Abstract
Objective
Patients with corticobasal syndrome (CBS) present with heterogeneous clinical features, including asymmetric parkinsonism, dyspraxia, aphasia, and cognitive impairment; to better understand the genetic etiology of this rare disease, we undertook a genetic analysis of microtubule-associated protein tau (MAPT).
Methods
We performed a genetic evaluation of MAPT mutations in 826 neurologically healthy controls and 173 cases with CBS using the Illumina NeuroChip genotyping array.
Results
We identified 2 patients with CBS heterozygous for a rare mutation in MAPT (p.V363I) that is located in the highly conserved microtubule-binding domain. One patient was pathologically confirmed and demonstrated extensive 4-repeat-tau-positive thread pathology, achromatic neurons, and astrocytic plaques consistent with corticobasal degeneration (CBD).
Conclusions
We report 2 CBS cases carrying the rare p.V363I MAPT mutation, one of which was pathologically confirmed as CBD. Our findings support the notion that this rare coding change is pathogenic.
Corticobasal syndrome (CBS) is a rare neurologic disease that presents with heterogeneous motor symptoms and cognitive impairment.1 A high misdiagnosis rate due to clinical heterogeneity limits efforts to extend disease-modifying therapy trials to this patient population. Improving the diagnostic accuracy of complex neurodegenerative syndromes is an important, yet unmet need in the research community.
Although understanding of the genetic underpinnings of CBS is limited, rare mutations in the microtubule-associated protein tau (MAPT) gene are implicated as a cause of CBS and related tauopathy spectrum disorders.2–6 One of these MAPT mutations is the variant p.V363I (rs63750869; c.1087G>A: NM_005910.5), located in the MAPT microtubule-binding domain. Previously described in a small number of patients with clinical tauopathy phenotypes (table 1), the mutation is present at a very low frequency in population databases and is hypothesized to be a disease-causing mutation with decreased penetrance rather than a rare polymorphism.6–10 The rare nature of the mutation makes it difficult to demonstrate disease segregation, and in silico prediction algorithms of this mutation are inconclusive (table e-1, links.lww.com/NXG/A162).
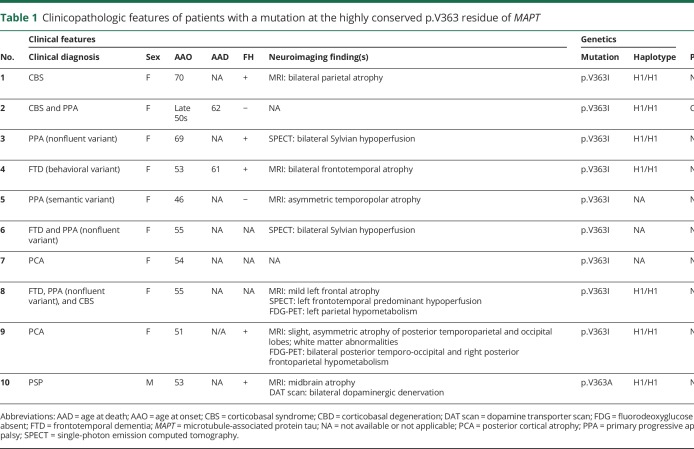
Table 1.
Clinicopathologic features of patients with a mutation at the highly conserved p.V363 residue of MAPT
We describe 2 CBS cases who were found to carry the rare p.V363I MAPT mutation. In addition, we summarize the clinicopathologic features of previously reported cases with a coding mutation at the MAPT p.V363 residue. One of our CBS cases had postmortem confirmation, which found abundant four-repeat tau accumulations consistent with corticobasal degeneration (CBD). As a pathologically confirmed case with this rare missense mutation, this case provides supporting evidence for the pathogenic nature of the p.V363I MAPT mutation.
Methods
Study population
Case 1 is a 73-year-old, right-handed, white woman who presented to the NIH Clinical Center in Bethesda, MD, for participation in genetic research. She was diagnosed with probable CBS based on the consensus criteria.11 A commercial genetic panel (Invitae, San Francisco, CA) that included screening of the genes CHCHD10, DCTN, FUS, GRN, TARDBP, VCP, UBQLN2, TBK1, PSEN1, PSEN2, APP, and MAPT had previously identified that she was a carrier of the MAPT p.V363I variant. Case 2 was identified by querying a research database for the presence of the MAPT p.V363I variant. This database contains genotype information on European-ancestry individuals, including 826 neurologically healthy controls and 961 patients with frontotemporal dementia (FTD) spectrum disorders (n = 772 cases with progressive supranuclear palsy [PSP], n = 173 patients with CBS/CBD, n = 41 patients with FTD; sample characteristics are summarized in table e-2 [links.lww.com/NXG/A162]; source of samples and number of samples per disease are described in table e-3). Case 2 was clinically diagnosed with hemiparkinsonism, primary progressive aphasia, and probable CBS.
Standard protocol approvals, registrations, and patient consents
The study was approved by the respective institutional review boards. Written informed consent for research participation was obtained from all participants.
Genetic analysis and validation
For each participant, DNA was extracted from blood or brain tissue using standard methods and followed by genotyping on the NeuroChip platform (Illumina, San Diego, CA). This affordable genotyping array contains a tagging single nucleotide polymorphism backbone combined with high-yield custom content that allows for rapid screening of ∼180,000 mutations and risk variants previously implicated in neurologic diseases, including the MAPT p.V363I variant. The detailed contents of this versatile genotyping platform have been described elsewhere.12 The MAPT p.V363I mutation was only present in 2 patients (henceforth referred to as case 1 and case 2), and we validated the mutation via direct Sanger sequencing using the following primers: forward 5′-GTGGCCAGGTGGAAGTAAAA, reverse 5′-ACATCCAGCCAGTCAACACA. To rule out other possible pathogenic mutations in these 2 patients, we assessed the NeuroChip data for damaging progranulin (PGRN) gene mutations. We also performed repeat-primed PCR screening of the C9orf72 repeat using methods described elsewhere.13 APOE genotypes were determined by extracting rs7412 and rs429358 as previously described.12 MAPT haplotype status was determined by imputation of the polymorphism rs1052553 (R2 = 0.99494), with the “A” allele determining the H1 haplotype and the “G” allele segregating with the H2 haplotype.14
Bioinformatic analysis
To better understand the effects of the p.V363I variant, a systematic literature review was conducted and summarized in table 1. In silico predictive tools (SIFT, PolyPhen2, FATHMM-XF, M-CAP, MutationTaster, CADD, ClinVar, and ClinPred) were applied to classify the MAPT p.V363I mutation.15–21 Sequence conservation analyses were performed in T-Coffee.22 A previously described, cryo-electron microscopy structure of the tau protofibril was used for 3-dimensional protein modeling (figure 1).23 Allele frequency differences between CBS cases and neurologically healthy controls were determined using a Fisher exact test with a significance threshold of 0.05.
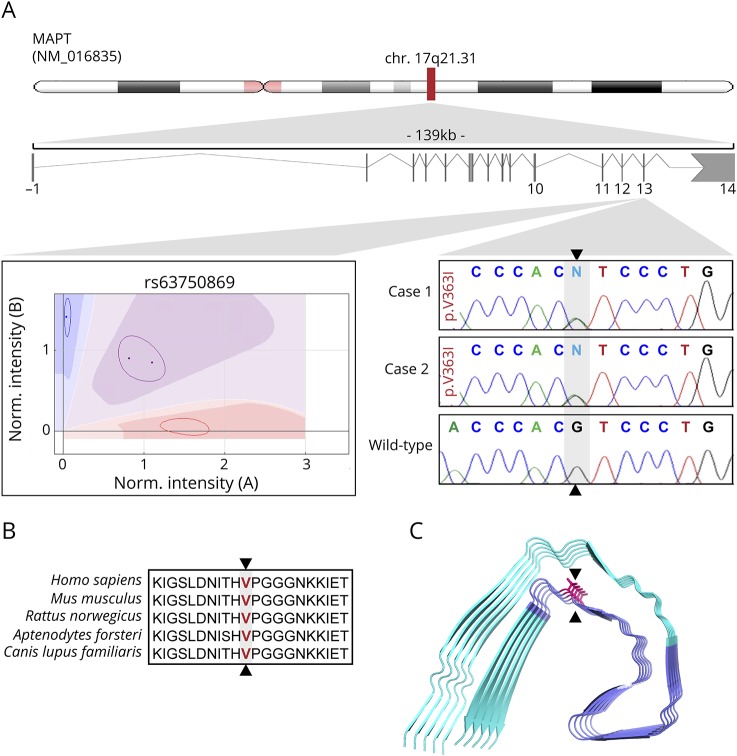
Figure 1. This schematic representation illustrates the location of the MAPT p.V363I mutation.
A Cartesian genotype plot and electropherograms of this mutation are shown for both CBS cases compared with a control (A). The mutation is localized within the highly conserved microtubule-binding domain (B). (C) Position of the p.V363I mutation (arrow heads) within the microtubule-binding domain (highlighted in purple) relative to the 3-dimensional reconstruction of the tau protofibril. MAPT = microtubule-associated protein tau.
Neuropathology
The brain of case 2 was pathologically evaluated at the Neurological Tissue Bank of the IDIBPAS Biobank in Barcelona, Spain, after obtaining written informed consent from the patient's relatives for use of tissue for diagnostic and research purposes. Hematoxylin and eosin staining was performed after standard formalin fixation and paraffin block sectioning of multiple cortical and subcortical brain areas. Immunohistochemistry was performed using phospho-tau (Ser202 and Thr205) monoclonal antibodies (AT8; 1:2000; Thermo Scientific, Rockford, IL) and anti-4R-tau (RD4) antibodies. In addition, selected areas were stained for ßA4-amyloid (6F/3D 1:400; Dako, Glostrup, Denmark), α-synuclein (KM51 2:200; Novocastra, Newcastle upon Tyne, UK), and TDP43 protein (2E2-E3 1:500; Abnova, Taipei, Taiwan) for identification of concomitant pathologies.
Data availability
Deidentified data are available upon request from qualified investigators.
Results
Genetic characteristics
In a cohort of 173 CBS cases, we identified 2 patients who were heterozygous for the rare p.V363I (c.1087G>A: NM_005910.5) mutation located in the highly conserved microtubule-binding domain of MAPT (Fisher exact test comparing CBS cases with neurologically healthy controls p = 0.0299). Both patients were homozygous for the H1 MAPT haplotype and carried no pathogenic mutations in PGRN or C9orf72. The patients' APOE genotypes were ε3/ε3. The MAPT p.V363I mutation was absent in ∼1,800 additional samples, including 826 neurologically healthy controls and 984 cases with diverse frontotemporal degeneration spectrum disorders. Bioinformatic predictions demonstrated that SIFT, PolyPhen2, MutationTaster, and ClinPred categorized the variant as tolerated and benign, whereas ClinVar, FATHMM-XF, M-CAP, and CADD predictions suggested a likely pathogenic mutation (table e-1, links.lww.com/NXG/A162).
Clinicopathologic features
Case 1 is a 73-year-old, right-handed, white woman with a medical history of hypertension, coronary artery disease, an old segmental left parietal stroke at age 54 years that resolved without residual neurologic deficits, and major depressive disorder. She was diagnosed with CBS at age 70 years after developing progressive right-sided impairment of her dexterity, slowed gait, and imbalance resulting in backward falls. A levodopa trial up to a maximum dose of 450 mg daily yielded no benefits. Over the course of 3 years, she gradually developed dysarthria, severe gait dysfunction rendering her wheelchair-bound, asymmetric parkinsonism, hand dystonia, apraxia, impaired word retrieval, and executive dysfunction. Her neurologic examination demonstrated bradyphrenia with a tendency to perseverate. She had severe ideomotor apraxia that was more prominent in her dominant hand. She was neglecting her right-sided space. Her speech was moderately dysarthric. Cranial nerve examination showed slowed, hypometric saccades (vertical more than horizontal), severe axial and right-sided rigidity with only mild rigidity on the left, bradykinesia, and dystonia with high-frequency/low-amplitude tremor in her right hand. She had agraphesthesia and astereognosis in her right hand. Reflexes were brisk throughout. Primitive reflexes, including grasp and palmomental reflexes, were present. She was unable to stand without assistance and would spontaneously fall without support. MRI of the brain demonstrated bilateral parietal atrophy with proportional, ex vacuo dilatation of the lateral ventricles. Her family history was notable for parkinsonism in her father (age at onset ∼65 years). No DNA was available from her father to test for segregation. The patient is alive after a 3-year disease duration.
Case 2 was a white woman who presented in her late 50s with primary progressive aphasia, left-sided parkinsonism, and CBS. The disease progressed to complete anarthria and severe dysphagia. She died at age 62 years. Clinical data on this case were limited. She had no known family history of dementia. The patient's neuropathologic findings were notable for widespread, 4-repeat-tau-positive inclusions in cortical and subcortical regions, including neurons and glial cells, consistent with CBD (figure 2). Frequent achromatic neurons were detected in frontal, parietal, and cingular cortices. These were associated with focal superficial spongiosis, diffuse neuronal loss, astrogliosis, and microglial activation in cortical areas, including the motor cortex, and diffuse gliosis of the underlying white matter. Prominent neuronal loss was noted in the globus pallidus and in the substantia nigra. Immunohistochemistry revealed astrocytic plaques, abundant pretangles, ballooned neurons, and neuropil threads in the cortex, abundant threads and pretangles in her basal ganglia, and prominent white matter pathology with widespread threads and coiled bodies involving also the brainstem. Remarkably, there was also prominent involvement of the hippocampus, including the granule cells of the dentate gyrus, without grain pathology. Co-comitant pathologies included a moderate amount of diffuse ßA4-amyloid deposits and few cored plaques in cortical areas, as well as few neuronal and glial cytoplasmic TDP43 protein inclusions in the globus pallidus, without frontal, temporal, or hippocampal involvement. No α-synuclein aggregates were identified.
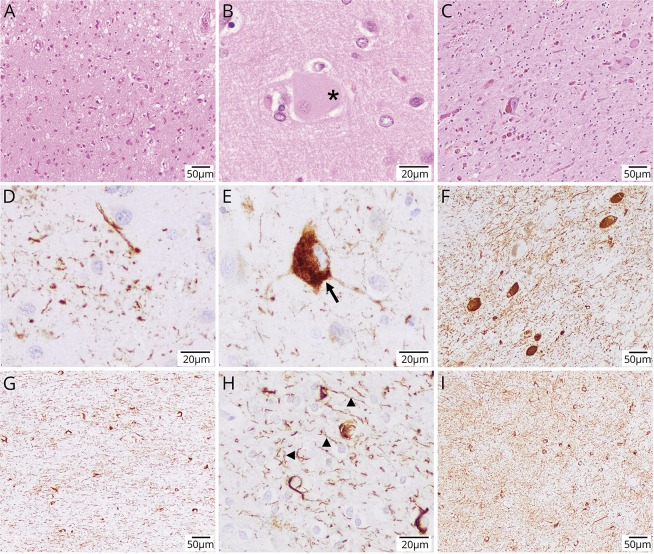
Figure 2. These images showcase the pertinent neuropathologic findings of case 2.
Hematoxylin and eosin staining shows superficial spongiosis in the postcentral region (A), a large achromatic or ballooned cell (highlighted by asterisk in B), and prominent nigral degeneration with severe neuronal loss and abundant extracellular neuromelanin pigment (C). (D–I) Abnormal pTau (AT8) and 4-repeat-tau-positive protein deposition on immunohistochemistry. Notable abnormal histopathologic findings included astrocytic plaques (D), frequent pretangles with some focal cytoplasmic condensations (E), tangles, pretangles, and abundant threads in the substantia nigra (F), very abundant threads (arrow heads) and coiled bodies (arrow) in the white matter (G and H), and abundant threads and pretangles in the striatum (I), overall consistent with the neuropathologic findings observed in corticobasal degeneration. Magnification scale bars are indicated in the bottom right corner of each panel.
Discussion
We describe 2 CBS cases carrying the rare p.V363I MAPT mutation located in the conserved microtubule-binding domain. One of the 2 cases was pathologically confirmed, demonstrating widespread, 4-repeat-tau-positive neuronal and glial pathology consistent with CBD. This report describes the pathology present in a p.V363I MAPT mutation carrier providing further support for the notion that this variant is likely disease causing. To date, this coding mutation has been described in 7 neurodegenerative disease cases with heterogeneous presentations, including FTD, primary progressive aphasia, and posterior cortical atrophy (table 1).6,8–10 Another mutation at the same residue (p.V363A) has been described in a single case with clinically diagnosed PSP.10 Of interest, all cases were female, had ε3/ε3 APOE genotypes, and were homozygous for the MAPT H1 haplotype. The average age at onset was 57 years, ranging from 46 to 70 years. The 2 CBS cases presented here extend the disease onset. This wide age spectrum is consistent with the pattern seen in tauopathies associated with MAPT mutations and could indicate a decreased, age-related penetrance.24
Among the cases, the initial disease manifestations were quite varied, including gait disturbances, memory deficits, and personality changes. This heterogeneity is not unusual for patients with MAPT mutations.25 The p.V363I mutation is present in 3 of 62,784 people in the NHLBI TopMed Bravo database (bravo.sph.umich.edu/freeze5/hg38/; allele frequency: 0.0000239; date accessed: October 14, 2018) and in 2 of 60,702 individuals in ExAC (allele frequency: 0.0000167, data accessed: October 14, 2018).26 The very rare presence within population databases might be explained by incomplete penetrance and late disease onset. In addition, limited in vitro analyses in 1 case demonstrated that this mutation leads to an increased propensity for microtubule polymerization and the formation of tau protein oligomers.10 Because of the lack of familial genetic data, we were not able to test for disease segregation, and this has not yet been reported by other investigators.
We present a pathologically confirmed patient with a p.V363I MAPT mutation. The neuropathologic findings of this case were consistent with CBD. An additional p.V363I carrier was identified with a CBS phenotype. This mutation was absent in neurologically healthy controls. Considering previous reports on mutation carriers with information about sequence conservation, functional studies, and pathologic confirmation, we nominate the MAPT p.V363I change as a likely disease-causing mutation. Identifying additional cases with this mutation will be important to understand the natural history and penetrance of this familial disease.
Acknowledgment
The authors thank all the subjects who donated their time and biological samples to be a part of this study. They thank the NIH NeuroBrainBank and the IDIBAPS Brain Bank, Barcelona, Spain, for contributing brain tissue samples. This study used tissue samples and data provided by the Michigan Brain Bank, the Michigan Alzheimer's Disease Center (5P30 AG053760), and the Protein Folding Disorders Program. This study used samples from the NINDS Repository at Coriell (catalog.coriell.org) and clinical data. The authors are grateful to the Banner Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona, for the provision of human brain tissue. The Brain and Body Donation Program is supported by the NINDS (U24 NS072026 National Brain and Tissue Resource for Parkinson's Disease and Related Disorders), the National Institute on Aging (P30 AG19610 Arizona Alzheimer's Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer's Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-0901, and 1001 to the Arizona Parkinson's Disease Consortium), and the Michael J. Fox Foundation for Parkinson's Research. The authors thank the Kathleen Price Bryan Brain Bank at Duke University Medical Center, the Harvard Brain Bank, and the Georgetown University Brain Bank for the provision of tissue and DNA samples. Tissue for this study was provided by the Newcastle Brain Tissue Resource, which is funded in part by a grant from the UK Medical Research Council (G0400074), by NIHR Newcastle Biomedical Research Centre and Unit awarded to the Newcastle upon Tyne NHS Foundation Trust and Newcastle University, and as part of the Brains for Dementia Research Programme jointly funded by Alzheimer's Research UK and Alzheimer's Society. The authors thank members of the North American Brain Expression Consortium for providing DNA samples on neurologically healthy controls. Tissue samples for genotyping were provided by the Johns Hopkins Morris K. Udall Center of Excellence for Parkinson's Disease Research (NIH P50 NS38377) and the Johns Hopkins Alzheimer Disease Research Center (NIH P50 AG05146).
Glossary
- CBD
corticobasal degeneration
- CBS
corticobasal syndrome
- FTD
frontotemporal dementia
- MAPT
microtubule-associated protein tau
- PGRN
progranulin
- PSP
progressive supranuclear palsy
Appendix. Authors
Study funding
This work was supported in part by the Intramural Research Programs of the NINDS and the National Institute on Aging (NIA) (project numbers Z01-AG000949 and 1ZIA NS003154). This study was partially supported by a CurePSP research grant (Cure PSP grant no.: 515-14; 2013-2015) to PP. EG received support from a grant from the Marató de TV3 (grant no. 20141610).
Disclosure
Disclosures available: Neurology.org/NG.
References
- 1.Kouri N, Murray ME, Hassan A, et al. . Neuropathological features of corticobasal degeneration presenting as corticobasal syndrome or Richardson syndrome. Brain 2011;134:3264–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poorkaj P, Bird TD, Wijsman E, et al. . Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol 1998;43:815–825. [DOI] [PubMed] [Google Scholar]
- 3.Hutton M, Lendon CL, Rizzu P, et al. . Association of missense and 5'-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998;393:702–705. [DOI] [PubMed] [Google Scholar]
- 4.Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci U S A 1998;95:7737–7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickering-Brown SM, Rollinson S, Du Plessis D, et al. . Frequency and clinical characteristics of progranulin mutation carriers in the Manchester frontotemporal lobar degeneration cohort: comparison with patients with MAPT and no known mutations. Brain 2008;131:721–731. [DOI] [PubMed] [Google Scholar]
- 6.Bessi V, Bagnoli S, Nacmias B, Tedde A, Sorbi S, Bracco L. Semantic dementia associated with mutation V363I in the tau gene. J Neurol Sci 2010;296:112–114. [DOI] [PubMed] [Google Scholar]
- 7.Rossi G, Conconi D, Panzeri E, et al. . Mutations in MAPT gene cause chromosome instability and introduce copy number variations widely in the genome. J Alzheimers Dis 2013;33:969–982. [DOI] [PubMed] [Google Scholar]
- 8.Anfossi M, Bernardi L, Gallo M, et al. . MAPT V363I variation in a sporadic case of frontotemporal dementia: variable penetrant mutation or rare polymorphism?. Alzheimer Dis Assoc Disord 2011;25:96–99. [DOI] [PubMed] [Google Scholar]
- 9.Munoz DG, Ros R, Fatas M, Bermejo F, de Yebenes JG. Progressive nonfluent aphasia associated with a new mutation V363I in tau gene. Am J Alzheimers Dis Other Demen 2007;22:294–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossi G, Bastone A, Piccoli E, et al. . Different mutations at V363 MAPT codon are associated with atypical clinical phenotypes and show unusual structural and functional features. Neurobiol Aging 2014;35:408–417. [DOI] [PubMed] [Google Scholar]
- 11.Armstrong MJ, Litvan I, Lang AE, et al. . Criteria for the diagnosis of corticobasal degeneration. Neurology 2013;80:496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blauwendraat C, Faghri F, Pihlstrom L, et al. . NeuroChip, an updated version of the NeuroX genotyping platform to rapidly screen for variants associated with neurological diseases. Neurobiol Aging 2017;57:247 e9–247 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renton AE, Majounie E, Waite A, et al. . A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011;72:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poorkaj P, Grossman M, Steinbart E, et al. . Frequency of tau gene mutations in familial and sporadic cases of non-Alzheimer dementia. Arch Neurol 2001;58:383–387. [DOI] [PubMed] [Google Scholar]
- 15.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 2003;31:3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jagadeesh KA, Wenger AM, Berger MJ, et al. . M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet 2016;48:1581–1586. [DOI] [PubMed] [Google Scholar]
- 17.Adzhubei IA, Schmidt S, Peshkin L, et al. . A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landrum MJ, Lee JM, Benson M, et al. . ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 2016;44:D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers MF, Shihab HA, Mort M, Cooper DN, Gaunt TR, Campbell C. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics 2018;34:511–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alirezaie N, Kernohan KD, Hartley T, Majewski J, Hocking TD. ClinPred: prediction tool to identify disease-relevant nonsynonymous single-nucleotide variants. Am J Hum Genet 2018;103:474–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rausch T, Emde AK, Weese D, Doring A, Notredame C, Reinert K. Segment-based multiple sequence alignment. Bioinformatics 2008;24:i187–192. [DOI] [PubMed] [Google Scholar]
- 23.Fitzpatrick AWP, Falcon B, He S, et al. . Cryo-EM structures of tau filaments from Alzheimer's disease. Nature 2017;547:185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irwin DJ. Tauopathies as clinicopathological entities. Parkinsonism Relat Disord 2016;22(suppl 1):S29–S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghetti B, Oblak AL, Boeve BF, Johnson KA, Dickerson BC, Goedert M. Invited review: frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging. Neuropathol Appl Neurobiol 2015;41:24–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lek M, Karczewski KJ, Minikel EV, et al. . Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Deidentified data are available upon request from qualified investigators.