Abstract
Phylogenomics is the practice of attempting to infer evolutionary relationships at a genome-level. This is becoming a standard step in the characterization of newly recovered genomes and to direct/constrain further research; yet the process from start to finish of building a de novo phylogenomic tree that is specific to the organisms of interest can still be computationally intractable for many biologists. GToTree is a recently published user-friendly workflow for phylogenomics intended to give more researchers the capability to generate phylogenomic trees to help guide their work. This commentary describes two common applications where GToTree can be helpful and then discusses some things to consider when using the program.
Keywords: phylogenomics, phylogenetics, bioinformatics
Comment on: Lee MD. GToTree: a user-friendly workflow for phylogenomics [published online ahead of print March 13, 2019]. Bioinformatics. doi:10.1093/bioinformatics/btz188. PMID: 30865266. https://www.ncbi.nlm.nih.gov/pubmed/30865266.
Introduction
The reconstruction of genomes from cultures and from culture-independent methods is now commonplace and continuing to accelerate. A standard step in the characterization of newly recovered genomes is placing them into a phylogenomic context. One of the reasons this is done is to assign a robust, genome-level taxonomic label, and there are excellent tools available designed that achieve this by inserting new genomes into a pre-constructed, reference phylogenomic tree (eg, CheckM1 and GTDB-Tk).2 GToTree differs from these in that it is not designed for assigning taxonomy, but rather for constructing de novo phylogenomic trees. Depending on the researcher’s goals, assigning taxonomy may be sufficient. But if the work being done would benefit from developing an understanding of the evolutionary landscape surrounding the organism(s) of interest in relation to other genomes—which is needed for purposes such as identifying appropriate relatives/groups for comparative genomics or pursuing ancestral reconstruction—then taxonomy alone is not sufficient and GToTree may be helpful. GToTree is also useful at broad-level scales, such as in generating a phylogenomic tree showing the distribution of a trait of interest across all bacterial genomes; for instance, see Example 1.
For a general summary of a typical workflow, GToTree3: (1) takes as input any combination of National Center for Biotechnology Information (NCBI) assembly accessions, GenBank files, nucleotide fasta files, and/or amino acid fasta files (compressed or uncompressed); (2) identifies4 single-copy genes (SCGs) suitable for phylogenomic analysis dependent on the breadth of the organisms being considered (using one of the 15-included SCG-sets or a user-provided one); (3) reports estimates of genome completion and redundancy; (4) handles filtering of genomes and target genes with adjustable parameters; (5) aligns5 and trims6 each group of target genes before concatenating all into one alignment (a partitions file describing the positions of individual genes is reported should the user wish to take the alignment and build a mixed-model tree elsewhere); (6) optionally replaces (or appends to) initial genome labels with taxonomy7 or user-specific information so the final alignment and tree can be more easily explored; (7) generates a phylogenomic tree.8,9 Here, two common applications of GToTree are presented.
Common Applications
Annotated code for these examples is available at the GToTree wiki on github (https://github.com/AstrobioMike/GToTree/wiki/example-usage), and complete data and log files can be downloaded from https://doi.org/10.6084/m9.figshare.8239763.
Example 1—visualization of flagellar-biosynthesis protein FliT across the bacterial domain
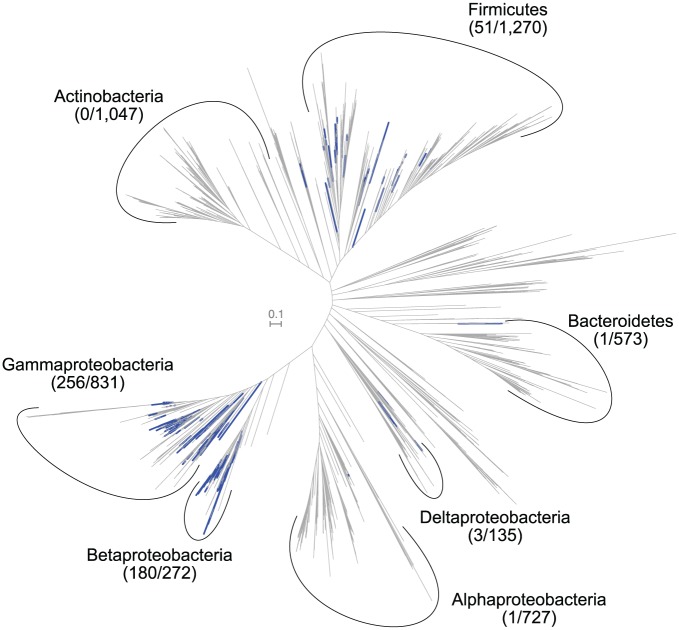
Depending on the scope of the question being pursued, it can sometimes be useful to visualize a trait or characteristic across an entire domain or large clade, as this may reveal patterns about its evolutionary distribution. The presence or absence of a particular trait in a given genome can be acquired in any fashion—eg, using annotations already available, using a program to identify a specific antibiotic resistance marker, etc. In this case, hidden Markov model (HMM) searches were performed with HMMER34 using a profile available from Pfam10 to identify the presence of the flagellar protein FliT11 (PF05400.13) in all bacterial genomes available from NCBI’s RefSeq12 (5550 genomes including “reference” or “representative” only accessed on June 3, 2019; see code linked above for exact retrieval and scanning). GToTree already uses HMMs to find target SCGs, but optionally can search for any additional target Pfam10 accessions. While searching all input genomes for the target SCGs for building the tree, if provided a file of additional PFam10 targets, GToTree will also search for these in each genome and: 1) pull out and store the identified sequences; 2) generate a count table of hits per genome; and 3) generate an Interactive Tree of Life13 (iToL)-compatible file for each target so all can be easily visualized at the iToL13 site. The results of this are presented in Figure 1. The distribution of this particular gene is highly concentrated within the Betaproteobacteria (with ~66% possessing at least 1 copy) and Gammaproteobacteria (~31%)—the class in which it was first studied.11 It is detected more sparsely in Firmicutes (~4%), while only 3 members of Deltaproteobacteria, 1 Alphaproteobacteria (Thalassospira profundimaris; GCF_000300275.1; WP_008890385.1), and 1 Bacteroidetes (Salinibacter ruber; GCF_000013045.1; YP_446723.1) had hits returned. Worth noting again is that all the sequences for these hits were already recovered and stored, so those are ready to be explored phylogenetically individually if wanted.
Figure 1.
Distribution of identified FliT genes across 5550 bacterial genomes (a total of 521 genomes had at least one copy of the gene identified within it, indicated by blue branches). Numbers in parentheses represent number of genomes of the total within each major clade that had at least one copy detected.
Example 2—placing a newly recovered genome into a phylogenomic context
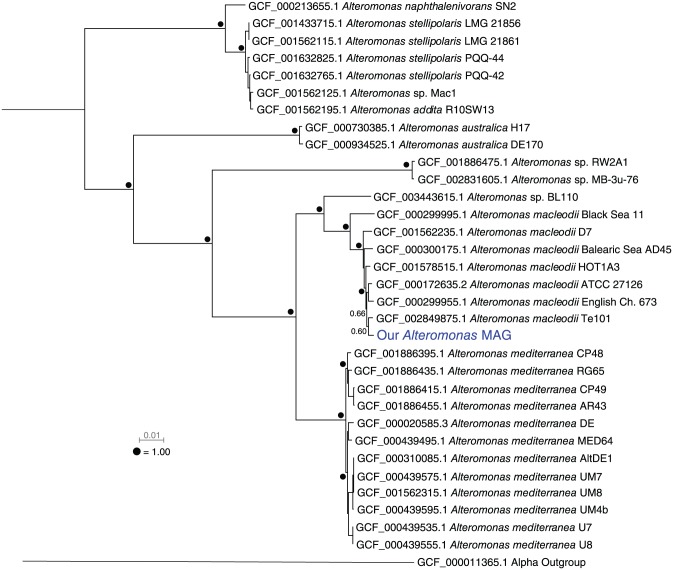
As mentioned above, sometimes getting a taxonomic label for a genome is just the start of things. If the goal is something like comparative genomics or ancestral reconstruction, then some understanding of the evolutionary relatedness between your genome(s) of interest and others is needed. Here is an example working with an Alteromonas metagenome-assembled genome (MAG; GCA_002271865.1) from a nitrogen-fixing, cyanobacterium enrichment culture.14 In most cases, a newly recovered genome will be in nucleotide fasta format. Part of the convenience of GToTree is its flexibility with regard to input format, so it is easy to combine new genomes with references. One way to retrieve references for this case is through a search at NCBI’s web interface with the search string “Alteromonas[ORGN] AND ‘latest refseq’ [filter] AND ‘complete genome’ [filter].” From the results, the accessions can be downloaded of all genomes that meet these criteria (this was 31 as accessed on January 1, 2019), and this accession list can be input into GToTree along with a fasta file of our newly recovered genome. Here, since we know we are working with all Gammaproteobacteria, we can select the Gammaproteobacteria SCG-set that comes packaged with GToTree that contains 172 SCGs specific to that class. And being constrained to this class makes it relatively straightforward to include an outgroup as well, so a member of Alphaproteobacteria was added to the accessions list (GCF_000011365.1). If wanted, GToTree also uses TaxonKit7 to add lineage information to genome labels, so the final tree is more easily explored. In this example, Genus, Species, and Strain were specified, but all ranks can be displayed. From the resultant tree in Figure 2, we see that based on the 172 gammaproteobacterial SCGs, our new Alteromonas MAG is of the species macleodii and is particularly closely related to strains Te101, English Channel 673, and ATCC27126. Beyond taxonomy and knowing the most similar reference genome available, having some idea of this layout can help guide further decisions. For instance, if comparative genomics were the goal, it would help to know at the start that the species A. macleodii and A. mediterranea are much more closely related to each other than either are to A. stellipolaris or A. naphthalenivorans (Figure 2). Otherwise, if the goal is pursuing pangenomics with an analysis and visualization program like anvi’o,15 knowing the evolutionary landscape like this can also help in deciding which reference genomes to include.
Figure 2.
Placing a newly recovered Alteromonas MAG into a phylogenomic context by providing GToTree solely NCBI accessions for the references and a fasta file of the newly recovered MAG. MAG indicates metagenome-assembled genome.
Things to Consider When Using GToTree
An important caveat on the idea of a workflow for phylogenomics
Phylogenetics is an incredibly complicated field, and things become even more complicated when working with many concatenated genes as is the case with phylogenomics. GToTree is meant to be a relatively high-throughput, user-friendly, and reproducible workflow. But anything designed this way needs to inherently sacrifice something in terms of flexibility and options. It is important that users new to this arena understand that many things impact the outcome of a phylogenetic/phylogenomic analysis, particularly including the alignment and the approach and model used for tree construction. Currently, GToTree employs only one alignment tool, and two options for tree construction, and not all options for these programs are available within the GToTree program. Users can take the concatenated alignment output by GToTree and use it with any tree assembly program and take full advantage of their entire functionality externally, including taking the partitions file GToTree generates that allows the use of mixed-model trees (where each gene can be assigned a different evolutionary model) in programs like IQ-TREE.9 It is also important that users new to this arena understand that no program or tool provides an “absolute answer” or the “truth.” Phylogenetic analysis is merely an attempt to model the evolutionary history and relatedness of some group of things.
Best-hit mode
By default, if a given genome has more than one hit to a specific HMM profile (target gene), GToTree will not include a sequence for that target gene from that genome in the final concatenated alignment. It will insert a gap-sequence just as would be the case if that genome had no hits to the target gene. This is a conservative approach, because if there are multiple copies of a target SCG within a genome, they may not all be under the same selective evolutionary pressures, which is a core assumption in phylogenetics. To override this, the user can provide the `-B` flag to tell GToTree to run in “best-hit” mode. In this case, when a given genome has more than one hit to a specific target gene, GToTree will take the best hit (based on HMMER34 output e-value).
Filtering hits by gene-length
GToTree is designed to automate the process of identifying and aligning many genes from many genomes, and it is not practical to manually inspect all gene hits. As any search method will occasionally return spurious hits, and these might then affect the alignment of that group of genes, GToTree has a built-in filtering parameter to try to conservatively address this by considering the length of any individual identified gene (hit) in relation to the median length of all hits of a given target gene. This is set with the `-c` option, and the default is 0.2. This means for target gene “A,” if the median length of all hits to target gene “A” is 100 amino acids, then any individual hit to target gene “A” that is greater in length than 120 amino acids or shorter in length than 80 will be removed from the analysis, and that genome will receive a gap-sequence for that target gene. This seems to work well, but only when there are enough genes in the gene set to give somewhat of a normal distribution. Meaning, at the extreme end, if there were only 3 genes to consider, and their lengths were 100, 100, and 102, the 102-length gene would be filtered out, even though it probably should not be in this case. If running GToTree with very few input genomes, you might consider increasing this threshold (set with the `-c` flag) and/or visually inspecting some of the alignments.
Filtering genomes by fraction of hits to target SCGs
To deal with cases where too few target genes were identified in a given input genome, GToTree has a parameter set with the `-G` flag that will filter out genomes with fewer hits than the specified proportion. The default value is 0.5, which means if searching for 100 target genes, if a genome has single-hits to fewer than 50 of those targets, it will be removed from analysis and reported.
Are eukaryotic genomes appropriate for use with GToTree?
If only using highly conserved ribosomal proteins that span all 3 domains, and/or if genes are already identified (eg, the input source is an NCBI accession with gene calls or a GenBank file with gene calls), then GToTree is suitable for working with Eukaryotes in addition to Bacteria and Archaea. If no gene calls are available, then GToTree is likely not suitable for eukaryotic genomes as the only gene-caller currently implemented is prodigal,16 which is designed for prokaryotes.
Conclusions
Phylogenomic analysis is becoming a standard step in the characterization of newly recovered genomes, but the computational barriers for the typical biologist can be much greater than for performing a phylogenetic analysis of an individual marker-gene (eg, the 16S ribosomal RNA gene). GToTree handles a lot of the computational steps involved in a typical phylogenomic workflow (eg, downloading genomes, handling different file formats, filtering genes and genomes, swapping labels, concatenating genes), making it feasible for more researchers to create phylogenomic trees to aid in their work.1 This commentary exemplified two common applications of the program and discusses some considerations when using GToTree.
Acknowledgments
The author would like to thank Jonathan A. Eisen, R. Craig Everroad, and Shengwei Hou for their thoughts and suggestions while putting this article together.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded in part by NASA Space Biology under grant NNH16ZTT001N-MOBE and by NASA’s Planetary Science Division (Astrobiology Program).
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: MD Lee is the sole contributor to this work.
ORCID iD: Michael D Lee  https://orcid.org/0000-0001-7750-9145
https://orcid.org/0000-0001-7750-9145
References
- 1. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM : assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Parks DH, Chuvochina M, Waite DW, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 3. Lee MD. GToTree: a user-friendly workflow for phylogenomics [published online ahead of print March 13, 2019]. Bioinformatics. doi: 10.1093/bioinformatics/btz188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eddy SR. Accelerated profile HMM searches. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformat. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gutíerrez SC, Martínez JMS, Gabaldón T. TrimAl: a tool for automatic alignment trimming. Bioinformatics. 2009;25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shen W, Xiong J. TaxonKit: a cross-platform and efficient NCBI taxonomy toolkit. bioRXiv. https://www.biorxiv.org/content/10.1101/513523v1.Up-dated2019. doi: 10.1101/513523. [DOI]
- 8. Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. El-gebali S, Mistry J, Bateman A, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bennett JCQ, Thomas J, Fraser GM, Hughes C. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Mol Microbiol. 2001;39:781–791. doi: 10.1046/j.1365-2958.2001.02268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O’Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Letunic I, Bork P. Interactive Tree of Life (iToL) v4: recent updates and new developments. Nucleic Acids Res. 2019. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee MD, Walworth NG, McParland EL, et al. The Trichodesmium consortium: conserved heterotrophic co-occurrence and genomic signatures of potential interactions. ISME J. 2017;11:1813–1824. doi: 10.1038/ismej.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eren AM, Esen ÖC, Quince C, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hyatt D, Locascio PF, Hauser LJ, Uberbacher EC. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics. 2010;28:2223–2230. doi: 10.1093/bioinformatics/bts429. [DOI] [PubMed] [Google Scholar]