Abstract
Background
Observational associations between asthma and obesity are well established, but inferring causality is challenging. We leveraged publicly available summary statistics to ascertain the causal direction between asthma and obesity via Mendelian randomization in European-ancestry adults.
Methods
We performed two-sample bi-directional Mendelian randomization analysis using publicly available genome-wide association studies summary statistics. Single nucleotide polymorphisms associated with asthma and body mass index at genome-wide significance were combined using a fixed effect meta-analysis in each direction. An extensive sensitivity analysis was considered.
Results
There was evidence in support of increasing causal effect of body mass index on risk of asthma (odds ratio 1.18 per unit increase, 95% confidence interval (CI) (1.11, 1.25), P = 2 × 10−8. No significant causal effect of asthma on adult body mass index was observed [estimate −0.004, 95% CI (−0.018, 0.009), P = 0.553].
Conclusions
Our results confirmed that in European-ancestry populations, adult body mass index is likely to be causally linked to the risk of asthma; yet the effect of asthma on body mass index is small, if present at all.
Keywords: Mendelian randomization, bi-directional causal estimation, asthma, body mass index, summary statistics
Key Messages
Epidemiological studies have established an observational association between asthma and obesity.
Mendelian randomization is a useful tool to estimate causal effect of a modifiable risk factor on a health outcome.
Current analyses on summary statistics from large cohorts suggested that increasing adiposity has a strong effect on the rising prevalence of asthma among European adults; yet the effect of asthma on body mass index is likely to be small, if present at all.
Introduction
Epidemiological studies have established an observational association between asthma and obesity, two major public health concerns across the globe. Biological dynamics between the two traits have been hypothesized as obesity is often considered a major risk factor and disease modifier for asthma, affecting individuals of all ages, and weight loss in obese asthmatics improves asthma control, whereas asthma exacerbations are observed more often among obese individuals.1–6 However, drawing causal inferences from observational epidemiology can be challenging because of residual confounding and potential reverse causation. Since its first appearance in inspecting whether low cholesterol causes cancer,7 Mendelian randomization (MR) has been a useful tool for examination of a causal link between a risk factor and a health outcome via human genetics.8–10 Conditional on parental genotype, genetic variants are randomly allocated at the time of conception and are unlikely to be confounded by environmental factors. As a result, these single nucleotide polymorphisms (SNPs) can robustly predict an exposure of interest and can be used as instrumental variables (IVs) to test whether an exposure has a causal effect on an outcome. Moreover, large, publicly available genome-wide association study (GWAS) summary statistics facilitate the examination of the bi-directional causal effects at lower cost with potentially better power and precision.
The causal link between asthma and obesity remains unclear and the effect of asthma on body mass index (BMI) has not been explored in a causal inference framework among adults.11–14 In this study, we use a two-sample MR analysis in which SNP-exposure associations and SNP-outcome associations are identified from independent previous studies and subsequently combined into a single causal estimate via meta-analysis.15 We perform sensitivity analyses to investigate directional pleiotropy to obtain effect estimates after bias- or outlier-corrections. In addition to showing the robustness of effect estimates, we performed a one-sample summary statistics based MR where SNP-exposure and SNP-outcome coefficients both come from a single large study.
Methods
Mendelian randomization
To assess the causal association between BMI and risk of asthma, we identified 77 SNPs associated with BMI at genome-wide significance in 322 154 European-descent individuals in the Genetic Investigation of ANthropometric Traits (GIANT) consortium.16 A manual search through the GWAS Catalog, Ensembl, and Phenoscanner showed that at genome-wide significance, none of these genetic variants associated with other phenotypes with a potential causal path to asthma. In a conservative fashion, rs11583200 and rs3888190 were excluded due to moderate linkage disequilibrium (LD) with rs657452 [r2 = 0.451 calculated by LDmatrix17 using a reference panel of Utah Residents from North and West Europe (CEU)] and rs2650492 (r2 = 0.524), respectively. See details of the remaining 75 BMI-SNPs in Supplementary Table S1, available as Supplementary data at IJE online. We extracted effect sizes and standard errors (s.e.) for the associations between the instrumental SNPs and BMI from GIANT.
The UK Biobank (UKB) is a population-based cohort that recruited half a million participants aged 40–69 years in 2006–2010 from sites across the UK.18 Details of its study design, quality control, genotyping and imputation are described elsewhere.19 Previous research has provided summary statistics of over 12 million variants in the UKB with BMI z-score and asthma.20 In both association analyses, the covariates include assessment centre, genotyping array, sex, age, age squared and 20 principal components to correct for ancestry. In UKB GWAS for asthma (cases defined as the participant responded to the touchscreen question that he/she was diagnosed of asthma by a doctor; UKB column codes 6152–0.0∼6152–0.4), we extracted summary statistics of 74 SNPs and a proxy SNP rs9581854 for the deprecated rs12016871. We transformed the effect sizes to get log odds ratio (OR) via a reasonable approximation logOR ≈ β/(µ(1 − µ)) for the genetic associations with asthma, where β denotes the reported effect size from the linear mixed model and µ is the case fraction for the binary trait. In UKB, the prevalence of asthma is roughly 11.57%, i.e. µ = 0.1157. Similarly, the corresponding s.e. were divided by µ(1 − µ).21 For the MR analysis, we first calculated instrumental variable estimates for individual SNPs, then pooled these estimates by including one SNP at a time (according to P-value in SNP-exposure association tests) using a fixed effect (inverse variance weighted) meta-analysis, and finally reported an overall causal effect estimate as the odds for asthma per-unit increase in BMI across all variants. A random effect meta-analysis was performed as a sensitivity analysis.
Effect of asthma on BMI
To examine the association of asthma with BMI, we started with 16 asthma-loci identified in the Trans-National Asthma Genetic Consortium (TAGC) of the USA22 at genome-wide significance. All SNPs are independent of each other and of any BMI-associated SNPs. The same criteria were applied to remove pleiotropic SNPs. Specifically, at genome-wide significance, rs1420101 and rs2033784 were associated with eosinophil counts23,24; rs20541 was associated with psoriasis,25,26 Hodgkin lymphoma,27 and IgE levels28; both rs9272346 and rs2855812 were reported to be associated with type 1 diabetes29,30 while the latter was also associated with diabetes mellitus type 131 and tuberculosis32; rs1233578 was associated with autism spectrum disorder33 and schizophrenia34; discovered rs7927894 and rs2033784 were associated with inflammatory bowel disease35; and rs2952156 was found to be associated with high-density lipoprotein (HDL) cholesterol.36 A total of eight asthma-SNPs were included in the subsequent analysis to assess the likelihood of causal effect of asthma on BMI. See detailed information of these SNPs listed in Supplementary Table S2, available as Supplementary data at IJE online. We extracted log ORs and s.e. for the associations between the eight instrumental SNPs with asthma from TAGC and with BMI z-score (coded in UKB 21001–0.0, constructed from height and weight measured during the initial assessment centre visit, and transformed into z-scores that were reasonably close to a standard normal distribution) from the UKB GWAS. The SNP-exposure and SNP-outcome coefficients were combined as described above in a fixed effect MR analysis yielding an estimate for the change in BMI z-score per-unit-log increase in asthma risk.
Sensitivity analyses
To gauge the potential impact of unmeasured directional pleiotropy, we used MR-Egger, a penalized weighted median estimator, and MR-PRESSO.37–39 All statistical analyses were performed using R version 3.5.0.40
As the UKB is a population-based study and larger than either TAGC or GIANT, we also performed one-sample summary statistics based MR analysis in which the SNP-exposure and SNP-outcome coefficients were both extracted from the UKB. To assess the causal effect of BMI on asthma, we first discovered 74 107 SNPs associated with BMI z-score in 457 824 European-ancestry individuals in UKB at P < 5 × 10−8, among which 75 SNPs were also associated with asthma at genome-wide significance. We excluded pleiotropic SNPs and variants that are in high LD with them, and pruned SNPs at a threshold r2 ≥ 0.05. A total of 1348 BMI-SNPs were used in the one-sample MR analysis. In the reverse analysis for the causal effect of asthma on BMI, 12 900 asthma-SNPs reached genome-wide significance among 458 699 individuals in UKB. After an aggressive pruning of SNPs associated with BMI or previously reported phenotypes, and the genetic variants in high LD, we included 234 independent asthma-SNPs in the analysis to examine the causal relationship between asthma and BMI. Similar to the two-sample MR analysis described earlier, we obtained causal estimates by meta-analyses and performed MR-Egger, penalized weighted median method, and MR-PRESSO to show robustness.
Results
Effect of BMI on risk of asthma
The 75 BMI-associated SNPs accounted for 1.55% of the variation in BMI within the European-ancestry population in GIANT (the proportion of variance in BMI was computed for each SNP based on the formula in41). F-statistics of these SNPs range from 29 to 696.3, indicating strong instruments.
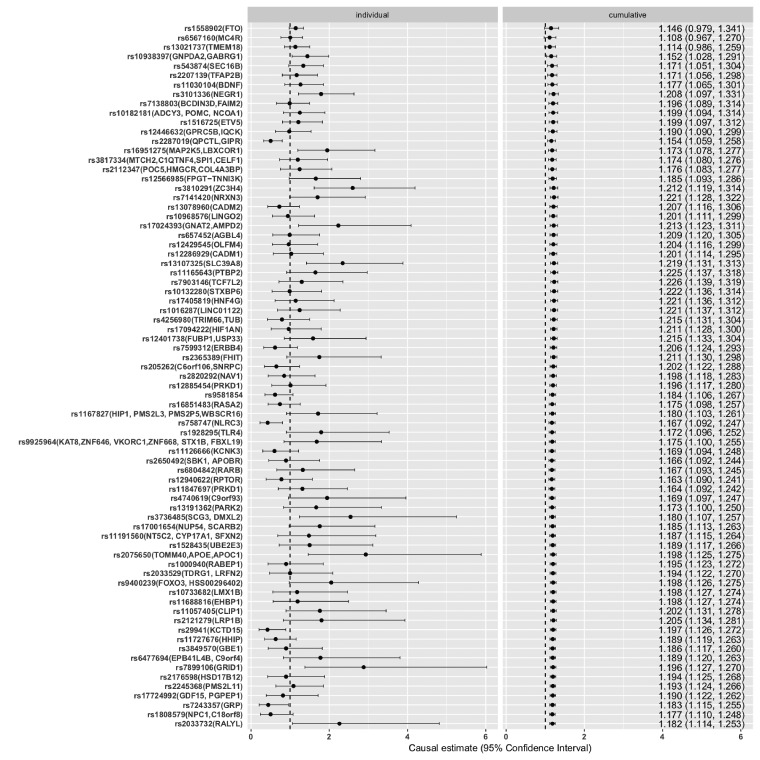
Supplementary Figure S1, available as Supplementary data at IJE online, displays the relationship between the genetic association with BMI in GIANT and genetic association with asthma in UKB. The increase in BMI was causally associated with high risk of asthma. OR per-unit increase in BMI by fixed effect meta-analysis yielded 1.18, 95% CI (1.11, 1.25), P = 2 × 10−8. In UKB, the BMI has mean 27.43 and standard deviation 4.785; thus on average, an increase of 4.785 in BMI is associated with 1.18 times the risk of asthma. Figure 1 displays forest plots of the instrumental variable estimates from each individual SNP (on the left) and cumulative effect sizes (on the right) where SNPs, sorted by ascending P-values of the genetic association with BMI in GIANT, were added one at a time. This provides a fuller picture showing both individually and cumulatively how each SNP, taking into account the strength of genetic association with the exposure, contributes to the effect estimates; an illustration of how sensitively the effect size is affected by individual SNPs. As shown, none of the top three BMI-SNPs are nominally associated with asthma; yet, as rs10938397 was included, the 95% CI of the cumulative point estimate no longer included 1. Random effect estimates were almost identical to that of fixed effect except for a slightly wider CI [OR: 1.18, 95% CI (1.08, 1.3), P = 5 × 10−4].
Figure 1.
Fixed effect meta-analysis of individual (left panel) and cumulative (right panel) genetically instrumented BMI and risk of asthma for the 75 SNPs under analysis. SNPs were sorted by ascending P-values of their associations with BMI in GIANT. Each SNP is marked by rs number, followed by it nearest genes in parentheses.
Effect of asthma on BMI
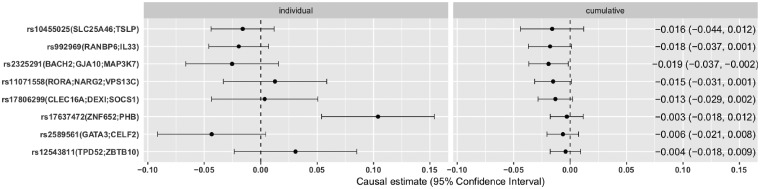
Instrumental SNPs for asthma overall are weaker than for BMI, with F-statistics ranging from 30.4 to 108.6 in TAGC. Altogether they explained 0.3% of the variation in asthma in the consortium. The relationship between effect sizes of SNPs-exposure and SNPs-outcome are shown in Supplementary Figure S2, available as Supplementary data at IJE online. Fixed effect meta-analysis yielded a non-significant estimate −0.004 per-1-log unit increase in risk of asthma, 95% CI (−0.018, 0.009), P = 0.553. The estimate from random effect meta-analysis was also small and with a slightly wider CI [estimate 0.004, 95% CI (−0.026, 0.034), P = 0.812]. Effect size was much smaller compared with that of BMI on asthma. Instrumental variable estimates of individual and cumulative asthma-SNPs are shown side by side in Figure 2 where the genetic variants are ordered by the significance level of their association with asthma in TAGC.
Figure 2.
Fixed effect meta-analysis of individual (left panel) and cumulative (right panel) genetically instrumented asthma and BMI z-score for the 8 SNPs under analysis. SNPs were sorted by ascending P-values of their associations with asthma in TAGC. Each SNP is marked by rs number, followed by its nearest genes in parentheses.
Sensitivity analysis
Effect of BMI on risk of asthma
MR-Egger regression suggested little vertical pleiotropy in the association between BMI and asthma [intercept OR 0.999, 95% CI (0.993, 1.005), P = 0.771]. The resulting estimated causal effect was slightly altered with a noticeably wider CI [OR 1.216, 95% CI (0.985, 1.5), P = 0.069]. Penalized weighted median yielded the most conservative estimate [OR 1.154, 95% CI (1.038, 1.282), P= 8 × 10−3]. MR-PRESSO, however, signalled pleiotropy (P < 2 × 10−4) and identified one outlier SNP rs2287019. After removing the outlier, the point estimate was slightly higher than that from meta-analysis [OR 1.198, 95% CI (1.104, 1.301), P = 5 × 10−5], however with no sign of a significant distortion in the causal estimate before and after MR-PRESSO correction. Point estimates from both methods agree with findings from the primary analysis.
Effect of asthma on BMI
For the asthma to BMI association, MR-Egger reported little evidence of pleiotropy [intercept 0.009, 95% CI (−0.0005, 0.018), P= 0.063], although the effect estimate did become more negative [estimate −0.08, 95% CI (−0.163, 0.003), P = 0.059]. In line with results from MR-Egger, the estimate from the penalized weighted median approach yielded estimate −0.017, 95% CI (−0.036, 0.001), P = 0.059. MR-PRESSO reported some presence of pleiotropy (P = 0.002) and suggested that rs17637472 be an outlier. The point estimate after correction remained insignificant [estimate −0.013, 95% CI (−0.027, 0.002), P = 0.136]. Results are listed in Table 1.
Table 1.
Causal estimates of BMI z-score on asthma and the reverse direction reported by fixed effect and random effect meta-analyses, MR-Egger, penalized weighted median, and MR-PRESSO
| Causal Link | Method | Estimate | 95% CI | P |
|---|---|---|---|---|
| BMI → asthma | Fixed effect | 1.182 | (1.114, 1.253) | 2e-08 |
| Random effect | 1.182 | (1.075, 1.300) | 5e-04 | |
| MR-Egger | 1.216 | (0.985, 1.500) | 0.069 | |
| Penalized | 1.154 | (1.038, 1.282) | 8e-03 | |
| MR-PRESSO | 1.198 | (1.104, 1.301) | 5e-05 | |
| Asthma → BMI | Fixed effect | −0.004 | (–0.018, 0.009) | 0.553 |
| Random effect | 0.004 | (–0.026, 0.034) | 0.812 | |
| MR-Egger | −0.080 | (–0.163, 0.003) | 0.059 | |
| Penalized | −0.017 | (–0.036, 0.001) | 0.059 | |
| MR-PRESSO | −0.013 | (–0.027, 0.002) | 0.136 |
One-sample MR analysis
The relationship between the genetic association with BMI z-score and that with asthma is shown in Supplementary Figure S3, available as Supplementary data at IJE online, for the 1348 independent SNPs associated with BMI z-score at genome-wide significance in UKB. The one-sample MR provided stronger evidence of an association between BMI and prevalence of asthma [OR 1.21, 95% CI (1.18, 1.25), P = 6 × 10−38] under fixed effect meta-analysis. Random effect estimates were almost identical [OR 1.21, 95% CI (1.17, 1.26), P = 6 × 10−23]. We also showed the individual (the top panel) and cumulative (the bottom panel) instrumental variable estimates of BMI on asthma in Supplementary Figure S4, available as Supplementary data at IJE online, where individual BMI-SNPs were sequentially added into the fixed effect meta-analysis as the priority was given to the variant most associated with BMI z-score in UKB. No pleiotropy was detected by MR-Egger [intercept OR 1.000, 95% CI (0.999, 1.002), P = 0.734] and its estimate was attenuated [OR 1.19, 95% CI (1.05, 1.34), P = 0.006]. The penalized weighted median yielded similar results with narrower CI [OR 1.19, 95% CI (1.14, 1.25), P = 2 × 10−13]. MR-PRESSO however signalled pleiotropy (P < 2 × 10−4), identified 7 outliers, and reported a slightly larger point estimate after correction [OR 1.22, 95% CI (1.18, 1.26), P = 3 × 10−25].
The one-sample MR using 234 independent asthma-SNPs in 458 699 UKB individuals suggested that asthma have a small and weak protective effect on BMI [estimate −0.011, 95% CI (−0.017, −0.005), P = 3 × 10−4] under fixed effect meta-analysis; see Supplementary Figure S5, available as Supplementary data at IJE online, for relationship between SNP-exposure and SNP-outcome. Random effect meta-analysis was consistent with the fixed effect [estimate −0.013, 95% CI (−0.024, −0.003), P = 0.018]. We showed individual and cumulative instrumental variable estimates of asthma on BMI in Supplementary Figure S6, available as Supplementary data at IJE online, where individual asthma-SNPs were sequentially included into the fixed effect meta-analysis in the order determined by P-values from association tests. Additionally, MR-Egger reported no pleiotropy and the estimate was no longer statistically significant [estimate −0.013, 95% CI (−0.041, 0.015), P = 0.369; intercept 0.0001, 95% CI (−0.001, 0.002), P = 0.893]. The estimate reported by the penalized weighted median method was similar to the fixed effect [−0.011, 95% CI (−0.021, −0.001), P = 0.034]. After removing 14 outliers, MR-PRESSO yielded a similar point estimate −0.013, 95% CI (−0.022, −0.004), P = 0.004. Overall, the point estimates from one-sample MR analysis for either direction were in line with findings from two-sample MR and the strength of the evidence is stronger, possibly due to larger sample size and stronger instrumental variables. Results are listed in Supplementary Table S3, available as Supplementary data at IJE online.
Discussion
The two-sample summary statistics based Mendelian randomization analysis, including 75 independent BMI-SNPs at genome-wide significance, supports a causal link that adult BMI is associated with the prevalence of asthma in individuals of European ancestry. The magnitude is smaller than the effect of BMI on type-2 diabetes, however on a par with that on cardiovascular diseases; see Supplementary Table S4, available as Supplementary data at IJE online for details. This echoes the findings using 26 BMI-SNPs in a subset of UKB (n = 162 124) and a two-stage least squares (2SLS) approach13 and that BMI is partially genetically causal for asthma from recent results.42 The effect sizes here are larger and the strength of the evidence is stronger due to larger sample sizes and inclusion of stronger instrumental variables. Similar results of the effect of body mass on asthma have been reported among European-ancestry children at age 7.5 y11 and Taiwanese children at age 10 y14 despite the use of different sets of BMI-SNPs. In,12 the authors used five BMI-genes to perform an MR analysis in the Copenhagen population and concluded that high BMI was associated with high risk of wheezing without asthma, but not with high risk of asthma without wheezing. We could not differentiate asthmatics by wheezing due to limited data access/availability. However, a two-sample (GIANT-UKB) MR fixed effect meta-analysis using the same set of genes (FTO, MC4R, TMEM18, GNPDA2 and BDNF) yielded an associated estimate of BMI on asthma OR 1.161, 95% CI (1.041, 1.295), P = 0.007.
The underlying pathophysiological mechanisms of how obesity could lead to asthma remains unclear and is beyond the scope of this discussion, yet we note that recent literature has been focused on a common denominator–inflammation–and advocated the adoption of aggressive management of obesity in asthma care.43–45 As weight loss improves increased airway closure, and noticeable exacerbations reduced at stable weight or weight loss, and weight gain increases asthma severity,46,47 the obese state could lead to the development of a novel form of late-onset asthma by affecting the tidal volumes mechanically and increasing metabolic inflammation markers.48–50
No causal effect of asthma on BMI was observed via meta-analysis approaches in the two-sample (TAGC-UKB) MR analysis. This does not contradict findings that obesity was observationally associated with asthma exacerbations, because the instrumental variables were selected here based on the genetic association with asthma diagnosed by a doctor, not with asthma exacerbation. In fact, promising asthma exacerbation loci51 were not significantly associated with asthma in UKB: specifically, rs7915695 OR 0.9987, P = 0.19 and rs993312 OR 0.9873, P = 0.88. When examining the effect of asthma on adiposity among Chinese-ancestry children, the authors14 noted that the effect of childhood asthma is likely to be small on adiposity accumulation.
Our investigation suggests that the co-occurrence of obesity and asthma is probably driven by obesity. However, the instruments used for BMI and for asthma are not necessarily equivalent. The instrumental genetic variants for asthma are at a disadvantage in both the quantity and proportion of the variation explained. This may result from disease heterogeneity and misclassification as individuals with asthma vary widely in clinical presentation, severity and pathobiology.52–54 In addition to increasing sample size, enhancing the quality of cases and controls may help the discovery of novel asthma loci, in turn increasing the accuracy of estimates in MR analysis.
As MR analysis makes strong assumptions, the present analysis is performed in a rather conservative fashion and accompanied with extensive sensitivity analyses. First, the analysis was restricted to an adult European-ancestry population. Each sub-study of TAGC and GIANT properly accounted for potential confounding due to population structure prior to the meta-analysis. Second, none of the BMI-SNPs overlapped with asthma-SNPs and no pairs of the instrumental BMI- and asthma-SNPs are in LD, defined very conservatively as r2 ≥ 0.05. Thus, little bias was likely to occur due to LD structure. Third, prior to entering into the main analysis, exposure-associated SNPs that were previously reported to be associated at genome-wide significance with any other potentially relevant phenotypes were excluded. Fourth, we investigated potential pleiotropic effects, as BMI and asthma are both complex polygenic phenotypes and thus likely to be influenced by multiple genetic and environmental risk factors. As a result, MR-Egger, penalized weighted median, and MR-PRESSO, each making different assumptions about the presence of directional pleiotropy, were tested for sensitivity analysis. Consistent results from these complementary methods suggest rather robust causal estimates. To further show the robustness of the estimates, we performed one-sample MR analysis where gene-exposure summary statistics also came from the UKB samples, larger than TAGC and GIANT. As both genetic associations with BMI z-score and asthma adjusted for the same set of covariates, the risk of introducing heterogeneity and potential confounding is lower than in the two-sample MR,55,56 whereas summary statistics in TAGC and GIANT are from different meta-analyses with different quality control procedures and variation in controlling for relevant variables. With the one-sample analysis, bias (≈ 1/E[F]) due to overlapping samples will be small as F-statistics for BMI-SNPs and asthma-SNPs are large, ranging from 25.9 to 569.5 and 27.1 to 226.2, respectively.57 Causal estimates in both directions in the one-sample MR analysis are stronger and the effect sizes are larger than in the two-sample MR. We note that one-sample MR is more likely prone to winner's curse;56 moreover, as the response rate for UKB is less than 5%, one-sample MR might introduce additional bias. In the one-sample analysis, the cumulative effect estimate of asthma on BMI is negative and close to zero. This agrees with the primary results and suggests that even if the effect of asthma on BMI exists among adults, it is likely to be small.
Focusing on European-ancestry adult populations aside, limitations of the current analysis are due to the use of summary statistics. We were unable to test whether the confounders of the exposure–outcome association are unrelated to the genetic instrument. It is also important in association testing to adjust for appropriate measured risk factors, for instance, diet, physical activity, smoking status and socio-economic status, among others, to obtain accurate effect estimates. In addition, we are limited by the availabilities of the phenotypes that the summary statistics were released on. It would be of great interest to infer causality using classes of obesity directly, or using waist–hip-ratio for better representation of the body mass distribution; and to differentiate asthma with and without wheezing in the inference. Furthermore, testing subgroups is only possible when individual-level data are available. For example, gender differences exist in both BMI and asthma development and progression; thus, it may be necessary to perform MR analysis in females and males separately and report gender-specific estimates.
Our results provide evidence that, genetically, adult BMI is causally associated with the prevalence of asthma. The strength of the evidence in support of the reverse causal link is weak, suggesting that the effect of asthma on adult BMI is likely to be small, if present at all. As the global epidemic of obesity continues to impact populations, downstream impacts need to be considered. This suggests that the rising prevalence of obese patients with asthma is probably driven by obesity; thus interventions to reduce obesity might help alleviate the adverse health effects inflicted by asthma.
Funding
This work was supported by the National Institute of Environmental Health Sciences at the National Institutes of Health (P01ES022845, 5P30ES007048, R01ES023262, R01ES025786, R01HL118455); the National Cancer Institute at the National Institutes of Health (P01CA196569); the Environmental influences on Child Health Outcomes at the National Institutes of Health (4UH3OD023287-03); the Environmental Protection Agency (RD-83544101–0); and the Hastings Foundation.
Supplementary Material
Acknowledgements
We express our gratitude to the participants and research teams from TAGC, GIANT, UK Biobank, and BOLT-LMM that made the GWAS results publicly accessible, and especially to Po-Ru Loh, Alkes Price, and Steven Gazal for providing more details of BOLM-LMM GWAS results. We thank the referees for helpful comments.
Conflict of interest: None declared.
References
- 1. Holguin F, Bleecker ER, Busse WW. et al. Obesity and asthma: an association modified by age of asthma onset. J Allergy Clin Immunol 2011;127:1486–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Juel CTB, Ali Z, Nilas L, Ulrik CS.. Asthma and obesity: does weight loss improve asthma control? A systematic review. J Asthma Allergy 2012;5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ali Z, Ulrik CS.. Obesity and asthma: a coincidence or a causal relationship? A systematic review. Respir Med 2013;107:1287–300. [DOI] [PubMed] [Google Scholar]
- 4. Sutherland E. Linking obesity and asthma. Ann N Y Acad Sci 2014;1311:31–41. [DOI] [PubMed] [Google Scholar]
- 5. Sivapalan P, Diamant Z, Ulrik CS.. Obesity and asthma: current knowledge and future needs. Curr Opin Pulm Med 2015;21:80–85. [DOI] [PubMed] [Google Scholar]
- 6. Peters U, Dixon AE, Forno E.. Obesity and asthma. J Allergy Clin Immunol 2018;141:1169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Katan M. Apoupoprotein E isoforms, serum cholesterol, and cancer. Lancet 1986;1:507–508. [DOI] [PubMed] [Google Scholar]
- 8. Davey Smith G, Ebrahim S.. Mendelian randomization: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 9. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G.. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–63. [DOI] [PubMed] [Google Scholar]
- 10. Emdin CA, Khera AV, Kathiresan S.. Mendelian randomization. JAMA 2017;318:1925–26. [DOI] [PubMed] [Google Scholar]
- 11. Granell R, Henderson AJ, Evans DM. et al. Effects of BMI, fat mass, and lean mass on asthma in childhood: a Mendelian randomization study. PLoS Med 2014;11:e1001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Çolak Y, Afzal S, Lange P, Nordestgaard BG.. Obese individuals experience wheezing without asthma but not asthma without wheezing: a Mendelian randomisation study of 85 437 adults from the Copenhagen General Population Study. Thorax 2016;71:247–54. [DOI] [PubMed] [Google Scholar]
- 13. Skaaby T, Taylor AE, Thuesen BH. et al. Estimating the causal effect of body mass index on hay fever, asthma and lung function using Mendelian randomization. Allergy 2018;73:153–64. [DOI] [PubMed] [Google Scholar]
- 14. Chen YC, Fan HY, Huang YT, Huang SY, Liou TH, Lee YL.. Causal relationships between adiposity and childhood asthma: bi-directional Mendelian Randomization analysis. Int J Obes 2019;43:73. [DOI] [PubMed] [Google Scholar]
- 15. Burgess S, Scott RA, Timpson NJ. et al. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol 2015;30:543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Locke AE, Kahali B, Berndt SI. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Machiela MJ, Chanock SJ.. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015;31:3555–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sudlow C, Gallacher J, Allen N. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bycroft C, Freeman C, Petkova D. et al. Genome-wide genetic data oñ 500, 000 UK Biobank participants. BioRxiv 2017;166298. [Google Scholar]
- 20. Loh PR, Kichaev G, Gazal S, Schoech AP, Price AL.. Mixed-model association for biobank-scale datasets. Nat Genet 2018;50:906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lloyd-Jones LR, Robinson MR, Yang J, Visscher PM.. Transformation of summary statistics from linear mixed model association on all-or-none traits to odds ratio. Genetics 2018;208:1397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Demenais F, Margaritte-Jeannin P, Barnes KC. et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet 2018;50:42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gudbjartsson DF, Bjornsdottir US, Halapi E. et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet 2009;41:342.. [DOI] [PubMed] [Google Scholar]
- 24. Astle WJ, Elding H, Jiang T. et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 2016;167:1415–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nair RP, Duffin KC, Helms C. et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat Genet 2009;41:199.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stuart PE, Nair RP, Tsoi LC. et al. Genome-wide association analysis of psoriatic arthritis and cutaneous psoriasis reveals differences in their genetic architecture. Am J Hum Genet 2015;97:816–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Urayama KY, Jarrett RF, Hjalgrim H. et al. Genome-wide association study of classical Hodgkin lymphoma and Epstein–Barr virus status–defined subgroups. J Natl Cancer Inst 2012;104:240–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Granada M, Wilk JB, Tuzova M. et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J Allergy Clin Immunol 2012;129:840–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cooper JD, Smyth DJ, Smiles AM. et al. Meta-analysis of genomewide association study data identifies additional type 1 diabetes risk loci. Nat Genet 2008;40:1399–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hakonarson H, Grant SF, Bradfield JP. et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature 2007;448:591.. [DOI] [PubMed] [Google Scholar]
- 31. Barrett JC, Clayton DG, Concannon P. et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tian C, Hromatka BS, Kiefer AK. et al. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat Commun 2017;8:599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16, 000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol Autism 2017;8:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee SH, Byrne EM, Hultman CM. et al. New data and an old puzzle: the negative association between schizophrenia and rheumatoid arthritis. Int J Epidemiol 2015;44:1706–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu JZ, van Sommeren S, Huang H. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Teslovich T. Genome Wide Associations Scans for Total Cholesterol, HDL-C, LDL-C and triglycerides 2010. http://csg.sph.umich.edu/willer/public/lipids2010/.
- 37. Bowden J, Davey Smith G, Burgess S.. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bowden J, Davey Smith G, Haycock PC, Burgess S.. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Verbanck M, Chen CY, Neale B, Do R.. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018;50:693.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria, 2018. https://www.R-project.org/ (23 April 2018, date last accessed). [Google Scholar]
- 41. Vaucher J, Keating BJ, Lasserre AM. et al. Cannabis use and risk of schizophrenia: a Mendelian randomization study. Mol Psychiatry 2017;23:1287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O’Connor LJ, Price AL.. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet 2018;50:1728–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sideleva O, Black K, Dixon AE.. Effects of obesity and weight loss on airway physiology and inflam- mation in asthma. Pulm Pharmacol Ther 2013;26:455–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Farzan S. The asthma phenotype in the obese: distinct or otherwise? J Allergy 2013;2013:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mohanan S, Tapp H, McWilliams A, Dulin M.. Obesity and asthma: pathophysiology and im- plications for diagnosis and management in primary care. Exp Biol Med (Maywood) 2014;239:1531–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mahadev S, Farah CS, King GG, Salome CM.. Obesity, expiratory flow limitation and asthma symptoms. Pulm Pharmacol Ther 2013;26:438–43. [DOI] [PubMed] [Google Scholar]
- 47. Nystad W, Meyer HE, Nafstad P, Tverdal A, Engeland A.. Body mass index in relation to adult asthma among 135, 000 Norwegian men and women. Am J Epidemiol 2004;160:969–76. [DOI] [PubMed] [Google Scholar]
- 48. Jensen ME, Wood LG, Gibson PG.. Obesity and childhood asthma–mechanisms and manifestations. Curr Opin Allergy Clin Immunol 2012;12:186–92. [DOI] [PubMed] [Google Scholar]
- 49. Sideleva O, Suratt BT, Black KE. et al. Obesity and asthma: an inflammatory disease of adipose tissue not the airway. Am J Respir Crit Care Med 2012;186:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sideleva O, Dixon A.. The many faces of asthma in obesity. J Cell Biochem 2014;115:421–26. [DOI] [PubMed] [Google Scholar]
- 51. McGeachie MJ, Wu AC, Tse SM. et al. CTNNA3 and SEMA3D: Promising loci for asthma exacerbation identified through multiple genome-wide association studies. J Allergy Clin Immunol 2015;136:1503–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carr TF, Bleecker E.. Asthma heterogeneity and severity. World Allergy Organ J 2016;9:41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vicente CT, Revez JA, Ferreira MA.. Lessons from ten years of genome-wide association studies of asthma. Clin Transl Immunol 2017;6:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Scherzer R, Grayson MH.. Heterogeneity and the origins of asthma. Ann Allergy Asthma Immunol 2018;121:400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boef AG, Dekkers OM, Le Cessie S.. Mendelian randomization studies: a review of the approaches used and the quality of reporting. Int J Epidemiol 2015;44:496–511. [DOI] [PubMed] [Google Scholar]
- 56. Lawlor DA. Commentary: two-sample Mendelian randomization: opportunities and challenges. Int J Epidemiol 2016;45:908.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Burgess S, Davies NM, Thompson SG.. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 2016;40:597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.