Abstract
Over the last years, many microRNAs (miRNAs) have been identified that regulate the formation of bioactive lipid mediators such as prostanoids and leukotrienes. Many of these miRNAs are involved in complex regulatory circuits necessary for the fine-tuning of biological functions including inflammatory processes or cell growth. A better understanding of these networks will contribute to the development of novel therapeutic strategies for the treatment of inflammatory diseases and cancer. In this review, we provide an overview of the current knowledge of miRNA regulation in eicosanoid pathways with special focus on novel miRNA functions and regulatory circuits of leukotriene and prostaglandin biosynthesis.
Keywords: microRNA, new miRNA functions, eicosanoids, inflammation, prostaglandins
Introduction
MicroRNAs (miRNAs) are a family of small non-coding RNAs that regulate a wide range of biological processes including cancer development (Garzon et al., 2006; Croce, 2009; Ochs et al., 2014). In 1993, lin-4 was the first miRNA to be discovered in the nematode Caenorhabditis elegans (C. elegans) and it was found to regulate the gene lin-14 on the post-transcriptional level during C. elegans development (Wightman et al., 1993). Later, a second small miRNA involved in worm development, let-7, was identified (Reinhart et al., 2000). However, at the time of discovery, it was assumed that these RNAs were rare exceptions and only present in nematodes. In 2001, three independent publications reported the existence of several hundreds of these small non-coding RNAs not only in nematodes but also in murine and human cells (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001). Currently, more than 2,500 human miRNAs have been identified (http://www.mirbase.org), although the functions of many of them are still unknown. In this review, we will give a short overview of novel miRNA functions involved in inflammatory processes. Moreover, we summarize the recent findings on miRNAs regulating key enzymes of the eicosanoid signaling pathway.
Biogenesis and Functions of MIRNAs
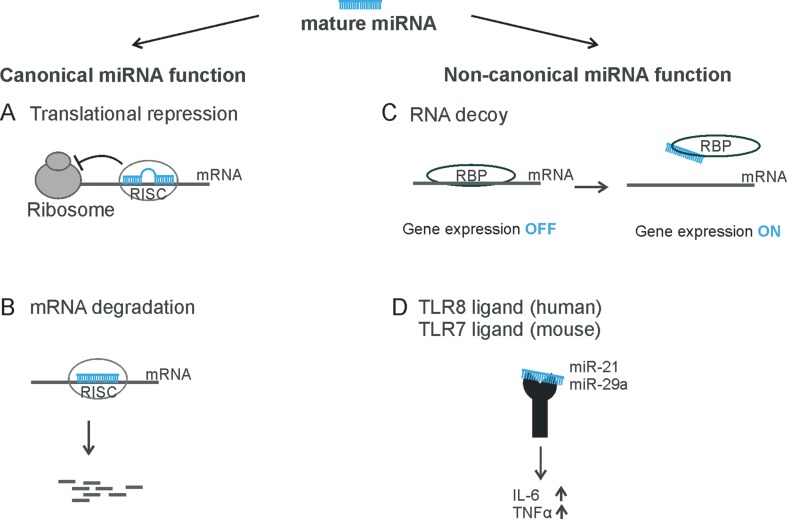
MiRNAs are transcribed from genes as long primary transcripts (pri-miRNAs) mostly by RNA polymerase II. In the nucleus, these pri-miRNAs are subsequently cleaved by the endonuclease Drosha. Drosha generates about 70 nucleotide (nt) long precursors (pre-miRNAs) that form imperfect stem-loop structures. Pre-miRNAs are transported out of the nucleus by exportin-5. In the cytoplasm, they are subsequently processed by a multiprotein complex including the RNAse III Dicer, Argonaute 2 (AGO2), and trans-activation-responsive RNA-binding protein (TRBP) to cleave the RNA into short 21–24 nt miRNA duplexes. Only one strand of the duplex is incorporated into a ribonucleoprotein complex, known as RNA-induced silencing complex (RISC). The passenger strand is mostly rapidly degraded. The miRNAs are then directed to their binding sites, which are usually located in the 3′ untranslated region (UTR) of their target messenger RNA (mRNA). Subsequently, they mediate endonucleotic cleavage, translational repression, or deadenylation of the mRNA transcript, followed by decapping and degradation of the target mRNA (Filipowicz et al., 2008) ( Figures 1A, B ). Of note, 40% of the currently known miRNAs are located within introns (mirtrons) and are processed by the spliceosome in an alternative way (Kim and Kim, 2007; De Rie et al., 2017).
Figure 1.
Overview of miRNA functions. The mature miRNA can be incorporated into the RNA-induced silencing complex (RISC) and binds to its target mRNA (A) to repress translation or (B) to induce mRNA degradation—for example, endonucleolytic cleavage. (C) miRNAs can act as RNA decoy to RNA-binding proteins, such as miR-328 to hnRNP E2 or miR-574-5p to CUGBP1. (D) miRNAs can be recognized by toll-like receptors 7/8, like miR-21 and miR-29a.
Traditionally, it has been assumed that miRNAs are loaded into RISC and bind to their target mRNA through specific base pairing and that they reduce gene expression at the post-transcriptional level as their sole canonical function. However, there is recent evidence that miRNAs are also able to activate gene expression via a non-canonical mechanism ( Figures 1C, D ). miRNAs can bind to RNA-binding proteins (RBPs) and sequester them from their target mRNAs in a RISC-independent manner. This was shown for miR-328, which acts as an RNA decoy for the heterogeneous nuclear ribonucleoprotein E2 (hnRNP E2), a global post-transcriptional regulator (Eiring et al., 2010; Saul et al., 2016) ( Figure 1C ). As a consequence, hnRNP E2 target gene expression is activated.
Recent findings demonstrate that cells can secrete bioactive molecules (like proteins, lipids, or nucleic acids) from cell to cell via extracellular vesicles. Those vesicles can be divided into small extracellular vesicles (sEVs, also known as exosomes), microvesicles or apoptotic vesicles distinguished by their size (Thery et al., 2018). Although these extracellular particles were traditionally considered to be a “disposal system” for unnecessary membrane proteins (Johnstone et al., 1991), they have now captured the interest of researchers as part of the cell to cell communication (Wen et al., 2017).
In fact, sEVs (<200 nm) are highly concentrated with miRNAs (Goldie et al., 2014). Different cell types, including immune and cancer cells, are capable of secretion and uptake of extracellular miRNAs from sEVs. This suggests that sEVs could be part of the intercellular communication and carry out novel biological functions. Since the extracellular miRNA content does not necessarily reflect the cellular miRNA profile of the recipient cell (Zhang et al., 2015), the functional analysis of sEV-delivered miRNAs is an interesting subject. In a study focusing on non-canonical miRNA functions, which are receptor-mediated, it was demonstrated that two sEV-delivered miRNAs, miR-21 and miR-29a, are able to bind to the murine toll-like receptor (TLR) 7 and the human TLR8 and induce cytokine expression (Fabbri et al., 2012). Similar results were found for miRNA let-7b in the context of the nervous system. Extracellular let-7b activates murine TLR7 and induces neurodegeneration. This observation is of particular interest because let-7b has been detected in the cerebrospinal fluid of patients with Alzheimer´s disease (Lehmann et al., 2012). Thus, sorting of miRNAS into EV and the interaction of EV-delivered miRNAs with target cell components such as certain receptors seems to be an important way of cell–cell communication.
An interesting aspect for further investigations in this context is to explore how miRNAs are sorted into EVs. Initial studies demonstrated that miRNAs are specifically recognized by RBPs, such as hnRNP A2/B1 and Y-box protein 1 (YBX1). Those RBPs bind sequence-specifically to miRNAs and load them selectively into sEVs (Villarroya-Beltri et al., 2013; Shurtleff et al., 2016). These data suggest that the loading of miRNAs into EV is a specific process that might control biological processes such as immune functions.
miRNAs and Eicosanoids
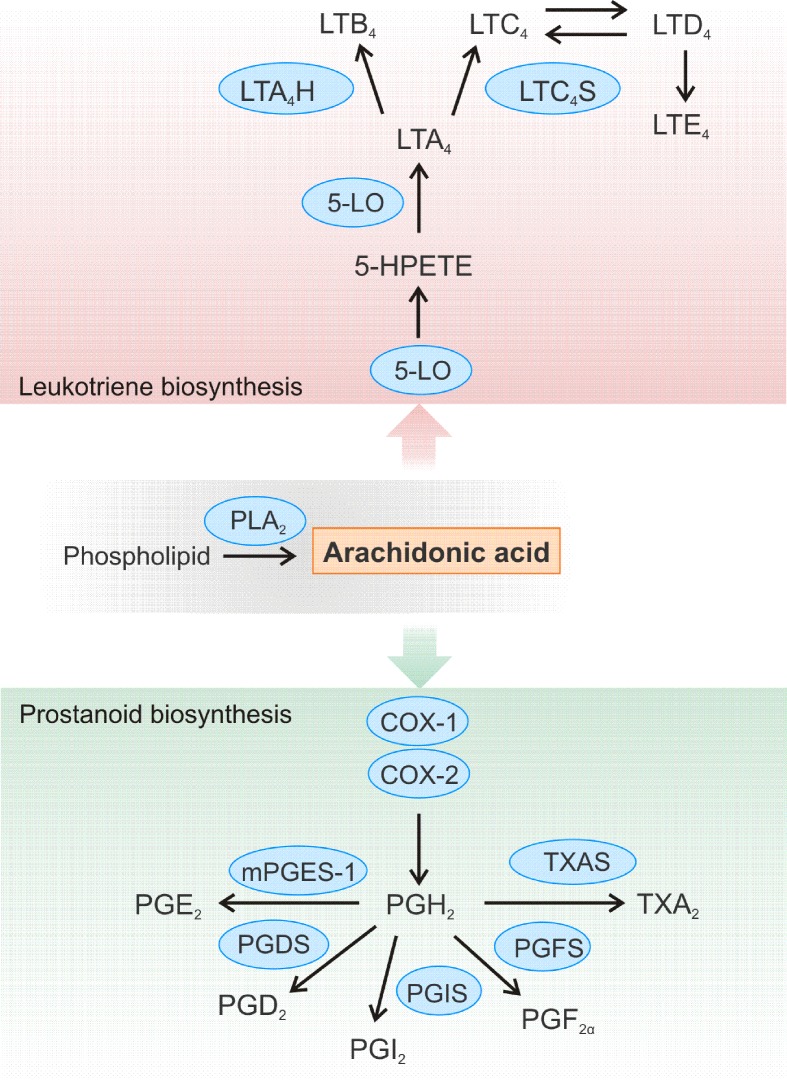
Eicosanoids such as prostaglandins and leukotrienes are biologically active lipid mediators that are products of a local cell type-specific arachidonic acid (AA) metabolism ( Figure 2 ). Such lipid mediators play a critical role in different pathological processes like inflammation and cancer (Zeldin, 2001; Wang and Dubois, 2010; Radmark et al., 2015). The synthesis of eicosanoids begins with the release of AA from the cell membrane by phospholipase A2 which is followed by the metabolism of the AA through cyclooxygenases (COXs), lipoxygenases (LOXs), and cytochrome P450 enzymes (Wang and Dubois, 2010). Due to the key role of these enzymes in the formation of bioactive lipid mediators, it is not surprising that 5-LO and COX enzymes are prominent miRNA targets (for previous reviews, see also Ochs et al., 2011; Ochs et al., 2014).
Figure 2.
Schematic overview of the leukotriene and prostanoid biosynthesis pathway. Arachidonic acid (AA) is released from cellular membranes by cytosolic phospholipase A2 (PLA2). The free AA can further be converted to different leukotrienes (LT). 5-LO is crucial for the conversion of AA to 5(S)-hydroperoxyeicosatetraenoic acid (5-HPETE) and LTA4. LTA4 is further converted to LTB4 by LTA4 hydrolase (LTA4H) or to the cysteinyl-containing LTC4 by LTC4 synthase (LTC4S), which can be metabolized to LTD4 and LTE4. Biosynthesis of prostanoids begins with the enzyme cyclooxygenase (COX)-1 or COX-2 converting AA to prostaglandin (PG) H2, which is further converted to a variety of other prostanoids. Thromboxane A2 (TXA2) is generated by TXA synthase (TXAS), while the synthases PGxS produce the certain PGs: PGF2α, PGI2, and PGD2. The microsomal PGE synthase-1 (mPGES-1) catalyzes formation of PGE2.
miRNA Regulation in Prostanoid Biosynthesis
The group of prostanoids consists of prostaglandins, thromboxane A2, as well as prostacyclin. Their common denominator is that they can all be formed from AA, which is converted by COX enzymes in a two-step process to PGH2 ( Figure 2 ). This intermediate is then converted to the different prostaglandins by the respective synthases. Prostanoids belong to the most important inflammatory signaling molecules. These lipid mediators exert their multiple biological effects in an autocrine and paracrine manner by binding to their specific cell surface G protein-coupled receptors. For example, prostaglandin E2 (PGE2) is a bioactive lipid that can elicit a wide range of biological effects associated with inflammation and cancer (Jakobsson et al., 1999; Pettersson et al., 2005; Nakanishi and Rosenberg, 2013). It contributes to the development of inflammation and plays a predominant role in promoting cancer progression by induction of cellular proliferation and tumor angiogenesis, inhibition of apoptosis, and suppression of immune responses (Wang and Dubois, 2006; Larsson and Jakobsson, 2015). PGE2 is formed by the conversion of AA to PGH2 by the cyclooxygenases COX-1 or COX-2, followed by processing by PGE synthases, of which microsomal prostaglandin E synthase 1 (mPGES-1) is the key enzyme. The formed PGE2 is then rapidly secreted to act on their specific receptors on recipient cells. These receptors are not only present on immune cells, but also on a variety of cells of the cardiovascular system like cardiomyocytes, smooth muscle cells, or vascular endothelial cells (Suzuki et al., 2011). Thus, PGE2 can be involved in the development of different cardiovascular diseases. It was shown that deletion of mPGES-1 impairs the left ventricular contractile function after acute myocardial infarction and leads to overall remodeling of the left ventricle (Degousee et al., 2008).
Non-Canonical miRNA Regulation of Prostaglandin E2 Biosynthesis
A variety of cancer types exhibit increased levels of mPGES-1 and its enzymatic product PGE2—for example, colon (Yoshimatsu et al., 2001b; Wang and Dubois, 2010), prostate (Jain et al., 2008; Hanaka et al., 2009), lung (Yoshimatsu et al., 2001a), and breast cancer (Olesch et al., 2015). Interestingly, there were marked differences in the extent of upregulation of mPGES-1 and PGE2 in individual lung tumors (Yoshimatsu et al., 2001a; Wu et al., 2010). This effect can be attributed to individual expression variations of COX-2 and mPGES-1 (Wang et al., 2006; Hughes et al., 2008). However, the observed variability cannot be explained solely by different transcriptional regulation mechanisms [such as nuclear factor κB (NFκB)] (Yamamoto et al., 1995; Kang et al., 2007), suggesting that additional—namely, post-transcriptional mechanisms—might be involved—for instance, miRNA regulation.
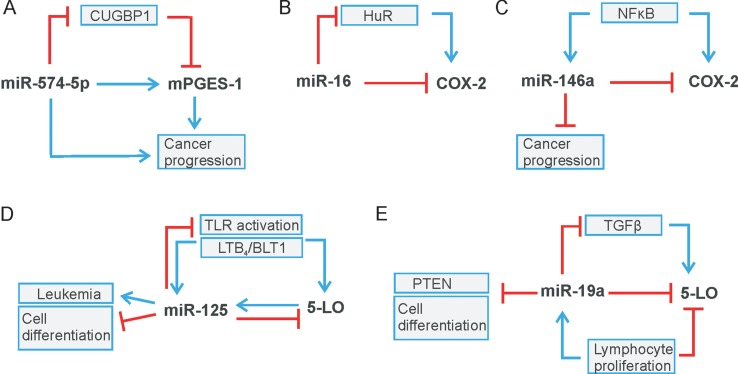
Recently, we demonstrated that miR-574-5p activates mPGES-1-derived PGE2 synthesis in human non-small cell lung cancer (NSCLC). Moreover, this miRNA promotes tumor growth in vivo, which is completely blocked by the administration of the mPGES-1 inhibitor CIII (Leclerc et al., 2013). Mechanistically, it could be shown that miR-574-5p acts as RNA decoy to CUG-RNA-binding protein 1 (CUGBP1) ( Figures 1C , 3A ) and that it antagonizes the CUGBP1 function as a post-transcriptional regulator (Mukhopadhyay et al., 2003; Subbaramaiah et al., 2003; Gao et al., 2015). In human, NSCLC as well as under inflammatory conditions, miR-574-5p is strongly upregulated and induces mPGES-1 expression by preventing CUGBP1 binding to the mPGES-1-3´UTR. This leads to an enhanced alternative splicing and the generation of a 3´UTR splice variant. Importantly, the nuclear localization of miR-574-5p and CUGBP1 is in line with its regulatory function on mPGES-1 mRNA splicing (Saul et al., 2019). The newly discovered association between miR-574-5p overexpression and PGE2-mediated growth of lung cancer cells in vivo suggests that pharmacological inhibition of PGE2 formation with mPGES-1 or COX inhibitors might be of considerable therapeutic value for NSCLC patients with high miR-574-5p levels (Saul et al., 2019). In this respect, mPGES-1 inhibition might be of particular interest since inhibition of mPGES-1, instead of COX-2, may be associated with fewer side effects as other prostanoids would not be affected.
Figure 3.
Regulatory circuits of leukotriene and prostaglandin biosynthesis. (A) miR-574-5p/CUGBP1 regulates mPGES-1 level, (B) miR-16/HuR and (C) miR-146a modulate COX-2 expression. (D) miR-125 and (E) miR-19a regulate 5-LO level. Stimulation is indicated by blue arrows, whereas red lines indicate inhibition.
Canonical miRNA Regulation of Prostaglandin Biosynthesis
There are two known isoenzymes responsible for the generation of prostanoids: cyclooxygenase-1 (COX-1) and cyclooxygenase-2 (COX-2), both of which catalyze the same enzymatic reaction. COX-1 represents a housekeeping enzyme responsible for maintaining basal prostanoid levels, which are important for tissue homeostasis. In contrast, COX-2 is barely detectable in most normal tissues, but is strongly induced in response to inflammatory cytokines, hypoxia, and other stressors (Samuelsson et al., 2007; Wang and Dubois, 2010).
COX-2 expression is regulated at different levels (Dixon et al., 2000; Harper and Tyson-Capper, 2008). Various transcription factors such as NFκB, activator protein 1 (AP1), or the cAMP-responsive element-binding protein (CREB) are involved in its transcriptional regulation (Yamamoto et al., 1995; Kang et al., 2007).
At the post-transcriptional level, COX-2 expression is regulated by mRNA stability and translation efficiency mostly including trans- and cis-acting factors (Dixon et al., 2000; Young and Dixon, 2010). For example, RNA-binding proteins can interact with AU-rich elements (AREs) within the COX-2 3´UTR like CUG triplet repeat-RNA-binding protein 2 (CUGBP2) (Mukhopadhyay et al., 2003) or the mRNA-stability factor HuR (Erkinheimo et al., 2003; Subbaramaiah et al., 2003). Tristetraprolin (TTP), another RNA-binding protein that promotes mRNA instability, has been shown to bind to the 3´UTR of COX-2 and thus contributes to the post-transcriptional regulation of COX-2 (Sawaoka et al., 2003). In addition, the use of an alternative polyadenylation signal may regulate COX-2 mRNA stability and translation (Hall-Pogar et al., 2005; Hall-Pogar et al., 2007).
In recent years, miRNAs have been identified as additional players in the post-transcriptional control of COX-2 expression (see Table 1 ). Interestingly, miR-16 is complementary to sequences in the AU-rich regions of the COX-2 3’UTR, which allows direct binding of miR-16, and in turn, alters COX-2 mRNA stability (Jing et al., 2005) ( Figure 3 ). In association with TTP, miR-16 can promote mRNA decay. However, TTP does not bind directly to miR-16 but interacts through the association with RISC components to form a complex with miR-16 and promote mRNA degradation (Jing et al., 2005). Furthermore, miR-16 silences COX-2 expression in hepatoma cells via two mechanisms: by binding directly to the COX-2 3′-UTR and by decreasing the levels of the RNA-binding protein HuR (Agra Andrieu et al., 2012) ( Figure 3B ). MiR-16 also competes with the RNA-binding protein heterogeneous nuclear ribonuclear protein K (hnRNP K) for binding to COX-2 3´UTR and thus modulates COX-2 expression on the post-transcriptional level (Shanmugam et al., 2008).
Table 1.
Summary—miRNAs influencing the prostaglandin pathway by binding to mRNAs of key enzymes or interfering with RNA-binding protein CUGBP1 (see mPGES-1).
| miRNA | Target | Tissue/disease | Reference |
|---|---|---|---|
| Hsa-miR-144 | COX-2 | Premature labor | (Li et al., 2016) |
| Hsa-miR-144-5p | COX-2 | Periodontitis | (Li et al., 2019) |
| Hsa-miR-144 | COX-2 | Esophageal squamous cell cancer | (Shao et al., 2016) |
| Hsa-miR-26a | |||
| Hsa-miR-26b | COX-2 | Nasopharyngeal epithelial cancer | (Ji et al., 2010) |
| Hsa-miR-26b | COX-2 | Oral lichen planus | (Danielsson et al., 2012) |
| Hsa-miR-26b | COX-2 | Breast cancer | (Li et al., 2013) |
| Hsa-miR-216a-3p | COX-2 | Colorectal cancer | (Wang et al., 2018) |
| Hsa-miR-30a | COX-2 | Gastric cancer | (Liu et al., 2017) |
| Hsa-miR-146a | mPGES-2 | Bone marrow-derived mesenchymal stem cells | (Matysiak et al., 2013) |
| Hsa-miR-146a | COX-2 | COPD | (Sato et al., 2010) |
| Hsa-miR-146a | COX-2 | Astrocytes | (Iyer et al., 2012) |
| Hsa-miR-146a | COX-2 | Colon cancer | (Zhang et al., 2019) |
| Hsa-miR-146a | COX-2 | Lung cancer | (Cornett and Lutz, 2014) |
| Hsa-miR-146a | COX-2 | COPD | (Wang et al., 2015) |
| Mmu-miR-199a | COX-2 | Mouse uterus, endometrial cancer | (Chakrabarty et al., 2007; Daikoku et al., 2008) |
| Mmu-miR-101a | |||
| Hsa-miR-101 | COX-2 | Colon cancer | (Strillacci et al., 2009) |
| Hsa-miR-101 | COX-2 | Endometrial serous carcinoma | (Hiroki et al., 2010) |
| Hsa-miR-101a | COX-2 | Mammary gland | (Tanaka et al., 2009) |
| Hsa-miR-101 | COX-2 | Gastric cancer | (He et al., 2012) |
| Hsa-miR-101 | COX-2 | Prostate cancer | (Hao et al., 2011) |
| Hsa-miR-101-3p | COX-2 | Esophageal squamous cell cancer | (Gong et al., 2016) |
| Hsa-miR-16 | COX-2 | Cervical cancer | (Jing et al., 2005) |
| Hsa-miR-16 | COX-2 | Hepatocellular carcinoma | (Agra Andrieu et al., 2012) |
| Hsa-miR-16 | COX-2 | Monocytes (THP-1 cells) | (Shanmugam et al., 2008) |
| Hsa-miR-137 | COX-2 | Glioma | (Chen et al., 2012) |
| Hsa-miR-143 | COX-2 | Amnion mesenchymal cells | (Kim et al., 2011) |
| Hsa-miR-542-3p | COX-2 | Colon cancer | (Moore et al., 2012) |
| Hsa-miR-574-5p | mPGES-1 | Lung cancer (NSCLC) | (Saul et al., 2019) |
| Hsa-miR-21 | 15-PGDH | Cholangiocarcinoma | (Lu et al., 2014) |
| Hsa-miR-21 | 15-PGDH | Tongue squamous cell cancer | (He et al., 2016) |
| Hsa-miR-21 | 15-PGDH | Gastric cancer | (Li et al., 2019) |
| Hsa-miR-21 | 15-PGDH | Colon cancer | (Monteleone et al., 2019) |
| Hsa-miR-26a/b | 15-PGDH | Cholangiocarcinoma | (Yao et al., 2015) |
| Hsa-miR-620 | 15-PGDH | Prostate adenocarcinoma cell line (DU145); breast cancer cell line (MDA-MB-231) | (Huang et al., 2015) |
| Hsa-miR-218 | 15-PGDH | Synovial mesenchymal stem cells (SMSCs) | (Cong et al., 2017) |
| Hsa-miR146b-3p | 15-PGDH | Cervical cancer | (Yao et al., 2018) |
In addition, a considerable number of reports focus on miR-144, which directly targets COX-2 mRNA to downregulate its protein level. For example, the balance between miR-144, COX-2, and c-fos regulates PGE2 synthesis in the amnion of pregnant humans and mice. The amnion is the major source of PGE2 and plays a central role in the process of premature labor. The transcription factor c-fos induces expression of COX-2 and miR-144. The latter in turn generates a negative feedback loop by directly and indirectly inhibiting both COX-2 and c-fos. This inhibition has a negative effect on PGE2 generation and thus prevents premature contractions (Li et al., 2016). A negative correlation of miR-144-5p and COX-2 was also observed in human inflamed gingival tissue from periodontitis patients (Li et al., 2019). Moreover, a significant downregulation of miR-144 and miR-26a compared to healthy surrounding tissue was found in the tumor tissue of patients with esophageal squamous cell carcinoma (ESCC). The two miRNAs were confirmed to bind to COX-2 mRNA, thus downregulating the protein level of COX-2. It has been shown that, due to downregulation of miR-144 and miR-26a in tumor tissue, COX-2 activity significantly increased and subsequently promoted cell proliferation and metastasis (Shao et al., 2016). COX-2 is also a canonical target of miR-101-3p that is downregulated in ESCC cells stimulated with cigarette smoking extract (CSE). It has been shown that the promoting effect of CSE on ESCC is due to COX-2 upregulation. It was further demonstrated that the mechanism by which CSE regulates COX-2 expression is mediated by miR-101-3p to promote cell proliferation (Gong et al., 2016). Another miRNA that regulates COX-2 via its canonical function is miR-146a, a miRNA that is regulated like COX-2 by NFκB signaling (Poligone and Baldwin, 2001; Taganov et al., 2006). In human lung cancer, miR-146a directly regulates COX-2 mRNA and thus the protein level of COX-2 in lung cancer cells (Cornett and Lutz, 2014) ( Figure 3C ). In human fibroblasts of smokers with chronic obstructive pulmonary disease, it was found that single nucleotide polymorphisms in the miR-146a precursor caused several patients to have reduced miRNA level, which significantly improved baseline lung function (Wang et al., 2015). Very recently, it was demonstrated that the polymorphisms of miR-146a (rs2910164) and plasmacytoma variant translocation 1 (PVT1; rs13281615) affect the prognosis of colon cancer by regulating COX-2 expression and cell apoptosis. The presence of PVT1 decreased the expression level of miR-146a, which in turn increased the COX-2 level (Zhang et al., 2019). Another miRNA that suppresses inflammation-related tumors is miR-30a. MiR-30a is crucial for regulation of growth and migration of Heliobacter pylori–infected gastric cancer via targeting COX-2 and B cell CLL/lymphoma 9 (BCL 9) (Liu et al., 2017). In the same way, colorectal cancer is influenced by miR-216a-3p, which directly targets COX-2 via its canonical function (Wang et al., 2018).
In addition to the miRNA regulation of enzymes involved in PGE2 formation like COX-2 and mPGES-1, it is important to explore how the PGE2-metabolizing enzyme is regulated by miRs. The key enzyme that converts PGE2 to its biologically inactive metabolite is the NAD+-dependent 15-hydroxyprostaglandin dehydrogenase (15-PGDH/HPGD) (Ensor and Tai, 1995). In cholangiocarcinoma cells, 15-PGDH was identified as a direct target of miR-21. In addition, COX-2 overexpression and PGE2 treatment increase the level of miR-21 associated with enhanced miR-21 promoter activity (Lu et al., 2014). The finding that miR-21 directly regulates 15-PDGH was further confirmed by recent publications on tongue squamous cell carcinoma (He et al., 2016), gastric cancer (Li et al., 2019) and colon cancer (Monteleone et al., 2019). Interestingly, epithelial growth factor (EGF) signaling in colorectal cancer cells reduces the level of 15-PGDH and simultaneously increases the miR-21 level (Monteleone et al., 2019).
In addition to this, other miRNAs are known to modulate 15-PGDH expression. Omega-3 polyunsaturated fatty acids (ω-3 PUFA) upregulate 15-PGDH expression by inhibiting miR-26a and miR-26b. This directly contributes to ω-3 PUFA-induced inhibition of human cholangiocarcinoma cell growth, providing a preclinical justification for the evaluation of ω-3 PUFA in the treatment of cholangiocarcinoma (Yao et al., 2015). MiR-620 directly targets 15-PGDH, which results in an increase in PGE2 promoting radioresistance in cancer cells like the human prostate adenocarcinoma cell line DU145 and the breast cancer cell line MDA-MB-231 (Huang et al., 2015). Furthermore, miR-218 directly regulates the expression level of 15-PGDH during differentiation of synovial mesenchymal stem cells (SMSCs) into cartilage and subsequently inhibits osteogenesis during chondrogenesis (Cong et al., 2017). Finally, 15-PDGH was identified as the canonical target of miR-146b-3p that promotes proliferation, migration, and anchorage-independent growth of cervical cancer cells (Yao et al., 2018).
Influence of the Prostaglandin Pathway on miRNA Expression
In addition to the regulation of the enzymes of the prostaglandin pathway by miRNAs, the influence of prostaglandin signaling on miRNA expression has also been investigated. It was found that COX-2 signaling increases the oncogenic miR-526b and miR-655 levels in human breast cancer by activating the EP4 receptor. Thus, COX-2 signaling strongly influences the phenotype of the tumor by promoting cellular migration, invasion, or proliferation (Majumder et al., 2015; Majumder et al., 2018).
In addition, it was found that cancer-associated fibroblasts (CAFs) develop a senescence-associated secretory phenotype (SASP) that contributes to cancer progression. Interestingly, senescent CAFs have increased levels of PGE2 and COX-2. Moreover, miR-335 is upregulated in the senescent normal fibroblasts as well as CAFs. This modulates the secretion of SASP factors and induces the mobility of cancer cells in co-cultures, at least partially by suppressing the expression of the phosphatase and tensin homologue (PTEN). With the application of the COX-2 inhibitor celecoxib, expression of miR-335 was strongly reduced, suggesting a new miR-335/COX-2/PTEN axis that modulates the inflammasome in senescent fibroblasts (Kabir et al., 2016).
In line with these results, it was demonstrated that the mPGES-1/PGE2 pathway affects the expression level of miR-15a and miR-186 in prostate cancer. High PGE2 levels reduced Dicer expression and consequently miRNA biogenesis in prostate cancer cells. It is noteworthy that PGE2-mediated downregulation of miR-15a and miR-186 is directly associated to vascular endothelial growth factor (VEGF) expression and angiogenesis. This suggests that these miRNAs may be potential candidates for mitigating the aggressive properties of prostate cancer. This alternative approach could overcome the chemo-resistance, which is common for drugs targeting VEGF and/or VEGF receptors (Terzuoli et al., 2016).
miRNA Regulation in Leukotriene Biosynthesis
The 5-lipoxygenase (5-LO) is the key enzyme of the leukotriene pathway. It interacts with the 5-LO-activating protein (FLAP) and catalyzes the conversion of AA into 5(S)-hydroperoxyeicosatetraenoic acid (5-HPETE) and leukotriene (LT) A4. The LTA4 is then subsequently converted either into biologically active LTB4 by LTA4 hydrolase or into LTC4 by LTC4 synthase and the LTC4 synthase isoenzyme MGST2. Over the last decades, different studies have shown that 5-LO-derived AA metabolites play an important role in inflammatory reactions like inflammatory disorders and allergic diseases such as asthma, allergic rhinitis, cardiovascular diseases, as well as in different types of cancer (Steinhilber and Hofmann, 2014).
Canonical miRNA Regulation on Leukotriene Pathway
In contrast to COX-2, the knowledge about post-transcriptional regulation of 5-LO is rather limited. It is known that the combination of alternative splicing and RNA decay modulates 5-LO gene expression (Ochs et al., 2012). Furthermore, several publications demonstrated that 5-LO is a canonical target for miR-19a-3p, miR-125-5p (Busch et al., 2015), miR-216-3p (Wang et al., 2018), and miR-674-5p (Su et al., 2016) (see Table 2 ). Specifically, miR-19a-3p and miR-125-5p regulate 5-LO expression in the human myeloid cell line MonoMac 6. In contrast, only miR-19a-3p modulates the 5-LO protein level in human T-lymphocytes stimulated with phytohemagglutinin (PHA). Overall, it reveals that miR-19a-3p and miR-125b-5p target 5-LO in a cell type and stimulus-specific manner (Busch et al., 2015). Interestingly, miR-125 and miR-19a seem to be parts of regulatory circuit-controlling immune reactions and cell proliferation ( Figures 3D, E ). 5-LO expression is induced by TLR/NFκB activation (Lee et al., 2015) or during cell stimulation by transforming growth factor (TGF)-β (Steinhilber et al., 1993). The same signals were also reported to induce miR-125 (Curtale et al., 2018; Hildebrand et al., 2018) as well as miR-19a, which in turn downregulates components of NFκB or TGF-β signaling, respectively, and also 5-LO ( Figures 3D, E ) (Dews et al., 2010; Busch et al., 2015). Thus, upregulation of miR-19a and downregulation of 5-LO expression are associated with cell proliferation in T-lymphocytes (Busch et al., 2015). Furthermore, it is well known that upregulation of miR-19a inhibits cell differentiation and promotes cell growth and cancer development—for instance, by suppression of PTEN (Lewis et al., 2003). MiR-674-5p was identified as a direct regulator of 5-LO mRNA in mice. It is further discussed as a negative regulator in 5-LO-mediated autoimmune diseases of the liver, thus representing a promising approach to future therapeutic measures (Su et al., 2016). Recently, it was shown that miR-216a-3p regulates not only COX-2 but also 5-LO expression in colon cancer, thus affecting colon cancer cell proliferation. These data indicate that miR-216-3p might represent a novel target for colorectal cancer treatment (Wang et al., 2018). Interestingly, not only 5-LO but also FLAP is targeted by miRs, like miR-146a in human lung cancer (Iacona et al., 2018). Moreover, miR-146a is also known to regulate COX-2 expression in lung cancer (Cornett and Lutz, 2014), which indicates a role of miR-146a as an endogenous dual inhibitor of AA metabolism in lung cancer by regulating prostaglandins and LTs, similar to miR-216a-3p (Iacona et al., 2018).
Table 2.
Summary—miRNAs influencing the leukotriene pathway by binding to mRNAs of key enzymes.
| miRNA | Target | Tissue/disease | Reference |
|---|---|---|---|
| Hsa-miR-19a-3p | 5-LO | Monocytes (MM6 cells), T-lymphocytes | (Busch et al., 2015) |
| Hsa-miR-125-5p | |||
| Hsa-miR-674-5p | 5-LO | Acute mouse liver injury | (Su et al., 2016) |
| Hsa-miR-216a-3p | 5-LO | Colorectal cancer | (Wang et al., 2018) |
| Hsa-miR-146a | FLAP | Lung cancer | (Iacona et al., 2018) |
| Hsa-miR-135a | FLAP | Pulmonary microvascular endothelial cells | (Gonsalves and Kalra, 2010) |
| Hsa-miR-199a |
Influence of the Leukotriene Pathway on miRNA Expression
It is well known that 5-LO interacts with the C-terminal domain of human Dicer. The interaction between 5-LO and Dicer leads to an enhanced 5-LO enzymatic activity as well as Dicer activity in vitro. These results suggest that the processing of specific miRNAs by Dicer might be regulated by the 5‐LO/Dicer interaction (Dincbas-Renqvist et al., 2009). In addition to the direct 5-LO/Dicer interaction, 5-LO products can also modulate miRNA expression. Thus, LTB4 induces the expression of inflammatory miRNAs including miR-155, miR-146a and miR125b in macrophages via the LT B4 receptor-1 (BLT1) and Gα1 signaling (Wang et al., 2014).
Conclusions
The number of miRNAs that were discovered in recent years to be involved in the regulation of the expression of key enzymes in prostanoid and LT biosynthesis is steadily increasing. Tables 1, 2 give a comprehensive overview of all currently known miRNAs involved in inflammatory processes. There is growing evidence that many of these miRNAs are involved in complex regulatory cascades and networks with multiple layers of integrated signals and feedback loops required for fine-tuning of biological functions such as inflammatory responses or cell growth. Besides the direct, canonical regulation of mRNA stability and translation of enzymes of the AA cascade by miRNAs, it was found that miRNAs that interfere with HUR or TTP function are involved in the regulation of COX-2 expression. Furthermore, a novel non-canonical mechanism was found for miR-574-5p, which acts as a decoy for CUGBP1 and strongly stimulates PGE2 formation. It becomes clear that miRNAs can also be packed into EVs so that they are involved in cell–cell communication. This suggests that miRNAs not only regulate cellular functions in an autocrine/intracrine manner but also can affect processes such as the formation of lipid mediators in a paracrine fashion. Of note, 5-LO was identified as a binding protein for Dicer, which suggests that 5-LO can modulate the Dicer function and interferes with miRNA generation. Thus, it becomes increasingly clear that there are multiple links between the miRNA network and lipid signaling. The progress in the understanding of these interactions will help to develop new therapeutic strategies for the treatment of inflammatory diseases and cancer.
Author Contributions
MS wrote the manuscript. AE, DS, and BS contributed to writing and editing the manuscript.
Funding
This project was supported by the Else Kröner-Fresenius Stiftung (Else Kröner-Fresenius-Graduiertenkolleg), the Deutsche Forschungsgemeinschaft (SFB902 and ECCPS), Athene Young Investigator program (Technische Universität Darmstadt; grant no: n/a), and BMBF (KMU-innovativ-22: miRTumorProst; 031B0768B).
References
- Agra Andrieu N., Motino O., Mayoral R., Llorente Izquierdo C., Fernandez-Alvarez A., Bosca L., et al. (2012). Cyclooxygenase-2 is a target of microRNA-16 in human hepatoma cells. PLoS One 7, e50935. 10.1371/journal.pone.0050935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch S., Auth E., Scholl F., Huenecke S., Koehl U., Suess B., et al. (2015). 5-lipoxygenase is a direct target of miR-19a-3p and miR-125b-5p. J. Immunol. 194, 1646–1653. 10.4049/jimmunol.1402163 [DOI] [PubMed] [Google Scholar]
- Chakrabarty A., Tranguch S., Daikoku T., Jensen K., Furneaux H., Dey S. K. (2007). MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc. Natl. Acad. Sci. U. S. A. 104, 15144–15149. 10.1073/pnas.0705917104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wang X., Wang H., Li Y., Yan W., Han L., et al. (2012). miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur. J. Cancer 48, 3104–3111. 10.1016/j.ejca.2012.02.007 [DOI] [PubMed] [Google Scholar]
- Cong R., Tao K., Fu P., Lou L., Zhu Y., Chen S., et al. (2017). MicroRNA218 promotes prostaglandin E2 to inhibit osteogenic differentiation in synovial mesenchymal stem cells by targeting 15hydroxyprostaglandin dehydrogenase [NAD(+)]. Mol. Med. Rep. 16, 9347–9354. 10.3892/mmr.2017.7795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornett A. L., Lutz C. S. (2014). Regulation of COX-2 expression by miR-146a in lung cancer cells. RNA 20, 1419–1430. 10.1261/rna.044149.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce C. M. (2009). Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704–714. 10.1038/nrg2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtale G., Renzi T. A., Mirolo M., Drufuca L., Albanese M., De Luca M., et al. (2018). Multi-step regulation of the TLR4 pathway by the miR-125a∼99b∼let-7e Cluster. Front. Immunol. 9, 2037. 10.3389/fimmu.2018.02037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daikoku T., Hirota Y., Tranguch S., Joshi A. R., Demayo F. J., Lydon J. P., et al. (2008). Conditional loss of uterine Pten unfailingly and rapidly induces endometrial cancer in mice. Cancer Res. 68, 5619–5627. 10.1158/0008-5472.CAN-08-1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson K., Ebrahimi M., Wahlin Y. B., Nylander K., Boldrup L. (2012). Increased levels of COX-2 in oral lichen planus supports an autoimmune cause of the disease. J. Eur. Acad. Dermatol. Venereol. 26, 1415–1419. 10.1111/j.1468-3083.2011.04306.x [DOI] [PubMed] [Google Scholar]
- De Rie D., Abugessaisa I., Alam T., Arner E., Arner P., Ashoor H., et al. (2017). An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 35, 872–878. 10.1038/nbt.3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degousee N., Fazel S., Angoulvant D., Stefanski E., Pawelzik S. C., Korotkova M., et al. (2008). Microsomal prostaglandin E2 synthase-1 deletion leads to adverse left ventricular remodeling after myocardial infarction. Circulation 117, 1701–1710. 10.1161/CIRCULATIONAHA.107.749739 [DOI] [PubMed] [Google Scholar]
- Dews M., Fox J. L., Hultine S., Sundaram P., Wang W., Liu Y. Y., et al. (2010). The myc-miR-17∼92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res. 70, 8233–8246. 10.1158/0008-5472.CAN-10-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dincbas-Renqvist V., Pepin G., Rakonjac M., Plante I., Ouellet D. L., Hermansson A., et al. (2009). Human Dicer C-terminus functions as a 5-lipoxygenase binding domain. Biochim. Biophys. Acta 1789, 99–108. 10.1016/j.bbagrm.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D. A., Kaplan C. D., Mcintyre T. M., Zimmerman G. A., Prescott S. M. (2000). Post-transcriptional control of cyclooxygenase-2 gene expression. The role of the 3’-untranslated region. J. Biol. Chem. 275, 11750–11757. 10.1074/jbc.275.16.11750 [DOI] [PubMed] [Google Scholar]
- Eiring A. M., Harb J. G., Neviani P., Garton C., Oaks J. J., Spizzo R., et al. (2010). miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 140, 652–665. 10.1016/j.cell.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensor C. M., Tai H. H. (1995). 15-Hydroxyprostaglandin dehydrogenase. J. Lipid Mediat. Cell Signal 12, 313–319. 10.1016/0929-7855(95)00040-W [DOI] [PubMed] [Google Scholar]
- Erkinheimo T. L., Lassus H., Sivula A., Sengupta S., Furneaux H., Hla T., et al. (2003). Cytoplasmic HuR expression correlates with poor outcome and with cyclooxygenase 2 expression in serous ovarian carcinoma. Cancer Res. 63, 7591–7594. [PubMed] [Google Scholar]
- Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., et al. (2012). MicroRNAs bind to toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 109, E2110–E2116. 10.1073/pnas.1209414109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114. 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- Gao C., Yu Z., Liu S., Xin H., Li X. (2015). Overexpression of CUGBP1 is associated with the progression of non-small cell lung cancer. Tumour Biol. 36 (6), 4583–4589. 10.1007/s13277-015-3103-1 [DOI] [PubMed] [Google Scholar]
- Garzon R., Fabbri M., Cimmino A., Calin G. A., Croce C. M. (2006). MicroRNA expression and function in cancer. Trends Mol. Med. 12, 580–587. 10.1016/j.molmed.2006.10.006 [DOI] [PubMed] [Google Scholar]
- Goldie B. J., Dun M. D., Lin M., Smith N. D., Verrills N. M., Dayas C. V., et al. (2014). Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 42, 9195–9208. 10.1093/nar/gku594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Chu Y., Xu M., Huo J., Lv L. (2016). Esophageal squamous cell carcinoma cell proliferation induced by exposure to low concentration of cigarette smoke extract is mediated via targeting miR-101-3p/COX-2 pathway. Oncol. Rep. 35, 463–471. 10.3892/or.2015.4379 [DOI] [PubMed] [Google Scholar]
- Gonsalves C. S., Kalra V. K. (2010). Hypoxia-mediated expression of 5-lipoxygenase-activating protein involves HIF-1alpha and NF-kappaB and microRNAs 135a and 199a-5p. J. Immunol. 184, 3878–3888. 10.4049/jimmunol.0902594 [DOI] [PubMed] [Google Scholar]
- Hall-Pogar T., Liang S., Hague L. K., Lutz C. S. (2007). Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3’-UTR. RNA 13, 1103–1115. 10.1261/rna.577707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Pogar T., Zhang H., Tian B., Lutz C. S. (2005). Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res. 33, 2565–2579. 10.1093/nar/gki544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaka H., Pawelzik S. C., Johnsen J. I., Rakonjac M., Terawaki K., Rasmuson A., et al. (2009). Microsomal prostaglandin E synthase 1 determines tumor growth in vivo of prostate and lung cancer cells. Proc. Natl. Acad. Sci. U. S. A. 106, 18757–18762. 10.1073/pnas.0910218106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Gu X., Zhao Y., Greene S., Sha W., Smoot D. T., et al. (2011). Enforced expression of miR-101 inhibits prostate cancer cell growth by modulating the COX-2 pathway in vivo . Cancer Prev. Res. (Phila) 4, 1073–1083. 10.1158/1940-6207.CAPR-10-0333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper K. A., Tyson-Capper A. J. (2008). Complexity of COX-2 gene regulation. Biochem. Soc. Trans. 36, 543–545. 10.1042/BST0360543 [DOI] [PubMed] [Google Scholar]
- He Q., Chen Z., Dong Q., Zhang L., Chen D., Patel A., et al. (2016). MicroRNA-21 regulates prostaglandin E2 signaling pathway by targeting 15-hydroxyprostaglandin dehydrogenase in tongue squamous cell carcinoma. BMC Cancer 16, 685. 10.1186/s12885-016-2716-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. P., Shao Y., Li X. L., Xu W., Chen G. S., Sun H. H., et al. (2012). Downregulation of miR-101 in gastric cancer correlates with cyclooxygenase-2 overexpression and tumor growth. FEBS J. 279, 4201–4212. 10.1111/febs.12013 [DOI] [PubMed] [Google Scholar]
- Hildebrand D., Eberle M. E., Wolfle S. M., Egler F., Sahin D., Sahr A., et al. (2018). Hsa-miR-99b/let-7e/miR-125a cluster regulates pathogen recognition receptor-stimulated suppressive antigen-presenting cells. Front. Immunol. 9, 1224. 10.3389/fimmu.2018.01224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroki E., Akahira J., Suzuki F., Nagase S., Ito K., Suzuki T., et al. (2010). Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. 101, 241–249. 10.1111/j.1349-7006.2009.01385.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Taeb S., Jahangiri S., Korpela E., Cadonic I., Yu N., et al. (2015). miR-620 promotes tumor radioresistance by targeting 15-hydroxyprostaglandin dehydrogenase (HPGD). Oncotarget 6, 22439–22451. 10.18632/oncotarget.4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D., Otani T., Yang P., Newman R. A., Yantiss R. K., Altorki N. K., et al. (2008). NAD+-dependent 15-hydroxyprostaglandin dehydrogenase regulates levels of bioactive lipids in non-small cell lung cancer. Cancer Prev. Res. (Phila) 1, 241–249. 10.1158/1940-6207.CAPR-08-0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacona J. R., Monteleone N. J., Lutz C. S. (2018). miR-146a suppresses 5-lipoxygenase activating protein (FLAP) expression and Leukotriene B4 production in lung cancer cells. Oncotarget 9, 26751–26769. 10.18632/oncotarget.25482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A., Zurolo E., Prabowo A., Fluiter K., Spliet W. G., Van Rijen P. C., et al. (2012). MicroRNA-146a: a key regulator of astrocyte-mediated inflammatory response. PLoS One 7, e44789. 10.1371/journal.pone.0044789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Chakraborty G., Raja R., Kale S., Kundu G. C. (2008). Prostaglandin E2 regulates tumor angiogenesis in prostate cancer. Cancer Res. 68, 7750–7759. 10.1158/0008-5472.CAN-07-6689 [DOI] [PubMed] [Google Scholar]
- Jakobsson P. J., Thoren S., Morgenstern R., Samuelsson B. (1999). Identification of human prostaglandin E synthase: a microsomal, glutathione-dependent, inducible enzyme, constituting a potential novel drug target. Proc. Natl. Acad. Sci. U. S. A. 96, 7220–7225. 10.1073/pnas.96.13.7220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., He Y., Liu L., Zhong X. (2010). MiRNA-26b regulates the expression of cyclooxygenase-2 in desferrioxamine-treated CNE cells. FEBS Lett. 584, 961–967. 10.1016/j.febslet.2010.01.036 [DOI] [PubMed] [Google Scholar]
- Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., et al. (2005). Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120, 623–634. 10.1016/j.cell.2004.12.038 [DOI] [PubMed] [Google Scholar]
- Johnstone R. M., Mathew A., Mason A. B., Teng K. (1991). Exosome formation during maturation of mammalian and avian reticulocytes: evidence that exosome release is a major route for externalization of obsolete membrane proteins. J. Cell. Physiol. 147, 27–36. 10.1002/jcp.1041470105 [DOI] [PubMed] [Google Scholar]
- Kabir T. D., Leigh R. J., Tasena H., Mellone M., Coletta R. D., Parkinson E. K., et al. (2016). A miR-335/COX-2/PTEN axis regulates the secretory phenotype of senescent cancer-associated fibroblasts. Aging (Albany NY) 8, 1608–1635. 10.18632/aging.100987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. J., Mbonye U. R., Delong C. J., Wada M., Smith W. L. (2007). Regulation of intracellular cyclooxygenase levels by gene transcription and protein degradation. Prog. Lipid Res. 46, 108–125. 10.1016/j.plipres.2007.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. K., Kim V. N. (2007). Processing of intronic microRNAs. Embo J. 26, 775–783. 10.1038/sj.emboj.7601512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., Romero R., Tarca A. L., Bhatti G., Lee J., Chaiworapongsa T., et al. (2011). miR-143 regulation of prostaglandin-endoperoxidase synthase 2 in the amnion: implications for human parturition at term. PLoS One 6, e24131. 10.1371/journal.pone.0024131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- Larsson K., Jakobsson P. J. (2015). Inhibition of microsomal prostaglandin E synthase-1 as targeted therapy in cancer treatment. Prostaglandins Other Lipid Mediat. 120, 161–165. 10.1016/j.prostaglandins.2015.06.002 [DOI] [PubMed] [Google Scholar]
- Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862. 10.1126/science.1065062 [DOI] [PubMed] [Google Scholar]
- Leclerc P., Idborg H., Spahiu L., Larsson C., Nekhotiaeva N., Wannberg J., et al. (2013). Characterization of a human and murine mPGES-1 inhibitor and comparison to mPGES-1 genetic deletion in mouse models of inflammation. Prostaglandins Other Lipid Mediat. 107, 26–34. 10.1016/j.prostaglandins.2013.09.001 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Ambros V. (2001). An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864. 10.1126/science.1065329 [DOI] [PubMed] [Google Scholar]
- Lee S. J., Seo K. W., Kim C. D. (2015). LPS Increases 5-LO Expression on monocytes via an activation of Akt-Sp1/NF-kappaB pathways. Korean J. Physiol. Pharmacol. 19, 263–268. 10.4196/kjpp.2015.19.3.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S. M., Kruger C., Park B., Derkow K., Rosenberger K., Baumgart J., et al. (2012). An unconventional role for miRNA: let-7 activates toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 15, 827–835. 10.1038/nn.3113 [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003). Prediction of mammalian microRNA targets. Cell 115, 787–798. 10.1016/S0092-8674(03)01018-3 [DOI] [PubMed] [Google Scholar]
- Li H., Zhou J., Wei X., Chen R., Geng J., Zheng R., et al. (2016). miR-144 and targets, c-fos and cyclooxygenase-2 (COX2), modulate synthesis of PGE2 in the amnion during pregnancy and labor. Sci. Rep. 6, 27914. 10.1038/srep27914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Kong X., Zhang J., Luo Q., Li X., Fang L. (2013). MiRNA-26b inhibits proliferation by targeting PTGS2 in breast cancer. Cancer Cell. Int. 13, 7. 10.1186/1475-2867-13-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang R., Ge Y., Chen D., Wu B., Fang F. (2019). Assessment of microRNA-144-5p and its putative targets in inflamed gingiva from chronic periodontitis patients. J. Periodontal Res. 54 (3), 266–277. 10.1111/jre.12627 [DOI] [PubMed] [Google Scholar]
- Liu X., Ji Q., Zhang C., Liu X., Liu Y., Liu N., et al. (2017). miR-30a acts as a tumor suppressor by double-targeting COX-2 and BCL9 in H. pylori gastric cancer models. Sci. Rep. 7, 7113. 10.1038/s41598-017-07193-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Byrnes K., Han C., Wang Y., Wu T. (2014). miR-21 targets 15-PGDH and promotes cholangiocarcinoma growth. Mol. Cancer Res. 12, 890–900. 10.1158/1541-7786.MCR-13-0419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder M., Dunn L., Liu L., Hasan A., Vincent K., Brackstone M., et al. (2018). COX-2 induces oncogenic micro RNA miR655 in human breast cancer. Sci. Rep. 8, 327. 10.1038/s41598-017-18612-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder M., Landman E., Liu L., Hess D., Lala P. K. (2015). COX-2 elevates oncogenic miR-526b in breast cancer by EP4 Activation. Mol. Cancer Res. 13, 1022–1033. 10.1158/1541-7786.MCR-14-0543 [DOI] [PubMed] [Google Scholar]
- Matysiak M., Fortak-Michalska M., Szymanska B., Orlowski W., Jurewicz A., Selmaj K. (2013). MicroRNA-146a negatively regulates the immunoregulatory activity of bone marrow stem cells by targeting prostaglandin E2 synthase-2. J. Immunol. 190 (10), 5102–5109. 10.4049/jimmunol.1202397 [DOI] [PubMed] [Google Scholar]
- Monteleone N. J., Moore A. E., Iacona J. R., Lutz C. S., Dixon D. A. (2019). miR-21-mediated regulation of 15-hydroxyprostaglandin dehydrogenase in colon cancer. Sci. Rep. 9, 5405. 10.1038/s41598-019-41862-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A. E., Young L. E., Dixon D. A. (2012). A common single-nucleotide polymorphism in cyclooxygenase-2 disrupts microRNA-mediated regulation. Oncogene 31, 1592–1598. 10.1038/onc.2011.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Houchen C. W., Kennedy S., Dieckgraefe B. K., Anant S. (2003). Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell 11, 113–126. 10.1016/S1097-2765(03)00012-1 [DOI] [PubMed] [Google Scholar]
- Nakanishi M., Rosenberg D. W. (2013). Multifaceted roles of PGE2 in inflammation and cancer. Semin. Immunopathol. 35, 123–137. 10.1007/s00281-012-0342-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs M. J., Sorg B. L., Pufahl L., Grez M., Suess B., Steinhilber D. (2012). Post-transcriptional regulation of 5-Lipoxygenase mRNA expression via alternative splicing and nonsense-mediated mRNA decay. PLoS One 7, e31363. 10.1371/journal.pone.0031363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs M. J., Steinhilber D., Suess B. (2011). MicroRNA involved in inflammation: control of eicosanoid pathway. Front. Pharmacol. 2, 39. 10.3389/fphar.2011.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs M. J., Steinhilber D., Suess B. (2014). MicroRNAs-novel therapeutic targets of eicosanoid signalling. Basic Clin. Pharmacol. Toxicol. 114, 92–96. 10.1111/bcpt.12116 [DOI] [PubMed] [Google Scholar]
- Olesch C., Sha W., Angioni C., Sha L. K., Acaf E., Patrignani P., et al. (2015). MPGES-1-derived PGE2 suppresses CD80 expression on tumor-associated phagocytes to inhibit anti-tumor immune responses in breast cancer. Oncotarget 6, 10284–10296. 10.18632/oncotarget.3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson P. L., Thoren S., Jakobsson P. J. (2005). Human microsomal prostaglandin E synthase 1: a member of the MAPEG protein superfamily. Methods Enzymol. 401, 147–161. 10.1016/S0076-6879(05)01009-8 [DOI] [PubMed] [Google Scholar]
- Poligone B., Baldwin A. S. (2001). Positive and negative regulation of NF-kappaB by COX-2: roles of different prostaglandins. J. Biol. Chem. 276, 38658–38664. 10.1074/jbc.M106599200 [DOI] [PubMed] [Google Scholar]
- Radmark O., Werz O., Steinhilber D., Samuelsson B. (2015). 5-Lipoxygenase, a key enzyme for leukotriene biosynthesis in health and disease. Biochim. Biophys. Acta 1851, 331–339. 10.1016/j.bbalip.2014.08.012 [DOI] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Samuelsson B., Morgenstern R., Jakobsson P. J. (2007). Membrane prostaglandin E synthase-1: a novel therapeutic target. Pharmacol. Rev. 59, 207–224. 10.1124/pr.59.3.1 [DOI] [PubMed] [Google Scholar]
- Sato T., Liu X., Nelson A., Nakanishi M., Kanaji N., Wang X., et al. (2010). Reduced miR-146a increases prostaglandin E(2)in chronic obstructive pulmonary disease fibroblasts. Am. J. Respir. Crit. Care Med. 182, 1020–1029. 10.1164/rccm.201001-0055OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul M. J., Baumann I., Bruno A., Emmerich A. C., Wellstein J., Ottinger S. M., et al. (2019). miR-574-5p as RNA decoy for CUGBP1 stimulates human lung tumor growth by mPGES-1 induction. Faseb J. 33 (6), 6933–6947. 10.1096/fj.201802547R [DOI] [PubMed] [Google Scholar]
- Saul M. J., Stein S., Grez M., Jakobsson P. J., Steinhilber D., Suess B. (2016). UPF1 regulates myeloid cell functions and S100A9 expression by the hnRNP E2/miRNA-328 balance. Sci. Rep. 6, 31995. 10.1038/srep31995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaoka H., Dixon D. A., Oates J. A., Boutaud O. (2003). Tristetraprolin binds to the 3’-untranslated region of cyclooxygenase-2 mRNA. A polyadenylation variant in a cancer cell line lacks the binding site. J. Biol. Chem. 278, 13928–13935. 10.1074/jbc.M300016200 [DOI] [PubMed] [Google Scholar]
- Shanmugam N., Reddy M. A., Natarajan R. (2008). Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. J. Biol. Chem. 283, 36221–36233. 10.1074/jbc.M806322200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Li P., Zhu S. T., Yue J. P., Ji X. J., Ma D., et al. (2016). MiR-26a and miR-144 inhibit proliferation and metastasis of esophageal squamous cell cancer by inhibiting cyclooxygenase-2. Oncotarget 7, 15173–15186. 10.18632/oncotarget.7908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff M. J., Temoche-Diaz M. M., Karfilis K. V., Ri S., Schekman R. (2016). Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 5, e19276. 10.7554/eLife.19276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhilber D., Hofmann B. (2014). Recent advances in the search for novel 5-lipoxygenase inhibitors. Basic Clin. Pharmacol. Toxicol. 114, 70–77. 10.1111/bcpt.12114 [DOI] [PubMed] [Google Scholar]
- Steinhilber D., Radmark O., Samuelsson B. (1993). Transforming growth factor beta upregulates 5-lipoxygenase activity during myeloid cell maturation. Proc. Natl. Acad. Sci. U. S. A. 90, 5984–5988. 10.1073/pnas.90.13.5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strillacci A., Griffoni C., Sansone P., Paterini P., Piazzi G., Lazzarini G., et al. (2009). MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp. Cell. Res. 315, 1439–1447. 10.1016/j.yexcr.2008.12.010 [DOI] [PubMed] [Google Scholar]
- Su K., Wang Q., Qi L., Hua D., Tao J., Mangan C. J., et al. (2016). MicroRNA-674-5p/5-LO axis involved in autoimmune reaction of Concanavalin A-induced acute mouse liver injury. Toxicol. Lett. 258, 101–107. 10.1016/j.toxlet.2016.06.010 [DOI] [PubMed] [Google Scholar]
- Subbaramaiah K., Marmo T. P., Dixon D. A., Dannenberg A. J. (2003). Regulation of cyclooxgenase-2 mRNA stability by taxanes: evidence for involvement of p38, MAPKAPK-2, and HuR. J. Biol. Chem. 278, 37637–37647. 10.1074/jbc.M301481200 [DOI] [PubMed] [Google Scholar]
- Suzuki J., Ogawa M., Watanabe R., Takayama K., Hirata Y., Nagai R., et al. (2011). Roles of prostaglandin E2 in cardiovascular diseases. Int. Heart J. 52, 266–269. 10.1536/ihj.52.266 [DOI] [PubMed] [Google Scholar]
- Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006). NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U. S. A. 103, 12481–12486. 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Haneda S., Imakawa K., Sakai S., Nagaoka K. (2009). A microRNA, miR-101a, controls mammary gland development by regulating cyclooxygenase-2 expression. Differentiation 77, 181–187. 10.1016/j.diff.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Terzuoli E., Donnini S., Finetti F., Nesi G., Villari D., Hanaka H., et al. (2016). Linking microsomal prostaglandin E Synthase-1/PGE-2 pathway with miR-15a and -186 expression: novel mechanism of VEGF modulation in prostate cancer. Oncotarget 7, 44350–44364. 10.18632/oncotarget.10051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C., Witwer K. W., Aikawa E., Alcaraz M. J., Anderson J. D., Andriantsitohaina R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C., Gutierrez-Vazquez C., Sanchez-Cabo F., Perez-Hernandez D., Vazquez J., Martin-Cofreces N., et al. (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 4, 2980. 10.1038/ncomms3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Dubois R. N. (2006). Prostaglandins and cancer. Gut 55, 115–122. 10.1136/gut.2004.047100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Dubois R. N. (2010). Eicosanoids and cancer. Nat. Rev. Cancer 10, 181–193. 10.1038/nrc2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Li Y., Zhang C., Li X., Yu J. (2018). MiR-216a-3p inhibits colorectal cancer cell proliferation through direct targeting COX-2 and ALOX5. J. Cell. Biochem. 119, 1755–1766. 10.1002/jcb.26336 [DOI] [PubMed] [Google Scholar]
- Wang H. W., Hsueh C. T., Lin C. F., Chou T. Y., Hsu W. H., Wang L. S., et al. (2006). Clinical implications of microsomal prostaglandin e synthase-1 overexpression in human non-small-cell lung cancer. Ann. Surg. Oncol. 13, 1224–1234. 10.1245/s10434-006-9001-4 [DOI] [PubMed] [Google Scholar]
- Wang R., Li M., Zhou S., Zeng D., Xu X., Xu R., et al. (2015). Effect of a single nucleotide polymorphism in miR-146a on COX-2 protein expression and lung function in smokers with chronic obstructive pulmonary disease. Int J. Chron. Obstruct. Pulmon. Dis. 10, 463–473. 10.2147/COPD.S74345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Filgueiras L. R., Wang S., Serezani A. P., Peters-Golden M., Jancar S., et al. (2014). Leukotriene B4 enhances the generation of proinflammatory microRNAs to promote MyD88-dependent macrophage activation. J. Immunol. 192, 2349–2356. 10.4049/jimmunol.1302982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C., Seeger R. C., Fabbri M., Wang L., Wayne A. S., Jong A. Y. (2017). Biological roles and potential applications of immune cell-derived extracellular vesicles. J. Extracell. Vesicles 6, 1400370. 10.1080/20013078.2017.1400370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- Wu Y. C., Su L. J., Wang H. W., Jeff Lin C. F., Hsu W. H., Chou T. Y., et al. (2010). Co-overexpression of cyclooxygenase-2 and microsomal prostaglandin E synthase-1 adversely affects the postoperative survival in non-small cell lung cancer. J. Thorac Oncol. 5, 1167–1174. 10.1097/JTO.0b013e3181e2f4f5 [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Arakawa T., Ueda N., Yamamoto S. (1995). Transcriptional roles of nuclear factor kappa B and nuclear factor-interleukin-6 in the tumor necrosis factor alpha-dependent induction of cyclooxygenase-2 in MC3T3-E1 cells. J. Biol. Chem. 270, 31315–31320. 10.1074/jbc.270.52.31315 [DOI] [PubMed] [Google Scholar]
- Yao L., Han C., Song K., Zhang J., Lim K., Wu T. (2015). Omega-3 polyunsaturated fatty acids upregulate 15-PGDH expression in cholangiocarcinoma cells by inhibiting miR-26a/b expression. Cancer Res. 75, 1388–1398. 10.1158/0008-5472.CAN-14-2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S., Xu J., Zhao K., Song P., Yan Q., Fan W., et al. (2018). Down-regulation of HPGD by miR-146b-3p promotes cervical cancer cell proliferation, migration and anchorage-independent growth through activation of STAT3 and AKT pathways. Cell. Death Dis. 9, 1055. 10.1038/s41419-018-1059-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimatsu K., Altorki N. K., Golijanin D., Zhang F., Jakobsson P. J., Dannenberg A. J., et al. (2001. a). Inducible prostaglandin E synthase is overexpressed in non-small cell lung cancer. Clin. Cancer Res. 7, 2669–2674. [PubMed] [Google Scholar]
- Yoshimatsu K., Golijanin D., Paty P. B., Soslow R. A., Jakobsson P. J., Delellis R. A., et al. (2001. b). Inducible microsomal prostaglandin E synthase is overexpressed in colorectal adenomas and cancer. Clin. Cancer Res. 7, 3971–3976. [PubMed] [Google Scholar]
- Young L. E., Dixon D. A. (2010). Posttranscriptional regulation of cyclooxygenase 2 expression in colorectal cancer. Curr. Colorectal Cancer Rep. 6, 60–67. 10.1007/s11888-010-0044-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldin D. C. (2001). Epoxygenase pathways of arachidonic acid metabolism. J. Biol. Chem. 276, 36059–36062. 10.1074/jbc.R100030200 [DOI] [PubMed] [Google Scholar]
- Zhang J., Li S., Li L., Li M., Guo C., Yao J., et al. (2015). Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinf. 13, 17–24. 10.1016/j.gpb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Xiao J., Lu X., Liu T., Jin X., Xiao Y., et al. (2019). PVT1 (rs13281615) and miR-146a (rs2910164) polymorphisms affect the prognosis of colon cancer by regulating COX2 expression and cell apoptosis. J. Cell. Physiol. 234 (10), 17538–17548. 10.1002/jcp.28377 [DOI] [PubMed] [Google Scholar]