IC80 concentration as determined for each cell line) for 4, 8 and 24 h and DNA content was assessed by FACS analysis. For all the cell lines, there was a significant increase in cells with sub-G1 DNA content with a corresponding decrease in cells in the G1 phase, consistent with apoptosis (Fig 3A and B). There was no evidence of cell cycle arrest in the G1 or G2/M phases of the cell cycle as has been seen in some human tumor cell lines.9, 12 In all the NB cell lines, there was a demonstrable increase in apoptotic cells by 8 h. To determine if the cell death induced by treatment of NB cells with romidepsin is caspase-dependent, cells were pretreated with 20 μM Z-VAD-FMK, a broad-spectrum caspase-3 inhibitor, and were incubated with or without romidepsin for 48 hours. Cell viability was assessed using the MTS assay. Z-VAD-FMK significantly inhibited romidepsin-induced cell death in all NB cell lines tested (Fig 3C). Although Z-VAD-FMK alone did not have any effect on cell viability, prior exposure to Z-VAD-FMK rescued NB cells from romidepsin induced cell death, indicating that romidepsin induces apoptosis in a caspase-dependent manner. Consistent with this result, an increased level of the caspase dependent cleaved product of PARP was detected in all cell lines by 24 h following treatment with romidepsin (Fig 3D).
Figure 3-.
Cell cycle analysis and apoptosis in NB cells treated with romidepsin. Panel A. Representative cell cycle analysis of KCNR cell line after treatment with IC80 concentration of romidepsin for 4, 8 and 24 h. Panel B. Graph of various cell cycle phases in all NB cell lines after exposure to IC80 concentrations of romidepsin for 4, 8 and 24 h. Panel C. PARP cleavage by western blot analysis in different NB cell lines after treatment with IC80 concentration of romidepsin for 4, 8 and 24 h. Panel D. Z-VAD-FMK, a pan-caspase inhibitor, rescues NB cells from romidepsin induced cell death. Pretreatment of cells with Z-VAD-FMK for 2 h followed by incubation with control media or IC80 concentration of romidepsin for 48 h reversed cell death seen by MTS assay.
Romidepsin decreases N-myc levels and induces p21 in NB
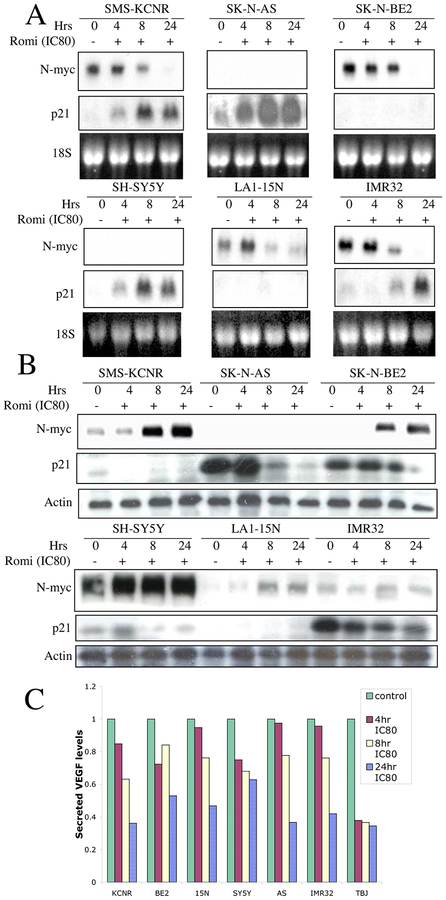
Patients with NB tumors that contain MYCN amplification have a poor prognosis.2 We evaluated the expression of N-myc in NB cells treated with romidepsin for various times. Within 4–8 h of romidepsin treatment, there was a decrease in N-myc mRNA (Fig 4A) and protein (Fig 4B) levels in the N-myc amplified cell lines SMS-KCNR, SK-N-BE and LA1–15N. N-myc expression was not detected in single-copy MYCN cell lines, SY5Y and AS at 24 h. In IMR32, the decrease in N-myc levels was evident at 24 h which coincided with the increase in histone acetylation. The time course observed for reduction in N-myc RNA levels was significantly correlated with increasing accumulation of Ac-H3 (Spearman r = −0.9254, p value- <0.0001).
Figure 4-.
Analysis of romidepsin-induced changes in gene expression. Panel A. Cell lines were incubated with IC80 concentration of romidepsin for various periods of time; total RNA was isolated and evaluated by Northern analysis for expression of N-myc and p21 mRNA. 18S RNA serves as a loading control. Panel B. Cell lines were incubated with IC80 concentration of romidepsin for various periods of time; protein was extracted and evaluated by Western analysis for expression of N-myc and p21. Blots were reprobed with actin as a loading control. Panel C. Effect of romidepsin on secreted VEGF in NB cell lines. NB cell lines were exposed to IC80 concentrations of romidepsin for 4, 8 and 24h, supernatant collected, centrifuged and analyzed by ELISA for VEGF.
Romidepsin11 and other HDAC inhibitors such as CHBA9 and MS-27–27516 have been shown to induce p21 in tumor cell lines. In vitro treatment of NB cells with romidepsin resulted in an increase in p21 in most of the cell lines at 4–8 h (Fig 4A and B). Notably, increases in p21 were not observed in cell lines with mutant p53 (SK-N-BE and LA1–15N) after treatment with romidepsin, suggesting that the increase in p21 in NB cell lines may be influenced by p53 status.
Romidepsin decreases VEGF levels in NB
High levels of VEGF and the extent of angiogenesis in neuroblastoma tumors are both associated with poor prognosis.17–19 Our previous study using MS-27–27516 and more recent studies using romidepsin indicate that HDAC inhibitors inhibit angiogenesis.20, 21 To determine the effect of romidepsin on VEGF levels, cells were incubated with romidepsin or a media control for the indicated times and the levels of VEGF in the culture supernatants were evaluated by ELISA. There was a time-dependent decrease in secreted VEGF levels in all NB cell lines (Fig 4C). Reductions in VEGF production were noted in all cell lines by 24 h with the reductions ranging from 34–62%.
Romidepsin alters growth of KCNR neuroblastoma tumors
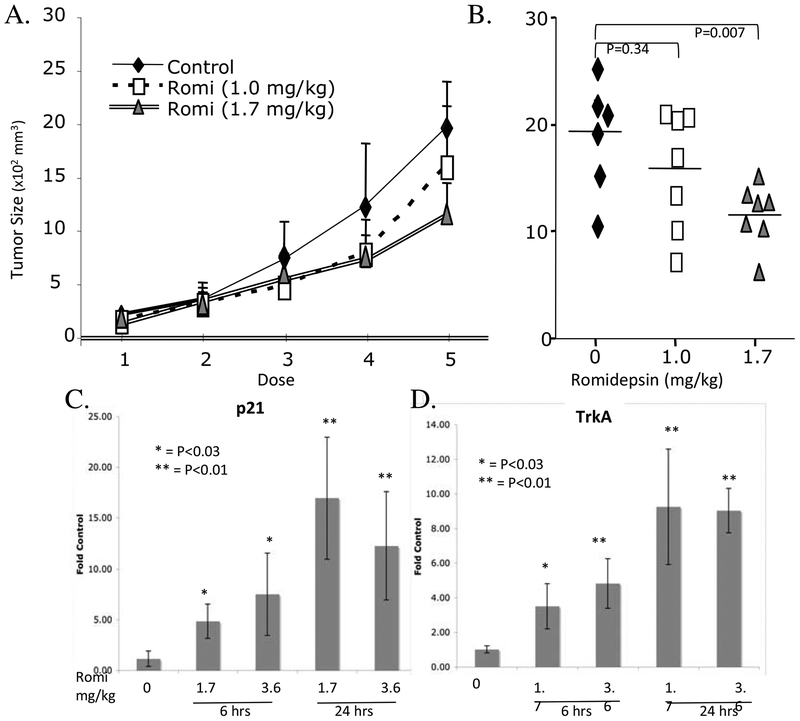
To test the biologic effects of romidepsin on neuroblastoma cells in vivo cohorts of nude mice bearing subcutaneous KCNR tumors were treated with romidepsin or vehicle alone as described in Methods. Romidepsin significantly inhibited the growth of KCNR tumors in mice (Fig 5A, B). Mice were given a single dose of romidepsin and tumors evaluated for changes in gene expression. Within 6 h of treatment, p21 levels in tumors increased almost 7-fold and reached 25-fold by 24 h compared to the levels detected in tumors from control treated mice (Fig 5C). Unlike the in vitro studies, the levels of MYCNmRNA or VEGFmRNA did not decrease in romidepsin treated tumors at this time or even at the end of the experiment (data not shown). However, TrkA which is a gene more highly expressed in tumors from patients with a good prognosis, increased in the tumors from romidepsin treated mice (Fig 5D).
Figure 5.
Romidepsin inhibits growth of subcutaneous KCNR cells. Panel A. Two million KCNR cells were injected subcutaneously into SCID-beige mice. When tumor size reached approximately 200 mm3 (width2 × length/4), mice received 5 intraperitoneal injections of romidepsin (1 or 1.7mg/kg) or vehicle control on days 13, 16, 20, 23 and 27. Panel B. Tumor size from mice at the end of the treatment receiving control solvent (black diamonds) or romidepsin at 1.0mg/kg (open squares) or 1.7mg/kg (grey triangles). Panels C & D. Quantitative PCR assessment of p21 (Panel C) or TrkA (Panel D) mRNA levels in tumors taken from mice 6 or 24 h after receiving a single dose of 1.7mg/kg or 3.6mg/kg romidepsin. (*p<0.05; **p<0.01, romidepsin treated groups compared to control group).
Discussion
Histone deacetylase inhibitors constitute a diverse group of compounds that promote histone acetylation, chromatin uncoiling, and transcription of a variety of genes involved in multiple cellular processes, including differentiation and apoptosis.22 Previous work with neuroblastoma cell lines, including our previous preclinical study with MS-27–27516 and other studies using BL-152123 and HKI46F0824, indicated that these tumors may be sensitive to transcriptionally based therapies using HDAC inhibitors. Phase I trials with various HDAC inhibitors including MS-27525, SAHA26 and romidepsin27–29 have shown that these HDAC inhibitors are relatively well tolerated at different dose schedules. Although cardiac toxicity was anticipated based on preclinical data with romidepsin, there was no evidence of myocardial damage.27–29 Reversible ECG changes with ST/T wave flattening or diffuse T wave inversions were regularly observed yet no changes were seen in the cardiac enzymes with romidepsin.30 in the pediatric phase I trial, only 1 of the 23 patients treated had neuroblastoma and while there were no objective responses there were 3 patients with prolonged disease stabilization in other solid tumors.14
In our study, cell cycle analysis of all NB cell lines treated with romidepsin showed an increase in cells in the sub-G1 fraction indicative of apoptosis. Moreover, the findings that romidepsin induced PARP cleavage and that the cell death could be blocked by Z-VAD-FMK is consistent with induction of apoptosis as a mechanism of action of romidepsin in NB. Glick et al31 showed that Z-VAD-FMK completely abrogated M-carboxycinnamic acid bishydroxamide (CBHA- a hybrid polar HDAC inhibitor) induced apoptosis in the 3 NB cell lines tested. Piekarz et al observed partial blockage of apoptosis with Z-VAD-FMK in a human T-cell lymphoma cell line treated with romidepsin.12 Despite the ability of HDACs to induce histone hyperacetylation in both tumor and normal cells, we like others found that tumor cells are preferentially sensitive to HDAC inhibition induced cell death compared to normal cells.7, 32
While HDAC inhibitors frequently induce p21 and cause a G1 arrest of cell growth in malignant epithelial cell lines13, our results are more consistent with those of Piekarz et al which demonstrated apoptosis in a human T-cell lymphoma cell line treated with romidepsin, sodium butyrate, TSA, or MS-27–275 without associated cell cycle arrest.12 Induction of p21 and marked apoptosis without concomitant cell cycle arrest was also previously observed in the A549 lung cancer cell line with HDAC inhibitor, trapoxin.33 Previous studies using the HDAC inhibitor CHBA and MS-27–275 have shown induction of p21 in NB cell lines and inhibition of NB tumor growth in vitro and in vivo.16, 34 The expression of p21 (WAF1/Cip1) is usually controlled at the transcriptional level by both p5 3-dependent and independent mechanisms.35 In our study, the increase in p21 with romidepsin appears to be p5 3-dependent since there was an increase in p21 in cell lines with wild type 53 but not in those with mutant p53. While some studies indicate HDAC inhibitors can restore p53 function in pseudo-null p53 cells,36 in some models p53 is not required.37 In this study, all NB cell lines underwent apoptosis irrespective of their p53 status. Thus, induction of p21 or wild type p53 is not essential for the anti-proliferative or the apoptosis induced by romidepsin in NB.
Consistent with the observations noted in our MS-27–275 study, similar apoptotic and anti-proliferative effects at nanomolar concentrations of romidepsin were noted in all NB cell lines irrespective of their MYCN status. Moreover there were no major differences in cell cycle changes, PARP cleavage or rescue by Z-VAD-FMK between romidepsin treated N-myc amplified and non-amplified cell lines. MS-27–275 induced decreases in MYCN and VEGF both in vitro and in vivo,16 yet we were not able to detect similar decreases in VEGF and MYCN in vivo in romidepsin treated tumors. This may have been due to the marked differences in the tolerability of the murine models to MS-275 which was well-tolerated by mice and completely blocked tumor growth in vivo16, while the doses of romidepsin that were better tolerated by mice only partially inhibited xenograft growth in vivo.
Although differentiation with HDAC inhibitors including romidepsin has been observed in various tumors, particularly leukemia14, 38 we did not observe any morphologic evidence of differentiation with romidepsin in the NB cell lines tested. However the marked induction of TrkA by romidepsin both in vitro and in vivo indicates that suppressed developmental pathways are induced upon inhibition of HDACs. Recent studies have elegantly shown that silencing expression of HDAC8 induces differentiation,39 while silencing of HDAC2 induces apoptosis39, and HDAC1 sensitizes neuroblastoma cells to chemotherapy.40 Romidepsin inhibits class I HDACs, which includes HDAC1, 2, 3 & 8; thus it appears that induction of apoptosis may be the dominant default pathway when multiple HDACs are inhibited by compounds such as romidepsin.
The challenge with HDAC inhibitors is to better translate the marked effects that are seen in preclinical in vitro and in vivo models to the clinical arena. In neuroblastoma, HDAC inhibitors in combination with TRAIL,41 interferon-a42, and retinoic acid43 have enhanced activity while combinations with gemcitabine are antagonistic.44 While combinations of phenyl butyrate (HDAC inhibitor) with 5 azacytadine (an inhibitor of DNA methylation) were not significantly different from the effects of either drug alone,45 such combinations in leukemia have enhanced activity.46 Recently key enzyme complexes (Polycomb Repression complex (PRC)) regulating chromatin biology have been identified and found to be over-expressed in tumors.47 It is possible that the combination of HDAC inhibitors with inhibitors of PRC may be a more effective therapy with which to control the dynamic epigenetic changes in tumor cells.
Materials and Methods
Cell culture and materials
Seven human NB cell lines, SMS-KCNR48, SK-N-BE249, SH-SY5Y50, SK-N-AS22, LA1–15N51, SH-SHEP52 and IMR3253 were used in this study (Table 1). All cell lines were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mM glutamine, 50 units/ml penicillin and 50 ug/ml streptomycin at 37°C with 5% CO2 as described previously.17 Romidepsin, provided by Fujisawa Pharmaceuticals (Osaka, Japan), was dissolved to 5 mg/mL in 4:1 propylene glycol and ethanol and then diluted to 100 μg/mL in dimethyl sulfoxide (DMSO) and stored at −20°C. For experiments, romidepsin was further diluted in cell culture media.
MTT and MTS assay
NB cells (5,000 to 10,000 cells/well) were seeded into 96 flat-bottomed well plates, and six replicate wells were incubated with different concentrations of romidepsin for 72 h. In some experiments, cell growth was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay (Sigma Chemical Company, St. Louis, MO) or an MTS/PMS colorimetric assay (Promega, Madison, WI) after 72 hrs of romidepsin treatment, as previously described.18 Samples were assessed according to manufacturer’s recommendations. The inhibition of viability was represented as a percentage of control cells. Each experiment was repeated 2–3 times and variability among the experiments on the same cell line and between MTT and MTS was less than 10%.
Cell cycle analysis
To assess cell cycle changes, control and romidepsin-treated cells were mechanically detached at the indicated times, washed twice with ice-cold phosphate-buffered saline (PBS) and incubated with RNAse at 100 μg/mL and PI at 50 μg/mL (Sigma Chemical Company) for 30 minutes in a dark environment at room temperature. The stained cells were analyzed for DNA content with a FACScan apparatus (Becton- Dickinson & Co., Franklin Lakes, NJ, USA) as previously described.16
Effect of Z-VAD-FMK
The caspase inhibitor Z-VAD-FMK was purchased from R&D Systems, Inc. (Minneapolis, MN). Five to 10,000 cells were seeded into 96 flat-bottomed well plates, and three replicate wells were incubated with 20 μmοl/L of the broad-spectrum caspase inhibitor Z-VAD-FMK for 2 h followed by incubation with romidepsin (IC80 concentration determined for each cell line) or control media for 48 hours. Cell viability was assessed by MTS assay and plotted as a percentage of control.
Acetylated H3 analysis by immunoblot assay
For acetylated histone analysis by immunoblot assay, control and romidepsin treated cells were detached using trypsin, washed twice with PBS, lysed in buffer (0.02 M Tris, 0.2 mM Triton X-100, 0.02% 2-mercaptoethanol, with 2 ng/mL aprotinin) and sonicated at 50% power to solubilize the protein. Protein lysate (12 μg total protein) was then added to 400 μL denaturing buffer (50% PBS 50% 4M GTC) and serially diluted (1:1) five times in denaturing buffer. Protein extracts were then analyzed by immunoblot using anti-GAPDH (American Research Products, 1:500 dilution) and anti-acetyl-Histone H3 (Upstate Biotechnology, 1:2200 dilution) antibodies as described previously.16
RNA preparation and northern blot analysis
Control and romidepsin-treated cells were detached mechanically from the 150-cm2 plate at the indicated time points, washed twice with ice-cold PBS, and processed for RNA extraction with the RNAeasy kit (Qiagen, Inc., Chatsworth, CA) according to the manufacturer’s instructions and analyzed by Northern blot analysis as described previously.16
Protein isolation and western blot analysis
Cells were cultured and harvested as above, and proteins extracts were prepared and anlayzed by Western blot as previously described.16 The following primary antibodies were utilized at a 1:1000 dilution except where noted; N-myc antibody (Santa Cruz Biotechnology), p21 antibody (1:250), poly (ADP-ribose) polymerase (PARP) antibody (Pharmingen, San Diego, CA); or anti-actin antibody (Oncogene Research Products, Darmstadt, Germany). For secondary antibodies, the appropriate species-specific horseradish peroxidase (HRP)–conjugated secondary antibodies (1: 2000 dilution; Santa Cruz Biotechnology) were utilized and bound antibodies were detected by chemiluminescence with the use of a LumiGlo detection system (Kirkegaard and Perry Laboratories, Gaithersburg, MD).
Analysis of secreted VEGF by ELISA
Cells were incubated for 4, 8 and 24 hours with or without romidepsin (IC80 concentration determined for each cell line), and the concentration of mouse (TBJ cell line) or human (other cell lines) VEGF was measured using ELISA (Quantikine VEGF immunoassay, R&D Systems, Minneapolis, MN).
Animals and treatment of established subcutaneous primary tumors.
Two million KCNR NB tumor cells were injected subcutaneous into the right backside of nude mice. Tumor size was measured with calipers. Tumor volume (V) was calculated as follows: V = a2 × b/4, where a is the width (small diameter) and b the length (large diameter) of the tumor in millimeters. When the tumor size reached 200 mm3, mice received intraperitoneal injections of vehicle control, 1.0, or 1.7 mg/kg romidepsin every 3 or 4 days for a total of 5 doses. To assess in treatment related changes in tumor gene expression, mice were given a single dose of romidepsin (1.7 mg/kg or 3.6 mg/kg) and tumors were harvested at 6 or 24 h. The 3.6 mg/kg dose of romidepsin could not be used in treatment protocols, as this dose was not tolerated by the mice using this schedule.
Acknowledgements
We are thankful to Robert Robey in the Cancer Therapeutics Branch, NCI for his insightful help. We are grateful to Kelly Martin, Reema Wahdan and other members of our lab, as well as the Cell and Molecular Biology Section for their immense help. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Maris J, Matthay K. Molecular biology of neuroblastoma. J Clin Oncol 1999; 17:2264–79. [DOI] [PubMed] [Google Scholar]
- 2.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 2003; 3:203–16. [DOI] [PubMed] [Google Scholar]
- 3.Mossé Y, Wood A, Maris J. Inhibition of ALK signaling for cancer therapy. Clin Cancer Res 2009; 15:5609–14. [DOI] [PubMed] [Google Scholar]
- 4.Allfrey V, Faulkner R, Mirsky A. Acetylation and méthylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A 1964; 51:786–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grunstein M Histone acetylation in chromatin structure and transcription. Nature 1997; 389:349–52. [DOI] [PubMed] [Google Scholar]
- 6.Struhl K Histone acetylation and transcriptional regulatory mechanisms. Genes Dev 1998; 12:599–606. [DOI] [PubMed] [Google Scholar]
- 7.Johnstone R, Licht J. Histone deacetylase inhibitors in cancer therapy: is transcription the primary target? Cancer Cell 2003; 4:13–8. [DOI] [PubMed] [Google Scholar]
- 8.Mulholland N, Soeth E, Smith C. Inhibition of MMTV transcription by HDAC inhibitors occurs independent of changes in chromatin remodeling and increased histone acetylation. Oncogene 2003; 22:4807–18. [DOI] [PubMed] [Google Scholar]
- 9.Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nature Rev Cancer 2001; 1:194–202. [DOI] [PubMed] [Google Scholar]
- 10.Nakajima H, Kim YB, Terano H, Yoshida M, Horinouchi S. FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp Cell Res 1998; 241:126–33. [DOI] [PubMed] [Google Scholar]
- 11.Piekarz RL, Robey R, Sandor V, Bakke S, Wilson WH, Dahmoush L, et al. Inhibitor of histone deacetylation, depsipeptide (FR901228), in the treatment of peripheral and cutaneous T-cell lymphoma: a case report. Blood 2001; 98:2865–8. [DOI] [PubMed] [Google Scholar]
- 12.Piekarz RL, Robey RW, Zhan Z, Kayastha G, Sayah A, Abdeldaim AH, et al. T-cell lymphoma as a model for the use of histone deacetylase inhibitors in cancer therapy: impact of depsipeptide on molecular markers, therapeutic targets, and mechanisms of resistance. Blood 2004; 103:4636–43. [DOI] [PubMed] [Google Scholar]
- 13.Blagosklonny MV, Robey R, Sackett DL, Du L, Traganos F, Darzynkiewicz Z, et al. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Mol Cancer Ther 2002; 1:937–41. [PubMed] [Google Scholar]
- 14.Fouladi M, Furman W, Chin T, Freeman Br, Dudkin L, Stewart C, et al. Phase I study of depsipeptide in pediatric patients with refractory solid tumors: a Children’s Oncology Group report. J Clin Oncol 2006; 24:3678–85. [DOI] [PubMed] [Google Scholar]
- 15.Graham C, Tucker C, Creech J, Favours E, Billups C, Liu T, et al. Evaluation of the antitumor efficacy, pharmacokinetics, and pharmacodynamics of the histone deacetylase inhibitor depsipeptide in childhood cancer models in vivo. Clin Cancer Res 2006; 12:223–34. [DOI] [PubMed] [Google Scholar]
- 16.Jaboin J, Wild J, Hamidi H, Khanna C, Kim CJ, Robey R, et al. MS-27–275, an inhibitor of histone deacetylase, has marked in vitro and in vivo antitumor activity against pediatric solid tumors. Cancer Res 2002; 62:6108–15. [PubMed] [Google Scholar]
- 17.Eggert A, Ikegaki N, Kwiatkowski J, Zhao H, Brodeur G, Himelstein B. High-level expression of angiogenic factors is associated with advanced tumor stage in human neuroblastomas. Clin Cancer Res 2000; 6:1900–8. [PubMed] [Google Scholar]
- 18.Meister B, Grünebach F, Bautz F, Brugger W, Fink F, Kanz L, et al. Expression of vascular endothelial growth factor (VEGF) and its receptors in human neuroblastoma. Eur J Cancer 1999;35:445–9. [DOI] [PubMed] [Google Scholar]
- 19.Meitar D, Crawford S, Rademaker A, Cohn S. Tumor angiogenesis correlates with metastatic disease, N-myc amplification, and poor outcome in human neuroblastoma. J Clin Oncol 1996; 14:405–14. [DOI] [PubMed] [Google Scholar]
- 20.Sasakawa Y, Naoe Y, Noto T, Inoue T, Sasakawa T, Matsuo M, et al. Antitumor efficacy of FK228, a novel histone deacetylase inhibitor, depends on the effect on expression of angiogenesis factors. Biochem Pharmacol 2003; 66:897–906. [DOI] [PubMed] [Google Scholar]
- 21.Mie Lee Y, Kim S, Kim H, Jin Son M, Nakajima H, Jeong Kwon H, et al. Inhibition of hypoxia-induced angiogenesis by FK228, a specific histone deacetylase inhibitor, via suppression of HIF-1alpha activity. Biochem Biophys Res Commun 2003; 300:241–6. [DOI] [PubMed] [Google Scholar]
- 22.White P, Maris J, Beltinger C, Sulman E, Marshall H, Fujimori M, et al. A region of consistent deletion in neuroblastoma maps within human chromosome lp36.2–36.3. Proc Natl Acad Sci U S A 1995; 92:5520–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Ruijter A, Kemp S, Kramer G, Meinsma R, Kaufmann J, Caron H, et al. The novel histone deacetylase inhibitor BL1521 inhibits proliferation and induces apoptosis in neuroblastoma cells. Biochem Pharmacol 2004; 68:1279–88. [DOI] [PubMed] [Google Scholar]
- 24.Wegener D, Deubzer H, Oehme I, Milde T, Hildmann C, Schwienhorst A, et al. HKI 46F08, a novel potent histone deacetylase inhibitor, exhibits antitumoral activity against embryonic childhood cancer cells. Anticancer Drugs 2008; 19:849–57. [DOI] [PubMed] [Google Scholar]
- 25.Ryan Q, Headlee D, Acharya M, Sparreboom A, Trepel J, Ye J, et al. Phase I and pharmacokinetic study of MS-275, a histone deacetylase inhibitor, in patients with advanced and refractory solid tumors or lymphoma. J Clin Oncol 2005; 23:3912–22. [DOI] [PubMed] [Google Scholar]
- 26.O’Connor O, Heaney M, Schwartz L, Richardson S, Willim R, MacGregor-Cortelli B, et al. Clinical experience with intravenous and oral formulations of the novel histone deacetylase inhibitor suberoylanilide hydroxamic acid in patients with advanced hematologic malignancies. J Clin Oncol 2006; 24:166–73. [DOI] [PubMed] [Google Scholar]
- 27.Byrd JC, Marcucci G, Parthun MR, Xiao JJ, Klisovic RB, Moran M, et al. A phase 1 and pharmacodynamic study of depsipeptide (FK228) in chronic lymphocytic leukemia and acute myeloid leukemia. Blood 2005; 105:959–67. [DOI] [PubMed] [Google Scholar]
- 28.Marshall J, Rizvi N, Kauh J, Dahut W, Figuera M, Kang M, et al. A phase I trial of depsipeptide (FR901228) in patients with advanced cancer. J Exp Ther Oncol 2002; 2:325–32. [DOI] [PubMed] [Google Scholar]
- 29.Sandor V, Bakke S, Robey RW, Kang MH, Blagosklonny MV, Bender J, et al. Phase I Trial of the Histone Deacetylase Inhibitor, Depsipeptide (FR901228, NSC 630176), in Patients with Refractory Neoplasms. Clin Cancer Res 2002; 8:718–28. [PubMed] [Google Scholar]
- 30.Piekarz RL, Frye AR, Wright JJ, Steinberg S, Liewehr DJ, Rosing DR, et al. Cardiac studies in patients treated with depsipeptide, FK228, in a phase II trial for T-cell lymphoma. Clin Cancer Res 2006. [DOI] [PubMed] [Google Scholar]
- 31.Glick R, Swendeman S, Coffey D, Rifkind R, Marks P, Richon V, et al. Hybrid polar histone deacetylase inhibitor induces apoptosis and CD95/CD95 ligand expression in human neuroblastoma. Cancer Res 1999; 59:4392–9. [PubMed] [Google Scholar]
- 32.Insinga A, Monestiroli S, Ronzoni S, Gelmetti V, Marchesi F, Viale A, et al. Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat Med 2005; 11:71–6. [DOI] [PubMed] [Google Scholar]
- 33.Gartel A, Tyner A. Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp Cell Res 1999; 246:280–9. [DOI] [PubMed] [Google Scholar]
- 34.Coffey D, Kutko M, Glick R, Butler L, Heller G, Rifkind R, et al. The histone deacetylase inhibitor, CBHA, inhibits growth of human neuroblastoma xenografts in vivo, alone and synergistically with all-trans retinoic acid. Cancer Res 2001; 61:3591–4. [PubMed] [Google Scholar]
- 35.Ito A, Kawaguchi Y, Lai C, Kovacs J, Higashimoto Y, Appella E, et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J 2002; 21:6236–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitazono M, Bates S, Fok P, Fojo T, Blagosklonny M. The histone deacetylase inhibitor FR901228 (desipeptide) restores expression and function of pseudo-null p53. Cancer Biol Ther 2002; 1:665–8. [DOI] [PubMed] [Google Scholar]
- 37.Vrana J, Decker R, Johnson C, Wang Z, Jarvis W, Richon V, et al. Induction of apoptosis in U937 human leukemia cells by suberoylanilide hydroxamic acid (SAHA) proceeds through pathways that are regulated by Bcl-2/Bcl-XL, c-Jun, and p21CIP1, but independent of p53. Oncogene 1999; 18:7016–25. [DOI] [PubMed] [Google Scholar]
- 38.Klisovic M, Maghraby E, Parthun M, Guimond M, Sklenar A, Whitman S, et al. Depsipeptide (FR 901228) promotes histone acetylation, gene transcription, apoptosis and its activity is enhanced by DNA methyltransferase inhibitors in AML1/ETO-positive leukemic cells. Leukemia 2003; 17:350–8. [DOI] [PubMed] [Google Scholar]
- 39.Oehme I, Deubzer H, Wegener D, Pickert D, Linke J, Hero B, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res 2009; 15:91–9. [DOI] [PubMed] [Google Scholar]
- 40.Keshelava N, Davicioni E, Wan Z, Ji L, Sposto R, Triche T, et al. Histone deacetylase 1 gene expression and sensitization of multidrug-resistant neuroblastoma cell lines to cytotoxic agents by depsipeptide. J Natl Cancer Inst 2007; 99:1107–19. [DOI] [PubMed] [Google Scholar]
- 41.Mühlethaler-Mottet A, Meier R, Flahaut M, Bourloud K, Nardou K, Joseph J, et al. Complex molecular mechanisms cooperate to mediate histone deacetylase inhibitors anti-tumour activity in neuroblastoma cells. Mol Cancer 2008; 7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michaelis M, Suhan T, Cinatl J, Driever P, Cinatl JJ. Valproic acid and interferon-alpha synergistically inhibit neuroblastoma cell growth in vitro and in vivo. Int J Oncol 2004;25:1795–9. [PubMed] [Google Scholar]
- 43.Hahn C, Ross K, Warrington I, Mazitschek R, Kanegai C, Wright R, et al. Expression-based screening identifies the combination of histone deacetylase inhibitors and retinoids for neuroblastoma differentiation. Proc Natl Acad Sci U S A 2008; 105:9751–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Ruijter A, Leen R, Hoebink J, Caron H, van Kuilenburg A. Antagonistic effects of sequential administration of BL1521, a histone deacetylase inhibitor, and gemcitabine to neuroblastoma cells. Cancer Lett 2006; 233:240–6. [DOI] [PubMed] [Google Scholar]
- 45.Tang X, Robinson M, Riceberg J, Kim D, Kung B, Titus T, et al. Favorable neuroblastoma genes and molecular therapeutics of neuroblastoma. Clin Cancer Res 2004; 10:5837–44. [DOI] [PubMed] [Google Scholar]
- 46.Scott S, Dong W, Ichinohasama R, Hirsch C, Sheridan D, Sanche S, et al. 5-Aza-2’-deoxycytidine (decitabine) can relieve p21WAF1 repression in human acute myeloid leukemia by a mechanism involving release of histone deacetylase 1 (HDAC1) without requiring p21WAF1 promoter demethylation. Leuk Res 2006; 30:69–76. [DOI] [PubMed] [Google Scholar]
- 47.Tiwari V, McGarvey K, Licchesi J, Ohm J, Herman J, Schübeler D, et al. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol 2008; 6:2911–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reynolds C, Biedler J, Spengler B, Reynolds D, Ross R, Frenkel E, et al. Characterization of human neuroblastoma cell lines established before and after therapy. J Natl Cancer Inst 1986; 76:375–87. [PubMed] [Google Scholar]
- 49.Gilbert F, Feder M, Balaban G, Brangman D, Lurie D, Podolsky R, et al. Human neuroblastomas and abnormalities of chromosomes 1 and 17. Cancer Res 1984; 44:5444–9. [PubMed] [Google Scholar]
- 50.Brodeur G, Green A, Hayes F, Williams K, Williams D, Tsiatis A. Cytogenetic features of human neuroblastomas and cell lines. Cancer Res 1981; 41:4678–86. [PubMed] [Google Scholar]
- 51.Ciccarone V, Spengler B, Meyers M, Biedler J, Ross R. Phenotypic diversification in human neuroblastoma cells: expression of distinct neural crest lineages. Cancer Res 1989; 49:219–25. [PubMed] [Google Scholar]
- 52.Thiele CJ. Neuroblastoma In: Masters JRW, Palsson B, eds. Human Cell Culture. Dordrecht, Great Britain: Kluwer Academic Publishers, 1999:21–53. [Google Scholar]
- 53.Thiele C, Reynolds C, Israel M. Decreased expression of N-myc precedes retinoic acid-induced morphological differentiation of human neuroblastoma. Nature 1985; 313:404–6. [DOI] [PubMed] [Google Scholar]