Summary
Multistate cure models are multistate models in which transitions into one or more of the states cannot occur for a fraction of the population. In the study of cancer, multistate cure models can be used to identify factors related to the rate of cancer recurrence, the rate of death before and after recurrence, and the probability of being cured by initial treatment. However, the previous method for fitting multistate cure models requires substantial custom programming, making these valuable models less accessible to analysts. In this article, we present an Expectation–Maximization (EM) algorithm for fitting the multistate cure model using maximum likelihood. The proposed algorithm makes use of a weighted likelihood representation allowing it to be easily implemented with standard software and can incorporate either parametric or non-parametric baseline hazards for the state transition rates. A common complicating feature in cancer studies is that the follow-up times for recurrence and death may differ. Additionally, we may have missingness in the covariates. We propose a Monte Carlo EM (MCEM) algorithm for fitting the multistate cure model in the presence of covariate missingness and/or unequal follow-up of the two outcomes, we describe a novel approach for obtaining standard errors, and we provide some software. Simulations demonstrate good algorithmic performance as long as the modeling assumptions are sufficiently restrictive. We apply the proposed algorithm to a study of recurrence and death in patients with head and neck cancer.
Keywords: Cure models, EM algorithm, Monte Carlo EM algorithm, Multistate models
1. Introduction
In medical applications, multistate models describe the rates at which individuals move between various health states. Multistate models have many valuable uses in medical research. Firstly, multistate models allow us to incorporate information from multiple event time outcomes (e.g. recurrence and death) in a unified way. Secondly, multistate models allow us to study which patient characteristics are relevant to which aspects of disease progression. Finally, multistate models are useful for making predictions, which can be valuable for medical decision-making.
The illness-death model is a popular multistate model explored in the literature and consists of three states: healthy (or no event), illness, and death (Andersen and Keiding, 2002). All subjects start out in the “healthy” state at baseline and can then move into the illness or death states as they develop illness or die from other causes. Subjects that develop illness can also transition into the death state. One common application of the illness-death model is in the study of cancer recurrence and death. In this setting, the “healthy” state represents subjects who have been treated for their initial cancer. For the remainder of this article, we will focus on the scenario with outcomes cancer recurrence and death, but these may be different types of events in general.
While the illness-death model is useful in many applications, one limitation is that the model implicitly assumes that all subjects can experience the illness event. In the context of cancer recurrence and death, this is equivalent to assuming that all subjects can experience a recurrence. For many types of cancer, however, this may not be a reasonable assumption. In the case of head and neck cancer, it has been well established that some subjects can be completely cured of their initial disease, and these subjects will never experience a recurrence of their primary cancer (Taylor, 1995; Cognetti and others, 2008). We call the set of cured subjects the “cured fraction.”
In this article, we consider a generalization of the illness-death model called the multistate cure model that accounts for the cured fraction. The Bayesian multistate cure model developed in Conlon and others (2013) breaks the “healthy” state of the illness-death model into two baseline states: cured and non-cured. The non-cured subjects can then experience either cancer recurrence or death under an illness-death model. The cured subjects can only experience the death event.
One challenge to fitting the multistate cure model is that cure status is partially latent. Subjects with an observed recurrence are known to be non-cured, but subjects censored for recurrence have unknown cure status. Cure status is unknown for all subjects at baseline. A natural question that arises in the context of cure models is our ability to identify the cured population when the event time distribution in the non-cured subjects may have a long tail (Farewell, 1982; Conlon and others, 2013). We will assume that we have sufficient follow-up after the last observed event time. One indicator of sufficient follow-up in a cure setting is that a Kaplan–Meier estimator applied to the time to recurrence outcome should have a clear plateau, indicating that there is a time-point after which recurrence events are no longer being observed. In the illness-death model setting, we require further follow-up for death before and after recurrence, so a lack of sufficient follow-up may be less of a concern for data sets that are well-suited for illness-death models.
Another problem for illness-death models in general and multistate cure models in particular is that the follow-up may not be the same for both outcomes of interest (Conlon and others, 2013). We call this situation “unequal follow-up.” For example, death status may be more easily obtained through death records, while assessment of cancer recurrence status requires a clinic visit, so it may often be the case that death status is known and recurrence status is unknown at a particular time  . Conlon and others (2013) propose a Gibbs-sampling algorithm for fitting the multistate cure model in which values of cure status are drawn using a data augmentation approach, and unequal follow-up is handled through a modification to the likelihood involving an integral. Their proposed algorithm performs well, but it requires substantial custom programming, it requires specification of prior distributions and tuning parameters, and it can take a long time to reach convergence. Additionally, the algorithm discussed in Conlon and others (2013) assumes that covariates are fully observed, which may not be the case in practice.
. Conlon and others (2013) propose a Gibbs-sampling algorithm for fitting the multistate cure model in which values of cure status are drawn using a data augmentation approach, and unequal follow-up is handled through a modification to the likelihood involving an integral. Their proposed algorithm performs well, but it requires substantial custom programming, it requires specification of prior distributions and tuning parameters, and it can take a long time to reach convergence. Additionally, the algorithm discussed in Conlon and others (2013) assumes that covariates are fully observed, which may not be the case in practice.
The EM algorithm is an alternative, maximum likelihood-based method in the literature for fitting models with latent variables or other types of missing data (Dempster and others, 1977). One advantage of the EM algorithm over Bayesian methods is that, in some cases, an EM algorithm can be more readily implemented using standard software. Additionally, the EM algorithm does not require specification of prior distributions or tuning parameters. In some complicated missing data scenarios, however, the conventional EM algorithm can be difficult to implement. The Monte Carlo EM (MCEM) algorithm proposed in Wei and Tanner (1990) provides a convenient, imputation-based approach for handling more complex missing data within a modified EM algorithm. EM and MCEM algorithms have not been previously explored in the context of multistate cure models in the literature and development of such fitting algorithms could make the multistate cure model much more accessible to investigators.
In this article, we first propose a simple EM algorithm for fitting the standard multistate cure model. We then propose a MCEM algorithm for fitting the model in the presence of covariate missingness and/or unequal follow-up of the outcomes. The proposed algorithms can incorporate either parametric or non-parametric baseline hazards for the transitions between states and can incorporate different assumptions about the rate of death from other causes. The proposed EM algorithm makes use of a weighted likelihood representation, allowing it to be easily implemented using standard software. We provide software for implementing the EM and MCEM algorithms. We describe a novel approach for estimating standard errors for the MCEM algorithm. Simulations demonstrate the performance of the EM and MCEM algorithms under different modeling assumptions. We apply the proposed MCEM to a study of cancer recurrence and death of head and neck patients. We then derive expressions for estimating state occupancy probabilities, which can used to make predictions for individual patients.
In Section 2, we present details about the multistate cure model structure. In Sections 3 and 4, we propose an EM and MCEM algorithm for fitting the model. In Section 5, we discuss how to estimate standard errors. In Section 6, we derive state occupancy probabilities. We present a simulation study in Section 7, and we apply the proposed methods to head and neck cancer data in Section 8. In Section 9, we include a discussion.
2. Multistate cure model specification
Suppose we have two semi-competing events: recurrence and death. By semi-competing, we mean that we can observe death after cancer recurrence but cannot observe recurrence after death. We further suppose that there is some subset of the subjects that are cured of their initial cancer and will never experience a cancer recurrence (even with very long follow-up).
Let  and
and  be the underlying recurrence and death times for subject
be the underlying recurrence and death times for subject  . For cured subjects,
. For cured subjects,  . Let
. Let  be the censoring time for recurrence (loss to follow-up) and
be the censoring time for recurrence (loss to follow-up) and  be the censoring time for death. Initially, we assume that the follow-up for recurrence and death is equal and define
be the censoring time for death. Initially, we assume that the follow-up for recurrence and death is equal and define  . For all subjects, we observe censored recurrence time information,
. For all subjects, we observe censored recurrence time information,  and
and  , and censored death time information,
, and censored death time information,  and
and 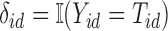 . Let
. Let 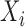 denote the covariates for subject
denote the covariates for subject 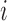 , which we will initially suppose are fully observed.
, which we will initially suppose are fully observed.
We assume that all subjects have been previously treated for their initial cancer and did not have observable cancer at baseline. For non-cured subjects, however, some unobservable cancer cells remain and will grow until they are eventually observable, called cancer recurrence. Subjects with an observed recurrence are known to have been non-cured at baseline, but all other subjects have unknown baseline cure status. Let  be a variable indicating baseline non-cure status:
be a variable indicating baseline non-cure status: 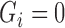 if cured,
if cured,  if not cured. While we never know for sure that subjects are cured, we may strongly believe subjects still at risk after some time
if not cured. While we never know for sure that subjects are cured, we may strongly believe subjects still at risk after some time  are cured and assign
are cured and assign 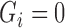 for these subjects. Assigning some subjects to be cured improves the stability of the proposed methods, and we use this approach in our simulations and data example. Similar assumptions are often implicitly made for standard cure rate models through restrictions on the event rate in the non-cured subjects (Peng and Dear, 2000; Cai and others, 2012).
for these subjects. Assigning some subjects to be cured improves the stability of the proposed methods, and we use this approach in our simulations and data example. Similar assumptions are often implicitly made for standard cure rate models through restrictions on the event rate in the non-cured subjects (Peng and Dear, 2000; Cai and others, 2012).
Figure 1 shows the conceptual structure of the model proposed in Conlon and others (2013). Solid arrows represent potential state transitions given baseline cure status. If we remove State 4, the multistate cure model would reduce to the popular Cox proportional hazards (CPH) mixture cure model (Kuk and Chen, 1992; Sy and Taylor, 2000). We will assume that the underlying transition times between states are conditionally independent given covariates. We will further assume that  and
and 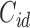 are independent of all underlying transition times given covariates.
are independent of all underlying transition times given covariates.
Fig. 1.

Diagram of the multistate cure model. States 1 and 2 present baseline cure status. State 3 (recurrence) can only be reached from baseline State 1. State 4 (death) can be reached from all other states.
We model the probability of not being cured by initial treatment using a logistic model:  , where
, where  is a subset of
is a subset of 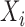 . We model the transition rate from state
. We model the transition rate from state  to state
to state 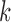 for all transitions except
for all transitions except 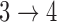 using proportional hazards model
using proportional hazards model  , where
, where 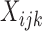 is the subset of
is the subset of  used in the model for transition
used in the model for transition 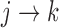 . One important decision in multistate modeling is whether we reset time back to zero upon entering a new state (Putter and others, 2007; Meira-Machado and others, 2009). In the model for the
. One important decision in multistate modeling is whether we reset time back to zero upon entering a new state (Putter and others, 2007; Meira-Machado and others, 2009). In the model for the 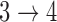 transition, we use the “clock reset” method in which time is reset to zero upon entering State 3, and use a proportional hazards regression to model the residual time in State 3 before entering State 4 as follows:
transition, we use the “clock reset” method in which time is reset to zero upon entering State 3, and use a proportional hazards regression to model the residual time in State 3 before entering State 4 as follows:  . We can incorporate the time spent in state 1,
. We can incorporate the time spent in state 1, 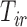 , as a covariate in
, as a covariate in 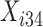 if desired.
if desired.
Let 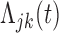 and
and 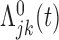 represent the cumulative hazard and cumulative baseline hazard for transition
represent the cumulative hazard and cumulative baseline hazard for transition 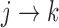 , and let
, and let 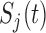 represent the probability of remaining in state
represent the probability of remaining in state  at time
at time 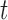 . We have that
. We have that 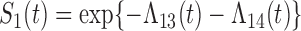 ,
,  , and
, and 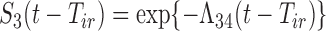 for
for  given
given  . We may use a parametric or non-parametric form for the baseline hazards.
. We may use a parametric or non-parametric form for the baseline hazards.
We may place additional assumptions on the hazards for the 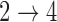 and
and  transitions. Since these two transitions represent typically death from other causes, it may be reasonable to assume that the hazards are identical (
transitions. Since these two transitions represent typically death from other causes, it may be reasonable to assume that the hazards are identical (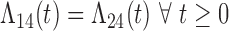 ). In this case, the multistate cure model reduces to a CPH cure model for recurrence time with two additional regressions for time to death with and without recurrence (Conlon and others, 2013). However, suppose we do not want to assume the hazards are equal. We may instead assume the hazards are proportional (
). In this case, the multistate cure model reduces to a CPH cure model for recurrence time with two additional regressions for time to death with and without recurrence (Conlon and others, 2013). However, suppose we do not want to assume the hazards are equal. We may instead assume the hazards are proportional (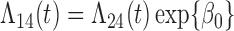 ) or that the baseline hazards are equal (
) or that the baseline hazards are equal (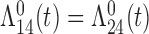 ,
, 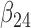 and
and  unrestricted), or we may make no equality assumptions (
unrestricted), or we may make no equality assumptions ( and
and  unrestricted).
unrestricted).
Let 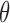 represent the set of model parameters.
represent the set of model parameters. 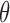 includes
includes  ,
,  , and possibly baseline hazard parameters (under parametric baselines). We will assume that
, and possibly baseline hazard parameters (under parametric baselines). We will assume that 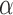 and
and 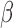 are distinct. Let
are distinct. Let 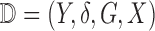 denote the complete data. The complete data log-likelihood for the multistate cure model takes the following form:
denote the complete data. The complete data log-likelihood for the multistate cure model takes the following form:
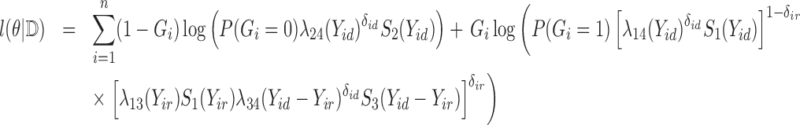 |
(1) |
3. EM algorithm
The EM algorithm is an approach for maximum likelihood estimation in the presence of missing data, which in our setting is the partially latent cure status (Dempster and others, 1977). Let  represent the observed data and
represent the observed data and 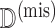 represent the missing data. The goal is to maximize the observed data log-likelihood,
represent the missing data. The goal is to maximize the observed data log-likelihood, 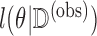 , with respect to
, with respect to  . The algorithm breaks the problem of maximizing
. The algorithm breaks the problem of maximizing 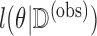 into iterations of two simpler steps: the E-step and the M-step. In the E-step, we calculate the expected value of the complete data log-likelihood conditioning on the observed data and the most recent parameter estimate,
into iterations of two simpler steps: the E-step and the M-step. In the E-step, we calculate the expected value of the complete data log-likelihood conditioning on the observed data and the most recent parameter estimate,  , to obtain
, to obtain
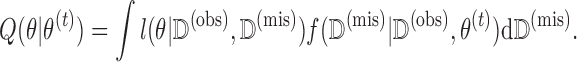 |
In the M-step, we maximize 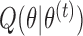 with respect to
with respect to  . We iterate these steps many times until convergence of the estimated
. We iterate these steps many times until convergence of the estimated  to the maximum likelihood estimate.
to the maximum likelihood estimate.
The EM algorithm is a common estimation method in the literature for models with latent classes. Frydman and Kadam (2004) proposed an EM algorithm for estimation for a continuous time multistate model in which the underlying population is split into movers and stayers such that only the movers are eligible to experience a state transition. This setting is very similar to our setting except that our model allows both the movers and stayers to experience death.
3.1. E-step
In the E-step for fitting the multistate cure model, we take the expectation of (1). Since (1) is linear in 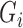 , we can obtain
, we can obtain 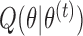 by replacing
by replacing 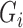 in (1) with
in (1) with 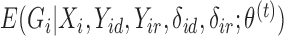 when
when 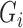 is unknown. Let
is unknown. Let 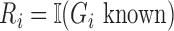 . For all subjects, we replace
. For all subjects, we replace 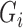 with
with
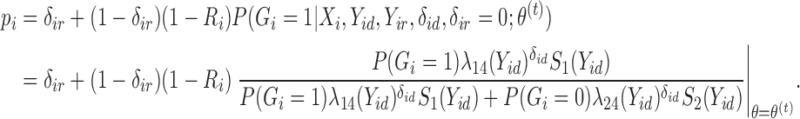 |
(2) |
In order to use the formula in (2), we need estimates of the baseline hazard functions for the  ,
,  , and
, and 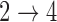 transitions. Under parametric assumptions, the complete form of the baseline hazards are determined in the M-step. When the baseline hazards are non-parametric, we estimate the baseline hazards prior to calculating (2). In the supplementary material available at Biostatistics online, we use the profile likelihood method to derive estimators for the baseline hazards. The form of the estimator depends on the estimate of
transitions. Under parametric assumptions, the complete form of the baseline hazards are determined in the M-step. When the baseline hazards are non-parametric, we estimate the baseline hazards prior to calculating (2). In the supplementary material available at Biostatistics online, we use the profile likelihood method to derive estimators for the baseline hazards. The form of the estimator depends on the estimate of 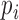 from the previous iteration and whether we assume that the baseline hazards for the
from the previous iteration and whether we assume that the baseline hazards for the  and
and 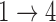 transitions are equal.
transitions are equal.
3.2. M-step
In the M-step, we maximize 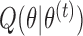 with respect to
with respect to  . After replacing
. After replacing 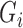 with
with 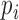 in (1) and reorganizing the terms, we have that
in (1) and reorganizing the terms, we have that
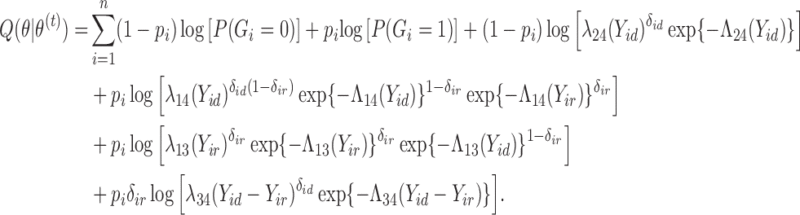 |
Since we assume that censoring times for recurrence and death are the same, 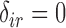 implies
implies  . Additionally, we note that
. Additionally, we note that 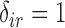 implies
implies 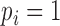 . We can therefore rewrite
. We can therefore rewrite 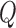 as:
as:
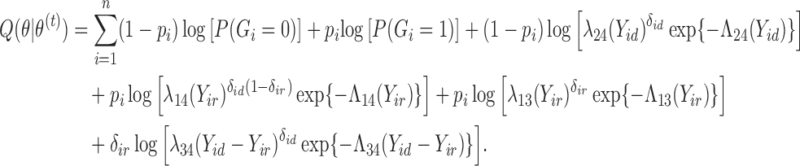 |
(3) |
The terms involving 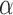 and
and 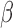 separate, so we can maximize (3) with respect to
separate, so we can maximize (3) with respect to  and
and 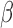 separately. The terms involving
separately. The terms involving 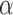 resemble the log-likelihood for a logistic model with
resemble the log-likelihood for a logistic model with 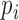 as the outcome. We can estimate
as the outcome. We can estimate  by fitting a logistic regression to
by fitting a logistic regression to 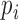 using predictors
using predictors  . We can estimate
. We can estimate 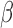 (and perhaps baseline parameters) by maximizing the last four terms of (3) with respect to the parameters. In the supplementary material available at Biostatistics online, we show how this maximization can be implemented by fitting a single, weighted proportional hazards regression model to an augmented version of the data. The proposed method for estimating
(and perhaps baseline parameters) by maximizing the last four terms of (3) with respect to the parameters. In the supplementary material available at Biostatistics online, we show how this maximization can be implemented by fitting a single, weighted proportional hazards regression model to an augmented version of the data. The proposed method for estimating 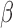 is similar to the methods used by mstate in R and other multistate modeling software except that it incorporates transition-specific weights and involves a different augmented data structure (de Wreede and others, 2011).
is similar to the methods used by mstate in R and other multistate modeling software except that it incorporates transition-specific weights and involves a different augmented data structure (de Wreede and others, 2011).
4. Extension to handle additional missing data
The EM algorithm in Section 3 assumes that cure status is the only source of missing data. However, additional missing data often arises in practice. One source of missing data is missingness in the covariates. Another common source of missingness occurs when the follow-up for recurrence is shorter than the follow-up for death. We call this phenomenon “unequal follow-up.” In order to follow-up for recurrence, patients must come into the clinic, while death status can be more easily obtained from death registries. In this case, recurrence status may only be known up to time 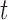 , while death status may be known up to time
, while death status may be known up to time 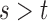 . This results in missing information about recurrence status on the interval
. This results in missing information about recurrence status on the interval  , which we will treat as missing data. This setting is similar to interval censoring and panel data for illness-death models (Jackson, 2011).
, which we will treat as missing data. This setting is similar to interval censoring and panel data for illness-death models (Jackson, 2011).
Conlon and others (2013) handles the problem of unequal follow-up by constructing a modified likelihood function involving an integral. Another potential solution is to censor death back to the follow-up time for recurrence for subjects with unequal follow-up. This approach is unappealing since it throws out valuable information about death. A third solution is to modify the conventional EM algorithm so that the E-step takes the expectation over all types of missing data. However, when we have complicated patterns of missing data, these expectations can be difficult to compute. We consider an alternative approach called the MCEM algorithm, which takes an imputation-based approach to dealing with missing data.
4.1. Monte Carlo EM algorithm
The MCEM algorithm proposed in Wei and Tanner (1990) provides a convenient approach to handling complicated missing data within a modified EM algorithm. The strategy is to replace the usual E-step from the EM algorithm with a step in which we obtain 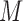 imputations
imputations 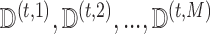 of
of 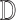 by drawing the missing data from
by drawing the missing data from 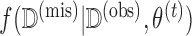 .
.
The M-step of the MCEM algorithm then involves maximizing the complete data log-likelihood mixed over the imputed values:
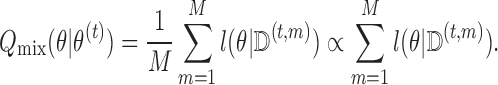 |
Suppose we create a stacked version of the data set, called 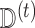 , obtained by stacking the imputed versions of the data such that each subject appears in the data set
, obtained by stacking the imputed versions of the data such that each subject appears in the data set 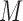 times. We then have that
times. We then have that 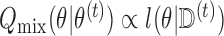 . We can estimate
. We can estimate  by maximizing
by maximizing  with respect to
with respect to  . We then iterate between the imputation and M-steps until “convergence,” where successive estimates of
. We then iterate between the imputation and M-steps until “convergence,” where successive estimates of  fall around the
fall around the 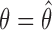 line with noise (Wei and Tanner, 1990). We can estimate
line with noise (Wei and Tanner, 1990). We can estimate  by taking the mean parameter estimate across the last few iterations of the MCEM algorithm.
by taking the mean parameter estimate across the last few iterations of the MCEM algorithm.
The imputation step involves drawing the missing data 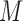 times from
times from 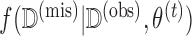 . Unlike conventional multiple imputation, missing data is drawn from the predictive distribution of the missing data evaluated at a single estimated parameter value,
. Unlike conventional multiple imputation, missing data is drawn from the predictive distribution of the missing data evaluated at a single estimated parameter value, 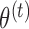 , rather than independent draws of the parameter
, rather than independent draws of the parameter  (Wei and Tanner, 1990; Neath, 2012). Therefore, the imputations produced are “improper” as described in Little and Rubin (2002). In addition to imputing missing covariate or outcome values, the MCEM algorithm will also involve imputing values for the partially latent cure status, and we will impute each type of missing data separately.
(Wei and Tanner, 1990; Neath, 2012). Therefore, the imputations produced are “improper” as described in Little and Rubin (2002). In addition to imputing missing covariate or outcome values, the MCEM algorithm will also involve imputing values for the partially latent cure status, and we will impute each type of missing data separately.
4.2. Imputation for unequal follow-up
Unequal follow-up is very common when the outcomes of interest are recurrence and death and occurs in many other semi-competing risks settings. In the supplementary material available at Biostatistics online, we present a derivation of the proposed imputation approach and provide recommendations for implementation. Here, we include a brief description of the general approach.
Let 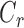 be the censoring time for recurrence and
be the censoring time for recurrence and 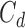 be the censoring time for death, but now assume that
be the censoring time for death, but now assume that 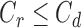 and for some subjects,
and for some subjects,  . For all subjects, we observe
. For all subjects, we observe  -censored recurrence information,
-censored recurrence information,  and
and 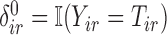 , and
, and  -censored death information,
-censored death information,  and
and 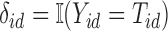 . Our goal is to impute values of
. Our goal is to impute values of 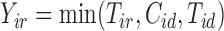 and
and  that would have been observed if we had followed subjects for recurrence as long as we followed them for death.
that would have been observed if we had followed subjects for recurrence as long as we followed them for death.
Suppose, we treat previously imputed 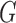 and
and  as known. Values of
as known. Values of  and
and 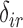 are only unknown for subjects with imputed
are only unknown for subjects with imputed 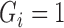 and observed
and observed  and
and  . Define
. Define  . We impute missing
. We impute missing  from a Bernoulli distribution with probability
from a Bernoulli distribution with probability
 |
If imputed  , we set
, we set  . Otherwise, we draw
. Otherwise, we draw 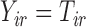 from
from
 |
4.3. Imputation of cure status
The cure status imputation approach will depend on whether we have unequal follow-up in the outcome. First, we will assume there is equal follow-up for all subjects. In this case, we can draw missing 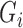 using
using 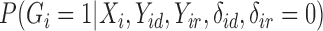 as shown in equation (2).
as shown in equation (2).
Suppose instead that we have unequal follow-up. We perform imputation of 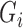 conditioning on the observed
conditioning on the observed 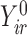 and
and  but not the imputed values of
but not the imputed values of  and
and  , which allows imputations to more easily move between
, which allows imputations to more easily move between 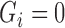 and
and 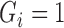 in successive iterations. We can impute missing
in successive iterations. We can impute missing 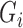 from a Bernoulli distribution using probability
from a Bernoulli distribution using probability  :
:
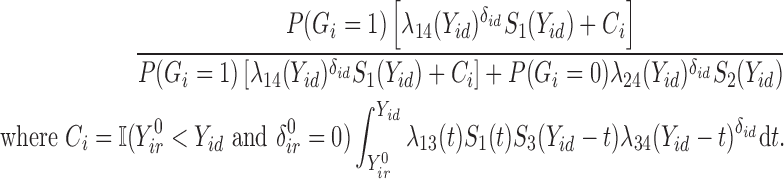 |
4.4. Imputation of missing covariates
Many methods can be used to perform the covariate imputation. For a detailed discussion of imputation methods, we refer the reader to Little and Rubin (2002). Chained equations is a popular approach in which we impute each covariate one-by-one using regression models (Van Buuren and others, 2006; Raghunathan, 2001). A method in Bartlett and others (2014) uses the structure of the outcome model in addition to a distribution for the covariates to obtain the imputation distributions. We use this approach in our simulations and data example. Let 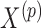 denote the
denote the 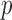 th covariate in
th covariate in 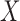 and
and 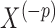 denote all but the
denote all but the 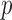 th covariate. Under missing at random assumptions (as defined in Little and Rubin, 2002), we impute each
th covariate. Under missing at random assumptions (as defined in Little and Rubin, 2002), we impute each  with missingness from a distribution proportional to
with missingness from a distribution proportional to 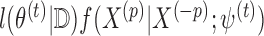 , where
, where 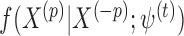 is the conditional distribution of
is the conditional distribution of 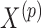 given
given 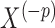 . This expression is viewed as a function of
. This expression is viewed as a function of 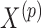 , treating all other imputed variables as fixed and using estimated values of
, treating all other imputed variables as fixed and using estimated values of 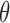 and
and 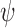 .
.
5. Estimating standard errors
The EM and MCEM algorithms provide an estimate of  , but they do not provide corresponding standard errors. Many methods have been explored in the literature for estimating standard errors for parameters estimated using an EM algorithm. Some methods involve
, but they do not provide corresponding standard errors. Many methods have been explored in the literature for estimating standard errors for parameters estimated using an EM algorithm. Some methods involve 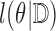 and its derivatives (e.g. Louis, 1982). Bootstrap methods are also popular. The bootstrap approach is commonly used to estimate standard errors for the CPH cure model (Sy and Taylor, 2000), and we use this approach for estimating standard errors for the EM algorithm in our simulations.
and its derivatives (e.g. Louis, 1982). Bootstrap methods are also popular. The bootstrap approach is commonly used to estimate standard errors for the CPH cure model (Sy and Taylor, 2000), and we use this approach for estimating standard errors for the EM algorithm in our simulations.
A similar bootstrap approach can be used to estimate the standard errors from the MCEM algorithm, but we do not recommend this approach due to the relative slowness of the MCEM fitting algorithm. The usual approach for estimating standard errors after an MCEM algorithm estimation of  is a generalization of Louis’s method proposed in Wei and Tanner (1990) with:
is a generalization of Louis’s method proposed in Wei and Tanner (1990) with:
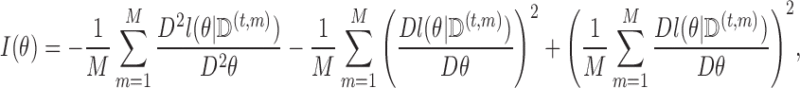 |
where 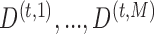 are the
are the 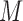 imputed versions of
imputed versions of 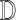 from the last iteration. The estimated covariance matrix for
from the last iteration. The estimated covariance matrix for 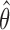 is
is  . This approach is usually implemented using large
. This approach is usually implemented using large 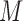 for the last few iterations of the fitting algorithm. This approach requires us to directly compute first and second derivatives of
for the last few iterations of the fitting algorithm. This approach requires us to directly compute first and second derivatives of 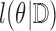 with respect to
with respect to 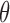 , which may not be convenient. Additionally, in the proposed MCEM algorithm, the
, which may not be convenient. Additionally, in the proposed MCEM algorithm, the 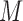 imputations at a given iteration depend on the
imputations at a given iteration depend on the  imputations from the previous iteration, so we cannot easily change the value of
imputations from the previous iteration, so we cannot easily change the value of 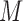 across iterations.
across iterations.
We propose a post-processing method (below) to obtain proper multiple imputations of  . After fitting the multistate cure model to each imputed data set separately, we can then use Rubin’s multiple imputation combining rules to obtain standard errors that correctly account for the uncertainty related to the missing data (Little and Rubin, 2002). This approach is convenient because (i) it does not require us to use large
. After fitting the multistate cure model to each imputed data set separately, we can then use Rubin’s multiple imputation combining rules to obtain standard errors that correctly account for the uncertainty related to the missing data (Little and Rubin, 2002). This approach is convenient because (i) it does not require us to use large  for any iterations and (ii) it does not require us to directly compute derivatives of the observed data log-likelihood. In simulations (not shown), we found similar performance under our proposed estimation method and the approach proposed in Wei 1990. However, the post-processing step is important, and skipping the post-processing resulted in undercoverage of multistate cure model parameters.
for any iterations and (ii) it does not require us to directly compute derivatives of the observed data log-likelihood. In simulations (not shown), we found similar performance under our proposed estimation method and the approach proposed in Wei 1990. However, the post-processing step is important, and skipping the post-processing resulted in undercoverage of multistate cure model parameters.
We propose the following method to obtain proper multiple imputations of  using the (improper) multiple imputations obtained within the MCEM algorithm. Our goal is to obtain
using the (improper) multiple imputations obtained within the MCEM algorithm. Our goal is to obtain  independent draws from
independent draws from  , which are our proper multiple imputations. At the end of the MCEM algorithm, we have
, which are our proper multiple imputations. At the end of the MCEM algorithm, we have 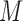 independent draws from
independent draws from 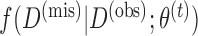 , where
, where 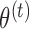 is the estimate of
is the estimate of  at the last iteration. Let
at the last iteration. Let 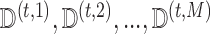 denote the imputations at the final iteration
denote the imputations at the final iteration  of the MCEM algorithm. We can obtain
of the MCEM algorithm. We can obtain  approximate draws from
approximate draws from  by performing the following for each
by performing the following for each 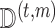 . [Step 1]: Estimate
. [Step 1]: Estimate 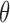 on a bootstrap sample of the most recent
on a bootstrap sample of the most recent  . Since
. Since 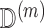 contains no missingness, this estimation is easy to perform. This results in an approximate draw of
contains no missingness, this estimation is easy to perform. This results in an approximate draw of  from
from 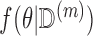 under a flat prior (Little and Rubin, 2002). [Step 2]: Using the draw of
under a flat prior (Little and Rubin, 2002). [Step 2]: Using the draw of  , obtain an updated imputation
, obtain an updated imputation 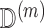 of
of  as described in Section 4. [Step 3]: Repeat Steps 1–2 several times. We can then use these proper multiple imputations of
as described in Section 4. [Step 3]: Repeat Steps 1–2 several times. We can then use these proper multiple imputations of 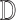 for variance estimation. We provide some theoretical justification for this approach in the supplementary material available at Biostatistics online.
for variance estimation. We provide some theoretical justification for this approach in the supplementary material available at Biostatistics online.
6. Prediction
We can use the multistate cure model fit to estimate the probability that a subject will be in a particular state at time 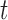 given that subject’s baseline predictors. These probabilities can be useful for predicting prognosis or exploring the potential impact of different treatments. Below, we provide expressions for estimating state occupancy probabilities over time given only baseline covariate information. In the supplementary material available at Biostatistics online, we provide examples and additional expressions for estimating these probabilities incorporating follow-up information post-baseline.
given that subject’s baseline predictors. These probabilities can be useful for predicting prognosis or exploring the potential impact of different treatments. Below, we provide expressions for estimating state occupancy probabilities over time given only baseline covariate information. In the supplementary material available at Biostatistics online, we provide examples and additional expressions for estimating these probabilities incorporating follow-up information post-baseline.
We are interested in estimating quantities related to the unmeasured (or incompletely measured) variables, 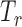 and
and 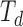 . Using the structure of the multistate cure model, we derive the following probabilities, which sum to 1 for a given
. Using the structure of the multistate cure model, we derive the following probabilities, which sum to 1 for a given 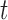 :
:
 |
These expressions can be calculated using numerical integration and a multistate cure model fit. We can estimate these probabilities for different values of 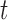 to create probability curves over time.
to create probability curves over time.
7. Simulation study
7.1. Simulation 1: bias, efficiency, and coverage of multistate cure model parameters
We simulate 500 data sets with 2000 subjects each under a multistate cure model with two covariates and Weibull baseline hazards. We simulate event times such that the hazards for the 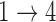 and
and 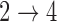 transitions are equal. All elements of
transitions are equal. All elements of  and
and 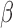 are set equal to 0.5. The baseline hazards for the event transitions were chosen to mimic a scenario we may expect to encounter in real data. Additional details can be found in the supplementary material available at Biostatistics online.
are set equal to 0.5. The baseline hazards for the event transitions were chosen to mimic a scenario we may expect to encounter in real data. Additional details can be found in the supplementary material available at Biostatistics online.
We consider three simulation scenarios: (i) no covariate missingness or unequal follow-up, (ii) covariate missingness, and (iii) unequal follow-up. For Scenarios 1 and 2, an outcome censoring time was generated from 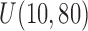 . This provides sufficient follow-up so that a clear plateau can be observed in the Kaplan–Meier estimator applied to time to recurrence, allowing the cure rate to be well estimated. For Scenario 3, censoring time for death was generated from
. This provides sufficient follow-up so that a clear plateau can be observed in the Kaplan–Meier estimator applied to time to recurrence, allowing the cure rate to be well estimated. For Scenario 3, censoring time for death was generated from  . For all but the first 750 subjects, we impose an earlier
. For all but the first 750 subjects, we impose an earlier  censoring time for recurrence. We use these simulated values to determine the observed data for each subject. This leads to roughly
censoring time for recurrence. We use these simulated values to determine the observed data for each subject. This leads to roughly 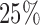 of the subjects needing imputation for unequal censoring. For Scenario 2, we impose
of the subjects needing imputation for unequal censoring. For Scenario 2, we impose 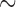 30% missing completely at random missingness in
30% missing completely at random missingness in 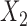 . In all scenarios, we assume subjects still at risk for recurrence after time 50 are cured. This value was chosen as to point in which the Kaplan–Meier estimator applied to recurrence shows a clear plateau.
. In all scenarios, we assume subjects still at risk for recurrence after time 50 are cured. This value was chosen as to point in which the Kaplan–Meier estimator applied to recurrence shows a clear plateau.
For each simulated data set, we fit a multistate cure model using the proposed EM (Scenario 1) or MCEM algorithm (Scenarios 2 and 3). Within each scenario, we consider different assumptions regarding baseline hazards (Weibull or Cox) and restrictions for the 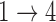 and
and  transition hazards. Variances from the MCEM algorithm are obtained using the Rubin’s rules-based approach described in Section 5 with five iterations of post-processing. We then compute the bias, empirical variance, and coverage rates of the multistate cure model parameter estimates across the 500 data sets. We also record the median run time and the number of simulations with numerical issues (non-converging M-step or difficulty with variance estimation) for each scenario.
transition hazards. Variances from the MCEM algorithm are obtained using the Rubin’s rules-based approach described in Section 5 with five iterations of post-processing. We then compute the bias, empirical variance, and coverage rates of the multistate cure model parameter estimates across the 500 data sets. We also record the median run time and the number of simulations with numerical issues (non-converging M-step or difficulty with variance estimation) for each scenario.
Tables 1 and 2 show the results. When we assume 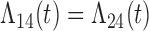 under Weibull or Cox baseline hazard assumptions, the proposed algorithms result in essentially unbiased parameter estimates with nominal coverage rates in all scenarios. When we assume
under Weibull or Cox baseline hazard assumptions, the proposed algorithms result in essentially unbiased parameter estimates with nominal coverage rates in all scenarios. When we assume 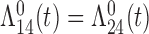 , we again see good bias and coverage properties under Weibull or Cox baseline hazards for Scenarios 1 and under Weibull baseline hazards for Scenario 2 and 3. For Scenarios 2 and 3 under Cox baseline hazards, we see increased bias and/or undercoverage for the parameters related to the
, we again see good bias and coverage properties under Weibull or Cox baseline hazards for Scenarios 1 and under Weibull baseline hazards for Scenario 2 and 3. For Scenarios 2 and 3 under Cox baseline hazards, we see increased bias and/or undercoverage for the parameters related to the 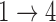 and
and  transitions and the logistic regression. When we assume
transitions and the logistic regression. When we assume 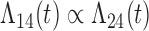 , we generally obtain good bias and coverage properties for all failure time model parameters. For the intercept in the logistic model, we tend to see some undercoverage, particularly when we assume Cox baseline hazards and when we have unequal follow-up. When we do not restrict
, we generally obtain good bias and coverage properties for all failure time model parameters. For the intercept in the logistic model, we tend to see some undercoverage, particularly when we assume Cox baseline hazards and when we have unequal follow-up. When we do not restrict 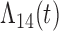 and
and  , we obtain good bias and covariate properties in Scenario 1 under Weibull baseline hazards, but we see increased bias and undercoverage in all other settings. Failing simulations provide additional evidence of numerical instability. Overall, the proposed algorithms can provide good numerical properties in all three scenarios when the assumptions for the
, we obtain good bias and covariate properties in Scenario 1 under Weibull baseline hazards, but we see increased bias and undercoverage in all other settings. Failing simulations provide additional evidence of numerical instability. Overall, the proposed algorithms can provide good numerical properties in all three scenarios when the assumptions for the  and
and  hazards are sufficiently restrictive. When the restrictions are relaxed (particularly when the baseline hazards are not equal), we can run into numerical problems, and these problems tend to be greater under Cox baseline hazard assumptions and when we have unequal follow-up.
hazards are sufficiently restrictive. When the restrictions are relaxed (particularly when the baseline hazards are not equal), we can run into numerical problems, and these problems tend to be greater under Cox baseline hazard assumptions and when we have unequal follow-up.
Table 1.
Bias, variance, and coverage of failure time model parameters corresponding to 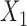 and
and 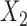 within the multistate cure model
within the multistate cure model
| Baseline hazard | 2 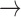 4, 1 4, 1 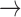 4 4 Assumption Assumption |
Failure time models | |||||||
|---|---|---|---|---|---|---|---|---|---|
1 
|
2 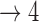
|
1 
|
3 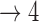
|
||||||

|
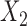
|
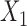
|
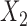
|
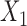
|
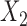
|

|

|
||
| Scenario 1: no covariate missingness or unequal follow-up | |||||||||
| Weibull |

|
0 (0.17) 94 | 0 (0.15) 97 | 0 (0.47) 94 | 0 (0.45) 96 | 0 (0.47) 94 | 0 (0.45) 96 | 0 (0.14) 95 | 0 (0.15) 95 |
| Weibull |
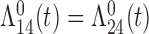
|
0 (0.18) 95 | 0 (0.16) 96 | 0 (1.03) 94 | 0 (0.97) 96 |
 1 (3.22) 93 1 (3.22) 93 |
0 (3.19) 94 | 0 (0.14) 94 | 0 (0.15) 94 |
| Weibull |
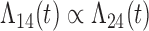
|
0 (0.17) 95 | 0 (0.15) 96 | 0 (0.48) 95 | 0 (0.47) 95 | 0 (0.48) 95 | 0 (0.47) 95 | 0 (0.14) 95 | 0 (0.15) 94 |
| Weibull | None | 0 (0.18) 95 | 0 (0.16) 96 | 0 (1.12) 95 | 1 (1.04) 96 | 0 (4.54) 95 | 0 (5.30) 95 | 0 (0.15) 95 | 0 (0.15) 95 |
| Cox |
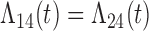
|
0 (0.18) 94 | 0 (0.15) 97 | 0 (0.48) 94 | 0 (0.45) 96 | 0 (0.48) 94 | 0 (0.45) 96 | 0 (0.15) 95 | 0 (0.16) 95 |
| Cox |

|
0 (0.18) 94 | 0 (0.16) 96 | 0 (1.05) 94 | 0 (0.97) 96 |
 1 (3.40) 94 1 (3.40) 94 |
0 (3.24) 93 | 0 (0.17) 95 | 0 (0.16) 94 |
| Cox |
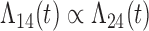
|
0 (0.18) 94 | 0 (0.16) 96 | 0 (0.48) 94 | 0 (0.47) 94 | 0 (0.48) 94 | 0 (0.47) 94 | 0 (0.15) 94 | 0 (0.16) 95 |
| Cox | None |
 1 (0.83) 70 1 (0.83) 70 |
 1 (0.82) 68 1 (0.82) 68 |
 6 (5.07) 60 6 (5.07) 60 |
 4 (5.19) 58 4 (5.19) 58 |
67 (15.1) 31 | 65 (16.7) 35 | 0 (0.15) 95 | 0 (0.16) 95 |
| Scenario 2: covariate missingness | |||||||||
| Weibull |

|
0 (0.17) 94 | 0 (0.22) 95 | 0 (0.45) 97 | 0 (0.63) 97 | 0 (0.45) 97 | 0 (0.63) 97 | 0 (0.15) 95 | 0 (0.17) 96 |
| Weibull |

|
0 (0.19) 95 | 0 (0.23) 95 | 0 (0.97) 96 | 0 (1.22) 96 |
 1 (3.36) 95 1 (3.36) 95 |
0 (4.20) 93 | 0 (0.15) 95 | 0 (0.17) 96 |
| Weibull |
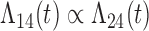
|
0 (0.17) 95 | 0 (0.22) 96 | 0 (0.47) 96 | 0 (0.64) 96 | 0 (0.47) 96 | 0 (0.64) 96 | 0 (0.47) 97 | 0 (0.64) 96 |
| Weibull | None | 0 (0.19) 94 | 0 (0.24) 94 | 1 (1.11) 94 | 0 (1.42) 95 |
 1 (3.81) 72 1 (3.81) 72 |
0 (5.28) 73 | 0 (0.14) 96 | 0 (0.18) 95 |
| Cox |
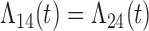
|
0 (0.18) 95 | 0 (0.22) 95 | 0 (0.46) 97 | 0 (0.64) 96 | 0 (0.46) 97 | 0 (0.64) 96 | 0 (0.16) 96 | 0 (0.19) 97 |
| Cox |
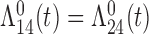
|
0 (0.20) 95 | 0 (0.24) 96 | 1 (1.07) 96 |
 3 (1.30) 95 3 (1.30) 95 |
0 (3.52) 94 | 0 (4.20) 94 | 0 (0.16) 95 | 0 (0.21) 96 |
| Cox |
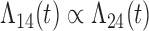
|
0 (0.18) 95 | 0 (0.23) 95 | 1 (0.49) 96 |
 2 (0.61) 95 2 (0.61) 95 |
1 (0.49) 96 |
 2 (0.61) 95 2 (0.61) 95 |
0 (0.16) 97 | 0 (0.19) 96 |
| Cox | None | 0 (0.53) 76 | 1 (0.52) 81 | 6 (6.13) 53 | 5 (5.97) 60 | 15 (28.8) 37 | 16 (26.7) 44 | 0 (0.15) 96 | 0 (0.18) 97 |
| Scenario 3: unequal follow-up | |||||||||
| Weibull |
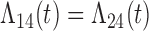
|
0 (0.19) 95 | 0 (0.21) 94 | 0 (0.50) 94 | 0 (0.50) 95 | 0 (0.50) 94 | 0 (0.50) 95 | 0 (0.16) 95 | 0 (0.16) 95 |
| Weibull |
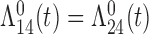
|
0 (0.21) 94 | 0 (0.22) 95 | 0 (1.05) 96 | 0 (1.03) 95 |
 1 (3.53) 94 1 (3.53) 94 |
 1 (3.39) 94 1 (3.39) 94 |
0 (0.16) 96 | 0 (0.16) 95 |
| Weibull |
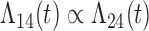
|
0 (0.19) 95 | 0 (0.21) 94 | 0 (0.50) 95 | 0 (0.51) 96 | 0 (0.50) 95 | 0 (0.51) 96 | 0 (0.16) 97 | 0 (0.16) 95 |
| Weibull | None | 0 (0.21) 94 | 0 (0.22) 95 | 1 (1.23) 95 | 0 (1.29) 93 |
 2 (4.30) 71 2 (4.30) 71 |
0 (3.63) 76 | 0 (0.17) 95 | 0 (0.17) 94 |
| Cox |
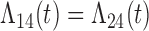
|
0 (0.20) 95 | 0 (0.21) 94 |
 1 (0.46) 94 1 (0.46) 94 |
 2 (0.49) 95 2 (0.49) 95 |
 1 (0.46) 94 1 (0.46) 94 |
 2 (0.49) 95 2 (0.49) 95 |
0 (0.16) 96 | 0 (0.16) 95 |
| Cox |
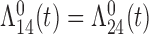
|
1 (0.23) 94 | 1 (0.24) 93 | 3 (1.39) 94 | 1 (1.40) 93 |
 3 (3.98) 92 3 (3.98) 92 |
 2 (4.15) 91 2 (4.15) 91 |
0 (0.16) 95 | 0 (0.16) 95 |
| Cox |
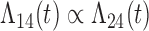
|
0 (0.21) 94 | 1 (0.23) 93 | 1 (0.55) 95 | 1 (0.55) 95 | 1 (0.55) 95 | 1 (0.55) 95 | 0 (0.16) 98 | 0 (0.16) 95 |
| Cox | None | 0 (0.65) 70 | 1 (0.61) 71 | 4 (6.49) 43 | 3 (5.75) 51 | 15 (30.5) 35 | 16 (25.0) 37 | 0 (0.16) 95 | 0 (0.17) 95 |
States 1 and 2 are the non-cured and cured baseline states. States 3 and 4 represent recurrence and death. Results across 500 simulations are presented using the following notation: bias (empirical variance) coverage of 95% confidence interval, each multiplied by 100.
Table 2.
Bias, variance, and coverage of logistic model parameters within the multistate cure model
| Baseline hazard | 2  4, 1 4, 1  4 Assumption 4 Assumption |
Logistic model | # Failed (out of 500) | Run time (min/sim) | ||
|---|---|---|---|---|---|---|
| Intercept |
|
|
||||
| Scenario 1: no covariate missingness or unequal follow-up | ||||||
| Weibull |
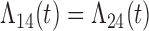
|
0 (0.33) 93 | 0 (0.42) 95 | 0 (0.44) 94 | 0 | 2.02 |
| Weibull |

|
0 (0.34) 94 | 0 (0.51) 94 | 0 (0.51) 94 | 0 | 2.12 |
| Weibull |
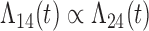
|
0 (0.59) 93 | 0 (0.41) 95 | 0 (0.45) 95 | 0 | 2.07 |
| Weibull | None | 1 (0.71) 93 | 0 (0.49) 96 | 0 (0.51) 97 | 46 | 2.16 |
| Cox |
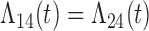
|
0 (0.33) 93 | 0 (0.42) 97 | 0 (0.44) 95 | 0 | 7.68 |
| Cox |
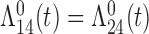
|
0 (0.34) 93 | 0 (0.51) 95 | 0 (0.52) 95 | 0 | 8.04 |
| Cox |
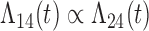
|
1 (0.89) 84 | 0 (0.43) 94 | 0 (0.48) 95 | 0 | 7.98 |
| Cox | None | 11 (0.37) 50 | 2 (2.64) 60 | 2 (2.83) 64 | 1 | 8.46 |
| Scenario 2: covariate missingness | ||||||
| Weibull |

|
0 (0.32) 96 | 0 (0.44) 97 | 0 (0.60) 95 | 0 | 5.65 |
| Weibull |

|
0 (0.33) 96 | 0 (0.54) 97 | 0 (0.70) 95 | 1 | 5.71 |
| Weibull |
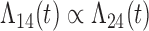
|
0 (0.63) 90 | 0 (0.47) 97 | 0 (0.60) 95 | 0 | 5.73 |
| Weibull | None | 2 (0.72) 89 | 0 (0.59) 95 | 0 (0.73) 93 | 141 | 5.57 |
| Cox |
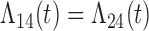
|
0 (0.32) 96 | 0 (0.46) 96 | 0 (0.60) 95 | 0 | 27.5 |
| Cox |
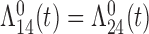
|
0 (0.33) 94 | 1 (0.58) 96 |
 2 (0.71) 93 2 (0.71) 93 |
0 | 27.3 |
| Cox |
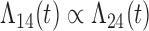
|
1 (1.59) 81 | 1 (0.50) 96 |
 3 (0.60) 94 3 (0.60) 94 |
0 | 27.4 |
| Cox | None | 6 (0.63) 80 |
 2 (2.15) 61 2 (2.15) 61 |
 2 (2.28) 67 2 (2.28) 67 |
82 | 26.7 |
| Scenario 3: unequal follow-up | ||||||
| Weibull |
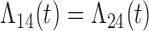
|
0 (0.35) 96 | 0 (0.51) 95 | 0 (0.48) 96 | 0 | 8.49 |
| Weibull |
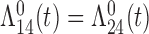
|
0 (0.35) 96 | 0 (0.64) 95 | 0 (0.63) 94 | 0 | 8.61 |
| Weibull |
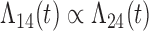
|
0 (0.63) 92 | 0 (0.54) 94 | 0 (0.51) 96 | 0 | 8.56 |
| Weibull | None | 3 (0.69) 92 | 0 (0.66) 95 | 0 (0.64) 93 | 102 | 8.58 |
| Cox |
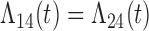
|
 2 (0.35) 94 2 (0.35) 94 |
1 (0.53) 93 | 1 (0.48) 95 | 0 | 18.3 |
| Cox |
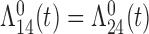
|
4 (0.43) 87 |
 2 (0.77) 93 2 (0.77) 93 |
 2 (0.81) 90 2 (0.81) 90 |
0 | 18.5 |
| Cox |
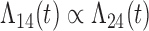
|
5 (1.95) 74 |
 2 (0.60) 94 2 (0.60) 94 |
 2 (0.57) 94 2 (0.57) 94 |
0 | 18.5 |
| Cox | None | 4 (0.70) 83 |
 1 (2.65) 53 1 (2.65) 53 |
 2 (2.44) 60 2 (2.44) 60 |
19 | 17.8 |
Results across 500 simulations are presented using the following notation: bias (empirical variance), coverage of 95% confidence interval, each multiplied by 100. The number of simulations (out of 500) with numerical issues and the median run time per simulation are also shown.
7.2. Simulation 2: multistate cure model estimates with more covariates
We simulate 500 data sets as in Scenario 1 in Simulation 1, but now we include 10 covariates in the model for each transition. All covariates are given effects of 0.5,  0.5, or 0 in
0.5, or 0 in 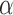 and
and  for all transitions. Additional details can be found in the supplementary material available at Biostatistics online. We then fit four different multistate cure models to the simulated data with different baseline hazards (Weibull and Cox) and different assumptions about the
for all transitions. Additional details can be found in the supplementary material available at Biostatistics online. We then fit four different multistate cure models to the simulated data with different baseline hazards (Weibull and Cox) and different assumptions about the 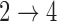 and
and 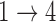 transitions.
transitions.
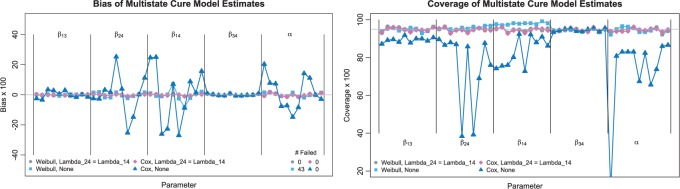
Figure 2 presents the bias and coverage for the estimated 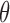 from each model fit. When we assume
from each model fit. When we assume 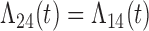 , we obtain essentially unbiased parameter estimates with nominal coverage under both Weibull and Cox baseline hazard assumptions. When we assume Weibull baseline hazards and no
, we obtain essentially unbiased parameter estimates with nominal coverage under both Weibull and Cox baseline hazard assumptions. When we assume Weibull baseline hazards and no 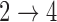 and
and 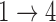 hazard restrictions, we see some increased bias and overcoverage for estimating
hazard restrictions, we see some increased bias and overcoverage for estimating 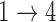 parameters, but otherwise we have good bias and coverage properties. However, we do see evidence of model instability as 119 out of the 500 simulations had numerical issues. When we assume Cox baseline hazards and no
parameters, but otherwise we have good bias and coverage properties. However, we do see evidence of model instability as 119 out of the 500 simulations had numerical issues. When we assume Cox baseline hazards and no 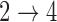 and
and 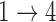 hazard restrictions, we see substantial bias and/or undercoverage, particularly in estimating
hazard restrictions, we see substantial bias and/or undercoverage, particularly in estimating  ,
, 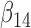 , and
, and  . As in Simulation 1, we see evidence of estimation instability in general when we place no restrictions on the hazards for the
. As in Simulation 1, we see evidence of estimation instability in general when we place no restrictions on the hazards for the 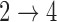 and
and 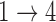 transitions.
transitions.
Fig. 2.
Bias and coverage of multistate model estimates with ten covariates. The plot on the left shows the bias (x100) of the multistate cure model parameters across 500 simulations, and the corresponding 95% confidence interval coverage rates are shown in the right plot. State 1 is the non-cured baseline state, State 2 is the cured baseline state, State 3 is the recurrence state, and State 4 is the death state.
8. Application to study of head and neck cancer recurrence and death
We consider data from a cohort study of 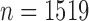 patients treated for head and neck squamous cell carcinoma (HNSCC). This study was conducted by the University of Michigan Head and Neck Specialized Program of Research Excellence (SPORE) and followed patients who were treated for HNSCC at the University of Michigan Cancer Center, the Henry Ford Hospital in Detroit, and the VA Hospital in Ann Arbor between 2003 and 2014. Details about this study can be found in Duffy and others (2008), Peterson and others (2016), and in the supplementary material available at Biostatistics online.
patients treated for head and neck squamous cell carcinoma (HNSCC). This study was conducted by the University of Michigan Head and Neck Specialized Program of Research Excellence (SPORE) and followed patients who were treated for HNSCC at the University of Michigan Cancer Center, the Henry Ford Hospital in Detroit, and the VA Hospital in Ann Arbor between 2003 and 2014. Details about this study can be found in Duffy and others (2008), Peterson and others (2016), and in the supplementary material available at Biostatistics online.
After treatment, patients were followed for recurrence and death. Covariate information was collected at baseline. We are interested in studying the association between these covariates and the time to HNSCC recurrence and death after treatment. Additionally, it is been well established that some head and neck cancer patients can be cured of their cancer through their primary treatment, and we are interested in identifying factors related to the underlying cure probability (Taylor, 1995; Grau and others, 1997; Cognetti and others, 2008). A previous analysis of an earlier version of these data explores time to recurrence and cure probability in a CPH cure model, but this analysis does not incorporate death information (Beesley and others, 2016).
Missing data, however, poses an additional complication. For many patients (62.3%), follow-up for recurrence was substantially shorter than for death. Human papillomavirus (HPV) status was unavailable for 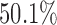 of the subjects, and a small amount of missingness was present in other variables. We restricted our analyzes to subjects with oropharynx, oral cavity, larynx, or hypopharynx cancer. We further restricted analyzes to subjects that appeared to clear their cancer through the initial treatment.
of the subjects, and a small amount of missingness was present in other variables. We restricted our analyzes to subjects with oropharynx, oral cavity, larynx, or hypopharynx cancer. We further restricted analyzes to subjects that appeared to clear their cancer through the initial treatment.
We fit a multistate cure model assuming Weibull baseline hazards and equal  and
and 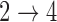 hazards using 100 iterations of the proposed MCEM algorithm. Subjects still at risk for recurrence and death after 80 months were assumed to be cured. Missing data were imputed using the method proposed in Bartlett and others (2014) as described in Section 4.4. Unequal follow-up was handled using the approach described in Section 4.2. Standard errors were estimated by treating the most recent imputations as proper imputations (after post-processing) and estimating variance within each imputed data set using 50 bootstrap samples. Rubin’s rules were then used to obtain the final estimates for the standard errors.
hazards using 100 iterations of the proposed MCEM algorithm. Subjects still at risk for recurrence and death after 80 months were assumed to be cured. Missing data were imputed using the method proposed in Bartlett and others (2014) as described in Section 4.4. Unequal follow-up was handled using the approach described in Section 4.2. Standard errors were estimated by treating the most recent imputations as proper imputations (after post-processing) and estimating variance within each imputed data set using 50 bootstrap samples. Rubin’s rules were then used to obtain the final estimates for the standard errors.
Figure 3 presents the results of the multistate cure model fit to the head and neck cancer data set. Higher cancer stage and HPV negativity were associated with higher rates of recurrence for non-cured subjects. Greater age, higher cancer stage, worse comorbidities, and increased smoking history were associated with higher rates of death from other causes for both cured and non-cured subjects. Higher cancer stage and increased smoking history were associated with higher rates of death after recurrence, and larynx site was associated with lower rates of death after recurrence compared to oral cavity cancer. Higher cancer stage and HPV negativity were associated with lower rates of cure. Additional details about the data set, this model fit, and a comparison to a standard illness-death model fit are included in the supplementary material available at Biostatistics online.
Fig. 3.
Results of applying MCEM algorithm to head and neck data. The corresponding 95% confidence intervals for each part of the multistate cure model are shown for each model covariate. Stars indicate confidence intervals that are significant at 0.05. State 1 is the non-cured baseline state, State 2 is the cured baseline state, State 3 is the recurrence state, and State 4 is the death state.
9. Discussion
In the study of cancer, multistate cure models can be used to identify factors related to the rate of cancer recurrence, the rate of death before and after recurrence, and the probability of being cured by initial treatment. Additionally, multistate cure models can be very useful for prediction. However, the previous method for fitting multistate cure models requires substantial custom programming, making multistate cure models less accessible to analysts. We are interested in developing methods for fitting multistate cure models that can be implemented more easily and can incorporate different modeling assumptions into the fitting procedure.
In this article, we proposed an Expectation–Maximization (EM) algorithm for fitting the multistate cure model using maximum likelihood. The proposed algorithm can be fit using standard software, can incorporate either parametric or non-parametric baseline hazards for the state transition rates, and can integrate parameter restrictions for the transitions to death from other causes for cured and non-cured subjects. We then propose a Monte Carlo EM (MCEM) algorithm for fitting the multistate cure model in the presence of covariate missingness and/or unequal follow-up of the two outcomes, and we provide some software.
In simulations, the proposed EM and MCEM algorithms demonstrate good bias and coverage properties when the modeling assumptions are sufficiently restrictive. Additionally, we can still see good model fitting performance when we include more covariates in the model. When the 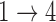 and
and 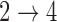 restrictions are relaxed (particularly when the baseline hazards are not restricted to be equal), we can run into numerical problems in some settings, suggesting that care should be taken to make reasonably restrictive modeling assumptions for these transitions. Additional exploration (not shown) suggests that these numerical issues stem from identifiability issues related to the
restrictions are relaxed (particularly when the baseline hazards are not restricted to be equal), we can run into numerical problems in some settings, suggesting that care should be taken to make reasonably restrictive modeling assumptions for these transitions. Additional exploration (not shown) suggests that these numerical issues stem from identifiability issues related to the 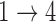 and
and 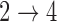 model parameters that occur when we have weaker restrictions for the two transitions. When applying the multistate cure model to a particular data set, we recommend fitting a model under different assumptions about the
model parameters that occur when we have weaker restrictions for the two transitions. When applying the multistate cure model to a particular data set, we recommend fitting a model under different assumptions about the 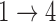 and
and 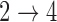 transitions and evaluating convergence properties and goodness of fit to determine if the
transitions and evaluating convergence properties and goodness of fit to determine if the 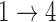 and
and 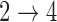 hazard restrictions can be reasonably relaxed. See Conlon and others (2013) for a discussion of goodness of fit diagnostics for multistate cure models.
hazard restrictions can be reasonably relaxed. See Conlon and others (2013) for a discussion of goodness of fit diagnostics for multistate cure models.
We applied the proposed MCEM algorithm to study cancer recurrence and death rates in subjects with head and neck cancer. Higher age, worse comorbidities, and increased smoking were associated with higher rates of death from other causes for both cured and non-cured subjects. Increased smoking was also associated with higher rates of death after recurrence, and larynx subsite was associated with lower rates of death after recurrence. HPV positive subjects had significantly lower rates of recurrence and higher rates of cure. Higher cancer stage was significantly associated with all transition rates and the probability of being cured.
Given the various types of imputation required, the relative advantages of the MCEM algorithm over the Bayesian MCMC algorithm should be considered. One disadvantage of the Bayesian approach (even if we apply our proposed imputation methods) is that we require accept–reject methods to draw the parameters, which involves careful consideration of parameter tuning, acceptance rates, and mixing. In contrast, the M-step of the MCEM algorithm is very simple to perform. As a result, the MCEM algorithm can perform estimation more quickly than the Bayesian Markov chain monte carlo. However, unlike with Bayesian approaches, the standard errors of 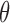 using MCEM are not readily available. Additionally, the Bayesian approach allows the user to more directly incorporate prior assumptions into the estimation. The relative merits of the two approaches may depend on the data and the experience of the analyst.
using MCEM are not readily available. Additionally, the Bayesian approach allows the user to more directly incorporate prior assumptions into the estimation. The relative merits of the two approaches may depend on the data and the experience of the analyst.
As illustrated by the head and neck cancer example, multistate cure models offer investigators a useful tool for identifying factors involved in different parts of the disease process, and they can be used for prediction and in medical decision-making. Multistate cure models can be applied in a variety of settings and are certainly not limited to the study of cancer. In this article, we developed methods to make multistate cure models easier to fit in practice. Previous work focused on the setting with fully parametric baseline hazards, and our proposed methods allow us to choose parametric or non-parametric baselines and incorporate different assumptions about the transitions to death from other causes. The novel imputation-based approach for dealing with unequal follow-up proposed as part of the MCEM algorithm can be applied in general semi-competing risks settings. Additionally, we propose a novel approach for obtaining standard errors for an MCEM algorithm, which can be applied in other MCEM settings. The proposed methods provide a convenient estimation method for fitting the multistate cure model and increased flexibility in model specification over existing methods.
10. Software
Software in the form of an R package called MultiCure is available on GitHub. This package provides functions for fitting the multistate cure model via the proposed EM and MCEM algorithms and for estimating corresponding standard errors. This package also includes functions for estimating the derived state occupancy probabilities.
Supplementary Material
Acknowledgments
The authors cite the many investigators (listed in Beesley and others, 2016) in the University of Michigan Head and Neck Specialized Program of Research Excellence for their contributions to patient recruitment, specimen collection, and study conduct. The authors gratefully acknowledge funding from NIH. Conflict of Interest: None declared.
Funding
National Institutes of Health (NIH) (CA83654 and CA129102).
References
- Andersen P. K. and Keiding N. (2002). Multi-state models for event history analysis. Statistical Methods in Medical Research 11, 91–115. [DOI] [PubMed] [Google Scholar]
- Bartlett J. W., Seaman S. R., White I. R. and Carpenter J. R. (2014). Multiple imputation of covariates by fully conditional specification: accommodating the substantive model. Statistical Methods in Medical Research 24, 462–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beesley L. J., Bartlett J. W., Wolf G. T. and Taylor J. M. G. (2016). Multiple imputation of missing covariates for the Cox proportional hazards cure model. Statistics in Medicine 35, 4701–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C., Zou Y., Peng Y. and Zhang J. (2012). smcure: an R-package for estimating semiparametric mixture cure models. Computer Methods and Programs in Biomedicine 108, 1255–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognetti D. M, Weber R. S. and Lai S. Y. (2008). Head and neck cancer: an evolving treatment paradigm. Cancer 113, 1911–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon A. S. C., Taylor J. M. G. and Sargent D. J. (2013). Multi-state models for colon cancer recurrence and death with a cured fraction. Statistics in Medicine 33, 1750–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wreede L. C., Fiocco M. and Putter H. (2011). mstate: an R package for the analysis of competing risks and multi-state models. Journal of Statistical Software 38, 1–30. [Google Scholar]
- Dempster A. P., Laird N. M. and Rubin D. B. (1977). Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society 39, 1–38. [Google Scholar]
- Duffy S., Taylor J. M. G., Terrell J., Islam M., Yuan Z., Fowler K., Wolf G. and Teknos T. (2008). IL-6 predicts recurrence among head and neck cancer patients. Cancer 113, 750–757. [DOI] [PubMed] [Google Scholar]
- Farewell V. T. (1982). The use of mixture models for the analysis of survival data with long-term survivors. Biometrics 38, 1041–1046. [PubMed] [Google Scholar]
- Frydman H. and Kadam A. (2004). Estimation in the continuous time mover stayer model with an application to bond ratings migration. Applied Stochastic Models in Business and Industry 20, 155–170. [Google Scholar]
- Grau J. J., Cuchi A., Traserra J., Arias C., Blanch J. L. and Estapé J. (1997). Follow-up study in head and neck cancer: cure rate according to tumor location and stage. Oncology 54, 38–42. [DOI] [PubMed] [Google Scholar]
- Jackson C. H. (2011). Multi-state models for panel data: the msm package for R. Journal of Statistical Software 38, 1–28. [Google Scholar]
- Kuk A. Y. C. and Chen C.-H. (1992). A mixture model combining logistic regression with proportional hazards regression. Biometrika 79, 531–541. [Google Scholar]
- Little R. J. A. and Rubin D. B. (2002). Statistical Analysis with Missing Data, 2nd edition Hoboken, NJ: John Wiley and Sons, Inc. [Google Scholar]
- Louis T. A. (1982). Finding the observed information matrix when using the EM algorithm. Journal of the Royal Statistical Society 44, 226–233. [Google Scholar]
- Meira-Machado L., De Uña-Álvarez J., Cadarso-Suárez C. and Andersen P. K. (2009). Multi-state models for the analysis of time-to-event data. Statistical Methods in Medical Research 18, 195–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neath R. C. (2012). On convergence properties of the Monte Carlo EM algorithm. Institute of Mathematical Statistics 10, 1–21. [Google Scholar]
- Peng Y. and Dear K. B G. (2000). A nonparametric mixture model for cure rate estimation. Biometrics 56, 237–243. [DOI] [PubMed] [Google Scholar]
- Peterson L. A., Bellile E. L., Wolf G. T., Virani S., Shuman A. G. and Taylor J. M. G. (2016). Cigarette use, comorbidities, and prognosis in a prospective head and neck squamous cell carcinoma population. Head and Neck 38, 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putter H., Fiocco M. and Geskus R. B. (2007). Tutorial in biostatistics: competing risks and multi-state models. Statistics in Medicine 26, 2389–2430. [DOI] [PubMed] [Google Scholar]
- Raghunathan T. E. (2001). A multivariate technique for multiply imputing missing values using a sequence of regression models. Survey Methodology 27, 85–95. [Google Scholar]
- Sy J. P. and Taylor J. M. G. (2000). Estimation in a Cox proportional hazards cure model. Biometrics 56, 227–236. [DOI] [PubMed] [Google Scholar]
- Taylor J. M. G. (1995). Semiparametric estimation in failure time mixture models. Biometrics 51, 899–907. [PubMed] [Google Scholar]
- Van Buuren S., Brand J. P. L., Groothuis-Oudshoorn C. G. M. and Rubin D. B. (2006). Fully conditional specification in multivariate imputation. Journal of Statistical Computation and Simulation 76, 1049–1064. [Google Scholar]
- Wei G. C. G. and Tanner M. A. (1990). A Monte Carlo implementation of the EM algorithm and the poor man’s data augmentation algorithms. Journal of the American Statistical Association 85, 699–704. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.