Abstract
Background
The transmission of methicillin-resistant Staphylococcus aureus (MRSA) between humans and animals has been identified in a number of countries. In this study, MRSA in urban rodents and shrews in a community was investigated. Further, comparisons of MRSA isolates from rodents, shrews, and humans were conducted to evaluate the relationships of these isolates from different origins.
Results
Between 2015 and 2016, 397 oropharynx samples from 212 rodents and 185 shrews, and 8 MRSA isolates from hospital patients were collected. Twelve MRSA were isolated from the small mammals (3.0, 95%CI: 1.3–4.7%), including 11 isolates from rodents and one from a shrew. Three MRSA isolates from Rattus norvegicus were PVL-positive, and seven isolates were IEC-negative (one from Suncus murinus, five from Rattus norvegicus, and one from a patient). The spa type, MLST, and antimicrobial resistance patterns showed that the MRSA retrieved from rodents and shrews are likely related to human strains.
Conclusion
MRSA derived from rodent shares similar antimicrobial resistance and molecular characteristics to those from humans, suggesting that urban rodents may play as maintenance host or vectors for MRSA which is important to human health.
Keywords: Urban rodent, House shrew, Methicillin-resistant Staphylococcus aureus, Antimicrobial resistance, Multilocus-sequence typing
Background
Staphylococcus aureus (S. aureus) is an important opportunistic pathogen, which causes a wide range of infections in humans and animals [1]. S. aureus has the capacity to rapidly develop resistance to most antibiotics used clinically [2]. For example, in 1961, 2 years after the clinical introduction of methicillin in the United Kingdom, the first methicillin-resistant S. aureus (MRSA) strain was identified [3]. Since then, MRSA infections have increased worldwide. In 2014 the World Health Organization reported that for all-cause mortality, MRSA had a Relative Risk (RR) of 1.61 compared to methicillin-susceptible S. aureus (MSSA) infections (95% Confidence Interval, CI: 1.43–1.82) [4]. Initially, MRSA was primarily hospital associated but in the late 1990s MRSA emerged as a community-associated infection (CA-MRSA) [5]. CA-MRSA caused serious infections in younger and healthier individuals and was unusually virulent with a marked capacity to disseminate throughout the community [6].
S. aureus methicillin resistance is due to the acquisition of the mecA gene that encodes a penicillin-binding protein (PBP 2a) with a low affinity for β-lactams [7]. A number of investigations have identified animals as potential reservoirs of mecA-containing staphylococci [8, 9]. Multiple animal-associated staphylococci have also been identified as the probable origin of methicillin resistance conferred by mecA [10, 11]. These reservoirs include companion animals [12, 13], livestock [14], and wildlife [15]. Transmission of MRSA between animals and humans, in both directions, has been well documented [16, 17].
In addition to domestic animals and livestock, urban wildlife such as rodents and shrews can also serve as reservoirs for zoonotic bacterial disease [18–20]. The first such report was livestock-associated MRSA (LA-MRSA) in rats living on pig farms in the Netherlands [21] in 2009, and more recently in urban Norway rats in Canada [22]. The Norway rat (Rattus norvegicus) [23] and the Asian house shrew (Suncus murinus) [24] are predominantly commensal animals in urban areas within regions of China. However, to the best of our knowledge, no information is available regarding the prevalence, antimicrobial resistance, or genetic characteristics of MRSA sampled from urban rodents in China. Additionally, there was no report on description of MRSA in shrews throughout the world.
The aims of this study were to determine the carriage rate, drug-resistance, and genetic characteristics of S. aureus and MRSA in urban rodents and shrews in Guangzhou City, Southern China. Further, in order to evaluate the relationships of MRSA isolates among these animals and humans, comparisons of molecular characterization and antibiotic susceptibility were conducted.
Results
Animal trapping and bacterial isolation
In total, 397 individual animals, including 212 rodents (197 Rattus norvegicus and 15 Rattus tanezumi) and 185 shrews (185 Suncus murinus), were captured between June 2015 and May 2016 (Table 1). They were captured near the garbage bin, on the lawn, and near house building in relation to human habitation.
Table 1.
Distribution of S. aureus and MRSA carriage in urban rodents and shrews trapped between 2015 and 2016 in Guangzhou, Southern China
| Species | No. of captures |
S. aureus no. (%) |
MRSA no. (%) |
|---|---|---|---|
| Rodents | 212 | 87/212 (41.0) | 11/212 (5.2) |
| Rattus norvegicus | 197 | 83/197 (42.1) | 11/197 (5.6) |
| Rattus tanezumi | 15 | 4/15 (26.7) | 0/15 (0) |
| Shrews | |||
| Suncus murinus | 185 | 14/185 (7.6) | 1/185 (0.5) |
| Total | 397 | 101/397 (25.4) | 12/397 (3.0) |
S. aureus Staphylococcus aureus;
MRSA Methicillin-resistant Staphylococcus aureus
Of the 397 animals trapped, S. aureus was isolated from 101 (25.4, 95% CI: 21.1–29.7%), of those 87 isolates were derived from rodents and 14 from shrews. S. aureus carriage rates in rodents (41.0, 95% CI: 34.4–47.7%) were higher than in shrews (7.6, 95% CI: 3.7–11.4%) (χ2 = 58.3, p < 0.001) (Table 1). Of the 101 S. aureus isolates, 12 were mecA-positive MRSA (3.0, 95% CI: 1.3–4.7%), including 11 from Rattus norvegicus and one from Suncus murinus. The MRSA carriage rate was 5.2% (95% CI: 2.2–8.2%) in rodents and 0.5% in shrews.
There was significant difference in seasonal carriage rates for S. aureus: summer (June to August, 17.2, 95% CI: 10.3–24.2%), fall (September to November, 12.3, 95% CI: 6.6–18.0%), winter (December to February, 46.1, 95% CI: 34.6–57.5%), and spring (March to May, 40.0, 95% CI: 28.7–51.3%) (χ2 = 41.336, p < 0.001). There was no difference in MRSA seasonal carriage rates: summer (5.2%), fall (0.8%), winter (3.9%), and spring (2.7%) (χ2 = 4.335, p = 0.227) (Fig. 1).
Fig. 1.
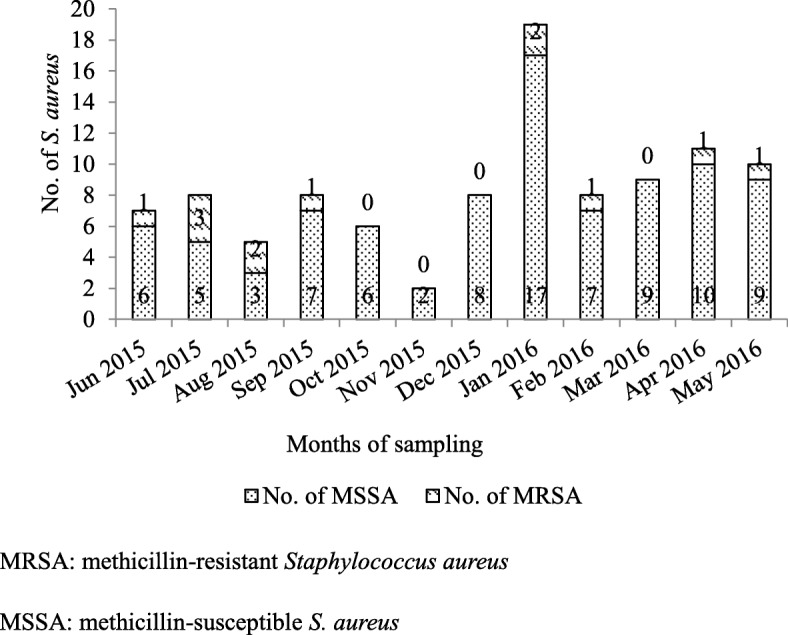
Distribution of MRSA and MSSA carriage in urban rodents and shrews by month between 2015 and 2016 in Guangzhou, Southern China. MRSA: methicillin-resistant Staphylococcus aureus. MSSA: methicillin-susceptible S. aureus
Antimicrobial susceptibility
All 101 S. aureus isolates were susceptible to minocycline, gentamicin, vancomycin, and trimethoprim-sulfamethoxazole. Of 89 MSSA isolates, the resistance rate to penicillin was the highest (50.6%), followed by azithromycin (20.2%), tetracycline (10.1%), clindamycin (3.4%), linezolid (1.1%), rifampin (1.1%) chloramphenicol (1.1%), and ciprofloxacin (1.1%). Four MSSA isolates showed intermediate resistance to clindamycin (n = 3) and nitrofurantoin (n = 1) (data are not shown).
Sixteen MDRSA isolates were detected among the S. aureus isolates. The proportion of MDRSA to S. aureus isolated from urban rodents and shrews was 16.1% (14/87) and 14.3% (2/14), respectively. Among these MDRSA isolates, four were MSSA (one from shrew and three from rodents).
The MRSA isolate derived from the shrew was susceptible to most antibiotics tested with the exception of penicillin, cefoxitin, and tetracycline. Among the 11 rodent MRSA isolates, resistance rates to penicillin and cefoxitin were highest (7/11), followed by azithromycin (3/11), clindamycin (3/11), tetracycline (3/11), rifampin (2/11), and ciprofloxacin (1/11). One rodent MRSA isolate showed intermediate susceptibility to tetracycline (Table 2).
Table 2.
Antimicrobial-resistant patterns of 20 MRSA isolates from rodents, shrews, and patients between 2015 and 2016 in Guangzhou, Southern China
| ID | Host | Antimicrobial resistance | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AZM | CM | CFX | P | SXT | LZD | MH | TE | RD | C | CIP | CN | F | VAc | ||
| 185 | SM | S | S | R | R | S | S | S | I | S | S | S | S | S | ≤1 |
| 213 | RN | S | S | R | R | S | S | S | R | R | S | R | S | S | ≤1 |
| 215 | RN | S | S | R | R | S | S | S | S | S | S | S | S | S | ≤1 |
| 239 | RN | R | R | R | R | S | S | S | R | S | S | S | S | S | ≤1 |
| 257 | RN | R | R | S | R | S | S | S | S | S | S | S | S | S | ≤1 |
| 263 | RN | S | S | R | R | S | S | S | S | R | S | S | S | S | ≤1 |
| 329 | RN | S | S | R | R | S | S | S | S | S | S | S | S | S | ≤1 |
| 65 | RN | S | S | S | S | S | S | S | S | S | S | S | S | S | ≤1 |
| 466 | RN | S | S | S | S | S | S | S | S | S | S | S | S | S | ≤1 |
| 484 | RN | S | S | R | S | S | S | S | S | S | S | S | S | S | ≤1 |
| 533 | RN | R | R | R | R | S | S | S | I | S | S | S | S | S | ≤1 |
| 564 | RN | S | S | S | S | S | S | S | R | S | S | S | S | S | ≤1 |
| 18 | PA | R | S | R | R | S | S | S | S | I | S | S | S | S | ≤1 |
| 07 | PA | R | S | R | R | S | S | S | R | R | S | R | R | S | ≤1 |
| 06 | PA | R | R | R | R | R | S | I | R | S | S | R | R | S | ≤1 |
| 73 | PA | R | R | R | R | S | S | S | I | I | R | S | S | S | ≤2 |
| 67 | PA | R | I | S | R | S | S | S | R | S | S | R | S | S | ≤1 |
| 418 | PA | S | S | S | S | S | S | S | S | S | S | S | S | S | ≤1 |
| 95 | PA | R | R | R | R | S | S | I | R | S | S | R | R | S | ≤2 |
| 76 | PA | R | R | R | R | S | S | S | S | S | S | S | S | S | ≤2 |
MRSA Methicillin-resistant Staphylococcus aureus
SM Suncus murinus, RN Rattus norvegicus, PA Patient, AZM azithromycin, CM clindamycin, CFX cefoxitin, P penicillin, SXT trimethoprim/sulfamethoxazole, LZD linezolid, MH minocycline, TE tetracycline, VA vancomycin, RD rifampin, C chloramphenical, CIP ciprofloxacin, CN gentamicin, F nitrofurantoin, R resistant, I intermediate, S susceptible
cMICs for vancomycin were determined by the agar dilution method
Drug resistance profiles for the patient MRSA isolates (n = 8) were similar to the profiles of isolates collected from animals in this study. Antibiotic-resistance rates were highest for β-lactams (penicillin/cefoxitin) and azithromycin (7/8). The resistant rates to clindamycin, tetracycline, ciprofloxacin, and gentamycin were 50%(4/8). One MRSA isolate was resistant to trimethoprim/sulfamethoxazole. Two isolates exhibited intermediate susceptibility to minocycline. All eight MRSA isolates were susceptible to linezolid, nitrofurantoin, and vancomycin (Table 2).
Molecular characterization of MRSA
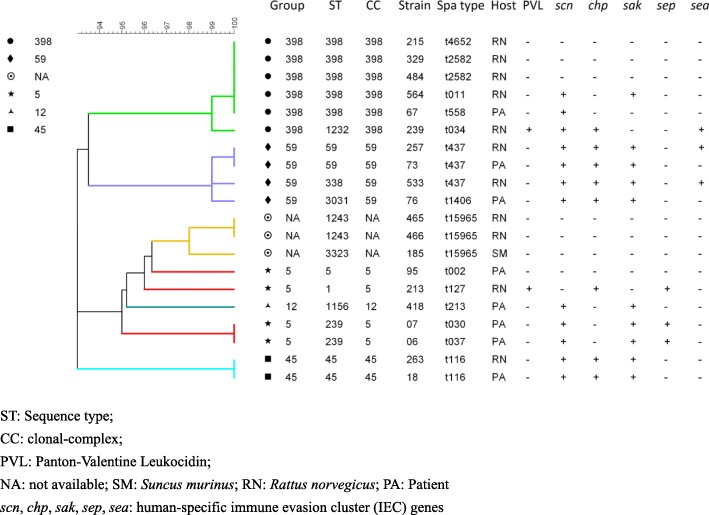
Among 11 MRSA isolates retrieved from rodents, 8 spa types were identified including t437 (n = 2), t2582 (n = 2), t127 (n = 1), t4652 (n = 1), t034 (n = 1), t116 (n = 1), t011 (n = 1), and t15965 (n = 2), which t15965 was a new spa type not observed previously. One MRSA isolate from shrew also belonged to t15965. Eight different spa types were detected among eight MRSA isolates from patients including t116, t030, t037, t437, t558, t213, t002, and t1406. The spa type t116 and t437 were found in both rodents and humans (Fig. 2).
Fig. 2.
Molecular characteristics of 20 MRSA isolates from rodents, shrews, and patients between 2015 and 2016 in Guangzhou, Southern China. ST: Sequence type; CC: clonal-complex; PVL: Panton-Valentine Leukocidin; NA: not available; SM: Suncus murinus; RN: Rattus norvegicus; PA: Patient, scn, chp, sak, sep, sea: human-specific immune evasion cluster (IEC) genes
After MLST analysis, seven sequence types (STs) were detected among 11 rodent MRSA isolates. The dominant type was ST398 (n = 4), followed by ST1243 (n = 2), ST59 (n = 1), ST45 (n = 1), ST1232 (n = 1), ST1 (n = 1), and ST338 (n = 1). A new type, ST3323, was identified from the shrew MRSA isolate. Seven STs were detected among the eight human MRSA isolates including ST45, ST239, ST59, ST398, ST1156, ST5, and ST3031. ST45, ST59, and ST398 were found in both rodents and humans (Fig. 2).
By eBURST analysis, 20 MRSA isolates (derived from the small animals and from humans) were divided into 6 groups based on Clonal Complexes (CCs) (Fig. 2). Five CCs were available, including CC5, CC398, CC59, CC45 and CC12. CC5 included ST1-t127, ST239-t030, ST239-t037, and ST5-t002. CC398 included ST398-t4652, ST1232-t034, ST398-t2582, ST398-t011, and ST398-t558. CC59 included ST59-t437, ST338-t437, and ST3031-t1406. CC45 included ST45-t116, and CC12 included ST1156-t213. Clonal Complexes of three isolates were not available (ST1243-t15965 and ST3323-t15965) (Fig. 2).
Among 101 S. aureus isolates from small animals only three MRSA isolates from Rattus norvegicus were PVL-positive, including CC398-ST1232-t034, CC59-ST59-t437, and CC59-ST338-t437. The PVL gene was not detected among eight MRSA isolates from patients (Fig. 2).
Among 11 MRSA isolates from rodents, five isolates were IEC-negative, including two isolates with a new type (ST1243-t15965) and another three isolates belonging to CC398 (ST398-t4652, ST1232-t034, and ST398-t2582). The shrew MRSA isolate belonged to a new type (ST3323-t15965) and was IEC-negative. Among eight patient MRSA isolates only one isolate (CC5-ST5-t002) was IEC-negative (Fig. 2).
Discussion
This study is the first to characterize S. aureus and MRSA in urban rodents in China. To the best of our knowledge, it is also the first report of carriage rates and bacterial characteristics of MRSA in house shrews worldwide. The most striking finding is that urban rodents and house shrews carry a diverse MRSA strains, including those found in humans and domestic animals. The house shrew, a small mole-like mammal, originated from the Indian subcontinent and are now found from southern Asia and Afghanistan to the Malay Archipelago and southern Japan [25]. As commensal animals, urban rodents and house shrews are commonly found near human households. The proximity to human is important in the transmission of pathogens.
The prevalence of S. aureus among urban rodents in this study was 25.4% (101/397, 95% CI: 21.1–29.7%), with a prevalence 41.0% (95% CI: 34.4–47.7%) in rodents and 7.6% (95% CI: 3.7–11.4%) in shrews. In total, the MRSA carriage rate among these animals was 3.0% (95% CI: 1.3–4.7%). In rodents and shrews, the MRSA carriage rate was 5.2% (95% CI: 2.2–8.2%) and 0.5%, respectively. The reported S. aureus carriage rate in rodents varies from 7.1% (43/608) [15] to 41.86% (18/43) [21]. The highest prevalence of MRSA was 11.6% (5/43) reported in rats living on pig farms in the Netherlands [21], followed by 3.5% (22/637) in urban rats in Canada [22]. These differences in carriage rates may be due to geographic differences, sample source, or sample size [26]. The carriage rates of S. aureus and MRSA in rodents and shrews could also be influenced by season. MRSA carriage among wild urban Norway rats was higher in the winter and spring as described in previous study [22]. A similar seasonality was noted for MRSA infection rates in humans in previous research, with CA-MRSA peaking in late summer and HA-MRSA peaking in the winter [27]. In this study, rodents and shrews caught in the summer and fall had a lower-carriage rate of S. aureus than in the winter and spring. While, no seasonal variation was observed among MRSA carriage rates. The seasonality of MRSA carriage rate among urban rodents and house shrews in Guangzhou, Southern China needs further research.
Antibiotic resistance reported here differs from a previous study looking at MRSA in urban-dwelling Rattus norvegicus [22]. In this study resistance to penicillin was 63.64% versus 100% (in the previously reported study), cefoxintin 63.64% versus 100%, rifampin 18.18% versus 9.09%, clindamycin 27.27% versus 0, and ciprofloxacin 9.09% versus 0. These differences may relate to geographic location and/or the genotypes of these MRSA isolates [28]. All MRSA isolates from the small animals were susceptible to trimethoprim/sulfamethoxazole, minocycline, gentamicin, and vancomycin, which is in agreement with a previous evaluation of human and animal MRSA isolates in China [29–31].
To differentiate human- from livestock-associated S. aureus isolates, clonal complexes (CCs) and resistance patterns have been used by other researchers [32, 33]. Absence of the human-specific IEC genes can be used to identify livestock-associated S. aureus [34], as those genes were lost with the evolution of LA-MRSA from its human-adapted ancestor [32]. As such, CCs, resistance patterns, and the presence of PVL or IEC genes (such as scn, chp, sak, sea, and sep) were used in this study to identify the potential origin of MRSA isolates.Current S. aureus typing methods include spa typing, MLST, SCCmec typing, pulsed-field gel electrophoresis (PFGE), and multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) [35]. However, no single-typing method is ideal [36, 37]. Based on international-standard nomenclature and flexibility, spa typing and MLST were used in this study.
MRSA CC398, which is also referred to as LA-MRSA, is the most frequent CC detected in this study, even though S. aureus CC9 has been identified as the main livestock-associated clone in most Asian countries [38, 39]. The CC398-t034 clone has been shown to be dominant among MSSA isolates from rodents [40]. MRSA ST398 has been detected in Rattus norvegicus from an inner-city neighborhood [22], suggesting that urban rats could be an important reservoir for S. aureus CC398 isolates. Generally, LA-MRSA ST398 is characteristically resistant to tetracycline, and lacks both PVL and IEC genes [41]. It is noteworthy that none of the five MRSA CC398 isolates in this study showed this typical pattern. Three MRSA CC398 isolates that lacked IEC genes were susceptible to tetracycline and were PVL-negative, suggesting that these isolates may have an animal origin. In contrast, the remaining two MRSA CC398 isolates were tetracycline resistant, IEC-positive, with one carrying the PVL gene, suggesting that these two isolates may have a human origin. Interestingly, one of patient-derived MRSA isolates shared the same pattern as that of rat isolates, PVL-negative, susceptible to tetracycline, and IEC-positive. In point of fact, LA-MRSA CC398 originated as a MSSA in humans and jumped to livestock by the acquisition of methicillin and tetracycline resistance and by the loss of phage-carried, human-virulence genes [32]. Recently, CC398 has been split into two distinct lineages, those that are of human origin and those that are livestock adapted [42].
S. aureus clones previously identified mainly in humans in Asian countries were also isolated from both Rattus norvegicus and patients in this study. These clones included MRSA CC59 (ST59-t437 and ST338-t437), the major CA-MRSA clones in China, and common HA-MRSA, CC5 (ST1-t127) and CC45 (ST45-t116) [38]. The molecular characterization of these MRSA clones conforms to the profiles of CA-MRSA and HA-MRSA [5]. More work needs to be done, such as long-read sequencing, to assess potential transmission events of CA-MRSA and/or HA-MRSA from humans to rodents or vice-versa.
In this study, three MRSA isolates were found to belong to a new-spa type (t15965) not previously identified. These isolates lacked PVL and IEC genes, suggesting derivation from animals. However, these isolates were from different MLST (ST3323 and ST1243) and had varying antimicrobial susceptibility patterns. Two of these were retrieved from Rattus norvegicus, belonging to ST1243, and susceptible to all antibiotics tested. The remaining one was retrieved from Suncus murinus, belonging to ST3323, and resistant to cefoxitin and penicillin, with intermediate resistance to tetracycline. Further analysis is required to confirm these resistance patterns, their associated host reservoir species, and their transmissibility.
Conclusion
This study is the first to report the carriage and characteristics of MRSA in urban house shrews. It is also the first to characterize MRSA isolates from urban rodents in China. The spa type, MLST, and antimicrobial resistance patterns suggest that the MRSA retrieved from rodents and shrews are likely related to humans and livestock associated strains. Urban rodents have high carriage rate of MRSA, with similar antimicrobial-resistance patterns and molecular characteristics to those of human isolates. These results suggest that urban rodents may play as maintenance host or vectors for MRSA which is important to human health.
Methods
Ethics statement
This study’s protocol was approved by the Animal Ethics and Welfare Committee of the School of Public Health, Southern Medical University, Guangzhou, China. Animals were cared for according to the Rules for the Implementation of Laboratory Animal Medicine (1998) by the Ministry of Health, China. All surgical procedures were performed under diethyl ether anesthesia to minimize suffering. Endangered or protected species were not involved in this study.
The protocol of collecting MRSA isolates from patients was approved by the ethics committee of Southern Medical University, and was performed in accordance with the ethical standards noted in the 1964 Declaration of Helsinki and its later amendments.
Sample collection
All samples were collected in a community of the Baiyun District of Guangzhou City, Southern China. Rodents and shrews were trapped on a single night each month from July 2015 to May 2016. The animals were trapped alive in iron cages (Yue-zong Co Ltd., Zhongshan, China). Trapped rodents and shrews were euthanized with diethyl ether and oropharynx swabs collected. During the operation, the trained personnel were protected from diethyl ether by wearing filtering facepiece respirators, safety chemical goggles, anti-static uniforms and chemical protective gloves. The swabs were soaked in 2 mL of 7.5% sodium-chloride broth (Land Bridge, Beijing, China) and incubated overnight at 37 °C for selective enrichment. Animal morphometric data were collected, including species (primary identification), sex, weight, body length (nose-to-rump), tail length, ear length, and sexual maturity. Subsequently, a full necropsy was performed. A small portion (~ 0.5 cm3) of the brain of each animal was retrieved and preserved in RNAlater® (Life Technologies, Grand Island, USA) and stored at − 80 °C for subsequent species identification by cytochrome B-gene sequencing [43, 44].
Bacterial isolation and identification
To isolate S. aureus, the sodium-chloride broth was streaked onto mannitol-salt agar plates (MSA) (Land Bridge, Beijing, China) and incubated at 37 °C for 24 h. Presumptive S. aureus colonies, which appeared yellow on MSA plates, were sub-cultured onto fresh MSA plates and incubated at 37 °C for an additional 24 h.
Microbiological tests of the isolated colonies included oxidation and fermentation of MSA, the catalase test (BioMerieux, France), the hemolysin test (5% sheep blood agar), the coagulase test (Land Bridge, Beijing, China), and Gram staining. S. aureus (ATCC® 25923) was used as a reference for each test.
Bacterial genomic DNA from each isolate was extracted using a TIANamp Bacteria-DNA kit (Tiangen Biotech Co., Ltd., Beijing, China). The presumptive isolates were confirmed as S. aureus by polymerase-chain reaction (PCR) using the 16S rRNA gene and the nuc gene for identification [45]. The mecA gene was detected using PCR as described previously [45].
Antibiotic susceptibility testing
S. aureus antibiotic susceptibility was conducted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [46]. A panel of 14 antimicrobials representing 13 different classes was used for susceptibility testing. The antibiotics were azithromycin, clindamycin, cefoxitin, penicillin, linezolid, rifampin, trimethoprim/sulfamethoxazole, minocycline, tetracycline, chloramphenicol, ciprofloxacin, gentamicin, nitrofurantoin, and vancomycin. The minimum-inhibitory concentrations (MICs) of S. aureus against vancomycin were determined by agar dilution as described previously [47]. Susceptibility to the remaining 13 agents was determined by the Kirby-Bauer disc-diffusion method [48]. S. aureus (ATCC® 29213) and S. aureus (ATCC® 25923) were used for quality control (QC). Results were interpreted according to CLSI guidelines. Cefoxitin-resistant isolates or mecA-positive isolates were identified as MRSA. Isolates that were not susceptible to ≥3 classes of antibiotics or were MRSA, were classified as multidrug-resistant S. aureus (MDRSA) [49, 50].
Molecular characterization of MRSA isolates
Spa typing for MRSA isolates was conducted as described previously [51]. Results were analyzed with BioNumerics Software version 7.6 (Applied Math, Sint-Matens-Latem, Belgium).
Multilocus-Sequence Typing (MLST) analysis of 7 housekeeping genes was performed as described previously [52]. Sequence types (STs) were assigned using the MLST database (https://pubmlst.org/saureus/). The eBURST algorithm was used to assign individual STs to specific clonal-complex (CC) types. Clustering analysis was used to infer relationships among the isolates from different origins using BioNumerics Software version 7.6 (Applied Math, Sint-Matens-Latem, Belgium).
The PVL gene, encoding the Panton-Valentine Leukocidin toxin, and the human-specific immune evasion cluster (IEC) genes (including scn, chp, sak, sep, and sea) were detected by PCR as described previously [53, 54].
MRSA isolates from humans
Eight MRSA isolates were obtained from hospital associated pneumonia inpatients in a teaching hospital, which was located within the rat-trapping region in the Baiyun District, were also assessed during the same period.
Statistical analysis
Statistical analysis was carried out using SPSS 20.0 (IBM Corp., Armonk, NY, USA). The distributions of S. aureus and MRSA positive samples by animal species and season were compared using the Chi-Square test. A two-sided p < 0.05 was considered statistically significant.
Acknowledgements
We are grateful to Shao-Wei Chen, Xue-Yan Zheng, Wen-Qiao He, Yu-Qi Wen for their work collecting animal samples.
Abbreviations
- AZM
Azithromycin
- C
Chloramphenical
- CA-MRSA
Community-associated infection
- CC
Clonal-complex
- CFX
Cefoxitin
- CI
Confidence Interval
- CIP
Ciprofloxacin
- CLSI
Clinical and Laboratory Standards Institute
- CM
Clindamycin
- CN
Gentamicin
- F
Nitrofurantoin
- I
Intermediate
- IEC
Immune evasion cluster
- LA-MRSA
Livestock-associated MRSA
- LZD
Linezolid
- MDRSA
Multidrug-resistant S. aureus
- MH
Minocycline
- MICs
Minimum-inhibitory concentrations
- MLST
Multilocus-Sequence Typing
- MLVA
Multilocus variable-number tandem-repeat analysis
- MRSA
Methicillin-resistant Staphylococcus aureus
- MSA
Mannitol-salt agar plates
- MSSA
Methicillin-susceptible S. aureus
- NA
Not available
- P
Penicillin
- PA
Patient
- PBP 2a
Penicillin-binding protein
- PCR
Polymerase-chain reaction
- PFGE
Pulsed-field gel electrophoresis
- PVL
Panton-Valentine Leukocidin
- QC
Quality control
- R
Resistant
- RD
Rifampin
- RN
Rattus norvegicus
- RR
Relative Risk
- S
Susceptible
- S. aureus
Staphylococcus aureus
- SM
Suncus murinus
- STs
Sequence types
- SXT
Trimethoprim/sulfamethoxazole
- TE
Tetracycline
- VA
Vancomycin
- VNTR
Variable-number tandem-repeat
Authors’ contributions
JG analyzed and interpreted the data, and was a major contributor in writing the manuscript. QC made substantial contributions to conception and design. XSZ made substantial contributions to interpreting the data. YQX, MQ and STH was involved in revising the manuscript critically. XJC, YM and MJC made substantial contributions to acquisition of data. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 81373051). The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The site where the rodents were collected belongs to Southern Medical University. This study’s protocol was approved by the Animal Ethics and Welfare Committee of the School of Public Health, Southern Medical University, Guangzhou, China. Animals were cared for according to the Rules for the Implementation of Laboratory Animal Medicine (1998) by the Ministry of Health, China. All surgical procedures were performed under diethyl ether anesthesia to minimize suffering. Endangered or protected species were not involved in this study.
The protocol of collecting MRSA isolates from patients was approved by the ethics committee of Southern Medical University, and was performed in accordance with the ethical standards noted in the 1964 Declaration of Helsinki and its later amendments.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jing Ge, Email: gjing120@163.com.
Xue-shan Zhong, Email: 759598106@qq.com.
Yi-quan Xiong, Email: 747756389@qq.com.
Min Qiu, Email: 453681264@qq.com.
Shu-ting Huo, Email: 441785869@qq.com.
Xue-jiao Chen, Email: 781413016@qq.com.
Yun Mo, Email: 390802971@qq.com.
Ming-ji Cheng, Email: 276131617@qq.com.
Qing Chen, Phone: +8602061648312, Email: qch.2009@163.com.
References
- 1.Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28(3):603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pantosti A, Sanchini A, Monaco M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Future Microbiol. 2007;2(3):323–334. doi: 10.2217/17460913.2.3.323. [DOI] [PubMed] [Google Scholar]
- 3.Eriksen KR. “Celbenin”-resistant staphylococci. Ugeskr Laeger. 1961;123:384–386. [PubMed] [Google Scholar]
- 4.WHO . Antimicrobial resistance global report on surveillance: 2014 summary. 2014. [Google Scholar]
- 5.Stefani S, Chung DR, Lindsay JA, Friedrich AW, Kearns AM, Westh H, Mackenzie FM. Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int J Antimicrob Agents. 2012;39(4):273–282. doi: 10.1016/j.ijantimicag.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 6.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375(9725):1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peacock SJ, Paterson GK. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu Rev Biochem. 2015;84:577–601. doi: 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- 8.Couto I, de Lencastre H, Severina E, Kloos W, Webster JA, Hubner RJ, Sanches IS, Tomasz A. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb Drug Resist. 1996;2(4):377–391. doi: 10.1089/mdr.1996.2.377. [DOI] [PubMed] [Google Scholar]
- 9.Schnellmann C, Gerber V, Rossano A, Jaquier V, Panchaud Y, Doherr MG, Thomann A, Straub R, Perreten V. Presence of new mecA and mph(C) variants conferring antibiotic resistance in Staphylococcus spp. isolated from the skin of horses before and after clinic admission. J Clin Microbiol. 2006;44(12):4444–4454. doi: 10.1128/JCM.00868-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 2010;54(10):4352–4359. doi: 10.1128/AAC.00356-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolo J, Worning P, Boye NJ, Sobral R, Bowden R, Bouchami O, Damborg P, Guardabassi L, Perreten V, Westh H, et al. Evidence for the evolutionary steps leading to mecA-mediated beta-lactam resistance in staphylococci. PLoS Genet. 2017;13(4):e1006674. doi: 10.1371/journal.pgen.1006674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loncaric I, Kunzel F, Licka T, Simhofer H, Spergser J, Rosengarten R. Identification and characterization of methicillin-resistant Staphylococcus aureus (MRSA) from Austrian companion animals and horses. Vet Microbiol. 2014;168(2–4):381–387. doi: 10.1016/j.vetmic.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 13.Vincze S, Stamm I, Kopp PA, Hermes J, Adlhoch C. Alarming proportions of methicillin-resistant Staphylococcus aureus (MRSA) in wound samples from companion animals, Germany 2010–2012. PLoS One. 2014;9:e85656. doi: 10.1371/journal.pone.0085656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald JR. Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 2012;20(4):192–198. doi: 10.1016/j.tim.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Porrero MC, Mentaberre G, Sánchez S, Fernández-Llario P, Gómez-Barrero S, Navarro-Gonzalez N, Serrano E, Casas-Díaz E, Marco I, Fernández-Garayzabal J, et al. Methicillin resistant Staphylococcus aureus (MRSA) carriage in different free-living wild animal species in Spain. Vet J. 2013;198(1):127–130. doi: 10.1016/j.tvjl.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Morris DO, Lautenbach E, Zaoutis T, Leckerman K, Edelstein PH, Rankin SC. Potential for pet animals to harbour methicillin-resistant staphylococcus aureus when residing with human MRSA patients. Zoonoses Public Health. 2012;59(4):286–293. doi: 10.1111/j.1863-2378.2011.01448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D, Voss A, Kluytmans J. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis. 2007;13(12):1834–1839. doi: 10.3201/eid1312.070384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bown KJ, Lambin X, Telford G, Heyder-Bruckner D, Ogden NH, Birtles RJ. The common shrew (Sorex araneus): a neglected host of tick-borne infections? Vector Borne Zoonotic Dis. 2011;11(7):947–953. doi: 10.1089/vbz.2010.0185. [DOI] [PubMed] [Google Scholar]
- 19.Firth C, Bhat M, Firth MA, Williams SH, Frye MJ, Simmonds P, Conte JM, Ng J, Garcia J, Bhuva NP, et al. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal rattus norvegicus in New York City. MBio. 2014;5(5):e1914–e1933. doi: 10.1128/mBio.01933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li K, Lin X, Wang W, Shi M, Guo W, Zhang X, Xing J, He J, Wang K, Li M, et al. Isolation and characterization of a novel arenavirus harbored by rodents and shrews in Zhejiang province, China. Virology. 2015;476:37–42. doi: 10.1016/j.virol.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 21.van de Giessen AW, van Santen-Verheuvel MG, Hengeveld PD, Bosch T, Broens EM, Reusken CBEM. Occurrence of methicillin-resistant Staphylococcus aureus in rats living on pig farms. Prev Vet Med. 2009;91(2–4):270–273. doi: 10.1016/j.prevetmed.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Himsworth CG, Miller RR, Montoya V, Hoang L, Romney MG, Al-Rawahi GN, Kerr T, Jardine CM, Patrick DM, Tang P, et al. Carriage of methicillin-resistant Staphylococcus aureus by wild urban Norway rats (Rattus norvegicus) PLoS One. 2014;9(2):e87983. doi: 10.1371/journal.pone.0087983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo XG, Dong WG, Men XY, Qian TJ, Wu D, Ren TG, Qin F, Song WY, Yang ZH, Fletcher QE. Species abundance distribution of Ectoparasites on Norway rats (Rattus norvegicus) from a localized area in Southwest China. J Arthropod Borne Dis. 2016;10(2):192–200. [PMC free article] [PubMed] [Google Scholar]
- 24.Guo WP, Lin XD, Wang W, Zhang XH, Chen Y, Cao JH, Ni QX, Li WC, Li MH, Plyusnin A, et al. A new subtype of Thottapalayam virus carried by the Asian house shrew (Suncus murinus) in China. Infect Genet Evol. 2011;11(8):1862–1867. doi: 10.1016/j.meegid.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Guan D, Li W, Su J, Fang L, Takeda N, Wakita T, Li T, Ke C. Asian musk shrew as a reservoir of rat hepatitis E virus, China. Emerg Infect Dis. 2013;19(8):1341–1343. doi: 10.3201/eid1908.130069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erickson MC. Overview: Foodborne Pathogens in Wildlife Populations. In: Jay-Russell MT, Doyle MP, editors. Food Safety Risks from Wildlife: Challenges in Agriculture, Conservation, and Public Health. Cham: Springer International Publishing; 2016. p. 1-30.
- 27.Klein EY, Sun L, Smith DL, Laxminarayan R. The changing epidemiology of methicillin-resistant Staphylococcus aureus in the United States: a national observational study. Am J Epidemiol. 2013;177(7):666–674. doi: 10.1093/aje/kws273. [DOI] [PubMed] [Google Scholar]
- 28.Stryjewski ME, Corey GR. Methicillin-resistant Staphylococcus aureus: an evolving pathogen. Clin Infect Dis. 2013;58(suppl 1):S10–S19. doi: 10.1093/cid/cit613. [DOI] [PubMed] [Google Scholar]
- 29.Cui S, Li J, Hu C, Jin S, Li F, Guo Y, Ran L, Ma Y. Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J Antimicrob Chemother. 2009;64(4):680–683. doi: 10.1093/jac/dkp275. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Kong F, Zhang X, Brown M, Ma L, Yang Y. Antimicrobial susceptibility of Staphylococcus aureus isolated from children with impetigo in China from 2003 to 2007 shows community-associated methicillin-resistant Staphylococcus aureus to be uncommon and heterogeneous. Br J Dermatol. 2009;161(6):1347–1350. doi: 10.1111/j.1365-2133.2009.09376.x. [DOI] [PubMed] [Google Scholar]
- 31.Chu H, Zhao L, Zhang Z, Gui T, Han L, Ni Y. Antibiotic resistance and molecular epidemiology of methicillin-resistant Staphylococcus aureus from lower respiratory tract: multi-resistance and high prevalence of SCCmec III type. Cell Biochem Biophys. 2013;67(2):795–801. doi: 10.1007/s12013-013-9542-7. [DOI] [PubMed] [Google Scholar]
- 32.Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, et al. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio. 2012;3(1):e305-e311. [DOI] [PMC free article] [PubMed]
- 33.Uhlemann AC, Porcella SF, Trivedi S, Sullivan SB, Hafer C, Kennedy AD, Barbian KD, McCarthy AJ, Street C, Hirschberg DL, et al. Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. MBio. 2012;3(2):e12-e27. [DOI] [PMC free article] [PubMed]
- 34.Ye X, Fan Y, Wang X, Liu W, Yu H, Zhou J, Chen S, Yao Z. Livestock-associated methicillin and multidrug resistant S. aureus in humans is associated with occupational pig contact, not pet contact. Sci Rep. 2016;6:19184. doi: 10.1038/srep19184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Struelens MJ, Hawkey PM, French GL, Witte W, Tacconelli E. Laboratory tools and strategies for methicillin-resistant Staphylococcus aureus screening, surveillance and typing: state of the art and unmet needs. Clin Microbiol Infect. 2009;15(2):112–119. doi: 10.1111/j.1469-0691.2009.02698.x. [DOI] [PubMed] [Google Scholar]
- 36.Struelens MJ. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin Microbiol Infect. 1996;2(3):2–11. doi: 10.1111/j.1469-0691.1996.tb00193.x. [DOI] [PubMed] [Google Scholar]
- 37.Van BA, Niesters HG, Mackay WG, van Leeuwen WB. Quality control of direct molecular diagnostics for methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2007;45(8):2698–2700. doi: 10.1128/JCM.00759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CJ, Huang YC. New epidemiology of Staphylococcus aureus infection in Asia. Clin Microbiol Infect. 2014;20(7):605–623. doi: 10.1111/1469-0691.12705. [DOI] [PubMed] [Google Scholar]
- 39.Chuang Y, Huang Y. Livestock-associated meticillin-resistant Staphylococcus aureus in Asia: an emerging issue? Int J Antimicrob Agents. 2015;45(4):334–340. doi: 10.1016/j.ijantimicag.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Ho PL, Chow KH, Lai EL, Law PY, Chan PY, Ho AY, Ng TK, Yam WC. Clonality and antimicrobial susceptibility of Staphylococcus aureus and methicillin-resistant S. aureus isolates from food animals and other animals. J Clin Microbiol. 2012;50(11):3735–3737. doi: 10.1128/JCM.02053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pantosti A. Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Front Microbiol. 2012;3:127. [DOI] [PMC free article] [PubMed]
- 42.Smith TC, Wardyn SE. Human infections with Staphylococcus aureus CC398. Curr Environ Health Rep. 2015;2(1):1–11. doi: 10.1007/s40572-014-0034-8. [DOI] [PubMed] [Google Scholar]
- 43.Schlegel M, Ali HS, Stieger N, Groschup MH, Wolf R, Ulrich RG. Molecular identification of small mammal species using novel cytochrome B gene-derived degenerated primers. Biochem Genet. 2012;50(5–6):440–447. doi: 10.1007/s10528-011-9487-8. [DOI] [PubMed] [Google Scholar]
- 44.Kang HJ, Kadjo B, Dubey S, Jacquet F, Yanagihara R. Molecular evolution of Azagny virus, a newfound hantavirus harbored by the west African pygmy shrew (Crocidura obscurior) in cote d’Ivoire. Virol J. 2011;8:373. doi: 10.1186/1743-422X-8-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang K, Sparling J, Chow BL, Elsayed S, Hussain Z, Church DL, Gregson DB, Louie T, Conly JM. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J Clin Microbiol. 2004;42(11):4947–4955. doi: 10.1128/JCM.42.11.4947-4955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cockerill FR, Patel JB, Alder J, Bradford PA, Dudley MN, Eliopoulos GM. Performance standards for antimicrobial susceptibility testing: twenty-third informational supplement; M100-S23. 2013. [Google Scholar]
- 47.Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standards—Ninth Edition; CLSI document M07-A9. Wayne: Clinical Laboratory Standards Institute; 2012.
- 48.Bauer AW, Kirby W, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- 49.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 50.Neyra RC, Frisancho JA, Rinsky JL, Resnick C, Carroll KC, Rule AM, Ross T, You Y, Price LB, Silbergeld EK. Multidrug-resistant and methicillin-resistant Staphylococcus aureus (MRSA) in hog slaughter and processing plant workers and their community in North Carolina (USA) Environ Health Perspect. 2014;122(5):471–477. doi: 10.1289/ehp.1306741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strommenger B, Braulke C, Heuck D, Schmidt C, Pasemann B, Nubel U, Witte W. Spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J Clin Microbiol. 2008;46(2):574–581. doi: 10.1128/JCM.01599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McClure J, Conly JM, Lau V, Elsayed S, Louie T, Hutchins W, Zhang K. Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from-resistant staphylococci. J Clin Microbiol. 2006;44(3):1141–1144. doi: 10.1128/JCM.44.3.1141-1144.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, van Strijp JA. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J Bacteriol. 2006;188(4):1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.