SUMMARY
Type VI CRISPR-Cas systems contain programmable single-effector RNA-guided RNases, including Cas13b, one of the four known family members. Cas13b, which has been used for both RNA editing and nucleic acid detection, is unique among type VI CRISPR effectors in its linear domain architecture and CRISPR RNA (crRNA) structure. Here we report the crystal structure of Prevotella buccae Cas13b (PbuCas13b) bound to crRNA at 1.65 Å resolution. This structure combined with biochemical experiments assaying the stability, kinetics, and function of Cas13b provide a mechanistic model for Cas13b target RNA recognition and identify features responsible for target and cleavage specificity. Based on these observations, we generated Cas13b variants with altered cleavage preferences, which may expand the utility of nuclease-based RNA detection assays and other applications of Cas13b in mammalian cells.
INTRODUCTION
A number of new Cas enzymes and CRISPR systems with novel properties have been discovered recently. Notable amongst these new discoveries are the Class 2 type VI CRISPR-Cas13 systems, which use a single enzyme to target RNA using a programmable CRISPR RNA (crRNA) guide (Abudayyeh et al., 2016; Konermann et al., 2018; Shmakov et al., 2015; 2017; Smargon et al., 2017; Yan et al., 2018). Cas13 binding to target single-stranded RNA activates a general RNase activity that cleaves the target and degrades surrounding RNA non-specifically, known as collateral activity or trans-cleavage (Abudayyeh et al., 2016; East-Seletsky et al., 2016). Type VI systems have been used for RNA knockdown, transcript labeling, splicing modulation, and ultra-sensitive virus detection (Abudayyeh et al., 2016; 2017; East-Seletsky et al., 2016; 2017; Gootenberg et al., 2018; 2017; Konermann et al., 2018; Smargon et al., 2017; Yan et al., 2018). In addition, nuclease dead Cas13 forms the basis for the RNA editing platform REPAIR (RNA Editing for Precise A-to-I Replacement), which relies on the targeting activity of Cas13 and the adenosine deaminase activity of ADAR to achieve precise base editing (Cox et al., 2017).
CRISPR-Cas13 systems are further divided into four subtypes based on the identity of the Cas13 protein (Cas13a – d) (Shmakov et al., 2017). All Cas13 protein family members contain two Higher Eukaryotes and Prokaryotes Nucleotide-binding (HEPN) domains. Unlike Cas13a, c, and d, however, in Cas13b the HEPN domains are at the extreme N- and C-termini of the linear protein, suggesting it may adopt a unique three-dimensional conformation (Shmakov et al., 2017; Smargon et al., 2017). Additionally, the direct repeat of the Cas13b crRNA is at the 3′ end, which is the opposite orientation found in other type VI systems (Abudayyeh et al., 2016; Cox et al., 2017; Konermann et al., 2018; Smargon et al., 2017; Yan et al., 2018). Here we report the structure of Cas13b from Prevotella buccae (PbuCas13b) in complex with a crRNA handle and partial spacer at 1.65 Å resolution. This structural information together with biochemical characterization of RNA targeting provide a model for target recognition and specificity by Cas13b which is distinct from other known programmable nucleic acid targeting systems.
RESULTS
High-resolution Crystal Structure of PbuCas13b
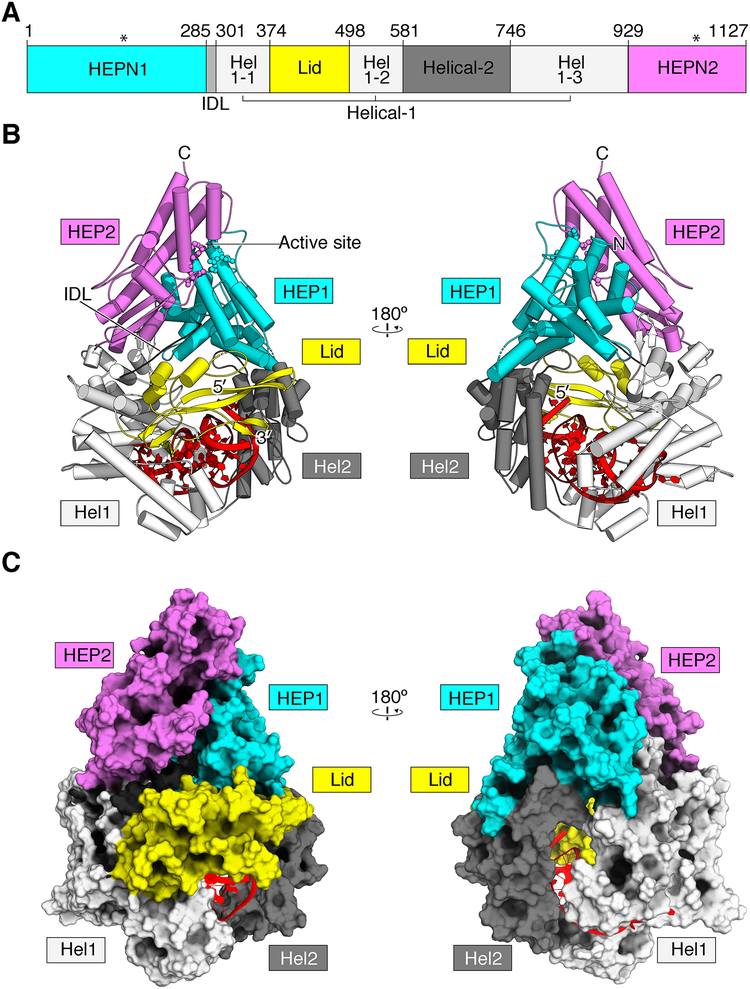
To understand the architecture of Cas13b, we solved a crystal structure of PbuCas13b complexed with a 36-nucleotide direct repeat sequence and a 5-nucleotide spacer at 1.65 Å (Figure 1, Table 1). The domains and organization are unique amongst other known structures, however the overall shape of PbuCas13b is bilobed, similar to other Class 2 CRISPR effectors (Jinek et al., 2014; Knott et al., 2017; Liu et al., 2017a; 2017b; Nishimasu et al., 2014; Yamano et al., 2016; Yang et al., 2016). Five domains are apparent within the structure: two HEPN domains (HEPN1 and HEPN2), two predominantly helical domains (Helical-1 and Helical-2), and a domain that caps the 3′ end of the crRNA with two beta hairpins (Lid domain) (Figure 1). The bound crRNA is coordinated within the Helical-1, Helical-2, and Lid domains, and the spacer protrudes into a large, positively charged central channel running through the center of the molecule, protected from the solvent (Figure S1A–D).
Figure 1. The crystal structure of PbuCas13b-crRNA Binary Complex.
A. Linear domain organization of PbuCas13b. Active site positioning is denoted by asterisks.
B. The overall structure of PbuCas13b. Two views are rotated 180 degrees from each other. Domains are colored consistent with the linear domain map and the crRNA is colored red. HEL1, Helical 1; HEL2, Helical 2; HEP1, HEPN1; HEP2, HEPN2; N, N-terminus; C, C-terminus
C. Space-filling model of PbuCas13b, each view rotated 180 degrees from each other.
Table 1.
Data collection and refinement statistics of PbuCas13b in complex with RNA.
| Data name | PbuCas13b-Se-peak | PbuCas13b-Se-inflection | PbuCas13b-Se-remote | PbuCas13b-native |
|---|---|---|---|---|
| Ligand | Citrate | Citrate | Citrate | Citrate |
| Data collection | ||||
| Space group | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 90.82, 124.65, 140.73 | 90.86 124.72 140.77 | 90.88, 124.76, 140.79 | 90.82, 124.65, 140.73 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 0.97934 | 0.97958 | 0.97204 | 1.03320 |
| Resolution (Å) | 140.73 – 2.32 (2.47 – 2.32)* | 140.77 – 2.35 (2.53 – 2.35)* | 140.79 – 2.40 (2.62 – 2.40)* | 43.9 – 1.65 (1.71 – 1.65)* |
| No. of unique reflections | 59,325 | 55,600 | 50,274 | 191,617 |
| Rsym | 0.203 (1.73)* | 0.207 (1.69)* | 0.214 (1.74)* | 0.118 (2.89)* |
| Rpim | 0.057 (0.494)* | 0.058 (0.474)v | 0.060 (0.490)* | 0.029 (1.028)* |
| I/σ(I) | 10.2 (1.4)* | 10.4 (1.5)* | 10.1 (1.5)* | 11.5 (0.57)* |
| CC1/2 | 0.996 (0.7)* | 0.996 (0.710)* | 0.996 (0.706)* | 0.998 (0.258)* |
| Completeness (%) | 94.2 (55.8)* | 94.0 (56.6)* | 93.8 (52.0)* | 99.9 (99.2) |
| Redundancy | 13.6 (12.3)* | 13.5 (12.5)* | 13.5 (12.7)* | 13.3 (10.7)* |
| Refinement | ||||
| No. of reflections used | 191,463 | |||
| Rwork / Rfree** | 0.1532/0.1850 | |||
| No. atoms | ||||
| Protein | 16,794 | |||
| RNA | 1,175 | |||
| Citrate | 126 | |||
| Water | 866 | |||
| Wilson B-factor | 31.02 | |||
| B-factors (Å2) | ||||
| Protein | 46 | |||
| RNA | 35 | |||
| Citrate | 57 | |||
| Water | 48 | |||
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.013 | |||
| Bond angles (°) | 1.36 | |||
| Rotamer outliers | 2.85 | |||
| Ramachandran analysis# (%) | ||||
| Favored | 97.88 | |||
| Allowed | 2.02 | |||
| Outliers | 0.10 |
Highest resolution shell is shown in parenthesis.
Rfree was calculated with 5% of the data.
Distribution of dihedral angles in Ramachandran diagram were calculated with MolProbity program (1).
, the Pearson’s correlation coefficient between subsets A and B.
The Domain Architecture of Cas13b
Both HEPN domains are largely alpha-helical: HEPN1 is made of twelve linearly connected α-helices with flexible loops in between the helices. HEPN2 is composed of nine α-helices, several short β-strands, and a β-hairpin with charged residues at the tip, which points towards the active site pocket. HEPN2 rests on HEPN1 such that the active site residues (R1068, N1069, H1073 and R156, N157, H161, respectively) are assembled into a canonical HEPN active site, despite being at the N- and C-terminal extremities of the linear protein (Figure 1) (Liu et al., 2017a; 2017b; Sheppard et al., 2016; Smargon et al., 2017). The HEPN1 domain is connected to the Helical-1 domain by a highly conserved inter-domain linker (IDL) that reaches across the center of a large, positively charged inner channel (Figure 1, Figure S1).
Helical-1 is broken up linearly into three segments by the Helical-2 and Lid domains, and makes extensive sugar-phosphate and nucleobase contacts with the direct repeat RNA (Figure 2). Helical-1 also makes minor interface contacts with both HEPN1/2 and the Lid domains. The Lid domain is mixed α and β secondary structure and caps the 3′ free end of the direct repeat RNA with two charged β-hairpins. The longer of the two β-hairpins reaches across the RNA loop to contact the Helical-1 domain, forming a lid over the free crRNA ends. Positively charged residues from the Lid domain point into a large central channel running through the center of the protein (Figure 1, S1). A side channel penetrating from the outer solvent to the inner channel is formed between a disordered loop (K431 to T438) of the Lid domain and the two HEPN domains (Figure S1). The Helical-2 domain is made of eleven α-helices and wraps under the body of the direct repeat RNA via its connection to Helical-1. Helical-2 interfaces extensively with the HEPN1 domain and makes minor contacts with the extended β-hairpin of the Lid domain. A second positively charged side channel oppositely oriented from the Lid domain between Helical-1 and Helical-2 provides bulk solvent accessibility to the crRNA in the unbound state (Figure S1A,B).
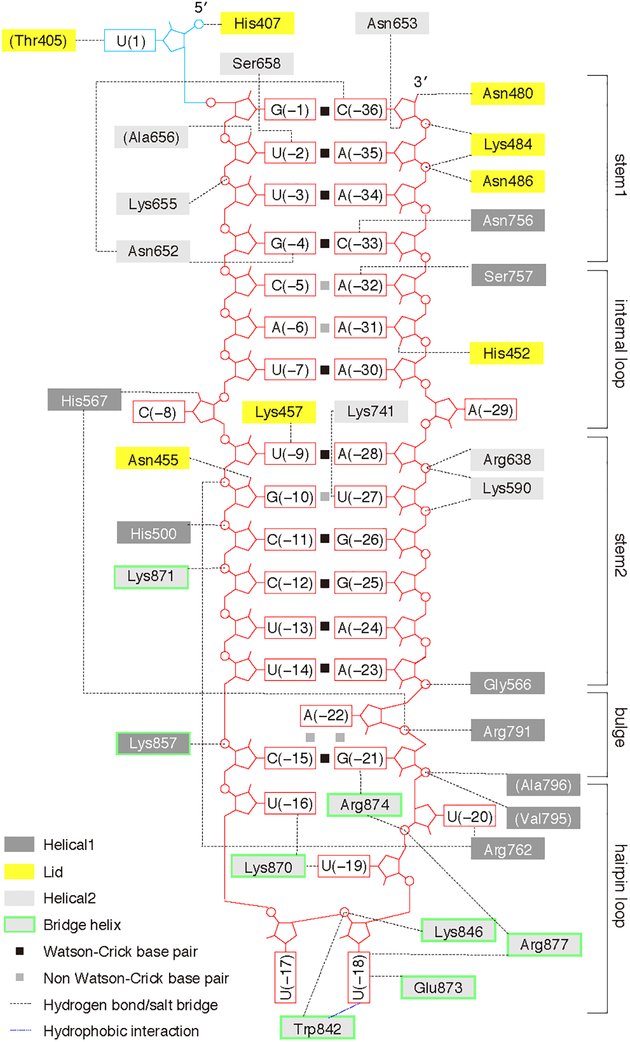
Figure 2. Map of crRNA—protein contacts in PbuCas13b.
A schematic diagram of the intermolecular contacts between PbuCas13b amino acids and crRNA bases.
crRNA Hydrogen Bonding Regulates Cas13b Nuclease Activity
The direct repeat of the crRNA forms a hairpin loop that is recognized by a network of protein interactions from highly conserved residues within the Helical-2 domain (Figure 2, Figure 3A). K870 coordinates with O4 from both U(−16) and U(−19), which indirectly flips U(−17) into the solvent at the hairpin turn, with no visible residue contacts. W842 stacks with the nucleobase of U(−18) while also interacting with the phosphate backbone together with K846. R877 and E873 further stabilize U(−18) though interactions with base N3 and O2 positions. R874 and R762 stabilize the U20 position though sugar O4` and base O2` interactions, respectively.
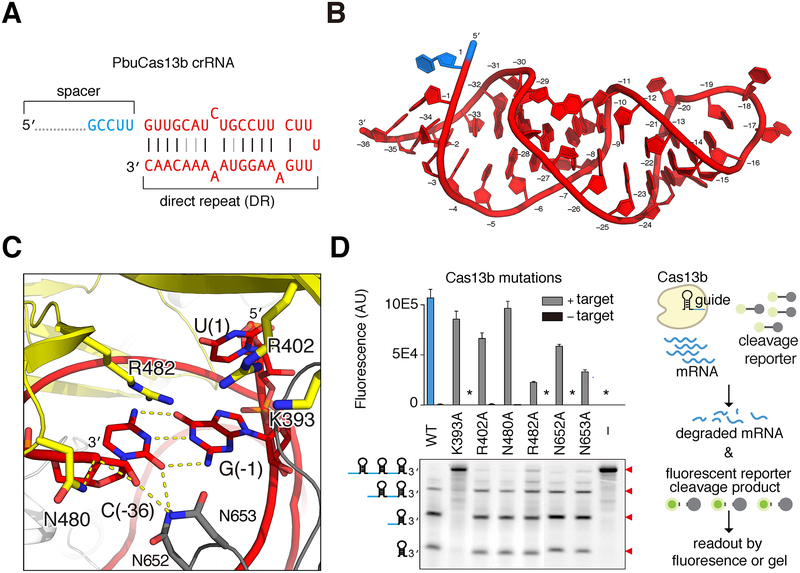
Figure 3. PbuCas13b crRNA recognition and processing.
A. Diagram of crRNA substrate co-crystallized with PbuCas13b. Direct repeat nucleotides are colored red and spacer nucleotides in light blue (full spacer is not shown). Watson-Crick base pairing denoted by black lines; non-Watson-Crick base pairing denoted by gray lines.
B. The structure of crRNA within the crystallized PbuCas13b complex. The coloring is consistent with panel (A) and individual bases are numbered (−1 to −36 in the crRNA, 1 for spacer).
C. Model of the 3′ end of the crRNA showing the catalytic residue K393 of the crRNA processing site and additional PbuCas13b residues that coordinate the crRNA.
D. Analysis of Lid domain residues predicted to coordinate and process crRNA within PbuCas13b. Right, schematic shows Cas13b-mediated RNA degradation. The upper panel shows collateral RNase activity in FLUORESCENT COLLATERAL RNA–CLEAVAGE assays with Lid domain mutants (asterisk indicates nearly undetectable levels of fluorescence); lower panel shows processing of crRNA by these mutants. Cartoons of the expected cleavage products are shown to the left of the gel; cleavage bands and expected sizes indicated by red triangles to the right of the gel.
Electron density for all RNA bases enabled complete model building of the direct repeat RNA. The direct repeat is mostly buried between the two Helical domains and the Lid domain but protrudes slightly into the solvent from Helical-1, explaining how Cas13b is able to utilize an alternate, longer crRNAs which would extend the RNA hairpin length. The overall crRNA structure is a deformed A-form duplex comprising a stem (bases G(−1)–G(−4), C(−33)–C(−36)), loop (C(−5)–U(−8), A(−29)–A(−32)), stem (U(−9)–U(−14), A(−23)–A(−28)), bulge (C(−15), G(−21)), and hairpin loop (U(−16)–U(−20)) architecture (Figure 2, Figure 3B, Figure S2I–N). Helical-1 and Helical-2 mediate direct and indirect recognition of the crRNA hairpin together with the Lid domain, which caps the 3′ free end. Three bases, C(−8), U(−20), and A(−29), are flipped out from the body of the RNA. The backbone carbonyl of T754 stabilizes the flipped out, highly conserved C(−8) base by interacting with the base N4 amine, holding the base in a hydrophobic pocket of highly conserved residues (Y540, 566–571, K751, 753–761) in the Helical-1 and Helical-2 domains. The base flip is further stabilized by interaction between the C(−8) N3` and the sugar (O2`) of U(−7). Changing C(−8) to G or U decreases nuclease activity and destabilizes the protein–RNA complex, as measured in a thermal stability assay (Figure S2P). U(−20) is absolutely conserved in Cas13b direct repeat sequences (Figure S2O) and is coordinated by completely conserved residues, most notably R762 which makes contacts with the nucleobase O2, and R874, which intercalates between G(−21) and U(−20), holding the base out and making contacts with the U(−20) sugar O4`. Mutating U(−20) to G decreases nuclease activity (Figure S2P). In contrast to C(−8) and U(−20), A(−29) is not conserved in Cas13b direct repeat sequences, and the nucleobase is not coordinated by any amino acids. Instead, A(−29) engages in multiplete base pairing with G(−26) and C(−11) (Figure 2K–M) (Lu et al., 2015). A(−29) is tolerant to identity changes to any other base (Figure S2P). Base changes that decrease the thermal stability of the Cas13+crRNA complex also decrease general nuclease activity. Furthermore, stabilizing the RNA hairpin by changing the wobble base pair between U(−27) and G(−10) to a Watson-Crick base pair increases general nuclease activity (Figure S2P). The hairpin loop distal end of the crRNA (−1 to −4 and −33 to −36) is helical and recognized by a combination of base and backbone interactions (Figure 2). Notably, N653 and N652 make critical minor groove direct contacts with U(−2) and C(−36) and coordinate the 5′ and 3′ ends of the hairpin (Figure 3C). Disruption of these base identities or mutation of N653 or N652 to alanine substantially decreases Cas13b activity in vitro and in mammalian interference assays (Figure 3D, Figure S2A and D, Figure S2P). C(−33) is coordinated by N756 via the nucleobase O2 and sugar O2`, and changing this C to A or G abrogates general RNase activity and decreases protein stability (Figure 2, Figure S2P). The crRNA hairpin end (nucleotides −17 to −20) is stabilized by extensive phosphate backbone hydrogen bonding and base interactions (Figure 2). Mutating U(−18) to G abolishes general nuclease activity. U(−19) and U(−20) are also intolerant of G mutations, though other base mutations are tolerated. This suggests that the G O6 or N2 nucleobase atoms disrupt nuclease activity (Figure S2P).
Cas13b Processes pre-crRNA into Mature crRNA Using a Second Nuclease Site
Several Class 2 CRISPR systems process long pre-crRNAs into mature crRNAs (East-Seletsky et al., 2016; Fonfara et al., 2016; Smargon et al., 2017). Cas13b has been shown to process its own crRNA at the 3′ end (Smargon et al., 2017). A number of highly conserved residues are in contact with or nearby the 3′ end of the RNA and potentially form a second, non-HEPN nuclease site (Figure 2, Figure 3C). We mutated to alanine six conserved residues (K393, R402, N480, R482, N652, and N653) nearby the 3′ RNA end and tested these mutants for crRNA processing and target-activated collateral nuclease activity (Figure 3D, Figure S2A–D). K393 when mutated to alanine abrogates RNA processing but retains targeted nuclease activity, confirming the location of a second nuclease site in the Lid domain responsible for crRNA processing (Figure 3C, D and Figure S2A–D).
Mechanism of RNA Targeting
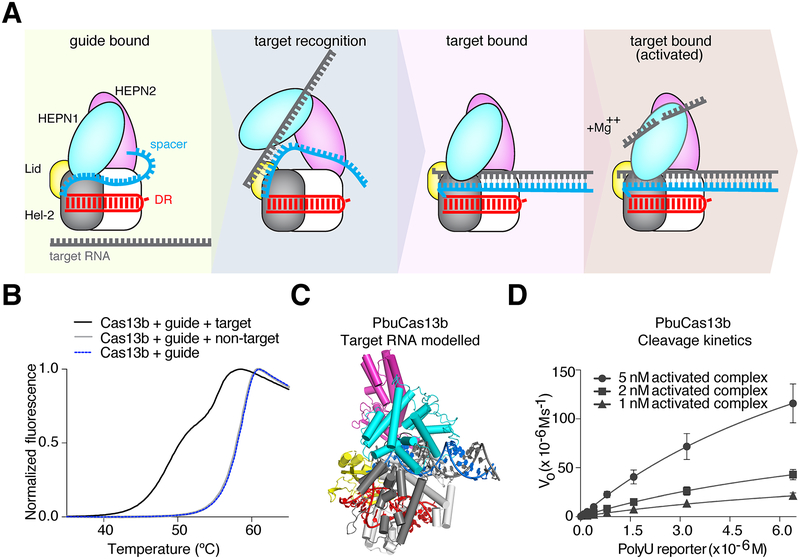
The crRNA is buried within the Cas13b protein in the crystal structure and inaccessible to solvent. Therefore, hybridization of crRNA to target RNA requires a major protein conformation change allowing the target RNA to enter the Cas13b central cavity. The only sterically permissible route from solvent to central channel is between the HEPN1 and Helical-2 domains, indicating the two domains must open to allow target RNA to hybridize with the crRNA. Loops bearing charged amino acids are poised to escort nucleic acid from the solvent into the central channel via this route, which is supported by the finding that mutagenesis of these residues decreases RNA knockdown efficiency (Figure S2H). Mutation of residue D397 on a Lid domain β-hairpin far from the predicted RNA binding pocket but at the interface between HEPN1 and Helical-2 also resulted in decreased RNA knockdown (Figure S2H). To see if residue D397 is also involved in RNA targeting, we tested this mutant with REPAIR RNA editing, which relies on successful targeting of Cas13b to RNA but does not require nuclease activity (Cox et al., 2017). As expected this mutation resulted in a dramatic decrease in RNA editing, suggesting a loss of PbuCas13b targeting and implicating the β-hairpin of the Lid domain in directing the RNA into the central channel (Figure S3E).
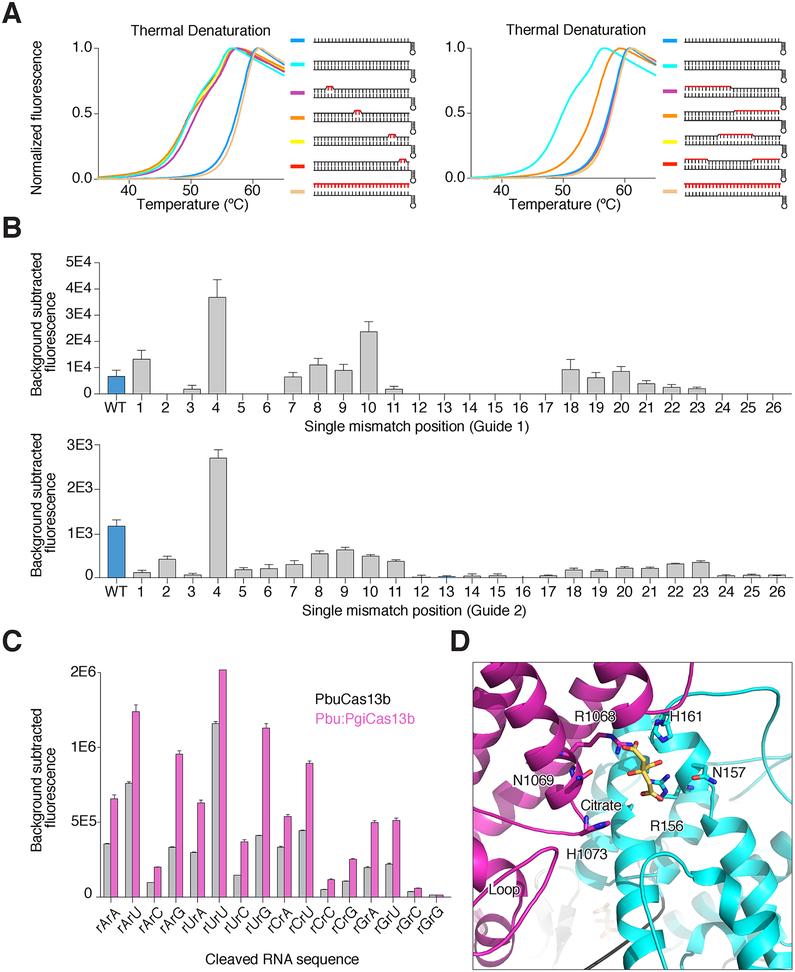
To further understand the mechanism of RNA targeting, we used a thermal denaturation assay to analyze changes in hydrogen bonding caused by altering PbuCas13b conformations (Figure S3G–I). Addition of target RNA to crRNA-bound PbuCas13b shifts the melting curve to a bimodal curve less stable than the binary crRNA-bound PbuCas13b complex, consistent with the structurally predicted central channel entrance between the HEPN1 and Helical domains (Figure S3G–I). The destabilization effect was also seen by limited proteolysis; PbuCas13b in the presence of target RNA was noticeably more susceptible to proteolysis by pepsin and chymotrypsin than with crRNA only (Figure S3J). We posited that the lower Tm curve represents the open conformation of Cas13b as it probes target RNA, whereas the more stable target-bound curve represents the closed state. To test this, we titrated target RNA to Cas13b-crRNA complex. As expected, lower molar ratios of target RNA decrease the presence of the lower Tm curve suggesting the alternate conformation of Cas13b is target dependent and may correspond to the predicted state with an opening between the HEPN1 and Helical domains (Figure S3K). Such an open conformation may actively chaperone target RNA from the solvent to the central channel after initial recognition of target RNA by crRNA. If so, we reasoned that partially matched RNA in the region responsible for initiating the conformational change would induce a corresponding partial decrease in thermal stability. We designed a target RNA with a mismatched 3′-half and repeated the melting experiments to test this hypothesis (Figure 4A). Target RNA with a 3′-half mismatched, but not other variants, partially shifted the Tm curve to the target bound conformation, indicating that the 3′ side of the spacer sequence initiates target recognition, most likely causing an opening between the HEPN1 and Helical-2 domains.
Figure 4. Biochemical characterization of RNA targeting by Cas13b.
A. Thermal melting curves of PbuCas13b complexed with crRNA and target RNA. To the right of the figure legends are cartoons of the substrate used with red lines indicating mismatches (tandem mismatches are shown on the left panel; larger mismatches are shown on the right panel). Curves shifted to the left indicate a destabilized complex.
B. The effect of single nucleotide mismatches between target and crRNA spacer on collateral nuclease activity activation in FLUORESCENT COLLATERAL RNA–CLEAVAGE assays. Two different guide sequences (upper, guide 1; lower, guide 2) are shown.
C. Dinucleotide cleavage preferences measured via FLUORESCENT COLLATERAL RNA–CLEAVAGE assays using wild-type PbuCas13b (gray) and PbuCas13b loop 938–951 swapped with the corresponding loop from PgiCas13b (Pbu:PgiCas13b, purple). See also Figure S5A.
D. Diagram of the active site of PbuCas13b in the crystal structure. Citrate (shown in yellow) is bound in the active site and loop 938–951 is shown in the lower left labeled as ‘Loop’.
Specificity and RNA Interactions within the Central Channel
We hypothesized the target RNA enters the central channel, and that the contacts with the protein are tighter at the 5′ end of the target and more open at the 3′ end (Figure 5C). If true, it would suggest that flexibility within the central channel is linked to the specificity of target binding and collateral activation. To identify the single base mismatch sensitivities along the length of PbuCas13b crRNA, we made single base mutations at each position along the spacer and tested their potential to activate general nuclease activity (Figure 4B). As expected the 3′ side of the crRNA spacer (positions 1 through 11), which is less tightly bound within the central channel, is tolerant of mismatches and even has increased activity with mismatches at some positions in the open part of the central channel. The narrowest region of the central channel, which corresponds to spacer positions 12–17, showed no detectable nuclease activity with single mismatches. Tandem mismatches are not tolerated along the length of the spacer, with the exception of slight tolerance at the first and second positions where the central channel is least constricted (data not shown). However, in denaturation assays, tandem mismatches between spacer and target did not lead to deviation from wild-type curves, indicating that targeting occurs prior to proof-reading within the central channel (Figure 4A). Taken together, these results suggest a model where crRNA-bound Cas13b initially probes the 3′ end of target RNA. If complementarity is found, the HEPN1 and Helical-2 domains open to enable access to the positively charged central channel within the protein, and the rest of the RNA is hybridized (Figure 5A). The bimodal thermal melting curves may reflect the target being repeatedly bound and released as well as present in several conformations in solution (Figure 5B). If binding is reversible, this also suggests a multi-turnover mechanism. Michaelis-Menten kinetics of PbuCas13b cleavage showed that indeed PbuCas13b is multi-turnover (987 turnovers/sec) (Figure 5D). Taken together these results establish a structural model for the mechanism for RNA targeting by Cas13b (Figure 5A).
Figure 5. Structural mechanism of RNA targeting by Cas13b.
A. A cartoon depicting the predicted dynamics of Cas13b. HEPN1 (blue oval) opens to accept the target RNA (gray) into the central channel which then base pairs with the guide RNA (blue). HEPN1 then closes, and cleavage occurs in a divalent metal ion-dependent fashion. Hel-2, Helical 2.
B. Thermal denaturation data showing bimodal curve shift in response to target RNA binding.
C. Model of PbuCas13b crystal structure with ideal A-form RNA docked into the central channel.
D. Kinetics data measuring turnover rate of PbuCas13b nuclease activity in FLUORESCENT COLLATERAL RNA–CLEAVAGE assays (987 turnovers/sec).
Structure Guided Engineering of Cas13b RNA Cleavage Specificity
Cas13 proteins have preference for cleaving specific dinucleotide RNA sequences (East-Seletsky et al., 2017; Gootenberg et al., 2018). We predicted that a β-hairpin loop (residues 938–951) positioned near the active site regulates RNA cleavage preferences (Figure 4D, Figure S4). The identities of the residues in this active site proximal loop and size of it are variable, but this loop is present within most known Cas13bs and positioned between highly conserved residues of the β-strands (Figure S4C, Data S1). To test the involvement of this loop in RNA cleavage specificity, we either deleted the loop or swapped it with amino acid loop sequences from other Cas13b orthologs to see if these mutations altered dinucleotide cleavage preference. The loop deletion mutant did not have any RNase activity. However, cleavage preferences for five of the nine loop swaps that retained activity had altered dinucleotide cleavage preferences, and three mutants had increased UU cleavage preference, demonstrating a role for this loop in cleavage activity and nucleotide recognition (Figure S4A, B). The ability to alter cleavage specificity will enable expansion of nuclease-based assays such as SHERLOCK (Gootenberg et al., 2017; 2018).
A General Strategy for Truncation of Cas13b for AAV packaging
AAV-mediated delivery is commonly used for gene therapies, but REPAIR exceeds the size limit of AAV’s cargo capacity (Cox et al., 2017; Wu et al., 2010). Because HEPN2 appears to be significantly less integrated into the overall structure than the other domains, we reasoned that a Cas13b enzyme with HEPN2 removed would retain RNA targeting activity for base-editing. We showed previously that C-terminal truncations of Prevotella sp. P5–125 (PspCas13b) did not decrease REPAIR activity, consistent with the idea that the HEPN2 domain is not critical (Cox et al., 2017). For additional validation, we used another ortholog of Cas13b, from Porphyromonas gulae (PguCas13b), which is stably expressed and shows high activity in mammalian cells, in contrast to PbuCas13b. Based on alignments between PbuCas13b and PguCas13b, we made truncations to remove the HEPN2 domain, fused it to ADAR, and tested its ability to carry out base editing with the REPAIR system. Not only did these truncated mutants retain RNA targeting, some were significantly more efficient at RNA editing, suggesting improved RNA binding (Figure S5A). HEPN2 partially obstructs this side channel, which may explain how deletion of HEPN2 improves RNA editing efficiency by enabling more sterically favorable binding.
Cas13b is Structurally and Mechanistically Distinct from other CRISPR Nucleases
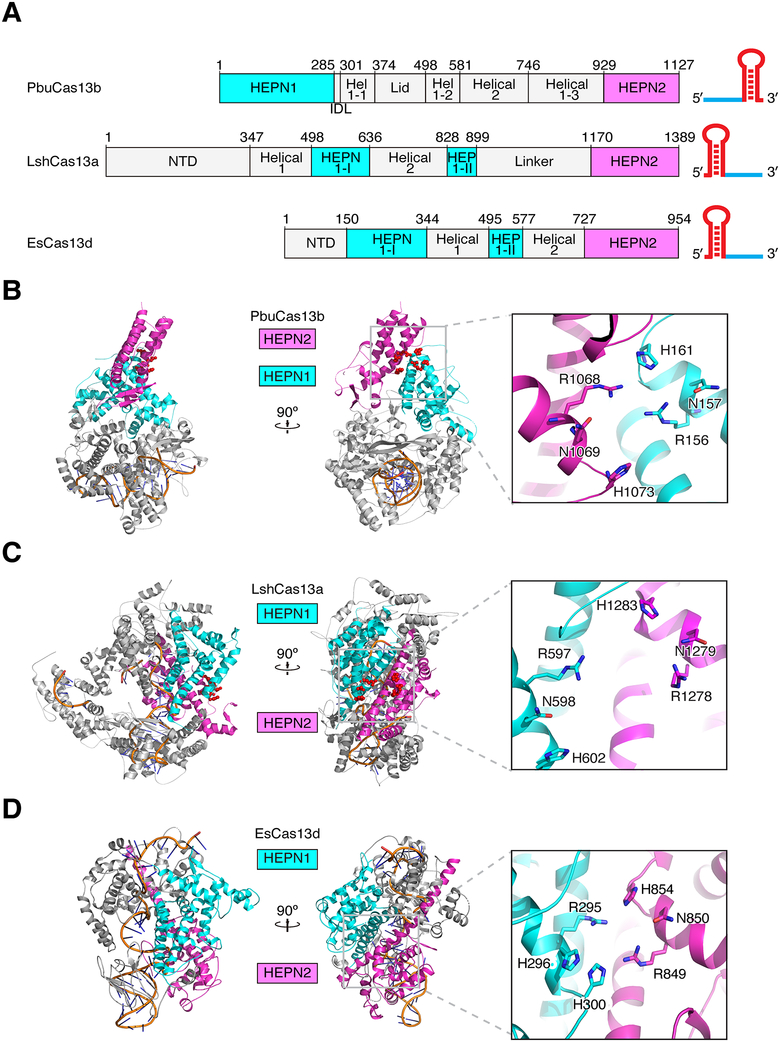
Lastly, we compared Cas13b to the structure of Cas13a and Cas13d (Figure 6) (Liu et al., 2017a; Zhang et al., 2018). Despite functional similarities between these family members, there are no identifiable structural similarities between PbuCas13b and other nucleases beyond the active site architecture. However, an SAS search provided a match to the crystal structure of LbCas12a (previously referred to as LbCpf1) and highlighted a bridge helix like sub-domain within Cas13b (Figure S8) (Milburn et al., 1998). Although this domain is poorly conserved within the Cas13b family, it appears to be a common structural feature with Cas12a that mediates essential nucleic acid contacts. Given the fundamental differences between Cas13b and Cas12a, we postulate that the bridge helix arose as a convergent feature and does not indicate a common ancestor for these two proteins. Nonetheless, we refer to this feature as the bridge helix for consistency with the nomenclature of other Class 2 effectors (Shmakov et al., 2015; Yamano et al., 2016). Finally, to identify similarities to other domains in the protein data bank, the complete PbuCas13b structure as well as isolated domains were queried using the DALI server (Holm and Laakso, 2016). HEPN1 matched to the HEPN2 domain of LshCas13a but no other domains had notable similarity (Liu et al., 2017a).
Figure 6. Comparison of Cas13b with Cas13a and Cas13d.
A. Comparison of the linear domain organization of PbuCas13b, LshCas13a, and EsCas13d. crRNA cartoons are shown to the right depicting architectural differences between type VI systems. Hel 1, Helical 1; HEP, HEPN; NTD, N-terminal domain.
B. – D. Two views of PbuCas13b (B), LshCas13a (PDBID:5WTK) (C), and EsCas13d (PDBID:6E9E) (D) rotated 90°. Insets show active site residues oriented with catalytic residues aligned.
DISCUSSION
The recent discovery of the Cas13 family of single-effector RNA-guided RNases has led to their rapid development as tools for manipulation of the mammalian genome as well as for the detection of nucleic acids. To date, four distinct Cas13 subtypes have been distinguished, Cas13a – d, of which Cas13a, b, and d have been characterized. These three subtypes share a number of features, such as crRNA processing and non-specific nuclease activity under some conditions following target recognition. However, there are clear differences between these enzymes, most notably at the structural level. For example, Cas13a and d share linear domain organization and overall structural similarities, whereas we show here that Cas13b is structurally and mechanistically distinct.
One of the major aspects of the Cas13b structure that distinguishes it from previously reported structures of Cas13a and d is the manner in which the target RNA is engaged. In Cas13a and d, there is a shared solvent-exposed cleft that grasps the target RNA. By contrast, PbuCas13b opens up to allow target RNA access to the central channel. This suggests the target probing dynamics are substantially different between these subtypes; whereas in Cas13a and d, the target binding cleft is persistently open (Liu et al., 2017a; Zhang et al., 2018), there is a conformational change in Cas13b that must precede target RNA binding. It will be interesting to determine if these mechanistic differences contribute to different enzymatic kinetics.
PbuCas13b nuclease activity is preferentially activated by RNA sequences with a protospacer flanking site (PFS) that can be any nucleotide other than C at the 5′ and a slight AA preference at the 3′ of the target (Cox et al., 2017; Smargon et al., 2017). The 5′ PFS nucleotides in our model of RNA binding protrude into the solvent, out of range for protein contacts, and therefore it is not surprising that targeting initiates at the 3′ preferential nucleotides, which are in range to make protein contacts. Higher-resolution structures of the target-bound complex as well as investigations into secondary structure of the RNA may provide deeper insight into why the 5′ PFS confers preferential nuclease activation.
Initial studies of Cas13 enzymes also revealed wide variation in the nucleotide cleavage preference amongst orthologs (Gootenberg et al., 2018). PbuCas13b shows a UU cleavage preference, which we show here can be reengineered by manipulating a single loop near the active site. Although no comparable loop on Cas13a or d is immediately apparent, further exploration of the protein region surrounding the active site may enable engineering of these systems with a similar mutagenic strategy.
The structure of PbuCas13b provides new information on the diversity of the type VI protein family and highlights the substantial differences between Cas13b and other CRISPR systems. We show the structural basis for crRNA recognition and processing and reveal key regulators of nuclease activity in both the guide RNA and protein. Our data suggest a dynamic targeting system that actively escorts the target RNA into the positively charged central channel by adopting an open configuration between the HEPN and Helical domains. Based on our structures of PbuCas13b, we identified important features that regulate RNA targeting and cleavage preference. Insights from this work will enable further rational engineering to improve functionality for RNA targeting specificity, base editing, and new enzymatic tools for nucleic acid detection (Cox et al., 2017; Gao et al., 2017; Gootenberg et al., 2018; Slaymaker et al., 2015).
Star Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Feng Zhang (zhang@broadinstitute.org). Additionally, all reagents are available to the academic community through Addgene and information about the protocols, plasmids, and reagents can be found at the Zhang lab website (http://zhanglab.bio).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HEK293FT cells
Mammalian cell culture experiments were performed in the HEK293FT line (American Type Culture Collection (ATCC)), which was grown in Dulbecco’s Modified Eagle Medium with high glucose, sodium pyruvate, and GlutaMAX (Thermo Fisher Scientific), additionally supplemented with 1× penicillin–streptomycin (Thermo Fisher Scientific) and 10% fetal bovine serum (VWR Seradigm). The sex of HEK293FT cells is female.
BL21 (DE3) cells
BL21 (DE3) cells were purchased from New England Biolabs.
METHOD DETAILS
Protein purification for crystallization
PbuCas13b was expressed in a pET28 based vector with a twin-strep-sumo tag fused at the N-terminal in chemically competent BL21 (DE3) cells purchased from New England Biolabs. Cells with the expression plasmid were grown at 37°C to OD 0.2, then the temperature was switched to 21°C. Growth continued until OD600 was 0.6, then cells were induced with 5 μM IPTG. Cultures were grown for 18–20 hours, and then cells were harvested by centrifugation at 5,000 rpm and frozen at −80°C. Frozen cell paste was homogenized in Buffer A (500 mM sodium chloride, 50 mM Hepes pH 7.5, 2 mM DTT) supplemented with benzonase and lysozyme. The cells were broken by two passes through a microfluidizer at 20,000 psi and cell debris was separated from the soluble fraction by centrifugation at 10,000 rpm. The soluble fraction was passed through Streptactin resin (GE life sciences) and washed with 10 column volumes of Buffer A, followed by 10 column volumes of wash buffer (1 M NaCl, 50 mM Hepes pH 7.5, 2 mM DTT), and finally by 10 column volumes of cleavage buffer (400 mM NaCl, 20 mM Hepes pH 7.5, 2 mM DTT). PbuCas13b was eluted from the resin by addition of 5 mM desthiobiotin (Sigma), then cleaved overnight by sumo protease after being supplemented with 20 mM DTT. After cleavage the protein was passed through a Heparin column, concentrated to 500 μL, and passed over a superdex 200 column (GE life sciences) equilibrated in storage buffer (500 mM NaCl, 10 mM Hepes pH 7.0, 2 mM DTT). Peak fractions were pooled and concentrated to at least 20 mg/ml. Seleno-methionine protein was similarly purified except with 5 mM DTT being supplemented in each buffer. Protein was quantified using Pierce reagent (Thermo).
Crystallization and data collection
RNA substrate was added to PbuCas13b protein at 2:1 molar ratio and dialyzed for 7 hours against dialysis buffer (50 mM NaCl, 10 mM Hepes pH 7.0, 2 mM TCEP). Complexed PbuCas13b+RNA was diluted to 10 mg/ml with dialysis buffer and set up at 20°C by hanging drop vapor diffusion against 165 mM sodium citrate pH 4.6, 5.5% PEG6000, and 2 mM TCEP at varying drop ratios. Rod-shaped crystals grew overnight and reached full size in 1–2 months. Crystals were transferred from the drop to cryo stabilization buffer (140 mM sodium citrate pH 4.6, 5% PEG6000, 35% PEG400), soaked for up to 24 hours, then flash cooled in liquid nitrogen. Selenium crystals for phasing were grown in similar conditions supplemented with 5 mM TCEP.
Native diffraction data from crystals of PbuCas13b and guide RNA were collected at the Advanced Photon Source, Argonne National Labs on beamlines 23-ID-B/D, and anomalous data at the Diamond light source on beamline I04. A small beam was used, either collimated (23-ID) or focused (Diamond) to 20μ, and multiple datasets were collected along the length of the crystal. Anomalous datasets were collected at 0.97934 Å (peak), 0.97958 Å (inflection) and 0.97204 Å (remote) wavelengths. Diffraction data were processed using XDS (Kabsch, 2010a; 2010b) and scaled in aimless (Evans and Murshudov, 2013) implemented in autoPROC toolbox (Vonrhein et al., 2011). The statistics are summarized in Table 1.
Crystal structure solution
The crystal structure of PbuCas13b was solved by multiwavelength anomalous diffraction (MAD) using selenium as anomalous scattering. The position of 27 SeMet sites were determined and refined using phenix.autosol (Adams et al., 2010; Terwilliger et al., 2009). A partial model was built by phenix.autobuild (Terwilliger et al., 2008) using a 3.5 Å resolution experimental map with a figure of merit of 0.35. Cycles of manual rebuilding in Coot (Emsley et al., 2010; Emsley and Cowtan, 2004) and refinement in phenix.refine (Afonine et al., 2012; Echols et al., 2014; Zwart et al., 2008) were done using the selenium experimental map. R-free flags and experimental phases were transferred from the selenium data to high-resolution native data using reflection file editor in PHENIX. These reflections were used for further cycles of rebuilding in Coot and refinement in phenix.refine. Anomalous difference maps were used ensure correct registry. Refinement in phenix.refine used TLS (translation, libration, and screw), positional, and individual B-factor refinement. Citrate geometry restraints were generated by phenix.elbow (Moriarty et al., 2009). The final model contains one polypeptide chain, one RNA nucleotide chain, seven citrates molecules, four tetraethylene glycol (PG4) molecule, two Cl atoms, two Na atoms, and 866 water molecules. Figures were created with PyMol Software (PyMol).
Structure Analysis
RNA structure was analyzed using DSSR (Lu et al., 2015). Protein conservation mapping to the structure was done using the Consurf server (Ashkenazy et al., 2016). Protein secondary structure was analyzed using the PDBSUM webserver (de Beer et al., 2014) (Figure S1E). APBS as part of the PyMol visualization program was used to calculate electrostatics (Jurrus et al., 2018).
Protein Alignment
Alignments of Cas13b enzymes were done using ClustalW or Muscle as implemented in Geneious (Ripma et al., 2014). Neighbor-joining trees were generated using a Jukes-Cantor distance model. Conservation alignments for structure analysis were done on a tree subgroup that successfully matched HEPN domain active site residues to other family members (Figure S5B–D).
Gel Filtration Experiments
For formation of guide complex, 100 μg of PbuCas13b were incubated with two molar equivalents of guide RNA for 20 minutes at room temperature, in 100 μL of buffer (125 mM NaCl, 10 mM HEPES pH 7.0, 2 mM TCEP). For formation of guide-target complex, 100 μg of PbuCas13b and two molar equivalents of guide RNA were incubated together for 20 minutes as above. Two molar equivalents of target RNA were then added to the solution and the mixture was incubated at room temperature for an additional 20 minutes (100 μL total, 125 mM NaCl, 10 mM HEPES pH 7.0, 2 mM TCEP). Apo protein was similarly diluted to 1 μg/μL in a buffer solution of 125 mM NaCl, 10 mM HEPES pH 7.0, 2 mM TCEP. Samples were injected from a 2 mL capillary loop onto a GE Superdex 200 Increase 10/300 GL column and run with 500 mM NaCl, 10 mM HEPES pH 7.0, 2 mM DTT buffer (Figure S7E).
ThermoFluor melting assay
ThermoFluor melting assays were adapted from a previously published protocol (Huynh and Partch, 2015). Samples were prepared to a final volume of 20 μL with 1 μg of PbuCas13b (apo, guide, or guide-target complex, as prepared above) in a solution with a final concentration of 50 mM NaCl, 10 mM HEPES pH 7.0, 6.25× SYPROTM Orange Dye. For MgCl2 cleavage and binding experiments, a final concentration of 6 mM Mg2+ was added to the buffer mix described. For control experiments with non-complementary RNA, 2 molar equivalents of RNA were incubated with the protein complex. Melting experiments were conducted in triplicate on a Roche LightCycler 480 II.
Limited proteolysis
For limited proteolysis experiments, 10 μg of PbuCas13b were incubated with 2 molar excess crRNA or crRNA and target for 30 min at room temperature. 400 μg of protease (buffer, trypsin, chymotrypsin or pepsin) were added, and the mix was incubated for 5 min at 37°C, then placed quickly on ice for 2 min before adding SDS loading buffer and running on a 4–12% acrylamide gel.
Protein expression and purification of PbuCas13b pre-crRNA processing mutants
Alanine mutants at each of the putative crRNA-processing catalytic residues were generated using PIPE-site-directed mutagenesis cloning from the TwinStrep-SUMO-PbuCas13b expression plasmid and transformed into BL21(DE3)pLysE E. coli cells. For each mutant, 2 L of Terrific Broth media (12 g/L tryptone, 24 g/L yeast extract, 9.4 g/L K2HPO, 2.2 g/L KH2PO4), supplemented with 100 μg/mL ampicillin, was inoculated with 15 mL of overnight starter culture and grown until OD600 0.4 −0.6. Protein expression was induced with the addition of 0.5 mM IPTG and carried out for 16 hours at 21°C with 250 RPM shaking speed. Cells were collected by centrifugation at 5,000 RPM for 10 minutes, and paste was directly used for protein purification (10–20 g total cell paste). For lysis, bacterial paste was resuspended via stirring at 4°C in 50 mL of lysis buffer (50 mM Tris-HCl pH 7.5, 500 mM NaCl, 1mM DTT) supplemented with 50 mg Lysozyme, 1 tablet of protease inhibitors (cOmplete, EDTA-free, Roche Diagnostics Corporation), and 500 U of Benzonase (Sigma). The suspension was passed through a LM20 microfluidizer at 25,000 psi, and lysate was cleared by centrifugation at 10,000 RPM, 4°C for 1 hour. Lysate was incubated with 2 mL of StrepTactin superflow resin (Qiagen) for 2 hours at 4°C on a rotary shaker. Resin bound with protein was washed three times with 10 mL of lysis buffer, followed by addition of 50 μL SUMO protease (in house) in 20 mL of IGEPAL lysis buffer (0.2% IGEPAL). Cleavage of the SUMO tag and release of native protein was carried out overnight at 4°C in Econo-column chromatography column under gentle mixing on a table shaker. Cleaved protein was collected as flow-through, washed three times with 5 mL of lysis buffer, and checked on a SDS-PAGE gel.
Protein was diluted two-fold with ion exchange buffer A containing no salt (50 mM Tris-HCl pH 7.5, 1mM DTT) to get the starting NaCl concentration of 250 mM. Protein was then loaded onto a 5 mL Heparin HP column (GE Healthcare Life Sciences) and eluted over a NaCl gradient from 250 mM to 1 M. Fractions of eluted protein (at roughly 700 mM) were analyzed by SDS-PAGE gel and Coomassie staining, pooled and concentrated to 1 mL using 50 MWCO centrifugal filters (Amicon). Concentrated protein was loaded onto a pre-equilibrated size exclusion column and eluted using S200 buffer containing 50 mM Tris-HCl pH 7.5, 500 mM NaCl, 2mM DTT. Monodisperse protein fractions were analyzed by SDS-PAGE gel and Coomassie staining, followed by concentrating and buffer exchange into protein storage buffer (600 mM NaCl, 50 mM Tris-HCl pH 7.5, 1 mM DTT).
Pre-crRNA processing assays
RNA for pre-crRNA processing and nuclease assays were ordered as Ultramers (IDT) and in vitro transcribed using the HiScribe T7 Quick High Yield RNA Synthesis kit (New England Biolabs). RNA was purified with AmpureXP RNA clean up beads and stored at −20°C for further use. For testing pre-crRNA processing, wild-type and mutant protein were incubated with pre-crRNA at four times molar excess of protein relative to the RNA. Pre-crRNA processing was carried out in Cas13b crRNA processing buffer (10 mM TrisHCl pH 7.5, 50 mM NaCl, 0.5 mM MgCl2, 20U SUPERase in (ThermoFisher Scientific), 0.1% BSA) for 30 minutes at 37°C, stopped by adding 2× TBE-Urea gel loading buffer and denatured for 5 minutes at 95°C. Samples were immediately put on ice for 10 minutes before running them on an 15% TBE-Urea gel in 1× TBE buffer at 200 V for 40 minutes. Gel staining was carried out in 1× Sybr Gold in 1× TBE for 15 minutes and imaged on a BioRad gel doc system.
Fluorescent collateral RNA-cleavage assay for pre-crRNA mutants
Detection assays were carried out as quadruplicates with equimolar ratios of PbuCas13b or PbuCas13b mutants, crRNA, and RNA target, in nuclease assay buffer (20 mM HEPES, 60 mM NaCl, 6 mM MgCl2, pH 6.8) with 0.5 μL murine RNase inhibitor (New England Biolabs) and 125 nM of poly-U homopolymer RNA sensor (Trilink). Samples were incubated for 3 hours at 37°C on a fluorescent plate reader equipped with a FAM filter set. Measurements were recorded at 5-minute intervals and data were normalized to the first time-point.
Kinetics and Data analysis
Varying amounts of activated protein complex (5 nM, 2 nM, and 1 nM) were diluted in a mixture containing murine RNase inhibitor (New England Biolabs, 0.5 μL per 20 μL of reaction volume), 1 × CutSmart® Buffer (New England Biolabs). Collateral cleavage was commenced by the addition of a fluorescent RNA reporter (FAM-(rU) 6-BHQ, TriLink) in a range of concentrations: 12.5 nM, 25 nM, 50 nM, 100 nM, 200 nM, 400 nM, 800 nM, 1600 nM, 3200 nM, and 6400 nM. Reactions (20 μL) were performed with four biological replicates on a 384-well plate (Corning®) and fluorescence was measured at 150-second intervals on a fluorescence plate reader (BioTek) for 180 minutes. The first 20 minutes of data at each substrate concentration was fitted by linear regression (GraphPad), and the initial velocity (nM s −1) was plotted against substrate concentration (nM). The data were fitted to the Michaelis-Menten equation (GraphPad) in the form where v is the velocity, Vmax is the maximum velocity, [S] is the substrate concentration, and Km is the Michaelis-Menten constant, or the substrate concentration at which .
Kcat was calculated by dividing the value obtained for Vmax by the enzyme concentration, and the three enzyme conditions (5 nM, 2 nM, and 1 nM of activated complex) were averaged to obtain a value of 987 turnovers per second.
Design and cloning of mammalian constructs for RNA editing
PguCas13b was made catalytically inactive (dPguCas13b) by mutating two arginine and two histidine residues in the catalytic sites of the HEPN domains to alanine (R146A/H151A/R1116A/H1121A). These catalytically inactivated Cas13bs were Gibson cloned into pcDNA-CMV vector backbones containing the deaminase domain of ADAR2(E488Q) fused to the C-terminal end of the Cas13b via a GS linker (Cox et al., 2017). To generate truncated versions, primers were designed to PCR amplify the dCas13b that truncated off 60 bp (20 amino acids) progressively up to 900 bp off of the C-terminal end (15 truncations in total), and these truncated Cas13b genes were Gibson cloned into the pcDNA-CMV-ADAR2 backbone described above. Guide RNAs targeting Cluc were cloned using golden gate cloning into a mammalian expression vector containing the direct repeat sequence for this ortholog at the 3’ end of the spacer sequence destination site, under the U6 promoter.
The luciferase reporter used was a CMV-Cluc (W85X) EF1alpha-Gluc dual luciferase reporter used by Cox et. al. (2017) to measure RNA editing (Cox et al., 2017). This reporter vector expresses functional Gluc as a normalization control, but a defective Cluc due to the addition of the W85X pretermination site.
Mammalian cell culture
Mammalian cell culture experiments were performed in the HEK293FT line (American Type Culture Collection (ATCC)), which was grown in Dulbecco’s Modified Eagle Medium with high glucose, sodium pyruvate, and GlutaMAX (Thermo Fisher Scientific), additionally supplemented with 1× penicillin–streptomycin (Thermo Fisher Scientific) and 10% fetal bovine serum (VWR Seradigm).
All transfections were performed with Lipofectamine 2000 (Thermo Fisher Scientific) in 96-well plates. Cells were plated at approximately 20,000 cells/well 16–18 hours prior to transfection to ensure 90% confluency at the time of transfection. For each well on the plate, transfection plasmids were combined with Opti-MEM I Reduced Serum Medium (Thermo Fisher) to a total of 25 μl. Separately, 24.5 μl of Opti-MEM was combined with 0.5 μl of Lipofectamine 2000. Plasmid and Lipofectamine solutions were then mixed and pipetted onto cells.
RNA knockdown in mammalian cells
To assess RNA targeting in mammalian cells with reporter constructs, 150 ng of Cas13 construct was co-transfected with 300 ng of guide expression plasmid and 45 ng of the dual luciferase reporter construct. Forty-eight hours post-transfection, media containing secreted luciferase was harvested and measured for activity with BioLux Cypridinia and Biolux Gaussia luciferase assay kits (New England Biolabs) on a plate reader (Biotek Synergy H4) with an injection protocol. Signal from the targeted Gluc was normalized to signal from un-targeted Cluc, and subsequently, experiments with PbCas13b mutant luciferase signal were normalized to experiments with guide–only luciferase signal (the average of three bioreplicates). All replicates performed are biological replicates.
REPAIR editing in mammalian cells
To assess REPAIR activity in mammalian cells, we transfected 150 ng of REPAIR vector, 300 ng of guide expression plasmid, and 45 ng of the RNA editing reporter. We then harvested media with the secreted luciferase after 48 hours and diluted the media 1:10 in Dulbecco’s phosphate buffered saline (PBS) (10 μl of media into 90 μl PBS). We measured luciferase activity with BioLux Cypridinia and Biolux Gaussia luciferase assay kits (New England Biolabs) on a plate reader (Biotek Synergy Neo2) with an injection protocol. All replicates performed are biological replicates.
Fluorescent cleavage assay for PbuCas13b loop mutants
Cloning for expression and purification of PbuCas13b loop mutants was carried out by mutagenic PIPE cloning (Klock and Lesley, 2009) using PbuCas13b expression plasmid as template. Protein expression and purification was carried out as described for pre-crRNA mutants. Detection assays were performed with 100 nM purified PbuCas13 and PbuCas13b loop mutants, 100 nM crRNA, quenched fluorescent RNA reporter (250 nM nucleotide reporters (IDT), 0.25 μL murine RNase inhibitor (New England Biolabs), and 100 nM of nucleic acid target in 1× CutSmart Buffer (New England Biolabs). Reactions were allowed to proceed for 3 hr at 37°C on a fluorescent plate reader (BioTek) with fluorescent kinetics measured every 5 min. Cleavage activity of four technical replicates is shown as mean + SEM, unless otherwise stated. For clustering of protein activity based on di-nucleotide motif preference, the mean cleavage activity matrix was clustered using Euclidean distance in R using the heatmap.2 function and exported for graphing in GraphPad Prism 7.
QUANTIFICATION AND STATISTICAL ANALYSIS
Crystallographic statistics were generated during refinement with Phenix and can be found in Table 1. RNA editing experiments are the average of four biological replicates, with the standard deviation shown. RNA knockdown experiments are the average of three biological replicates, normalized to the average of three replicates of guide alone condition. Fluorescence collateral RNA-cleavage assays were performed in technical quadruplicates. For each measurement, initial fluorescence was subtracted to adjust for background fluorescence (background – subtracted fluorescence). Unless otherwise indicated, mean cleavage activities ± S.E.M from four replicates of background-subtracted signals are reported. For clustering of protein activity based on di-nucleotide motif preference, the mean cleavage activity from four technical replicates was used to calculate a distance matrix based on Euclidean distance. This distance matrix was used for clustering with the default ‘complete agglomeration’ method. Distance matrix calculations and clustering were computed using the heatmap.2 function (gplots) in R. The final clustered matrix was exported for graphing in GraphPad Prism 7.
DATA AND SOFTWARE AVAILABILITY
The crystal structure and reflections for PbuCas13b are available from the Protein Data Bank: ID: 6DTD
Supplementary Material
Oligos used in this study.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and Virus Strains | ||
| Rosetta BL21(DE3) pLysE | EMD Millipore | 70956 |
| Biological Samples | ||
| pbCas13b protein | E coli produced | N/A |
| Dulbecco’s Modified Eagle Medium with high glucose, sodium pyruvate, and GlutaMAX | Thermo Fisher Scientific | 10566016 |
| fetal bovine serum | VWR Seradigm | 97068–088 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Twin-Strep-Sump-PbCas13b+flag | E coli produced | N/A |
| Sumo protease (ULP1) | E coli produced | N/A |
| Lipofectamine 2000 | Thermo Fisher Scientific | 11668500 |
| penicillin–streptomycin | Thermo Fisher Scientific | 15140122 |
| Benzonase | Sigma | E1014 |
| PEG6000 | Hampton | HR2–533 |
| Sodium Citrate pH 4.6 | Hampton | HR2-935-05 |
| TCEP | Sigma | 646547 |
| Streptactin resin | GE life sciences | 28935600 |
| IPTG | Sigma | I6758 |
| Opti-MEM I Reduced Serum Medium | Thermo Fisher | 31985062 |
| Critical Commercial Assays | ||
| BioLux Cypridinia and Biolux Gaussia luciferase assay kits | New England Biolabs | E3300 |
| RNA oligo clean and concentrator kit | Zymo research | D4060 |
| NEB Multiplex Small RNA sequencing kit | New England Biolabs | E7300 |
| Deposited Data | ||
| Crystal Structure of PbuCas13b | PDB | 6DTD |
| Experimental Models: Cell Lines | ||
| HEK293FT | American Type Culture Collection (ATCC) | PTA-5077 |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Guide RNA | IDT | N/A |
| nucleotide reporters for fluorescent cleavage assays | IDT | N/A |
| fluorescent RNA reporter (FAM-(rU) 6-BHQ) | TriLink | N/A |
| A list of oligonucleotide sequences are provided in the supplementary materials. | ||
| Recombinant DNA | ||
| CMV-Cluc (W85X) EF1alpha-Gluc dual luciferase reporter | Cox et. al. (2017) | N/A |
| pcDNA-CMV-ADAR2 | Cox et. al. (2017) | N/A |
| Software and Algorithms | ||
| ClustalW or Muscle as implemented in Geneious | Ripma, et al., 2014 | https://www.geneious.com/ |
| DSSR | Lu et al., 2015 | http://x3dna.org |
| XDS | Kabsch, 2010a; 2010b | http://xds.mpimf-heidelberg.mpg.de/ |
| aimless | Evans and Murshudov, 2013 | http://www.ccp4.ac.uk/ccp4/html/aimless.html |
| phenix.autosol | Adams et al., 2010; Terwilliger et al., 2009 | www.phenix-online.org |
| phenix.autobuild | Terwilliger et al., 2008 | www.phenix-online.org |
| Coot | Emsley et al., 2010; Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| phenix.refine | Afonine et al., 2012; Echols et al., 2014; Zwart et al., 2008 | www.phenix-online.org |
| phenix.elbow | Moriarty et al., 2009 | www.phenix-online.org |
| GraphPad Prism 7 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| autoPROC toolbox | Vonrhein et al., 2011 | https://www.globalphasing.com |
| PyMol | PyMol | |
| R version 3.5.1 for MacOSX | CRAN | https://cran.r-project.org/bin/macosx/ |
Acknowledgments
We wish to thank Craig Ogata and the staff at beamline 23ID-D/B at Argonne National Labs and the staff at the Diamond Synchrotron at beamlines I04 and I24 and the Skolkovo Institute of Science and Technology for support of crystallography. We also wish to thank Donald Raymond, Colin Garvie, Chris Lemke, and Doug Daniels at the Broad Institute as well as Robert Grant at MIT for assistance with crystal screening and data processing. Special thanks to Rich Belliveau and Rhiannon Macrae for experimental and manuscript assistance. We also thank the entire Zhang laboratory for support and advice. O.O.A. is supported by a Paul and Daisy Soros Fellowship and a NIH F30 NRSA 1F30-CA210382. F.Z. is a New York Stem Cell Foundation–Robertson Investigator. F.Z. is supported by NIH grants (1R01-HG009761, 1R01-MH110049, and 1DP1-HL141201); the Howard Hughes Medical Institute; the New York Stem Cell, Simons, Paul G. Allen Family, and Vallee Foundations; the Poitras Center for Affective Disorders Research at MIT; the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT; and J. and P. Poitras, and R. Metcalfe. The Novo Nordisk Foundation Center for Protein Research is supported financially by the Novo Nordisk Foundation (Grant NNF14CC0001). This work was also supported by the cryoEM (Grant NNF0024386) and cryoNET (Grant NNF17SA0030214) grants to GM. GM is a member of the Integrative Structural Biology Cluster (ISBUC) at the University of Copenhagen. All reagents are available to the academic community through Addgene and information about the protocols, plasmids and reagents can be found at the Zhang lab website (http://zhanglab.bio).
Declaration of Interests
Broad Institute has filed patent applications relating to CRISPR technology, including CRISPR-Cas13. F.Z. is a scientific founder of and advisor to Arbor Biotechnologies, Beam Therapeutics, Editas Medicine, and Pairwise Plants. F.Z. is a director of Beam Therapeutics.
References
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. (2017). RNA targeting with CRISPR–Cas13. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573–aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, and Ben-Tal N (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucl. Acids Res. 44, W344–W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, and Zhang F (2017). RNA editing with CRISPR-Cas13. Science 550, eaaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer TAP, Berka K, Thornton JM, and Laskowski RA (2014). PDBsum additions. Nucl. Acids Res. 42, D292–D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Burstein D, Knott GJ, and Doudna JA (2017). RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Molecular Cell 66, 373–383.e373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, and Doudna JA (2016). Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols N, Morshed N, Afonine PV, McCoy AJ, Miller MD, Read RJ, Richardson JS, Terwilliger TC, and Adams PD (2014). Automated identification of elemental ions in macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr 70, 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Evans PR, and Murshudov GN (2013). How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr 69, 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovič M, Le Rhun A, and Charpentier E (2016). The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521. [DOI] [PubMed] [Google Scholar]
- Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, and Zhang F (2017). Engineered Cpf1 variants with altered PAM specificities. Nature Biotechnology 163, 759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, and Zhang F (2018). Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science eaaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, et al. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science eaam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, and Laakso LM (2016). Dali server update. Nucl. Acids Res. 44, W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh K, and Partch CL (2015). Analysis of protein stability and ligand interactions by thermal shift assay. Curr Protoc Protein Sci 79, 28.9.1–.9.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. (2014). Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science 343, 1247997–1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurrus E, Engel D, Star K, Monson K, Brandi J, Felberg LE, Brookes DH, Wilson L, Chen J, Liles K, et al. (2018). Improvements to the APBS biomolecular solvation software suite. Protein Sci. 27, 112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010a). Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr 66, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010b). XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klock HE, and Lesley SA (2009). The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol. Biol 498, 91–103. [DOI] [PubMed] [Google Scholar]
- Knott GJ, East-Seletsky A, Cofsky JC, Holton JM, Charles E, O’Connell MR, and Doudna JA (2017). Guide-bound structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme. Nat Struct Mol Biol 24, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, and Hsu PD (2018). Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 173, 665–676.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li X, Ma J, Li Z, You L, Wang J, Wang M, Zhang X, and Wang Y (2017a). The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell 170, 714–726.e10. [DOI] [PubMed] [Google Scholar]
- Liu L, Li X, Wang J, Wang M, Chen P, Yin M, Li J, Sheng G, and Wang Y (2017b). Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities. Cell 168, 121–134.e12. [DOI] [PubMed] [Google Scholar]
- Lu X-J, Bussemaker HJ, and Olson WK (2015). DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucl. Acids Res. 43, e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn D, Laskowski RA, and Thornton JM (1998). Sequences annotated by structure: a tool to facilitate the use of structural information in sequence analysis. Protein Eng. 11, 855–859. [DOI] [PubMed] [Google Scholar]
- Moriarty NW, Grosse-Kunstleve RW, and Adams PD (2009). electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr 65, 1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, and Nureki O (2014). Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell 156, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PyMol. The PyMOL molecular graphics system, Version 1.8. Schrödinger, LLC. [Google Scholar]
- Ripma LA, Simpson MG, and Hasenstab-Lehman K (2014). Geneious! Simplified genome skimming methods for phylogenetic systematic studies: A case study in Oreocarya (Boraginaceae). Appl Plant Sci 2, 1400062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard NF, Glover CVC, Terns RM, and Terns MP (2016). The CRISPR-associated Csx1 protein of Pyrococcus furiosus is an adenosine-specific endoribonuclease. Rna 22, 216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. (2015). Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Molecular Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, et al. (2017). Diversity and evolution of class 2 CRISPR-Cas systems. Nature Reviews Microbiology 15, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, and Zhang F (2015). Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargon AA, Cox DBT, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P, Shmakov S, Makarova KS, et al. (2017). Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Molecular Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, and Hung LW (2009). Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr 65, 582–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, and Adams PD (2008). Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr 64, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, and Bricogne G (2011). Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr 67, 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang H, and Colosi P (2010). Effect of genome size on AAV vector packaging. Mol Ther 18, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y, Fedorova I, Nakane T, Makarova KS, Koonin EV, et al. (2016). Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell 165, 949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, and Scott DA (2018). Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Molecular Cell 70, 327–339.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gao P, Rajashankar KR, and Patel DJ (2016). PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease. Cell 167, 1814–1828.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Konermann S, Brideau NJ, Lotfy P, Wu X, Novick SJ, Strutzenberg T, Griffin PR, Hsu PD, and Lyumkis D (2018). Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d. Cell 175, 212–223.e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. (2008). Automated structure solution with the PHENIX suite. Methods Mol. Biol 426, 419–435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligos used in this study.
Data Availability Statement
The crystal structure and reflections for PbuCas13b are available from the Protein Data Bank: ID: 6DTD