Abstract
Purpose:
Cutaneous T cell lymphomas (CTCL), encompassing a spectrum of T-cell lymphoproliferative disorders involving the skin, have collectively increased in incidence over the last 40 years. Sézary syndrome (SS) is an aggressive form of CTCL characterized by significant presence of malignant cells in both the blood and skin. The guarded prognosis for SS reflects a lack of reliably effective therapy, due in part to an incomplete understanding of disease pathogenesis.
Methods:
Using single-cell sequencing of RNA and the machine-learning reverse graph embedding approach in the Monocle package, we defined a model featuring distinct transcriptomic states within SS. Gene expression used to differentiate the unique transcriptional states were further utilized to develop a boosted tree classification for early versus late CTCL disease.
Results:
Our analysis showed the involvement of FOXP3+ malignant T cells during clonal evolution, transitioning from FOXP3+ T cells to GATA3+ or IKZF2+ (HELIOS) tumor cells. Transcriptomic diversities in a clonal tumor can be used to predict disease stage, and we were able to characterize a gene signature that predicts disease stage with close to 80% accuracy. FOXP3 was found to be the most important factor to predict early disease in SS, along with another 19 genes used to predict CTCL stage.
Conclusions:
This work offers insight into the heterogeneity of SS, providing better understanding of the transcriptomic diversities within a clonal tumor. This transcriptional heterogeneity can predict tumor stage and thereby offer guidance for therapy.
Introduction
Cutaneous T cell lymphomas (CTCLs) are a group of heterogenous T cell neoplasms with skin involvement. Two predominant types of CTCL include mycosis fungoides (MF) and Sézary syndrome (SS), both of which are thought to be derived from mature skin-homing CD4+ T cells (1,2). Given this commonality and their often overlapping clinicopathologic features, MF and SS had historically been regarded as closely related entities on a spectrum; however, recent elucidation of distinct cells of origin (3) has favored MF and SS to represent distinct clinical entities (4-6). SS refers to a rare form of CTCL characterized by circulating malignant cells with widespread skin involvement and possesses a poor 5-year survival rate (1,7). In contrast, MF refers to a substantially more common CTCL with a skin-predominant, and usually a skin-limited, presentation. MF most often has an indolent course, with a 5-year survival of 70-80% (5,7); however, a subset of patients exhibit a progressive course such that malignant cells may be identified in the circulation, lymph nodes, and viscera. Treatments for advanced stage MF and SS ultimately become ineffective, contributing to the morbidity and mortality of this patient population. Methods to identify those patients who will progress to advanced and widespread disease may facilitate optimal transition from skin-directed therapies to more aggressive treatment, but such methods have not yet been established.
Despite a number of high-quality computational inquiries into the genomic makeup of CTCL (8-12), the development of differentiated T cells phenotypes and their relationship to disease pathogenesis represents a knowledge gap in the understanding of CTCL. In particular, the contribution of Treg-like cells to the malignant population in MF/SS has been controversial, with heterogeneous and sometimes conflicting results (13-17). Heterogeneity within SS has been suggested by a recent targeted gene sequencing of single cells (18). A deeper understanding of differences within the clonal malignant population in CTCL may yield insights into more effective treatment regimens and strategies.
Here, we use single-cell RNA sequencing and single-cell V-D-J sequencing to examine SS at a previously unrealized transcriptomic resolution by pairing isolated SS cells with matched normal CD4+ T cells. Using this unique dataset, we investigated the degree as well as trajectory of heterogenous transcriptional profiles within the malignant cell population to identify novel markers of SS that may aid in the detection, diagnosis, and staging of CTCL. We further validate the power of our methodology by applying our findings to a publicly available dataset consisting of a large cohort of CTCL patients and demonstrate that when used in conjunction with an artificial intelligence (AI)-based approach, transcripts can be identified that distinguish early and late stage disease.
Methods
Patient Recruitment
The current study was approved by the University of Iowa Institutional Review Board and conducted under the Declaration of Helsinki Principles. The patient was recruited from the University of Iowa Cutaneous Lymphoma clinic in the Department of Dermatology. Informed written consent was received from the participant before inclusion in the study. At the time of collection, the patient was a 61-year-old male with stage IVA SS (T4N1M0B2) being treated with photophoresis and vorinostat. Interferon and bexarotene had previously been ineffective and/or not well tolerated.
Flow Cytometry
A blood draw was performed, and peripheral blood mononuclear cells (PBMCs) were isolated using a Ficoll gradient. Cells were labeled with fluorescent antibodies specific for CD3, CD4, CD8, CD45RA, CD45RO, CD5, CD7, and CD26 and flow sorted on a Becton Dickinson Aria II.
Single-cell RNA sequencing
A malignant (CD3+CD4+CD5brightSSChi) and nonmalignant CD4 (CD3+CD4+CD5intSSCint) population were flow sorted in parallel. T cell receptor sequencing and 5’ gene expression sequencing was performed using the Chromium (10x Genomics, Pleasonton, CA) and Illumina (San Diego, CA) sequencing technologies. Amplified cDNA was used to construct both 5’ expression libraries and TCR enrichment libraries. Libraries were pooled together and run on sperate lanes of a 150 based-paired, paired-end, flow cell using the Illumina HiSeq 4000. Basecalls were converted into FASTQs using the Illumina bcl2fastq software by the University of Iowa Genomics Division. FASTQ files were aligned to human genome (GRCh38) using the CellRanger v2.2 pipeline as described by manufacturer. Single-cell immune profiling of the clonotypes of the CD4+ T cells was performed in conjunction with the single-cell RNA sequencing following the protocols described above. Single-cell data is available at the Gene Expression Omnibus at the accession number: GSE122703.
Single-Cell Data Processing and Analysis
Initial processing of peripheral (n=4,485) and malignant (n=3,526) CD4+ T cells was performed using the Seurat R Package (v2.3.4) (19). Individual cells filtered for total number of genes expressed and percentage of mitochondrial reads. This filtering was set to retain cells with greater than 200 genes, but less than 3500 genes, and percent mitochondrial reads less than 9%. Individual cells were then normalized using log-normalization with a scale factor of 10,000. After processing, clustering was performed using the Seurat package on peripheral (n=4,436) and malignant (n=3,443) CD4+ T cells. Dimensional reduction to form the tSNE plot utilized the top 10 calculated principal components and a resolution, or granularity of the clusters, of 1.2. The number of principal components utilized in the tSNE cluster was based on examining the standard deviations of the top 20 principal components and running a jackstraw analysis to quantify p-value distributions (19). Cluster markers and differential gene expression analyses were performed using the Wilcoxon rank sum test. In the context of the differential gene expression between malignant and normal T cells, the Wilcoxon rank test was performed without filtering or thresholding parameters. In contrast, the cluster markers utilized default threshold of 0.25 for log2-fold change and a filter for the minimum percent of cells in a cluster greater than 25%. The differential markers between the clusters were isolated by comparing significantly upregulated genes as defined as adjusted p-values <0.05 and unique, non-shared genes between clusters. Single-cell immune phenotype correlations utilized the SingleR (v0.2.0) R package on mean raw count data for clusters identified in Seurat (20), scores are reported in quantile-normalized Spearman ρ values. Cell types for the analysis were derived from the Human Primary Cell Atlas (21). Differential markers between peripheral and malignant CD4+ T cells utilized the percentage of cells that express the individual mRNA species and log2-fold change between the two cell populations. Cell trajectory and pseudo-time analysis was performed using the Monocle R package (v2.8.0) and the reverse-graph embedding machine learning algorithm (22). Differential gene testing for the pseudo-time analysis was based on the previously identified malignant cell clusters and a cut-off for significance q-value < 0.01. Single-sample gene set enrichment analysis (ssGSEA) was performed on malignant T cell clusters using the SingleR R package with recently reported T-cell-related gene sets (23).
Machine Learning Gene Signature Analysis
Raw TruSeq FASTQs of 152 CTCL and 29 normal/benign skin lesions were downloaded from SRP114956 (24,25). Additional clinical data on patient age and disease were downloaded from the SRA repository. Files were pseudo-aligned with kallisto using the GRCh38 build of the human transcriptome (26). Transcript-level quantifications were condensed to gene-level and scaled to transcripts-per-million (TPM) values. In total, 344 genes were sequenced and quantified across the 151 of the total 152 patients, a single patient sample, SRR5906152, was removed due to pseudo-alignment issues in which kallisto produced a domain error after quantifying abundances and running the Expectation-Maximization algorithm. Quantified genes were then cross-referenced to significant (q-value < 0.05) genes identified in the Monocle 2 algorithm to narrow down signature candidates, with a total of 93 genes used for classification prediction. Boosted classification trees were constructed with the gbm (v2.1.3) package using log TPM values. Boosting was performed with 10,000 classification trees with a multinomial distribution; interaction depth and shrinkage parameters were selected via 10-fold cross validation on the training dataset. Variable importance of each gene was assessed by quantifying the mean decrease in the Gini index of each predictor averaged over all splits.
Results
Separation of Malignant and Normal CD4+ T cells by expression profiling
We performed parallel single cell RNA-sequencing and T cell receptor V-D-J sequencing of sorted malignant CD4+ T cells paired with normal CD4+ T cells using a single cell droplet platform, as outlined in Figure 1A. Malignant CD4 T cells were identified in a patient with SS by high side scatter (3) as well as aberrantly high expression of CD5; these cells made up nearly 86% of circulating CD4+ T cells. Normal CD4+ T cells were sorted in parallel, with a normal side scatter profile and normal CD5 expression (Figure 1B).
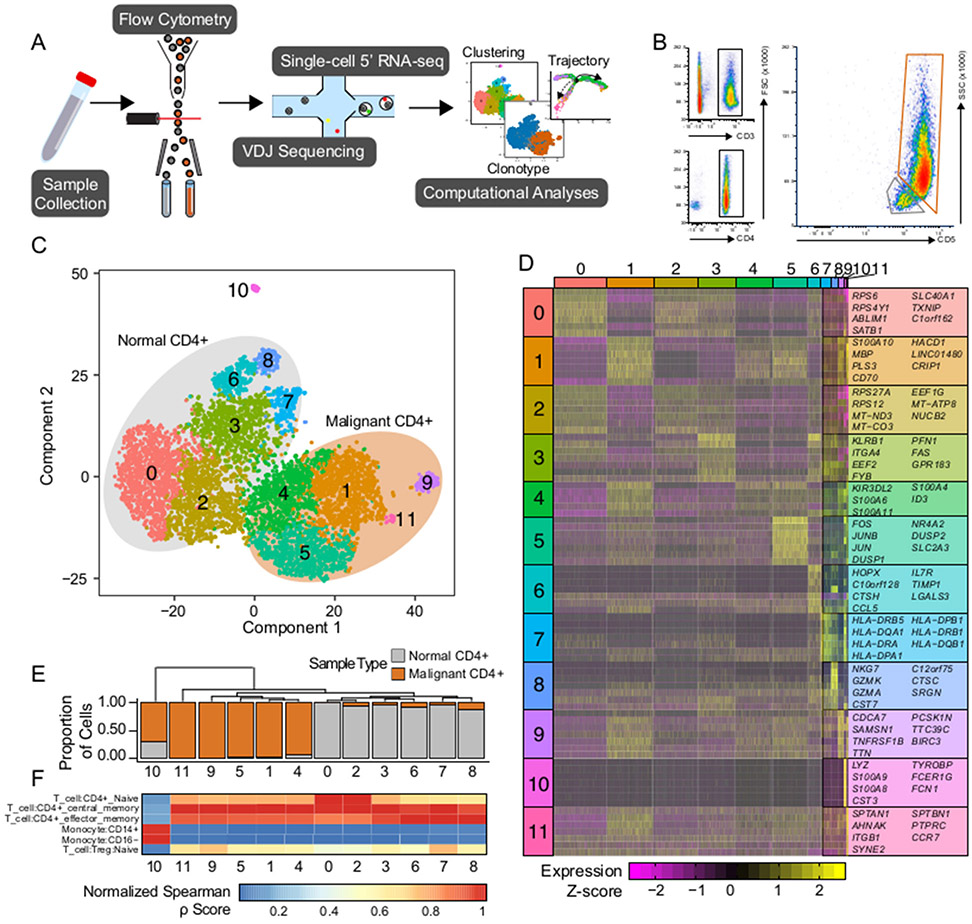
Figure 1. Single-cell isolation and sequencing of peripheral blood and Sézary Syndrome cells.
(A) Schematic of the isolation, sequencing, and analysis of the single-cells. (B) Flow cytometry gating of the patient sample to isolate peripheral blood and tumor cells. (C) tSNE projection of patient sample with normal peripheral blood samples (n=4436) outline in grey and tumorigenic CD4 cells (n=3443) in orange. (D) Unique significant cluster genes without overlap between clusters and based on the Wilcoxon rank sum test, adjusted P-value < 1e-50. (E) Phylogenic tree of cluster identities based on mean mRNA values in the cluster with corresponding cluster proportion of cells composition. (F) Quantile-normalized Spearman correlation values of predicted immune cell phenotype based on SingleR algorithm for each tSNE cluster.
After single-cell RNA and T cell receptor (TCR) sequencing of the isolated CD4+ T cells, data were filtered for low-quality cells and normalized. Assessing the collective heterogeneity of both normal and malignant CD4+ T cells, we observed 12 distinct clusters based on mRNA expression (Figure 1C). Accompanying the clustering, we also identified the top 5-7 genes that define each cluster (Figure 1D). Of the tSNE clusters, 6 were comprised of normal CD4+ T cells, while 5 consisted of the malignant SS cells. Using Euclidean hierarchical clustering, we found the tSNE clusters were most closely related to the normal versus malignant classification (Figure 1E), further confirming the separation within the tSNE itself. Using the mean mRNA expression of each cluster, we correlated the gene expression with known marker genes, with the majority of cells of both normal and malignant origin correlating with CD4+ central memory T cells (Tcm, Figure 1F). Notably, the normal CD4+ T cell Clusters 0 and 2 appeared to contain a naïve CD4+ T cell phenotype, and the clusters corresponding to the malignant SS cell population appeared to contain a phenotype consistent with Tcm (3). An additional cluster (Cluster 10) consisted of CD4+ myeloid cells and was excluded from further analysis.
SS cells are clonal and transcriptionally distinct from normal CD4+ T cells
In order to investigate the difference in malignant and normal CD4+ T cells, we confirmed the separation of normal and malignant cells using previously identified markers (Figure 2A). We verified that sequencing was performed on isolated CD4+ T cells, and that the malignant population exhibited a characteristic decrease in CD26 (DPP4) (27,28) and increase in CD70 (29) (Figure 2A). As previously mentioned, the patient’s malignant cells expressed an aberrant increase in CD5, which we could demonstrate at the mRNA level and a maintenance of CD7, both of which are unusual for SS (5) (Figure 2A). To further demonstrate the observable difference in the malignant SS cells, we filtered the VDJ sequencing results for the top TCR hits and matched 94.7% of sequenced cells with the corresponding VDJ sequencing information. Of the 3328 cells sorted for the SS phenotype (Figure 1B) with recoverable TCR sequencing information, 97.3% consisted of a single clonotype containing TRBV14 (CDR3 amino acid sequence: CASSPLQGTNSPLHF) and TRAV9-2 (CDR3 amino acid sequence: CALFPNTGFQKLVF) (Figure 2B). In contrast, the normal CD4+ T cells had 4007 unique clonotypes with 37 individual cells (0.9%) possessing the same malignant TRBV14/TRAV9-2 clonotype (Figure 2B), likely due to the close proximity of the flow sorting gates to each other.
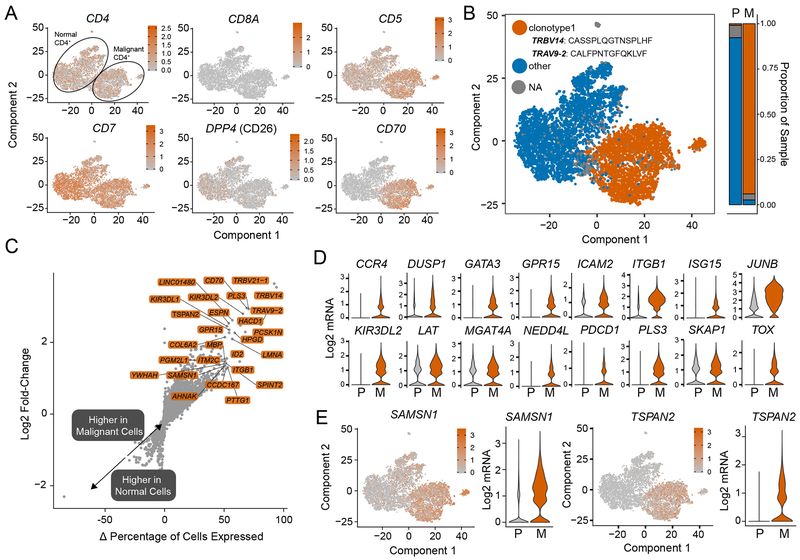
Figure 2. Transcriptomic comparison of malignant versus normal CD4+ T cells.
(A) tSNE projects of common markers used to diagnose CTCL (B) VDJ sequencing of malignant CD4+ T cells examining the distribution of a single prominent clonotype in the malignant T cells (orange) (C) Log2-fold change expression versus the difference in the percent of cell expressing the gene comparing malignant to normal peripheral blood CD4+ T cells (Δ percentage of cells expressed). Genes labeled have a Δ percentage of cells expressed > 50%, log2-fold change > 1 and an adjusted p-value < 0.05. (D) Potential novel markers of CTCL cells with a Δ percentage of cells expressed greater than 50% and adjusted p-values < 1e-100. (E) Violin plots of previously identified markers of CTCL (adjusted P < 1e-10).
In order to investigate potential novel markers and/or therapeutic targets of SS, we performed differential gene analysis comparing the malignant and normal CD4+ T cells. The complete differential expression results are available in Supplemental Table 1. We used this comparison analysis by contrasting the log2-fold change (y-axis) and the difference in the percentage of cell expressing the gene (Δ percentage of cells expressed, x-axis) (Figure 2C). By examining the difference in the percentage of malignant versus normal peripheral blood CD4+ T cells, this allows for the identification of specific markers of SS. As expected, of the genes with the highest log2-fold change and greatest discrimination between malignant versus normal cells were TRBV14 (log2-fold change = 3.53, Δ percentage = 94%) and TRAV9-2 (log2-fold change = 2.49, Δ percentage = 81%) (Figure 2B). Interestingly, 95.5% of malignant SS cells also expressed a second TRBV region variant, the pseudogeneTRBV21-1 (CDR3 amino acid sequence: CALFPNTGFQKLVF, Δ percentage = 92%) (Figure 2C). Using this analysis, we examined previously identified genes that relied on pooled SS RNA sequencing and found differential expression in CCR4 (30), DUSP1 (28,31), GPR15(32), ICAM2 (31), JUNB (31,33), KIR3DL2 (34), PLS3 (35), ITGB1 (10,28), GATA3 (28,31), NEDD4L (9,32), LAT (36), MGAT4A (36), PDCD1 (10,36,37), SKAP1 (11,36), and TOX (36,38) (Figure 2D). In addition to previously-identified markers, we report potential novel markers for SS (Figure 3E) as defined by a log2-fold change greater than one in SS cells versus normal cells and a Δ percentage > 50%. Both genes displayed similar expression differences to previously-reported gene markers (Figure 2D). SAM domain, SH3 domain and nuclear localization signals 1 (SAMSN1) has previously been reported to be involved in resistance to interferon α, a treatment modality for CTCL (39). Tetraspanin-2 (TSPAN2) is a cell-surface protein that has been implicated in cell migration in lung cancer (40).
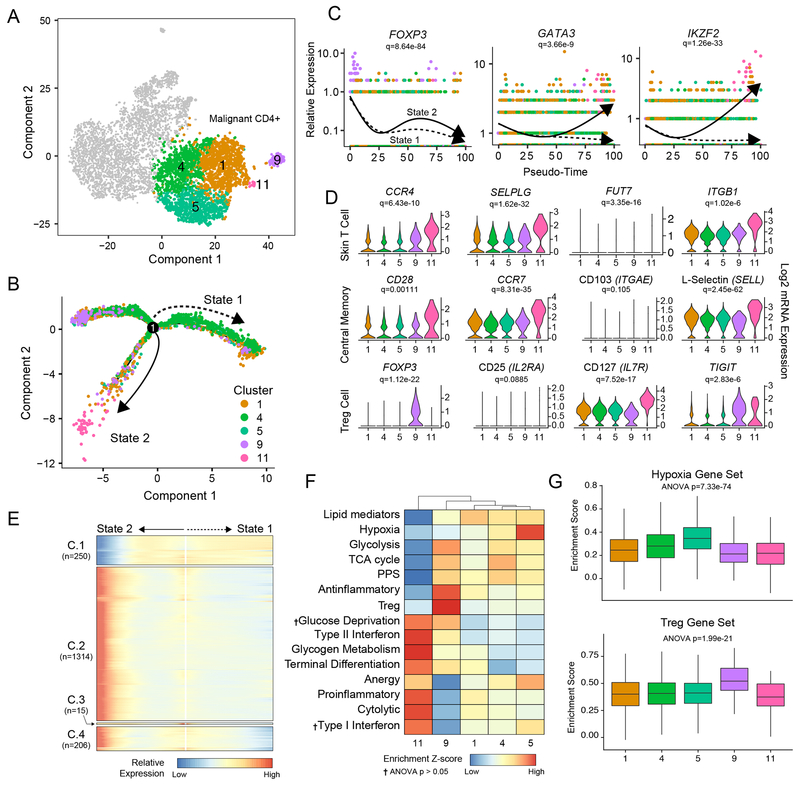
Figure 3. Transcriptional heterogeneity in malignant CD4+ T cells.
(A) tSNE projection of patient malignant CD4 cells (n=3,443). (B) Trajectory of malignant cells from clusters 1, 4, 5, 9, and 11 using the Monocle 2 algorithm, solid and dotted line represent distinct cell trajectories defined by single-cell transcriptomes (C) Pseudo-time projections of major immune transcriptional drivers in the malignant CD4+ T cells, demonstrating the change in relative expression over pseudo-time for the distinct transcriptional states, with each point representing a single cell. Significance based on differential testing by cluster identification used to generate pseudo-time and adjusted for multiple comparisons. (D) Selection of genes by cluster identity for skin-homing, central memory, and regulatory T cell phenotypes. Significance based on the pseudo-time generated by the Monocle 2 algorithm and correct for multiple comparisons. (E) Relative expression heatmap of significant (Q < 1e-4) genes based on branch expression analysis modeling comparing the two SS cell states and were used in the ordering of the pseudo-time variable. (F) Z-score transformed enrichment score for ssGSEA of T-cell-related gene sets in the malignant clusters. Pathways were significant with P < 0.05, as assessed by one-way ANOVA with multiple comparison adjustment unless indicated by †. (G) Hypoxia (upper panel) and Treg (lower panel) gene set enrichment across malignant SS clusters.
Heterogeneous transcriptional profiles of single cells in SS
Unlike previous genomic studies which have relied on pooled SS cells in comparison to normal CD4+ controls, we also were able to investigate the heterogeneity of the SS cells at a single-cell level (Figure 3A), and our previous analysis separated this malignant population into 5 clusters. Using the machine-learning reverse graph embedding for dimensional reduction available in the Monocle 2 algorithm, we constructed a manifold using the malignant SS cells (Figure 3B). This technique orders the single cells by expression patterns to represent distinct cellular fates or biological processes (22). Despite our finding of the clonal expansion of the SS cells (Figure 2B), we observed distinct bifurcated architecture of the cell trajectory, implying a divergence in transcriptional states (Figure 3B). Based on this ordering, SS cells appear to start principally from Cluster 9 and moved towards Clusters 1,4 and 5 (State 1, dotted line) or Cluster 11 (State 2, solid line) (Figure 3B).
In order to better understand the differential genes driving the ordinal construction of the manifold, we examined major immune transcription factors expression across the malignant CD4+ T cells, focusing on FOXP3, GATA3, IKZF2 (Figure 3C). Using the pseudo-time created by the reverse graph ordering, we produced pseudo-time projections in which we can compare the change in relative expression over pseudo-time for the distinct transcriptional states. From these projections, we observed a general decrease in FOXP3 in both directions of the bifurcation (Figure 3C). In contrast, both GATA3 and IKZF2 (HELIOS) had marked increased expression with the transcriptional state associated with cluster 11 (Figure 3C). An expanded analysis of major transcriptional factors relating to immune differentiation is available in Supplemental Figure 1A. Utilizing the differential expression analysis based on the pseudo-time construction, we next investigated the underlying differences of the malignant clusters by defined immune phenotypes. We separated the analysis into markers of skin-homing T cells, central memory T cells, and Tregs (Figure 3D). As expected we found consistent expression of skin-homing markers, CCR4, SELPLG (CLA) and ITGB1 (Figure 3D, upper row). Additionally, we found low expression of FUT7, a fucosyl-transferase required for the modification of CLA, across all clusters (41), however, FUT7 had significant differential expression between clusters (Figure 3D, upper row). Similarly, the malignant cells exhibited an expression pattern similar to Tcm cells, with sustained levels of CD28, CCR7 and SELL (L-Selectin/CD62L). Interestingly in both skin-homing and central memory T cell markers, Cluster 11 had consistent increased expression in both phenotype markers compared to the other SS clusters (Figure 3D). An additional analysis marker revealed a distinct FOXP3+ IL7Rlow TIGIT+ population in cluster 9, consistent with Treg or Treg-like cells (Figure 3D, lower row). All clusters had low and inconsistent expression of the Treg marker IL2RA (CD25), however, consistent with previous reports demonstrating lack of CD25+ Tregs in SS (14).
We next performed branched expression analysis modeling to discern the significant expression differences between the SS transcriptional state (Figure 3E). Differential genes between the two states were placed into four groups, C.1 through C.4, by expression pattern using the ward.D2 clustering algorithm (42). The complete list of genes in each group is available as Supplemental Table 2. A number of genes had increased expression in State 2 (C.2 and C.4) which were principally comprised of immune mediators (Figure 3E). In contrast C.1, which had maintained expression in State 1 had a number of ribosomal genes (Figure 3E). To better understand how these diverging gene patterns may play a role in SS, we performed single-sample gene set enrichment analysis (ssGSEA) (43). We found significant differences between SS clusters in T-cell-related gene sets (Figure 3F). Of note, along with the previously noted increase in Tcm and skin-homing gene markers, Cluster 11 (terminal portion of State 2) was significantly enriched for type II interferon signaling, terminal differentiation and cytolytic activity gene signatures. Clusters 1, 4, and 5 that form the major portion of Cell State 1, lacked distinct alterations in gene set enrichment with the exception of high levels of hypoxia in Cluster 5 (Figure 3F,G). In contrast to the other SS clusters, Cluster 9 was enriched for anti-inflammatory and Treg markers (Figure 3F,G), the latter fitting with our previous expression analysis. The pathway analysis with enrichment of the differentiated T cell signatures in cluster 11, lends support to our cell trajectory starting at FOXP3+ Cluster 9. Together these data suggest transcriptional and potentially functional heterogeneity among the malignant SS cell population and implya changing transcriptional profile within this clonal population.
Application of artificial intelligence-enabled genetic architecture to single cell SS pseudo-time scheme to predict disease stage
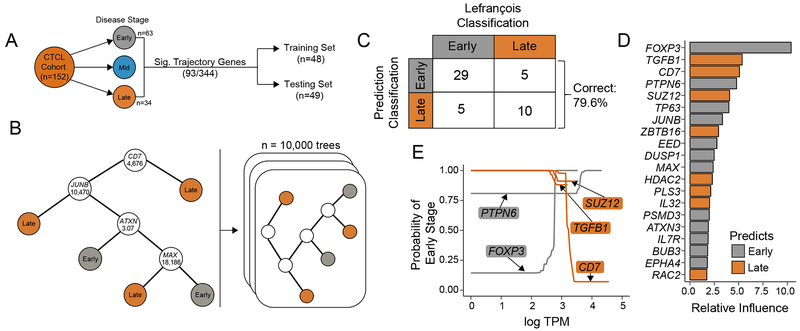
To better determine and validate if the observed heterogeneity had clinical significance, we downloaded raw sequencing reads from a cohort of CTCL patients (24,25). This cohort consisted of 152 CTCL patient samples from skin lesions of MF with targeted sequencing in 344 genes and 3 clinical CTCL classifications: early (Stage ≤ IIA), intermediate/mid (Stages IIB and III), and advanced/late (Stage IV, Figure 4A). Although often clinically distinct and derived from different T cells states (44), late-stage MF can have overlapping clinical features with SS and is often treated similarly (5). To improve the separation of the predictions, we isolated early (n=63) and late (n=34) CTCL patients and utilized 93 genes that were predictive of pseudo-time in our single cell data (Figure 4A). After splitting the cohort into training (n=48) and testing (n=49) sets, we constructed a series of boosted classification trees (n=10,000) using the training set (Figure 4B). We applied the boosted classification trees to the independent testing set and correctly classified 79.6% of samples into early versus late stages (Figure 4C). Variable importance was quantified for each gene across the boosted classification trees. The single gene with the largest relative influence in classification was FOXP3 at 10.39% (Figure 4D). Other genes with high relative influence in the classification model include TGFB1 (5.37%), CD7 (5.09%), PTPN6 (4.79%), and SUZ12 (4.07%). Partial dependence plots for the five most influential genes were constructed to illustrate the effect of each important gene’s expression on the probability of early disease stage classification while integrating out other variables (Figure 4E).
Figure 4. Predictive clinical correlates in CTCL using SS single-cell heterogeneity.
(A) Representative schematic of the composition of SRP114956 and the separation into training and testing sets for prediction of clinical stage. (B) A hypothetical classification decision tree is constructed to predict the CTCL stage based on RNA-seq expression data for each patient in the training set (n=48). At each branch in the tree, the patient’s transcripts per million (TPM) for a given gene are compared to a cutoff value. If the patient’s TPM are below the cutoff, the algorithm proceeds to the left and vice versa, until a terminal classification node is reached. A series of 10,000 boosted trees are grown in sequence utilizing information from previous trees, improving upon previous misclassifications. (C) The independent test patient data set (n=49) is applied to the 10,000 boosted classification trees and predicted disease states are compared to original classifications. Overall, the boosted decision trees correctly classify 79.6% of the disease states. (D) The 20 most important genes in generating the boosted classification trees are quantified and displayed in a ranked variable importance plot. Bar color logic is described below. (E) Partial dependence plots for the five most important variables represent how different levels of gene expression (log TPM) effect the probability of early-disease classification after integrating out the expression of all other genes. Genes with high expression predictive of early disease are colored in grey, while high gene expression more predictive of late stage disease are colored in orange.
A partial dependence plot was constructed for each of the 20 most important genes (data not shown), and the highest expression level of each gene was compared to the probability of early disease stage classification. Recent work has linked late-stage disease progression in MF to SS, specifically in the increased expression of TOX, FYB, CD52, and CCR4; however, based on the boosted classification tree, these genes did not have large relative influence in prediction (45). Genes with their highest expression predictive of early disease include FOXP3 and PTPN6, while genes with highest expression predictive of late stage disease include TGFB1, CD7, and SUZ12. We next examined the distribution of expression of selected genes from the top 20 genes based on the pseudo-time projection of the manifold (Supplemental Figure 1B-D). We found several genes that had diverging relative expression between the SS transcriptional states, like PLS3 and SUZ12 (Supplemental Figure 1C). Using the cell trajectory, we were unable to see clear expression trends in late-associated genes (Supplemental 1D, orange boxes), while early-associated genes had consistent decreases in at least one tail of the trajectory (Supplemental 1D, grey boxes). Larger single-cell datasets from patients at different stages of diseases may therefore increase the power of this technique and increase the precision of prognostic and predictive biomarkers.
Discussion
Beyond examining transcriptional states of clonal SS cells, this study examines the implications of predicting CTCL progression based on divergent gene drivers. The boosted classification trees demonstrated an efficacious prediction model for classifying early versus late disease stage (Figure 4). In contrast to the binary evaluation of differential expression of a gene between two different disease states, the boosted classification trees utilize combinations of continuous expression values associated with early versus late disease (46). Underscoring the value of the boosted classification tree approach was the nearly 80% prediction efficiency for CTCL stage (Figure 4C), particularly intriguing considering that feature selection was performed using data from a single patient. Although limited to a single patient, our analysis provides a framework for the application of single-cell-based high throughput technologies to analyze disease in a clinically meaningful way.
In particular, the expression of FOXP3 was the most influential predictor of CTCL stage identified from our analysis. FoxP3 is a master transcription factor for regulatory T cells (47,48). The observation of Treg or Treg-like malignant cells in SS and MF is controversial, with a number of conflicting results reported (13-17). Our work demonstrated decreasing FOXP3 over purported pseudo-time estimation, and this decrease was associated with an increase in the major Th2 immune driver, GATA3 (Figure 3C). Intriguingly, in the absence of adequate CD25 expression, bona fide Tregs retain developmental plasticity, allowing the cells to differentiate into helper T cells dependent on the microenvironment and cytokine milieu (49). Our data indicate that SS cells may initially express high FOXP3 and low CD25 (IL2RA) and retain similar mutability to FoxP3+CD25− Tregs.
The maintenance of FOXP3 expression in Tregs is required to maintain a suppressive phenotype, the loss of which is termed Treg fragility (50,51). A previous report in SS found a subset of patients with CD25− FOXP3+ tumor cells, similar to our RNA findings (Figure 3), that retain suppressive function (14). Instead of the malignant proliferation of regulatory T cells first suggested by Berger, et al, our data suggests the possibility of a FOXP3+ intermediate state of SS tumor cells (13). The association with increased TGFB1 expression with later stage disease, however, would indicate that loss of FOXP3 does not equate to loss of the ability to elaborate immunoregulatory/suppressive factors. Also of interest, HDAC inhibition, which is effective in treating 30% of SS, has been shown to drive FOXP3 expression and Treg suppressive function in vivo (52). Recent targeted single-cell sequencing of SS cells before and after treatment with HDAC inhibition demonstrated reduction in T cells of the Tcm transcriptional phenotype (18), while response to HDAC inhibitors in CTCL have also been associated with increase in open chromatin at a number of gene loci, including FOXP3 (53). Thus, the promotion of the early or intermediate FoxP3+ state in CTCL may be a mechanism of action for vorinostat or other histone deacetylase inhibitors (54) in disease treatment and the prevention of disease progression.
Newer single-cell methods, as used here, allow researchers to characterize and stratify common drivers and sources of heterogeneity in clonal tumors. Similar to our approach, two recent reports in CTCL using transcript-indexed ATAC-seq (55) and limited mRNA sequencing of 110 T-cell-related genes (18) found heterogeneity in malignant SS cells. Beyond the observations of transcriptional heterogeneity, this new level of data provides the opportunity for clinically-meaningful advances in SS and to other cancers. Although limited in scope, our supervised machine-learning approach to predicting CTCL disease state demonstrates the early stages of combining these new highthroughput approaches with predictive algorithms to move beyond simple observations.
Supplementary Material
Translational Relevance.
An analysis of Sézary syndrome (SS) using single cell RNA-sequencing revealed transcriptional heterogeneity among malignant SS cells. The current study is the first to show a shift in Treg-like to a more central memory CD4+ T cells phenotype at the transcriptional level in SS. From the heterogeneity of SS cells in a single patient, we were able to construct an artificial-intelligence-based algorithm that predicted early versus late disease state, implicating the role of the observed transcriptional dynamics in disease progression and potentially drug resistance.
Acknowledgements
Funding for this project was provided from the National Institute of Health under the K08 award AR069111 (PI: A.J.), from the F30 fellowship CA206255 (PI: N.B.; mentor: W.Z.) and from R01s CA200673 and CA203834 (PI: W.Z.). The data presented herein were obtained at the Flow Cytometry Facility, which is a Carver College of Medicine / Holden Comprehensive Cancer Center core research facility at the University of Iowa, funded through user fees and the generous financial support of the Carver College of Medicine, Holden Comprehensive Cancer Center, and Iowa City Veteran’s Administration Medical Center, as well as at the Genomics Division of the Iowa Institute of Human Genetics which is supported, in part, by the University of Iowa Carver College of Medicine and the Holden Comprehensive Cancer Center (National Cancer Institute of the National Institutes of Health under Award Number P30CA086862). We thank Sergei Syrbu for stimulating discussions and Julie McKillip for excellent clinical support.
Footnotes
Declaration of Interests
The authors declare no competing interests
References
- 1.Willemze R, Jaffe ES, Burg G, Cerroni L, Berti E, Swerdlow SH, et al. WHO-EORTC classification for cutaneous lymphomas. Blood. 2005. page 3768–85. [DOI] [PubMed] [Google Scholar]
- 2.Kirsch IR, Watanabe R, O’Malley JT, Williamson DW, Scott LL, Elco CP, et al. TCR sequencing facilitates diagnosis and identifies mature T cells as the cell of origin in CTCL. Sci Transl Med. 2015;7:308ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark RA, Shackelton JB, Watanabe R, Calarese A, Yamanaka KI, Campbell JJ, et al. High-scatter T cells: A reliable biomarker for malignant T cells in cutaneous T-cell lymphoma. Blood. 2011;117:1966–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ormsby A, Bergfeld WF, Tubbs RR, Hsi ED. Evaluation of a new paraffin-reactive CD7 T-cell deletion marker and a polymerase chain reaction-based T-cell receptor gene rearrangement assay: Implications for diagnosis of mycosis fungoides in community clinical practice. J Am Acad Dermatol. 2001;45:405–13. [DOI] [PubMed] [Google Scholar]
- 5.Wilcox RA. Cutaneous T-cell lymphoma: 2017 update on diagnosis, risk-stratification, and management. Am J Hematol. 2017;92:1085–102. [DOI] [PubMed] [Google Scholar]
- 6.Michie SA, Abel EA, Hoppe RT, Warnke RA, Wood GS. Discordant expression of antigens between intraepidermal and intradermal T cells in mycosis fungoides. Am J Pathol. 1990;137:1447–51. [PMC free article] [PubMed] [Google Scholar]
- 7.Agar NS, Wedgeworth E, Crichton S, Mitchell TJ, Cox M, Ferreira S, et al. Survival Outcomes and Prognostic Factors in Mycosis Fungoides/Sezary Syndrome: Validation of the Revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer Staging Proposal. J Clin Oncol. 2010;28:4730–9. [DOI] [PubMed] [Google Scholar]
- 8.Shin J, Monti S, Aires DJ, Duvic M, Golub T, Jones DA, et al. Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome. Blood. 2007;110:3015–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booken N, Gratchev A, Utikal J, Weiß C, Yu X, Qadoumi M, et al. Sézary syndrome is a unique cutaneous T-cell lymphoma as identified by an expanded gene signature including diagnostic marker molecules CDO1 and DNM3. Leukemia. 2008;22:393–9. [DOI] [PubMed] [Google Scholar]
- 10.Lee CS, Ungewickell a., Bhaduri a., Qu K, Webster DE, Armstrong R, et al. Transcriptome sequencing in Sezary syndrome identifies Sezary cell and mycosis fungoides-associated lncRNAs and novel transcripts. Blood. 2012;120:3288–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Doorn R, van Kester MS, Dijkman R, Vermeer MH, Mulder A a, Szuhai K, et al. Oncogenomic analysis of mycosis fungoides reveals major differences with Sezary syndrome. Blood. 2009;113:127–36. [DOI] [PubMed] [Google Scholar]
- 12.Choi J, Goh G, Walradt T, Hong BS, Bunick CG, Chen K, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015;47:1011–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger CL, Tigelaar R, Cohen J, Mariwalla K, Trinh J, Wang N, et al. Cutaneous T-cell lymphoma: Malignant proliferation of T-regulatory cells. Blood. 2005;105:1640–7. [DOI] [PubMed] [Google Scholar]
- 14.Heid JB, Schmidt A, Oberle N, Goerdt S, Krammer PH, Suri-Payer E, et al. FOXP3+CD25− tumor cells with regulatory function in Sezary syndrome. J Invest Dermatol. 2009;129:2875–85. [DOI] [PubMed] [Google Scholar]
- 15.Tiemessen MM, Mitchell TJ, Hendry L, Whittaker SJ, Taams LS, John S. Lack of suppressive CD4+CD25+FOXP3+ T cells in advanced stages of primary cutaneous T-cell lymphoma. J Invest Dermatol. 2006;126:2217–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krejsgaard T, Gjerdrum LM, Ralfkiaer E, Lauenborg B, Eriksen KW, Mathiesen AM, et al. Malignant Tregs express low molecular splice forms of FOXP3 in Sézary syndrome. Leukemia. 2008;22:2230–9. [DOI] [PubMed] [Google Scholar]
- 17.Gjerdrum LM, Woetmann a, Odum N, Burton CM, Rossen K, Skovgaard GL, et al. FOXP3+ regulatory T cells in cutaneous T-cell lymphomas: association with disease stage and survival. Leuk Off J Leuk Soc Am Leuk Res Fund, UK. 2007;21:2512–8. [DOI] [PubMed] [Google Scholar]
- 18.Buus TB, Willerslev-Olsen A, Fredholm S, Blümel E, Nastasi C, Gluud M, et al. Single-cell heterogeneity in Sézary syndrome. Blood Adv. American Society of Hematology; 2018;2:2115–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aran D, Agnieszka P, Looney AP, Liu L, Wu E, Fong V, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nature Immunol 2019;20:163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mabbott NA, Baillie JK, Brown H, Freeman TC, Hume DA. An expression atlas of human primary cells: Inference of gene function from coexpression networks. BMC Genomics. 2013;14:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell. 2018; 174:1293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefrançois P, Tetzlaff MT, Moreau L, Watters AK, Netchiporouk E, Provost N, et al. TruSeq-Based Gene Expression Analysis of Formalin-Fixed Paraffin-Embedded (FFPE) Cutaneous T-Cell Lymphoma Samples: Subgroup Analysis Results and Elucidation of Biases from FFPE Sample Processing on the TruSeq Platform. Front Med. 2017;4:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Litvinov IV, Tetzlaff MT, Thibault P, Gangar P, Moreau L, Watters AK, et al. Gene expression analysis in Cutaneous T-Cell Lymphomas (CTCL) highlights disease heterogeneity and potential diagnostic and prognostic indicators. Oncoimmunology. 2017;6:e1306618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 27.Bernengo MG, Quaglino P, Novelli M, Cappello N, Doveil GC, Lisa F, et al. Prognostic factors in Sezary syndrome: A multivariate analysis of clinical, haematological and immunological features. Ann Oncol. 1998;9:857–63. [DOI] [PubMed] [Google Scholar]
- 28.Kari L, Loboda A, Nebozhyn M, Rook AH, Vonderheid EC, Nichols C, et al. Classification and Prediction of Survival in Patients with the Leukemic Phase of Cutaneous T Cell Lymphoma. J Exp Med. 2003;197:1477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao X, Orchard G, Mitchell TJ, Oyama N, Russell-Jones R, Vermeer MH, et al. A genomic and expression study of AP-1 in primary cutaneous T-cell lymphoma: Evidence for dysregulated expression of JUNB and JUND in MF and SS. J Cutan Pathol. 2008;35:899–910. [DOI] [PubMed] [Google Scholar]
- 30.Ferenczi K, Fuhlbrigge RC, Pinkus JL, Pinkus GS, Kupper TS. Increased CCR4 expression in cutaneous T cell lymphoma. J Invest Dermatol. 2002;119:1405–10. [DOI] [PubMed] [Google Scholar]
- 31.Nebozhyn M, Loboda A, Kari L, Rook AH, Vonderheid EC, Lessin S, et al. Quantitative PCR on 5 genes reliably identifies CTCL patients with 5% to 99% circulating tumor cells with 90% accuracy. Blood. 2006;107:3189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Su M, Zhou LL, Tu P, Zhang X, Jiang X, et al. Deficiency of SATB1 expression in Sézary cells causes apoptosis resistance by regulating FasL/CD95L transcription. Blood. 2011;117:3826–35. [DOI] [PubMed] [Google Scholar]
- 33.Mao X, Orchard G, Lillington DM, Russell-Jones R, Young BD, Whittaker SJ. Amplification and overexpression of JUNB is associated with primary cutaneous T-cell lymphomas. Blood. 2003;101:1513–9. [DOI] [PubMed] [Google Scholar]
- 34.Poszepczynska-Guigné E, Schiavon V, D’Incan M, Echchakir H, Musette P, Ortonne N, et al. CD158k/KIR3DL2 is a new phenotypic marker of sezary cells: Relevance for the diagnosis and follow-up of sezary syndrome. J Invest Dermatol. 2004;122:820–3. [DOI] [PubMed] [Google Scholar]
- 35.Su MW, Dorocicz I, Dragowska WH, Ho V, Li G, Voss N, et al. Aberrant Expression of T-Plastin in Sezary Cells. Cancer Res. 2003;63:7122–7. [PubMed] [Google Scholar]
- 36.Zhang Y, Wang Y, Yu R, Huang Y, Su M, Xiao C, et al. Molecular markers of early-stage mycosis fungoides. J Invest Dermatol. 2012;132:1698–706. [DOI] [PubMed] [Google Scholar]
- 37.Samimi S, Benoit B, Evans K, Wherry EJ, Showe L, Wysocka M, et al. Increased Programmed Death-1 Expression on CD4+ T Cells in Cutaneous T-Cell Lymphoma. Arch Dermatol. 2010;146:1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y, Su MW, Jiang X, Zhou Y. Evidence of an oncogenic role of aberrant TOX activation in cutaneous T-cell lymphoma. Blood. 2015;125:1435–43. [DOI] [PubMed] [Google Scholar]
- 39.Tracey L, Villuendas R, Ortiz P, Dopazo A, Spiteri I, Lombardia L, et al. Identification of genes involved in resistance to interferon-α in cutaneous T-cell lymphoma. Am J Pathol. 2002;161:1825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otsubo C, Otomo R, Miyazaki M, Matsushima-Hibiya Y, Kohno T, Iwakawa R, et al. TSPAN2 Is Involved in Cell Invasion and Motility during Lung Cancer Progression. Cell Rep. 2014;7:527–38. [DOI] [PubMed] [Google Scholar]
- 41.Buffone A, Mondal N, Gupta R, McHugh KP, Lau JTY, Neelamegham S. Silencing 1,3-fucosyltransferases in human leukocytes reveals a role for FUT9 enzyme during e-selectin-mediated cell adhesion. J Biol Chem. 2013;288:1620–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murtagh F, Legendre P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J Classif. 2014; 2014;31:274–95. [Google Scholar]
- 43.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell JJ, Clark RA, Watanabe R, Kupper TS. Sézary syndrome and mycosis fungoides arise from distinct T-cell subsets: A biologic rationale for their distinct clinical behaviors. Blood. 2010;116:767–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lefrançois P, Xie P, Wang L, Tetzlaff MT, Moreau L, Watters AK, et al. Gene expression profiling and immune cell-type deconvolution highlight robust disease progression and survival markers in multiple cohorts of CTCL patients. Oncoimmunology. 2018;7:e1467856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dudoit S, Fridlyand J, Speed TP. Comparison of Discrimination Methods for the Classification of Tumors Using Gene Expression Data Comparison of Discrimination Methods for the Classi’ cation of Tumors Using Gene Expression Data. J Am Stat Assoc. 2002;97457:77–87. [Google Scholar]
- 47.Hori S, Nomura T, Sakaguchi S. Control of Regulatory T Cell Development by the Transcription Factor. Science 2003;299:1057. [PubMed] [Google Scholar]
- 48.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–6. [DOI] [PubMed] [Google Scholar]
- 49.Komatsu N, Mariotti-Ferrandiz ME, Wang Y, Malissen B, Waldmann H, Hori S. Heterogeneity of natural Foxp3 + T cells: A committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc Natl Acad Sci. 2009;106:1903–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–84. [DOI] [PubMed] [Google Scholar]
- 51.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–70. [DOI] [PubMed] [Google Scholar]
- 52.Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–307. [DOI] [PubMed] [Google Scholar]
- 53.Qu K, Zaba LC, Satpathy AT, Giresi PG, Li R, Jin Y, et al. Chromatin Accessibility Landscape of Cutaneous T Cell Lymphoma and Dynamic Response to HDAC Inhibitors. Cancer Cell. 2017; 2017;32:27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–52. [DOI] [PubMed] [Google Scholar]
- 55.Satpathy AT, Saligrama N, Buenrostro JD, Wei Y, Wu B, Rubin AJ, et al. Transcript-indexed ATAC-seq for precision immune profiling. Nat Med. 2018;24:580–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.