Abstract
Objective
Given the known strong relationship of DNA methylation with environmental exposure, we investigated whether brain regions affected in Parkinson disease (PD) were differentially methylated between PD cases and controls.
Methods
DNA chip arrays were used to perform a genome-wide screen of DNA methylation on the dorsal motor nucleus of the vagus (DMV), substantia nigra (SN), and cingulate gyrus (CG) of pathologically confirmed PD cases and controls selected using the criteria of Beecham et al. Analysis examined differentially methylated regions (DMRs) between cases and controls for each brain area. RNA sequencing and pathway analysis were also performed for each brain area.
Results
Thirty-eight PD cases and 41 controls were included in the analysis. Methylation studies revealed 234 significant DMR in the DMV, 44 in the SN, and 141 in the CG between cases and controls (Sidak p < 0.05). Pathway analysis of these genes showed significant enrichment for the Wnt signaling pathway (FDR < 0.01).
Conclusions
Our data suggest that significant DNA methylation changes exist between cases and controls in PD, especially in the DMV, one of the areas affected earliest in PD. The etiology of these methylation changes is not yet known, but the predominance of methylation changes occurring in the DMV supports the hypothesis that vagus nerve function, perhaps involving the gastrointestinal system, is important in PD pathogenesis. These data also give independent support that genes involved in Wnt signaling are a likely factor in the neurodegenerative processes of PD.
Parkinson disease (PD) is the second most common neurodegenerative disorder affecting older adults. The clinical presentation includes bradykinesia, resting tremor, and rigidity.1 Monogenic mutations for PD have been identified that greatly increase the risk of PD.2 However, 90% or more of PD cases are idiopathic.
Epigenetics is a potentially important factor contributing to PD risk, particularly since environmental factors have been associated with an increased risk of developing PD.3–5 However, little work has been done to explore the potential epigenetic contribution to PD. DNA methylation, the mostly studied form of epigenetic modification, has been primarily investigated in PD within select candidate genes.6,7 Several studies have found differential methylation in SNCA and MAPT.8–11 Furthermore, significant changes in DNA methylation were identified in multiple genes in both blood10,12,13 and brain.10 Relevant to the current study, dysregulation of Wnt signaling due to methylation was observed in the frontal cortex and midbrain sections of PD brains.14
Here, we report an initial analysis of the genome-wide methylation profile in the dorsal motor nucleus of the vagus (DMV), substantia nigra (SN), and cingulate gyrus (CG) of pathology-confirmed PD cases compared with age- and sex-matched, pathology-confirmed controls. Each of these brain regions represents the location of neuropathologic changes in PD at different stages of the disease. We found that patients with PD have significant DNA methylation changes in these 3 brain regions, and find the largest number of significant DNA methylation changes are found in the DMV. Furthermore, pathway analysis in the DMV of patients with PD supports the involvement of the Wnt pathway in the pathophysiology of PD.
Methods
Brain samples were obtained from the autopsy program of the University of Miami, Morris K. Udall Parkinson Disease Center of Excellence (n = 11), the NIH Neurobiobank (n = 12), and the Pacific Udall Center Neuropathology Core (n = 22). Our initial discovery sample set consisted of 22 PD pathology-confirmed cases and 24 pathology-confirmed controls. The replication data set had 16 PD pathology-confirmed cases and 17 pathology-confirmed controls, providing a total of 38 PD cases and 41 controls for the joint analysis. All brains were from non-Hispanic white men, aged >60 years at death with no premortem diagnosis of cancer, postmortem interval of less than 24 hours, and a Braak neurofibrillary tangle stage ≤IV. Neuropathologic evaluations were performed in every case confirming Lewy body (LB) PD, and followed the filtering criteria of Beecham et al.,15 for inclusion into the study. Staging was performed according to the Braak hypothesis of LB staging in PD. PD samples included brains with Braak stages ranging from 3 to 5 (36% stage 3, 37% stage 4, and 27% stage 5). Tissue punches of 0.3 cm in diameter were taken from fresh, quick-frozen 1 cm coronal sections from each region studied, e.g., the medulla oblongata containing the DMV, the SN, and the anterior CG. Tissue punches corresponded to approximately an average of 50% of the DMV, 30% of the SN, and 5% of the CG. These brain regions represent the location of neuropathologic changes in PD at different stages of the disease. One of the earliest regions to display LBs and Lewy neurites, the hallmarks of PD, is the DMV, with the characteristic motor symptoms occurring later, when over 50% of the dopaminergic neurons of the SN's pars compacta are lost.1 The DMV has a direct connection to the environment through the vagus nerve's innervation of the gastrointestinal tract, of which dysfunction in PD has been reported in multiple studies.16,17 The CG generally develops later pathologic changes in PD.18 A 40-µm section of the tissue punch containing the DMV was used for anatomic verification.
Standard protocol approvals, registrations, and patient consents
The authorization for retention of the brain, review of medical records, and informant interviews were approved by the respective institutional review boards.
Profiling of CpG methylation using 450k/850k array
Genomic DNA (500 ng) was bisulfite modified (EZ-96 DNA Methylation Kit; Zymo Research, Orange, CA) as per manufacturers' instructions. For analysis of CG dinucleotide (CpG) methylation, both the Illumina Infinium HumanMethylation450 BeadChip and the Infinium MethylationEPIC (850K array) Beadchip were used (Illumina, San Diego, CA) because of discontinuation of the 450 BeadChip by the manufacturers.
Statistical analysis of methylation data
Raw intensity files were processed using the methylation analysis software RnBeads.19 Because all samples were male, we did not filter sex-specific probes. Beta-Mixture Quantile normalization was used for intra-array normalization of beta values, which are the ratio of the methylated signal intensity to the sum of both methylated and unmethylated signals after background subtraction. The beta values were then logit transformed to attain M values for statistical analysis.20 Unsupervised hierarchical clustering of the methylome data revealed 1 outlier from the CG group, which was removed from further analysis.
For the analysis of individual CpGs, linear regression models were used to test differential methylation between case-control status adjusting for age at death (AAD) and methylation chip effects. To account for additional unmeasured confounding factors such as cellular composition, we included surrogate variables (SVs) estimated from independent surrogate variable analysis (iSVA) as covariate variables.21 iSVA estimates major independent components (ICs) in genome-wide DNA methylation patterns. We tested each IC with status using a T test. The significant ICs (IC3 for DMV and IC7 for SN), which could be confounders of the association between case-control status and differential methylation, were then included in the linear model: M value ∼PD + AAD + array + IC. We only considered CpGs that showed a difference in group means in methylation M values (|ΔM|) of at least 25% (|ΔM| ≥ 1.5) and false discovery rate (FDR) <0.05 in the 2 group comparison.
Differentially methylated region analysis
The majority of genome-wide methylation studies have focused on single CpG sites. However, modification of single CpG often produces weak correlations with gene expression data.22 Contextualizing the methylation level of an individual CpG by the status of neighboring CpG sites facilitates biological inferences. Clusters of neighboring CpGs with coordinated differential methylation are identified as differentially methylated regions (DMRs). Hypermethylated DMRs in promoters are usually associated with silencing, whereas in the gene body, they associate with upregulation of expression.23 We defined a DMR as a region including (1) 3 or more consecutive significantly differentially methylated (p < 0.05) sites between PD and control groups with the same direction of methylation change; (2) each differentially methylated CpG separated by less than 500 bp; and (3) a multiple-comparison corrected p value (Sidak p) less than 0.05 for the region.
DMR analysis was performed using comb-p.24 Comb-p takes as input unadjusted p values for each probe, identifies regions of enrichment (i.e., series of adjacent low p values), and computes statistical significance of the regions using the Sidak correction.
Levodopa
Levodopa (l-Dopa) has been shown to affect methylation levels25 and thus could be a contributor to the methylation changes observed. To address this, we used 2 approaches: (1) we examined the dose-response relationship with l-Dopa in a line of iPSC-derived dopaminergic neurons generated from a control non-Hispanic white male donor. (2) We also compared methylation changes we found to those reported in a model of 7-day l-Dopa administration to rats rendered hemiparkinsonian through unilateral injections of 6-hydroxydopamine (6-OHDA).25
We selected nontoxic concentrations ranging from 0 to 50 μM of l-Dopa, including treatment with 10 μM, a concentration proven to induce methylation changes in cultured human peripheral blood mononuclear cells.26 iPSC-derived dopaminergic neurons differentiated for 70 days were cultured with varying levels of l-Dopa (0, 5, 10, and 50 uM) in Neurobasal N2/B27 media (Gibco) supplemented with 1 ng/mL of transforming growth factor beta-3, 10 ng/mL of sonic hedgehog signaling molecule, 20 ng/mL of brain-derived neurotrophic factor, and 30 ng/mL of glial cell-derived neurotrophic factor. After 7 days, DNA was extracted and subjected to methylation analysis.
RNA sequencing and statistical analysis
RNA-seq (RNA integrity number ≥ 5) was performed using a paired-end 125 bp protocol on a HiSeq 2500. Reads were aligned to the human reference genome (hg19) using the STAR algorithm and analyzed using the “voom” method in the Limma package.27
A linear model with AAD was fitted to each gene, and empirical Bayes moderated t-statistics and p values were used to assess expression differences between PD and controls. To account for underlying unknown confounding factors, we used SVAseq with default parameters to estimate SVs.28 None of the estimated SVs differed significantly between case-control status, so we did not include them in the linear model: log (cpm) ∼ PD + AAD.
Pathway analysis
We used Enrichr (amp.pharm.mssm.edu/Enrichr/), which yields an FDR-adjusted p value for each pathway.29 The binding affinity of most transcription factors to DNA is altered by DNA CpG methylation.30 Thus, we analyzed the presence of transcription factor binding sites (TFBSs) using the R package Goldmine.31
Data availability
Raw data for the primary analyses are available upon reasonable request from the corresponding and senior author.
Results
Samples
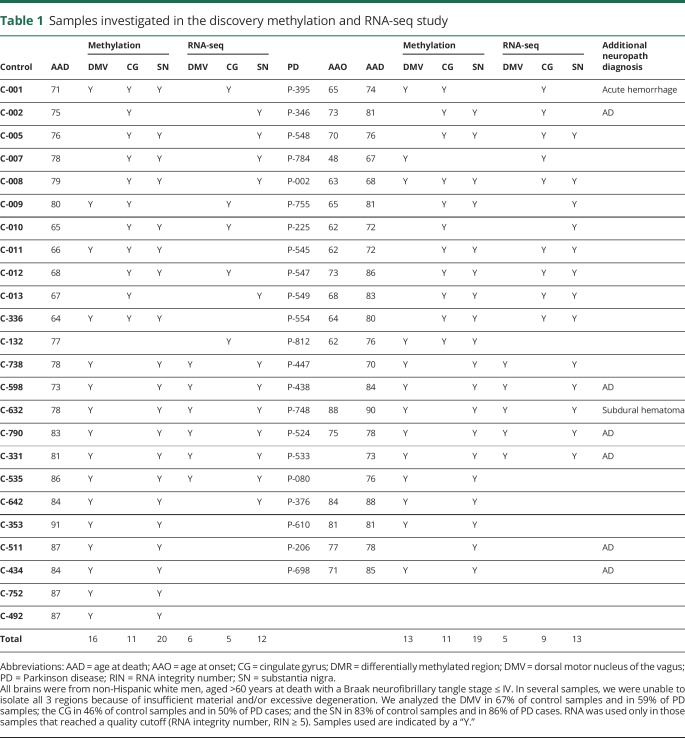
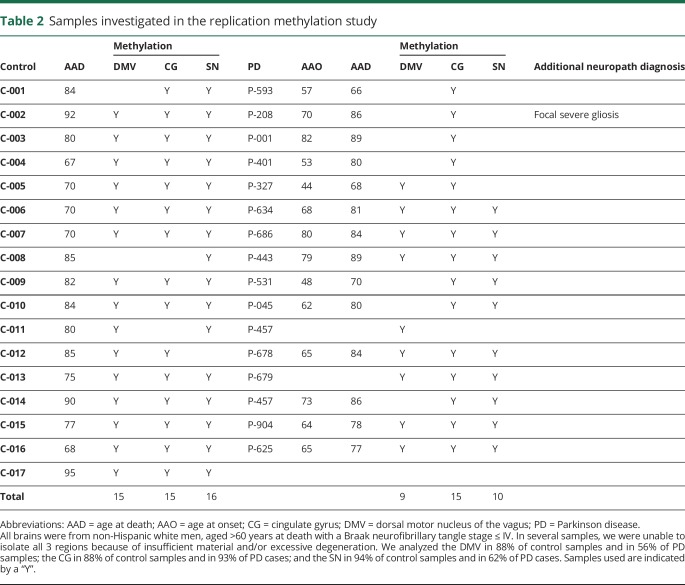
The average AAD for PD cases was 78.1 years (range 67–90 years) in the discovery data set and 79.8 years (range 66–89 years) in the replication data set. For controls, it was 77.7 years (range 64–91 years) in the discovery and 79.6 years (range 67–95 years) in the replication data set. In multiple samples, we were unable to isolate all 3 regions because of insufficient material or excessive degeneration. This is shown in tables 1 and 2, with age at onset, AAD, and the regions isolated from each donor.
Table 1.
Samples investigated in the discovery methylation and RNA-seq study
Table 2.
Samples investigated in the replication methylation study
DMR analysis
Discovery data set
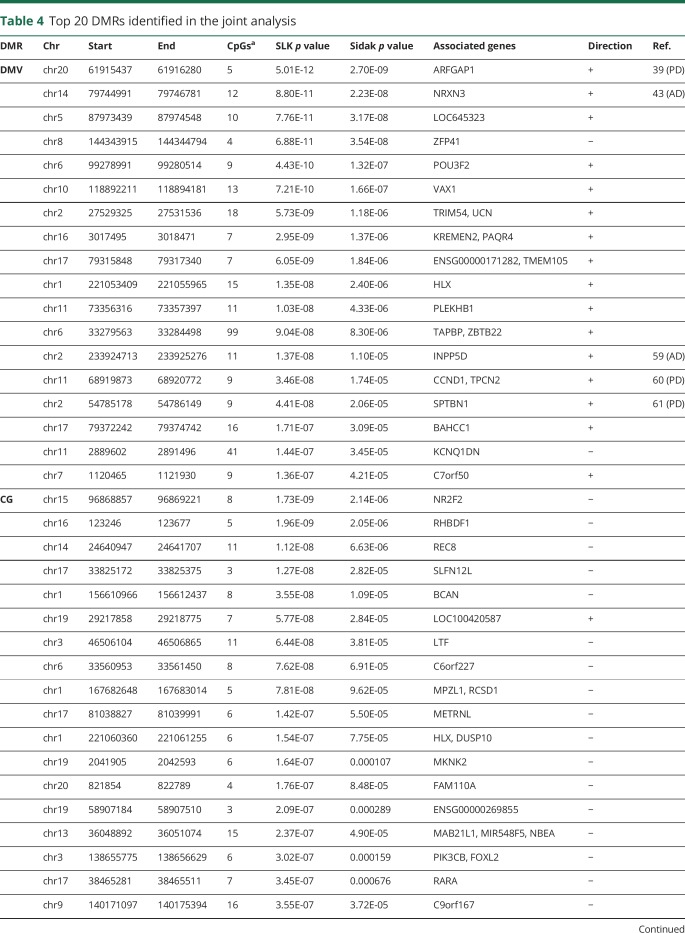
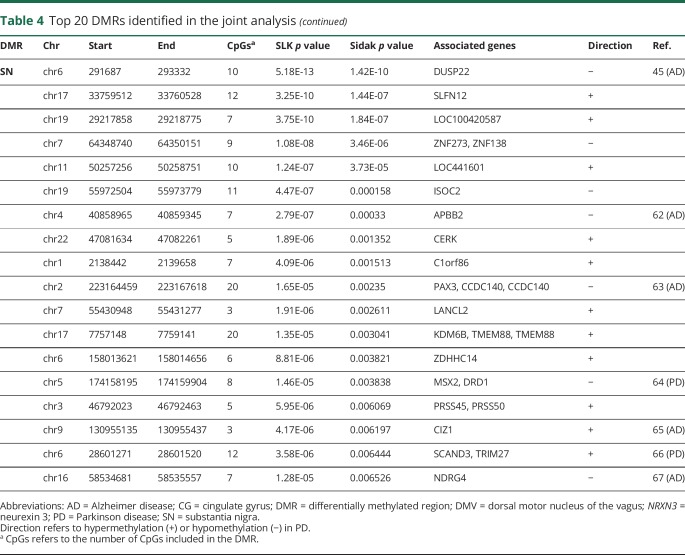
Analysis in the discovery data set identified 85 DMRs in the DMV, 65 in the CG, and 27 in the SN samples with Sidak p < 0.05 (table e-1, links.lww.com/NXG/A164). These DMRs were associated with 108, 84, and 31 genes in the DMV, CG, and SN, respectively. Within the discovery data set, comparison of the DMRs identified in the 3 tissues revealed that a region spanning the promoter region of dual-specificity phosphatase 22 (DUSP22) was hypomethylated in PD brains in the 3 tissues. Furthermore, another 7 genomic regions were differentially hypermethylated in both the DMV and the SN (RNF5, AGPAT1, LANCL2, LMTK3, SCAND3, SLFN12, and ZDHHC14).
Replication data set
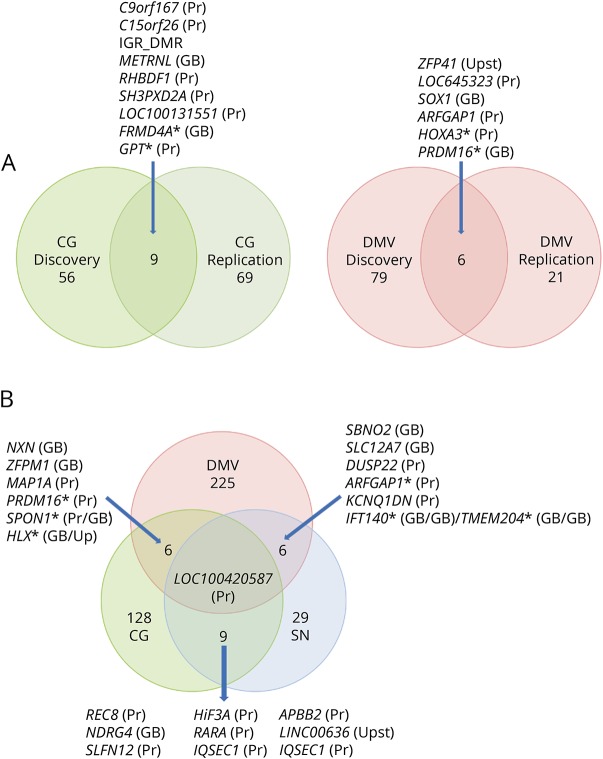
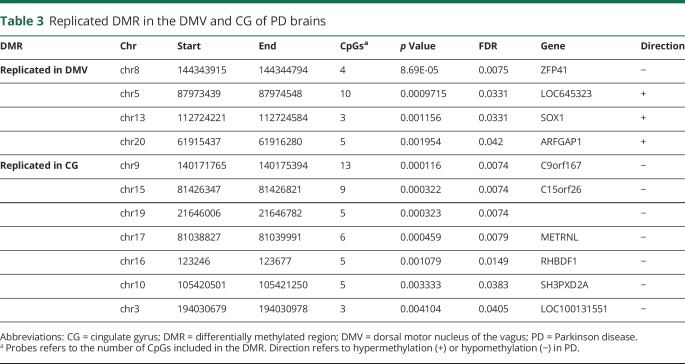
We tested whether any of the significant DMRs identified in the discovery data set were also differentially methylated in the replication data set (Sidak p < 0.05). We found that 7 of the discovery DMRs were reproduced in the replication data set in the CG, 4 in the DMV, and none in the SN (figure and table 3). We therefore performed a gene-based analysis that revealed genes that contained DMRs in both the DMV (FRMD4A and GPT) and the CG (HOXA3 and PRDM16) of patients with PD in which the location of the DMR is not the same in the discovery and the replication data set (figure). This analysis identified several genes, including ARFGAP1, a reported regulator of LRRK2 toxicity in PD.32,33
Figure. PD-associated DMRs.
(A) DMRs previously identified in the discovery data set were also significantly differentially methylated in the same direction in the replication data set. The SN is not included because we did not detect overlap at the DMR or gene level with any of the other 2 tissues. (B) Common PD-associated DMRs identified in the 3 analyzed tissues in the joint analysis. A DMR in LOC100420587, an SHC binding and spindle associated 1 pseudogene, is present in all 3 regions. If DMRs could be assigned to more than 1 gene, both genes are shown separated by a slash. Asterisks denote overlap at the gene level but not at the DMR level (distinct DMRs assigned to the same gene). In the Venn diagram, the numbers are the significant DMRs in the 2 data sets/brain regions and those shared between them. Brackets besides gene names indicate the genomic location of the DMR. CG = cingulate gyrus; DMR = differentially methylated region; DMV = dorsal motor nucleus of the vagus; GB = gene body; PD = Parkinson disease; Pr = promoter; SN = substantia nigra; Upst = upstream intergenic region.
Table 3.
Replicated DMR in the DMV and CG of PD brains
Joint data set
As both the discovery and replication autopsy data sets were limited in sample numbers and showed replication between them, to increase the power of detecting disease-associated methylation changes, we performed a single joint analysis. This provided us a total of 53 DMV (22 PD and 31 controls), 52 CG (26 PD and 26 controls), and 65 SN (29 PD and 36 controls) for the analysis. In the joint analysis, we identified 234 significant DMR in the DMV, 44 in the SN, and 141 in the CG (table e-2, links.lww.com/NXG/A165). These correspond to 266, 53, and 168 genes, respectively. The top 20 DMRs in the joint analysis from each region are shown in table 4. In the joint analysis, a DMR in the promoter area of LOC100420587 is hypermethylated in the 3 brain regions. It is interesting to note that an SNP in this noncoding gene of unknown function was identified as associated with the volume of the CG by neuroimaging and GWASs.34
Table 4.
Top 20 DMRs identified in the joint analysis
Pathway analysis identified Wnt signaling as epigenetically affected in the DMV of PD brains
We identified physiologic pathways overrepresented among the genes associated with DMRs identified in the joint analysis. Significant enrichment was observed in the KEGG “Hippo signaling pathway” (FDR = 0.007) and “Wnt signaling pathway” (FDR = 0.01) in the DMV (table e-3, links.lww.com/NXG/A166). No pathway enrichment was observed for CG or SN, even using FDR < 0.25 as the significance cutoff, as previously suggested for pathway analysis.35,36
Integrated analysis of differential methylation and RNA-Seq
We observed that ∼80% of the DMRs identified contain TFBS (table e-4, links.lww.com/NXG/A167), suggesting that the identified differences in DNA methylation are likely to have transcriptional consequences. We then performed transcriptome analysis by RNA-seq on a subset of samples (table 1). Analysis of the RNA-seq data identified 515 differentially expressed genes (DEG) in the CG, 390 DEG in the SN, and 3 DEG in the DMV associated with PD (FDR <0.05, adjusted for AAD). An overlap analysis of both methylation data and RNA-seq revealed 6 genes with DMRs that were also differentially expressed (NDRG4, PTPRN2, SYT7, IQSEC1, DLG4, and KCNIP1) in the CG and 1 (NDRG4) in the SN.
Levodopa
We found 14 DMRs that changed their methylation levels significantly (Sidak p < 0.05; table e-5, links.lww.com/NXG/A168) on l-Dopa treatment of iPSC-derived dopaminergic neurons. Comparison of the DMRs identified in the joint analysis in the different tissues with the methylation changes induced by l-Dopa treatment showed no overlap. Furthermore, comparison of our data (all genes containing DMR irrespective of the brain region) with the genes identified as differentially methylated in the dorsal striatum of 6-OHDA–treated rats receiving l-Dopa25 with FDR < 0.05 and absolute change of 10% (2703 genes) revealed an overlap of 82 genes. Thus, the rat model–human comparison data suggest that approximately 24.1% of the genes identified as differentially methylated in PD could be related to l-Dopa administration. To attempt to identify methylation changes that could be induced by the presence of cell death or hypofunctioning neurons, we determined which of the DMR-containing genes (from the joint analysis) were also differentially methylated in hemiparkinsonian rats not given l-Dopa. This revealed 60 genes with differential methylation shared between our human data and those responsive to 6-OHDA lesion in the rat striatum as previously identified.25
Discussion
This initial study of DNA methylation changes in the DMV, SN, and CG supports our hypothesis of an epigenetic contribution to PD risk. Whether the identified changes are inherited, acquired de novo during development, in part due to cellular composition changes or induced by environmental variables is currently unknown. However, it is certainly interesting to speculate that these methylation changes might be due to environmental influences through the vagus nerve. If this were the case, it would suggest that methylation is an early factor in the development of PD, as the DMV is thought to be one of the earliest regions to develop characteristic PD pathologic changes.
Epigenetic patterns are different between cell types, specifically neurons and glia.37 Unlike the DMV, which does not have extensive degeneration, cell loss in the SN is prominent, and thus, it is possible that a change in cellular composition between controls and patients with PD contributes to some of the changes we have seen despite the use of iSVA to correct for cellular heterogeneity. Furthermore, heterogeneity in the amount of cell death in the different cell areas studied here is anticipated because of variable Lewy pathology Braak staging (ranging from stage 3 to stage 5), potentially influencing the results.
We analyzed the overlap between our results and changes observed in l-Dopa–treated rats. The 6-OHDA–induced parkinsonism model has limitations in terms of progression and recapitulation of the age-related effects of PD. However, this parkinsonism model has been useful in the study of l-Dopa-related dyskinesia.38 The small overlap in l-Dopa–associated changes in DNA methylation and the changes we observed, with the fact that the DMV, SN, and CG displayed minimal overlap in DMRs, yet are all exposed to the drug, would suggest that l-Dopa is not the major contributor to the changes observed here.
The finding that most DMRs contain TFBS suggests that the PD-associated DNA methylation changes have regulatory potential. We, nevertheless, did not observe a correlated change in gene expression in the DMV and only one gene in the SN. Notably, in the CG, an area affected late in the course of PD, we did detect some transcriptional dysregulation accompanied by DNA methylation changes. This suggests the possibility that the identified DMRs constitute stable modifications of the epigenome, but their acute effects (transcriptional changes) are not stably maintained and compensated by other mechanisms. Furthermore, a subset of the differentially methylated/expressed genes in the CG has also DMRs in the SN, supporting the idea of stable DNA modifications reflecting previous transcriptional changes.
A DMR in ADP-ribosylation factor 1 GTPase activating protein 1 (ARF1GAP) is the most significant DMR in the joint analysis of the DMV (table 3). ARFGAP1 and LRRK2 interact and appear to regulate each other's expression.32,33 Thus, the methylation changes would suggest a wider role for ARFGAP1 in PD pathophysiology. Neurexin 3, thought to be involved in synaptic plasticity, has been associated with multiple psychiatric disorders including Alzheimer disease.39 Of interest, the promoter of DUSP22, associated with the most significant DMR in the SN in this study, was shown to be hypermethylated in the hippocampus,40 whereas a region upstream of DUSP22 was found to be hypomethylated in the superior temporal gyrus41 of patients with Alzheimer disease. It has been recently suggested that DUSP22 is involved in both the phosphorylation of tau and CREB signaling, both shown to be involved in Alzheimer disease.40 Furthermore, hypermethylation of the DUSP22 promoter was associated with schizophrenia.42 It is interesting that we did not find any DMR in SNCA that has been shown to have methylation changes previously in the cortex, SN, and blood in PD. The reasons for this are not clear, but may reflect tissue degeneration in the earlier affected regions, well as a larger data set reported here, as it is likely to vary between individuals.
Identifying enriched functional pathways revealed epigenetic perturbation in the interrelated pathways of “Wnt signaling” and “Hippo signaling” in the DMV. Wnt signaling has been previously implicated in PD via expression and methylation,14 genetic,43–50 and modeling46,47 approaches. Wnt proteins are critical mediators of cell-to-cell communication and intracellular signaling associated with CNS development.43 Particularly important for PD is their role in determining midbrain dopaminergic cell fate and functioning.48 Recently, Wnt signaling has been implicated in propagating the innate immune function in multiple tissues. Such regulation of inflammation, a process implicated in PD49 and in neurodegeneration in general, could be a primary source of Wnt's influence on PD.50
Thus, our data support an epigenetic component to the development of PD and fit well within the growing body of evidence involving the DMV and the vagus nerve in PD. Furthermore, our data support the previous studies suggesting deregulated Wnt signaling contributing to the pathogeneses of PD.
Acknowledgment
The authors are grateful to the families and staff who participated in this study. Some of the samples used in this study were collected while the Udall PDRCE was based at Duke University.
Glossary
- AAD
age at death
- CG
cingulate gyrus
- DEG
differentially expressed gene
- DMR
differentially methylated region
- DMV
dorsal motor nucleus of the vagus
- DUSP22
dual-specificity phosphatase 22
- IC
independent component
- iSVA
independent surrogate variable analysis
- LB
Lewy body
- PD
Parkinson disease
- SN
substantia nigra
- SV
surrogate variable
- TFBS
transcription factor binding site
Appendix. Authors
Study funding
This study was supported by NIH grants NS071674 and NS062684.
Disclosure
Disclosures available: Neurology.org/NG.
References
- 1.Kalia LV, Lang AE. Parkinson's disease. Lancet 2015;386:896–912. [DOI] [PubMed] [Google Scholar]
- 2.Mullin S, Schapira A. The genetics of Parkinson's disease. Br Med Bull 2015;114:39–52. [DOI] [PubMed] [Google Scholar]
- 3.Allen MT, Levy LS. Parkinson's disease and pesticide exposure: a new assessment. Crit Rev Toxicol 2013;43:515–534. [DOI] [PubMed] [Google Scholar]
- 4.Ammal Kaidery N, Tarannum S, Thomas B. Epigenetic landscape of Parkinson's disease: emerging role in disease mechanisms and therapeutic modalities. Neurotherapeutics 2013;10:698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock DB, Martin ER, Mayhew GM, et al. . Pesticide exposure and risk of Parkinson's disease: a family-based case-control study. BMC Neurol 2008;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wüllner U, Kaut O, deBoni L, Piston D, Schmitt I. DNA methylation in Parkinson's disease. J Neurochem 2016;139(suppl 1):108–120. [DOI] [PubMed] [Google Scholar]
- 7.Miranda-Morales E, Meier K, Sandoval-Carrillo A, Salas-Pacheco J, Vazquez-Cardenas P, Arias-Carrion O. Implications of DNA methylation in Parkinson's disease. Front Mol Neurosci 2017;10:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jowaed A, Schmitt I, Kaut O, Wüllner U. Methylation regulates alpha-synuclein expression and is decreased in Parkinson's disease patients' brains. J Neurosci 2010;30:6355–6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto L, Takuma H, Tamaoka A, et al. . CpG demethylation enhances alpha-synuclein expression and affects the pathogenesis of Parkinson's disease. PLoS One 2010;5:e15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masliah E, Dumaop W, Galasko D, Desplats P. Distinctive patterns of DNA methylation associated with Parkinson disease: identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics 2013;8:1030–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan YY, Wu L, Zhao ZB, et al. . Methylation of alpha-synuclein and leucine-rich repeat kinase 2 in leukocyte DNA of Parkinson's disease patients. Parkinsonism Relat Disord 2014;20:308–313. [DOI] [PubMed] [Google Scholar]
- 12.Su X, Chu Y, Kordower JH, et al. . PGC-1alpha promoter methylation in Parkinson's disease. PLoS One 2015;10:e0134087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsh AG, Cottrell MT, Goldman MF. Epigenetic DNA methylation profiling with MSRE: a quantitative NGS approach using a Parkinson's disease test case. Front Genet 2016;7:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Deng J, Pan Q, et al. . Targeted methylation sequencing reveals dysregulated Wnt signaling in Parkinson disease. J Genet Genomics 2016;43:587–592. [DOI] [PubMed] [Google Scholar]
- 15.Beecham GW, Dickson DW, Scott WK, et al. . PARK10 is a major locus for sporadic neuropathologically confirmed Parkinson disease. Neurology 2015;84:972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sampson TR, Debelius JW, Thron T, et al. . Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell 2016;167:1469–1480.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stokholm MG, Danielsen EH, Hamilton-Dutoit SJ, Borghammer P. Pathological alpha-synuclein in gastrointestinal tissues from prodromal Parkinson disease patients. Ann Neurol 2016;79:940–949. [DOI] [PubMed] [Google Scholar]
- 18.Uribe C, Segura B, Baggio HC, et al. . Patterns of cortical thinning in nondemented Parkinson's disease patients. Mov Disord 2016;31:699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods 2014;11:1138–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du P, Zhang X, Huang CC, et al. . Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teschendorff AE, Zhuang J, Widschwendter M. Independent surrogate variable analysis to deconvolve confounding factors in large-scale microarray profiling studies. Bioinformatics 2011;27:1496–1505. [DOI] [PubMed] [Google Scholar]
- 22.Vanderkraats ND, Hiken JF, Decker KF, Edwards JR. Discovering high-resolution patterns of differential DNA methylation that correlate with gene expression changes. Nucleic Acids Res 2013;41:6816–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet 2011;20:670–680. [DOI] [PubMed] [Google Scholar]
- 24.Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics 2012;28:2986–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figge DA, Eskow Jaunarajs KL, Standaert DG. Dynamic DNA methylation regulates levodopa-induced dyskinesia. J Neurosci 2016;36:6514–6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitt I, Kaut O, Khazneh H, et al. . L-dopa increases alpha-synuclein DNA methylation in Parkinson's disease patients in vivo and in vitro. Mov Disord 2015;30:1794–1801. [DOI] [PubMed] [Google Scholar]
- 27.Ritchie ME, Phipson B, Wu D, et al. . Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leek JT. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res 2014;42:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuleshov MV, Jones MR, Rouillard AD, et al. . Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin Y, Morgunova E, Jolma A, et al. . Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017;356;eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhasin JM, Ting AH. Goldmine integrates information placing genomic ranges into meaningful biological contexts. Nucleic Acids Res 2016;44:5550–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong Y, Yuan C, Chen R, Dawson TM, Dawson VL. ArfGAP1 is a GTPase activating protein for LRRK2: reciprocal regulation of ArfGAP1 by LRRK2. J Neurosci 2012;32:3877–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stafa K, Trancikova A, Webber PJ, Glauser L, West AB, Moore DJ. GTPase activity and neuronal toxicity of Parkinson's disease-associated LRRK2 is regulated by ArfGAP1. PLoS Genet 2012;8:e1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bi X, Yang L, Li T, Wang B, Zhu H, Zhang H. Genome-wide mediation analysis of psychiatric and cognitive traits through imaging phenotypes. Hum Brain Mapp 2017;38:4088–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bueno MJ, Gómez de Cedrón M, Gómez-López G, et al. . Combinatorial effects of microRNAs to suppress the Myc oncogenic pathway. Blood 2011;117:6255–6266. [DOI] [PubMed] [Google Scholar]
- 37.Kessler NJ, Van Baak TE, Baker MS, Laritsky E, Coarfa C, Waterland RA. CpG methylation differences between neurons and glia are highly conserved from mouse to human. Hum Mol Genet 2016;25:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duty S, Jenner P. Animal models of Parkinson's disease: a source of novel treatments and clues to the cause of the disease. Br J Pharmacol 2011;164:1357–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Mir A, Gonzalez-Perez A, Gayan J, et al. . Genetic study of neurexin and neuroligin genes in alzheimer's disease. J Alzheimers Dis 2013;35:403–412. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez-Mut JV, Aso E, Heyn H, et al. . Promoter hypermethylation of the phosphatase DUSP22 mediates PKA-dependent TAU phosphorylation and CREB activation in alzheimer's disease. Hippocampus 2014;24:363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watson CT, Roussos P, Garg P, et al. . Genome-wide DNA methylation profiling in the superior temporal gyrus reveals epigenetic signatures associated with Alzheimer's disease. Genome Med 2016;8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boks MP, Houtepen LC, Xu Z, et al. . Genetic vulnerability to DUSP22 promoter hypermethylation is involved in the relation between in utero famine exposure and schizophrenia. NPJ Schizophr 2018;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inestrosa NC, Arenas E. Emerging roles of Wnts in the adult nervous system. Nat Rev Neurosci 2010;11:77–86. [DOI] [PubMed] [Google Scholar]
- 44.Rawal N, Corti O, Sacchetti P, et al. . Parkin protects dopaminergic neurons from excessive Wnt/beta-catenin signaling. Biochem Biophys Res Commun 2009;388:473–478. [DOI] [PubMed] [Google Scholar]
- 45.Berwick DC, Harvey K. LRRK2 functions as a Wnt signaling scaffold, bridging cytosolic proteins and membrane-localized LRP6. Hum Mol Genet 2012;21:4966–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou T, Zu G, Zhang X, et al. . Neuroprotective effects of ginsenoside Rg1 through the Wnt/beta-catenin signaling pathway in both in vivo and in vitro models of Parkinson's disease. Neuropharmacology 2016;101:480–489. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Hao S, Yang B, et al. . Wnt/beta-catenin signaling plays an essential role in alpha7 nicotinic receptor-mediated neuroprotection of dopaminergic neurons in a mouse Parkinson's disease model. Biochem Pharmacol 2017;140:115–123. [DOI] [PubMed] [Google Scholar]
- 48.Prakash N, Brodski C, Naserke T, et al. . A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development 2006;133:89–98. [DOI] [PubMed] [Google Scholar]
- 49.Wang Q, Liu Y, Zhou J. Neuroinflammation in Parkinson's disease and its potential as therapeutic target. Transl Neurodegener 2015;4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderegg A, Lin HP, Chen JA, et al. . An Lmx1b-miR135a2 regulatory circuit modulates Wnt1/Wnt signaling and determines the size of the midbrain dopaminergic progenitor pool. PLoS Genet 2013;9:e1003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data for the primary analyses are available upon reasonable request from the corresponding and senior author.