Abstract
The ability to conserve energy in the presence or absence of oxygen provides a metabolic versatility that confers an advantage in natural ecosystems. The switch between alternative electron transport systems is controlled by the fumarate nitrate reduction transcription factor (FNR) that senses oxygen via an oxygen-sensitive [4Fe-4S]2+ iron-sulfur cluster. Under O2 limiting conditions, FNR plays a key role in allowing bacteria to transition from aerobic to anaerobic lifestyles. This is thought to occur via transcriptional activation of genes involved in anaerobic respiratory pathways and by repression of genes involved in aerobic energy production. The Proteobacterium Acidithiobacillus ferrooxidans is a model species for extremely acidophilic microorganisms that are capable of aerobic and anaerobic growth on elemental sulfur coupled to oxygen and ferric iron reduction, respectively. In this study, an FNR-like protein (FNRAF) was discovered in At. ferrooxidans that exhibits a primary amino acid sequence and major motifs and domains characteristic of the FNR family of proteins, including an effector binding domain with at least three of the four cysteines known to coordinate an [4Fe-4S]2+ center, a dimerization domain, and a DNA binding domain. Western blotting with antibodies against Escherichia coli FNR (FNREC) recognized FNRAF. FNRAF was able to drive expression from the FNR-responsive E. coli promoter PnarG, suggesting that it is functionally active as an FNR-like protein. Upon air exposure, FNRAF demonstrated an unusual lack of sensitivity to oxygen compared to the archetypal FNREC. Comparison of the primary amino acid sequence of FNRAF with that of other natural and mutated FNRs, including FNREC, coupled with an analysis of the predicted tertiary structure of FNRAF using the crystal structure of the related FNR from Aliivibrio fisheri as a template revealed a number of amino acid changes that could potentially stabilize FNRAF in the presence of oxygen. These include a truncated N terminus and amino acid changes both around the putative Fe-S cluster coordinating cysteines and also in the dimer interface. Increased O2 stability could allow At. ferrooxidans to survive in environments with fluctuating O2 concentrations, providing an evolutionary advantage in natural, and engineered environments where oxygen gradients shape the bacterial community.
Keywords: fumarate nitrate reductase, anaerobic regulation, transcriptional regulation, DNA binding, iron-sulfur cluster, biomining, microbial ecology
Introduction
A central challenge in microbial ecology is to understand how microorganisms interact in complex communities, including how they respond to dynamically changing environments. Answers to this challenge are important for addressing issues such as the role of biogeochemical reactions in nutrient and energy cycling and in understanding ecosystem functioning in earth, ocean, and atmospheric environments (Widder et al., 2016). However, it is difficult to model such systems because of their complexity as well as that their experimental investigation in the field may require long time scales, measured in years to centuries, e.g., ecological succession.
Hyperacidic environments (pH < 3) provide an advantage over neutral milieu for addressing these issues as they generally exhibit low microbial diversity (Mendez-Garcia et al., 2015; Teng et al., 2017). This facilitates data collection, observation, and experimental exploration of ecological models over periods measured in weeks or months and simplifies model building of microbial interactions. One such environment is bioleaching heaps (termed “bioheaps”) that exploit acidophilic microorganisms’ metabolism (Bonnefoy and Holmes, 2012; Dopson and Johnson, 2012) to catalyze commercial metal recovery from sulfide minerals in many parts of the world (Brierley and Brierley, 2013; Vera et al., 2013). Bioheaps offer additional advantages for studying microbial community function and dynamics as they are subjected to dynamically changing conditions including levels of heavy metals, acidity, CO2, temperature, nutrients, and available redox couples for growth (Dopson et al., 2009; Valdés et al., 2010; Riekkola-Vanhanen, 2013; Tupikina et al., 2013; Dopson and Holmes, 2014). In addition, bioheap microbes are challenged by a gradient of O2 availability when thick biofilms are formed (Baker-Austin et al., 2010), due to limitations of O2 gas–liquid mass transfer that are exacerbated at higher temperatures (Petersen, 2010), and decreasing O2 concentrations in the center of the bioheap (Yin et al., 2011).
Acidithiobacillus ferrooxidans is a keystone bioheap species that is especially prevalent during early stage bioleaching (Demergasso et al., 2005; Remonsellez et al., 2009; Halinen et al., 2012). This is likely as it is able to grow at higher pH values than e.g., Leptospirillum ferriphilum (Dopson, 2016) and as it fixes carbon that aids in the subsequent growth of heterotrophic acidophilic species, as has been demonstrated during co-culture of autotrophic, and heterotrophic biomining species (Nancucheo and Johnson, 2010). At. ferrooxidans is an acidophilic, obligatory chemolithoautotrophic mesophile that gains its energy from the oxidation of ferrous iron, elemental sulfur, inorganic sulfur compounds, and hydrogen (Bonnefoy and Holmes, 2012; Dopson and Johnson, 2012; Hedrich and Johnson, 2013). At. ferrooxidans is a facultative anaerobe that grows under aerobic and anaerobic conditions and the main electron transport components couple the aerobic oxidation of iron and sulfur to the reduction of O2 (Quatrini et al., 2009) and the anaerobic oxidation of elemental sulfur to reduction of ferric iron (Pronk et al., 1991; Ohmura et al., 2002; Osorio et al., 2013). This switch from aerobic to anaerobic growth is expected to require a regulator of gene expression, which has not been studied in detail in At. ferrooxidans.
The transition from oxic, to hypoxic (low concentrations of O2), and finally anoxic environments may require gene regulation systems to respond to the varying O2 concentrations (Osorio et al., 2009). In addition, At. ferrooxidans requires iron homeostasis systems (such as for the ferric iron utilized as electron acceptor) as its concentration can reach 1018-fold higher than in pH neutral environments (Osorio et al., 2008a, b). Bacterial O2 sensing systems include the direct interaction of O2 with membrane sensors such as FixL and the cytoplasmic transcription factor FNR (Fumarate and Nitrate Reduction), along with redox responsive regulatory systems that include, but are not limited to, ArcBA and Rex (reviewed in (Green et al., 2009; Bueno et al., 2012; Mettert and Kiley, 2018). The FNR transcription factor is a member of the cyclic AMP (cAMP) receptor protein (Crp) superfamily and plays a major role in altering gene expression between oxic and anoxic conditions (Constantinidou et al., 2006). The Escherichia coli FNR protein (termed FNREC) senses O2 via four cysteine residues that ligate an O2-sensitive [4Fe-4S]2+ iron-sulfur cluster in the N-terminal region and affects its regulatory function via the C-terminal helix-turn-helix (HTH) DNA binding domain (Fleischhacker and Kiley, 2011; Mettert and Kiley, 2018). In anoxic conditions, FNR is activated by Isc protein-dependent acquisition of the [4Fe-4S]2+ cluster that promotes dimerization. The dimer binds to target DNA sequences, and induces or represses transcription. As O2 levels increase, the FNR [4Fe-4S]2+ cluster is degraded and the protein is converted to a monomeric form, which is no longer active in gene regulation (Crack and Le Brun, 2018; Mettert and Kiley, 2018). At. ferrooxidans gene clusters predicted to be under the control of FNR are suggested to be involved in carbon and energy metabolism along with nitrogen fixation (Osorio et al., 2009). However, the predicted role of FNR and the mechanisms of adaption to changing O2 concentrations in At. ferrooxidans have not been experimentally tested.
Due to the importance of At. ferrooxidans as a model organism in natural and man-made acidic environments, it is important to understand the regulation of growth, adaptation, and extracellular electron transfer under anoxic and acidic conditions. Here, we characterized the At. ferrooxidans FNR master gene regulator and examined the effect of changing O2 concentrations on its Fe-S cluster ligand.
Materials and Methods
Bacterial Strains and Growth Conditions
Acidithiobacillus ferrooxidansT ATCC 23270 was obtained from the American Type Culture Collection (Table 1). The strain was maintained in sterile 9K basal salts medium (sterilized by Tyndallization) adjusted to pH 3.5 with H2SO4 (Silverman and Lundgren, 1959) containing 0.5% (wt/vol) S0 and incubated under aerobic conditions at 30°C with shaking. Anaerobic (S0/Fe3+) cultures of At. ferrooxidans were grown in identical medium with the exception of the addition of 25 mM ferric iron (sterile filtered through a 0.22 μm filter) as electron acceptor and the pH was adjusted to 1.8. At. ferrooxidans was pre-grown in aerobic conditions before transferring the cells to an anaerobic jar using the Anaerocult A system (Merck).
TABLE 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Genotype | References or sources |
| At. ferrooxidans | ||
| ATCC 23270 | Wild type | |
| Escherichia coli | ||
| JM109 | endA1, recA1, gyrA96, thi, hsdR17 (rk–, mk+), relA1, supE44, Δ(lac-proAB), [F′ traD36, proAB, laqIqZΔM15] | Promega |
| BL21 | F-, ompT, hsdSB (rB-mB-), gal, dcm, rne131, (DE3) | Invitrogen |
| PK22 | hsdS, gaL λDE53, Δcrp-bs990, rpsL, Δfnr, zcj-3061::Tn10 | Lazazzera et al., 1993 |
| RZ7350 | lacZ Δ145, narG234::MudI1734 | Kiley and Reznikoff, 1991 |
| RZ8480 | Δfnr, lacZ Δ145, narG234::MudI1734 | Lazazzera et al., 1993 |
| Plasmids | ||
| pET100/D-TOPO | Expression vector | Invitrogen |
| pET100/D-TOPO FNRAF | Expression vector, FNRAF protein | This study |
| pKK223-3 | Plasmid vector, Ampr | PL-Pharmacia |
| pKK223-3 fnrAF | fnrAF coding region cloned into pKK223-3 vector with a tac promoter | This study |
| pET11A | Expresion vector, Ampr with a T7 promoter | Novagen |
| pET11A fnrAF | fnrAF coding region cloned into pET11A vector with a T7 promoter | This study |
Escherichia coli strains (Table 1) were grown on a rotary shaker in sterile LB medium at 37°C. The following antibiotics were added as required: spectinomycin (Sp; 25 μg/mL), streptomycin (Sm; 25 μg/mL), ampicillin (Ap; 50 μg/mL), and tetracycline (Tc; 10 μg/mL). For β-galactosidase assays, E. coli strains were grown under aerobic or anaerobic conditions at 37°C in minimal medium M9 containing 0.1% glucose with the respective antibiotics. Anaerobic cultures were carried out in anaerobic jars (as described for At. ferrooxidans) on M9 minimal medium containing 0.1% (wt/vol) glucose.
Ferrous Iron Production During Anaerobic Cultures
The formation of ferrous iron in anaerobic cultures was determined by titration with 2,2′-dypyridyl. Samples (1 mL) of culture medium were passed through a 0.2 μm membrane filter and 160 μL aliquots of the filtrate were added to 40 μL of 5 mM 2,2′-dypyridyl. The ferrous iron concentration was determined via a calibration curve of FeSO4 × 7H2O at an absorbance of 510 nm in a spectrophotometer. At. ferrooxidans growth was quantified by counting in a Petroff-Hausser chamber. The cells in aerobic and anaerobic cultures were quantified in triplicate cultures to construct the respective growth curves [data presented are means (n = 3) ± standard deviations].
Bioinformatics
The amino acid sequence of FNR from E. coli K12 (FNREC; accession number, WP_000916335) was used in a BlastP search against the genome of At. ferrooxidans ATCC 23270. A potential FNR candidate (FNRAF; locus tag, AFE_0270) was identified with 28% identity. Using FNRAF in a reciprocal best Blast hit against the NCBI nr database recovered hits against the Crp-Fnr family of transcriptional regulators (domain architecture ID 11429533) from multiple organisms. A conserved domain analysis of FNRAF was carried out (Marchler-Bauer et al., 2013). Multiple sequence alignments of FNRAF, FNREC and FNR from Aliivibrio fisheri (FNRAFI, Q5E593) were carried out using Clustal Omega (Sievers and Higgins, 2018) and Swiss-Model (Waterhouse et al., 2018). Where there was a difference between the two alignment methods, the alignment by Swiss-Model was chosen.
Secondary structure analysis of FNRAF was carried out using homology modeling by comparing the predicted protein with the FNR crystal structure from A. fisheri (PDB 5e44) with a sequence identity of 27.35% and full coverage of the complete protein (Volbeda et al., 2015). The homology model was constructed using Modeler V.9 (Eswar et al., 2008) and validated using the ADIT! Validation Server from PDB (Richardson et al., 2013).
RNA Extraction and Real-Time PCR
Cells were harvested from At. ferrooxidans cultures (maximum 1 × 109 total cells) by centrifugation at 12000 × g for 10 min at 4°C. The pellet was washed with 10 mM H2SO4 and then with TE buffer pH 8.0 and finally resuspended in 100 μL of TE pH 8.0. To this mixture, 10 μL RNAse-free lysis buffer (0.5 M TrisHCl, 20 mM EDTA, 10% SDS, pH 6.8) was added and mixed gently. The tubes were incubated at 100°C for 3 min and allowed to cool to room temperature. The previous steps were sufficient to guarantee the rupture of the cells without damaging the RNA. RNA was isolated using RNeasy Mini Kit (Qiagen®) and contaminant DNA removed using RNase free DNase I (Fermentas) according to the manufacturer’s recommendations. The RNA was resuspended in five volumes of RNAlater solution (Qiagen) and subsequently frozen at −80°C until use. RNA samples were reverse-transcribed using Revertaid M-MuLV (Fermentas) and specific oligonucleotides (Table 2) according to the manufacturer’s recommendations and 0.5 μg of total RNA for each reaction. The real-time PCR reactions were performed using an iCycler thermal cycler (Bio-Rad) and the KAPA SYBR FAST qPCR kit (KAPABIOSYSTEMS). The 20 μL PCR reactions contained 2 μL of a 1:100 diluted cDNA sample, 200 nM of each primer (Table 2), and 1 × KAPA SYBR FAST qPCR Master Mix. The reference dye ROX was included at a final concentration of 5 nM. The cycling protocol was as follows: initial denaturation for 10 min at 95°C followed by 40 cycles of 30 s each at 95°C, 56°C, and 72°C. Fluorescence was measured after the extension phase at 72°C and specific amplification was confirmed by a single peak in the melting curve. For each experimental condition, total RNA was extracted from replicate At. ferrooxidans cultures and the real-time PCR reactions were performed in triplicate and thus, the data sets consist of six values per gene. Relative expression levels of At. ferrooxidans fnr (amplified with qPCR fnr FF and qPCR fnr REV primers; Table 2) were normalized with the expression of the stable reference gene rpoC (amplified with qPCR rpoC FF and qPCR rpoC REV primers; Table 2). The rpoC gene has previously been demonstrated to be expressed at a constant level and is a valid choice as a reference (Nieto et al., 2009). Stationary phase genomic DNA (10-fold dilutions ranging from 10 ng to 1 pg) was used to generate a five-point standard curve for every gene by using the Cycle Threshold (Ct) value vs. the logarithm of each dilution factor. Reaction efficiency {E = [10(−1/slope)]–1} for every gene was derived from the slope of the corresponding standard curves. A one-way Anova (multiple comparison analysis) or a two way ANOVA test swere carried out to test the statistical significance of gene expression results (McDonald, 2009) using the Graphpad Prism software1.
TABLE 2.
Oligonucleotides used in this study.
| Name | Sequence (5′–3′) | Function |
| pKK FF fnr-EcoRI | ATCGATGAATTCATGACTGCCAGGCACTCCG | Cloning |
| pKK REV fnr-EcoRI | ATCGATGAATTCTCAGGCGCGGGTGCC | Cloning |
| fnr FF-NdeI | CATATGACTGCCAGGCACTCC | Cloning |
| fnr REV-NdeI | CATATGGTCTGCATTGACAATTATCAA | Cloning |
| qPCR fnr FF | AAGCTGGTCAAGAGTCTGCCCAAT | RT-qPCR |
| qPCR fnr REV | TGCCGGTCAAGGTAATGGCACTAT | RT-qPCR |
| qPCR rpoC FF | AATGCGGTGTTGAGGTAACC | RT-qPCR |
| qPCR rpoC REV | AGGTACTGGTCTTCGGTAAG | RT-qPCR |
Cloning Procedures
Acidithiobacillus ferrooxidans genomic DNA was prepared using the Wizard® Genomic DNA Purification Kit (Promega Corp.). Plasmid DNA was prepared from E. coli JM109 cultures with the QIAprep® Spin MiniPrep Kit (Qiagen). PCR products for cloning were amplified using oligonucleotides in Table 2 and purified from agarose gels with the SpinPrepTM Gel DNA Kit (Novagen).
To carry out β-galactosidase assays, the coding region of the At. ferrooxidans fnr gene (termed fnrAF) was amplified with primers containing embedded EcoRI restriction sites: pKK FF fnr-EcoRI and pKK REV fnr-EcoRI (Table 2). Cloning and transformation was carried out using standard techniques as described by Miller (1972). The amplification product was cloned into the multiple cloning site of pKK223-3 carrying the Ptac promoter (Pharmacia Biotech), generating pKK223-3 fnrAF. pKK223-3 fnrAF was transformed into E. coli strain RZ8480 which is Δfnr and contains lacZ under control of the PnarG promoter (Lazazzera et al., 1993). Finally, the fnrAF coding region was cloned from PKK223-3 FNRAF into the pET100/D-TOPO expression vector to generate pET100/D-TOPO FNRAF that was subsequently used as a control in the Western blot analysis.
For the construction of the plasmid pET11A fnrAF,the At. ferrooxidans fnr gene coding region was amplified with primers containing embedded NdeI restriction sites fnr FF-NdeI and fnr REV-NdeI (Table 2). The product was cloned into pET11A (Novagen), generating plasmid pET11A fnrAF (Table 2). The construct was then transformed into E. coli strain PK22 (Lazazzera et al., 1993) for FNRAF purification experiments. E. coli strain PK22 was used because it lacks both FNREC and the structurally related CRP (cAMP-activated global transcriptional regulator) that could potentially contaminate the preparation of FNRAF (Lazazzera et al., 1993).
β-Galactosidase Assays
Escherichia coli was pre-grown aerobically overnight in M9-glucose medium (Sambrook et al., 1989). The medium (10 mL) was inoculated with 1% (vol/vol) seed culture and incubated in anaerobic jars until the cell density reached an optical density at 600 nm (OD600) of 0.8. The cultures had not yet achieved stationary phase since the cell mass increased at least twofold with further incubation. β-galactosidase assays were performed as described by (Miller, 1972) using chloroform and 0.1% sodium dodecyl sulfate to permeabilize the cells. All β-galactosidase assay results are the average of triplicate samples for each strain ± standard deviations.
Purification of FNRAF
FNR-like protein was purified from strain PK22 carrying pET11A-fnrAF using the anaerobic protocol developed for FNREC. The cells were grown aerobically in 4 L of M9 minimal medium plus 0.2% (wt/vol) glucose and ampicillin at 37°C to an OD600 ∼0.3, and IPTG was added to a final concentration of 400 μM for 1 h to induce FNR biosynthesis. After induction, cells were sparged overnight at 4°C with argon to remove the presence of O2. All subsequent steps in FNR purification were carried out under anaerobic conditions in a Coy anaerobic chamber with an atmosphere of 90% N2 and 10% H2 or in sealed tubes. The cells were harvested by centrifugation at 7,900 g for 15 min at 4°C and concentrated 200-fold in buffer A [50 mM potassium phosphate (pH 6.8), 0.1 mM EDTA, 0.1 M KC1, 10% glycerol, 1 mM dithiothreitol (DTT), and 0.1 mM phenylmethanesulfonylfloride (PMSF)], and passed once through a French press at 20,000 psi. The extracts were centrifuged at 139,000 g for 1 h to remove the membrane fraction. Cell extracts were passed over a 5 mL Bio-Rex 70 cation-exchange column (BioRad Laboratories) at a flow rate of 0.17 mL/min and eluted with a 70 mL linear gradient of 0.1 to 1 M KC1 in buffer A. Fractions containing a green color were pooled and diluted 1:4 with buffer C [50 mM phosphate (pH 6.8) plus 10% (vol/vol) glycerol] and loaded onto a 1 mL BioRex-70 gravity column, washed with 2 column volumes of buffer A, and eluted with 1 column volume of buffer B [50 mM phosphate (pH 6.8), 10% (vol/vol) glycerol, and 1 M KCl]. The purity of the FNR protein preparations was estimated from Coomassie-stained SDS-polyacrylamide gels. The protein concentration was estimated by a Bradford assay using the Coomassie Plus Protein Assay Reagent (Pierce).
Purification of His-tagged FNRAF was carried out by first transforming pET100/D-TOPO FNRAF into E. coli BL21 cells. Induction of FNRAF was carried out in cell cultures grown to an OD600 of 0.8 in LB supplemented with amp (100 μg/ml) by adding 1 mM IPTG to the culture medium for 1 h. The overexpressed FNRAF protein contained in the soluble extracts was purified by nickel-charged agarose resins (BIO-RAD) using 1M imidazole.
Western Blotting
Aliquots of total protein extract and purified FNR protein (approximately 10 μM of protein) were separated by SDS-PAGE with either 15 or 18% acrylamide (total acrylamide/bisacrylamide) and transferred onto nitrocellulose filters by standard methods with a Bio-Rad blotting apparatus.
The blotted proteins were subsequently screened using a polyclonal rabbit anti-FNR serum generated against FNREC. Filters were blocked overnight in blocking solution [5% skimmed milk, 0.05% Triton X-100, and Tris-buffered saline (TBS)] at 4°C with agitation, incubated for 1 h with a 1:500 dilution of the primary antibody in TBS/Tween 20 (0.05%) and further incubated in a 1:15,000 dilution of peroxidase-conjugated anti-rabbit immunoglobulin in TBS/Tween 20 (0.05%) for another hour. Immunoreactive proteins were detected using the Supersignal West Pico chemiluminescent substrate (Pierce). Pre-stained broad-range molecular mass protein standards from Bio-Rad were used. Protein concentrations were determined with Bio-Rad Protein Assay using BSA as standard.
UV-Visible Spectroscopy
To measure the absorbance of the FNRAF, 1 mL of the protein stored under anaerobic conditions was used at a concentration of 10 μM in 50 mM phosphate buffer pH 6.8 containing 0.4 M KCl and its absorbance was recorded between 200 to 700 nm in a Lambda 25 UV/Vis spectrophotometer (PerkinElmer). The impact of O2 on the spectral characteristics of FnrAF was evaluated by exposure of the sample to air for 0, 30, 90, 120, 150, 180, and 210 min and the absorbance spectra between 200 and 700 nm was recorded. As a control, the absorbance of the protein under anaerobic conditions was measured over the same time period in order to rule out other environmental factors causing a change in the protein’s spectral properties.
Iron determinations in FNRAF were performed by the TPTZ method which forms a deep blue-purple color with ferrous iron that is spectrophotometrically measured at 562 nm as previously described (Yan and Kiley, 2009).
Results and Discussion
Anaerobic Growth of At. ferrooxidans ATCC 23270
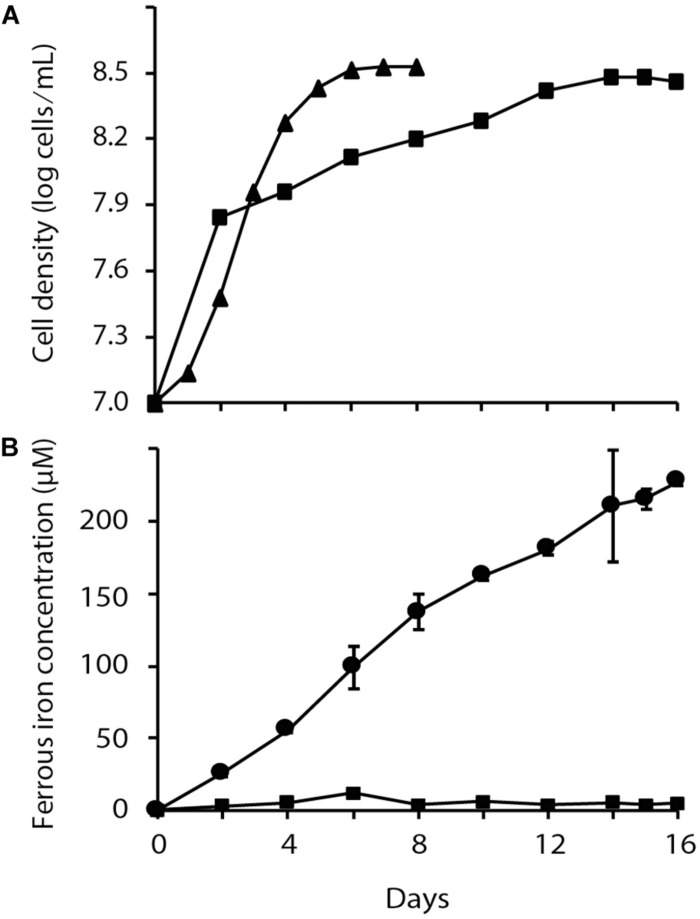
A comparison of At. ferrooxidans growth in oxic and anoxic conditions was performed in which elemental sulfur oxidation was coupled, respectively to reduction of O2 or Fe3+ as final electron acceptors. Despite the fact that both cultures reached similar levels of cell density, a reduced growth rate was observed when At. ferrooxidans used Fe3+ anaerobically instead of O2 as terminal electron acceptor (Figure 1). During anaerobic reduction of Fe3+, the amount of Fe2+ rose to a maximum of 228 ± 4 μM at stationary phase in the presence of At. ferrooxidans compared to 4 ± 4 μM Fe2+ in the un-inoculated control. These growth curves confirm and extend earlier observations (Osorio et al., 2013).
FIGURE 1.
Chemolithoautotrophic growth of Acidithiobacillus ferrooxidans via aerobic and anaerobic respiration with S0 as the electron donor. (A) Time dependent changes in cell density of aerobically (▲) and anaerobically (■) respiring cultures and (B) Fe2+ formation in aerobic (■) and anaerobic ( ) cultures. Data points are biological triplicates ± SD.
) cultures. Data points are biological triplicates ± SD.
Primary and Secondary Structure Analysis of FNRAF
The amino acid sequence of FNREC from E. coli K12 (accession number: WP_000916335) was used in a BlastP search against the genome of At. ferrooxidans ATCC 23270. A potential FNRAF candidate (AFE_0270) was identified with 28% identity. Using FNRAF in a reciprocal best Blast hit against the NCBI nr database recovered hits against the Crp-Fnr family of transcriptional regulators (domain architecture ID 11429533) from multiple organisms.
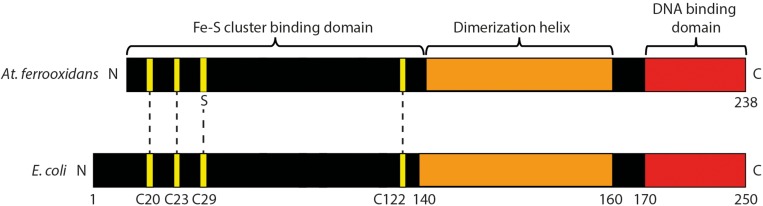
A conserved domain analysis of FNRAF (Marchler-Bauer et al., 2013) and a comparison of its primary amino acid sequence with FNREC [as reviewed in (Crack and Le Brun, 2018; Mettert and Kiley, 2018)] showed that it contained the following motifs characteristic of an FNR-like protein2 : (i) three of the four cysteines (positions 20, 23, and 122, using a numbering system based on the FNREC sequence) potentially forming part of an Fe-S cluster binding domain involved in coordinating an [4Fe-4S]2+ center; (ii) a dimerization helix; and (iii) a DNA binding domain (Figure 2).
FIGURE 2.
Cartoon of predicted domain structure of FNRAF compared to FNREC. The four cysteines that coordinate the [4Fe-4S]2+ cluster at positions 20, 23, 29, and 122 in FNREC (Mettert and Kiley, 2007) are highlighted in yellow. Three of these cysteines are conserved in FNRAF.
FNRAF Can Drive Expression From the FNR-Responsive E. coli Promoter PnarG
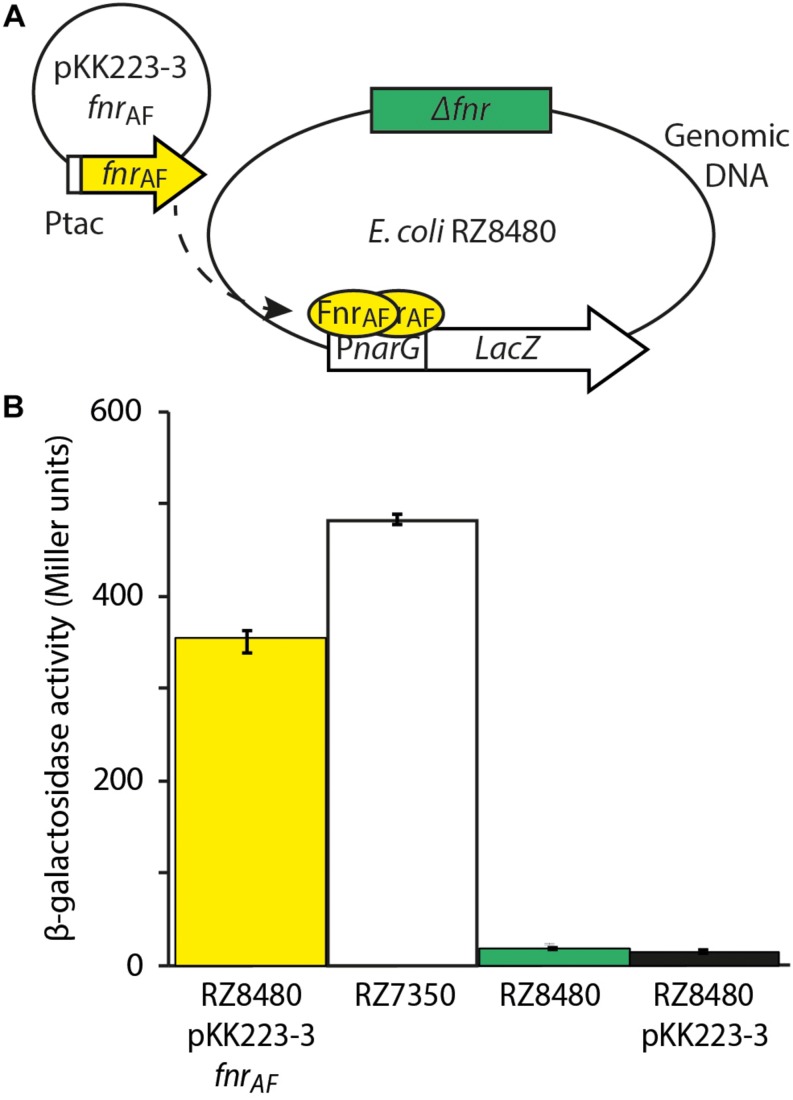
Although bioinformatic analyses strongly support the contention that FNRAF is an FNR-like protein, we investigated whether FNRAF could complement a mutant strain that lacked fnr, providing evidence for its function. It is difficult to generate a Δfnr mutant of At. ferrooxidans as the organism is challenging to manipulate genetically (Inaba et al., 2018) as reviewed in (Gumulya et al., 2018). Therefore, we chose to complement an E. coli strain RZ8480 lacking fnr (Δfnr). To accomplish this, a plasmid pKK223-3 fnrAF was constructed containing the predicted fnrAF coding sequence fused to the IPTG inducible promoter Ptac, and was transformed into RZ8480 (Δfnr, PnarG–lacZ). This strain contains lacZ, under the control of the FNR inducible PnarG promoter (Figure 3A). Therefore, when a functional FNR is cloned and expressed in E. coli strain RZ8480, it can induce the expression of lacZ, giving rise to a measurable β-galactosidase activity.
FIGURE 3.
Experimental design and results of FNRAF activity from the Escherichia coli promoter PnarG. (A) Experimental design to test the effect of FNRAF (encoded on plasmid pKK223-3 fnrAF) on in vivo expression of PnarG in E. coli Δfnr. (B) Data are reported as β-galactosidase activities (n = 3 ± SD, one way Anova p < 0.05) in strains RZ8480 A. f. [E. coli fnr (−) fnr A,f. (+)] in plasmid pKK223-3, PK7350 [E. coli fnr (+)], RZ8480 [E. coli fnr (−)], and RZ8480 PKK [E. coli fnr (–)] with pKK223-3 plasmid. The dotted arrow indicates the expression of FNRAF from fnrAF, its dimerization and binding to PnarG.
Escherichia coli strain RZ8480 transformed with pKK223-3 fnrAF was grown anaerobically until mid-log and β-galactosidase activity was measured after induction with 0.5 mM IPTG for 1 hr (Figure 3B). This activity was compared to that of E. coli strain RZ7350 that contains a native fnr and the PnarG-lacZ allele. We observed that β-galactosidase was expressed in the recombinant strain harboring FNRAF, indicating FNRAF is able to drive expression from the PnarG promoter. However, expression from PnarG is about 70% of that produced by FNREC. One-way ANOVA (multiple comparisons analysis) was used to test the statistical significance yielding p < 0.05. Possible explanations for the observed decrease in expression are that there are important amino acid and/or structural differences between the respective FNRs or the architecture of the respective FNR binding sites are different. Alternatively, since Ptac is a strong promoter, a certain amount of expressed FNRAF could be present in the cell in a non-soluble form potentially accounting, at least in part, for the lower activity.
As expected, little β-galactosidase activity was detected in the E. coli strain RZ8480 transformed with the vector only control (pKK223-3 lacking fnrAF). Thus, fnrAF is driving the expression of β-galactosidase and hence is capable of complementing ΔfnrEC.
The observation that fnrAF can regulate expression from the E. coli PnarG promoter provides evidence that it could potentially be involved in the regulation of anaerobic metabolism in At. ferrooxidans as has been observed in a number of organisms. However, as yet, there is no experimental evidence to test this hypothesis because of the difficulties involved in genetic manipulation of this organism.
Transcription Levels of At. ferrooxidans fnr in Different Growth Conditions
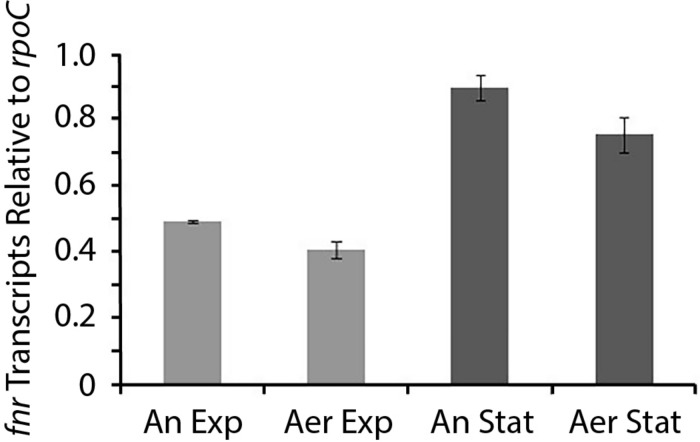
Having demonstrated that FNRAF is functional in a surrogate host, we wished to evaluate whether it was expressed in cell cultures of At. ferrooxidans and if so, under what conditions. Whole cell RNA was prepared from cells in four culture conditions: (i) anaerobic exponential growth; (ii) aerobic exponential growth; (iii) anaerobic stationary phase; and (iv) aerobic stationary phase and was quantified by RT-qPCR using the housekeeping gene rpoC mRNA as an internal standard (Figure 4). In anaerobic stationary phase, the number of RNA transcripts of FNRAF exceeded that observed in aerobic conditions with statistical support (two-way ANOVA, multiple t unpaired test, p < 0.05). Also, the number of RNA transcripts in the stationary phase in both anaerobic and aerobic conditions exceeded (p < 0.05) those detected in the equivalent exponential phase. These results suggest the existence of a mechanism for regulating the level of fnrAF transcripts depending on the growth phase and the presence or absence of O2. Using known transcription factor binding sites of fnrAF as models (Osorio et al., 2009), no FNR-type binding sites could be detected bioinformatically upstream of fnrAF, suggesting that it is not auto-regulated.
FIGURE 4.
Level of fnrAF transcripts in aerobic and anaerobic conditions. RT-qPCR data are expressed as relative values normalized to the housekeeping gene rpoC and presented as the average of triplicate samples ± the standard deviation, two-way Anova p < 0.05. An Exp, anaerobic exponential; Aer Exp, aerobic exponential; An Stat, anaerobic stationary; and Aer Stat, aerobic stationary.
Purification and Biochemical Characterization of FNRAF
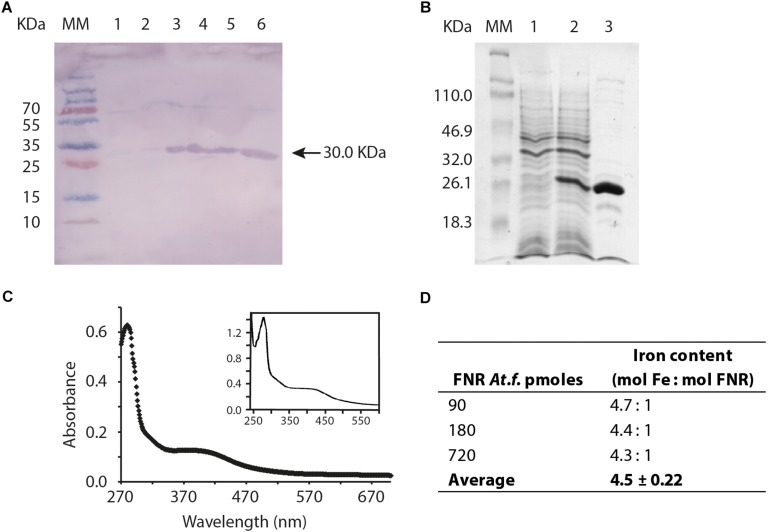
Antibodies prepared against FNREC were able to react with FNRAF prepared from plasmid pET100/D-TOPO fnrAF cloned into E. coli strain BL21 consistent with the observation that the two FNRs have similar structural regions that the antibody recognizes (Figure 5A). To determine if FNRAF contains an O2-sensitive metal cofactor, it was purified under anaerobic conditions (Figure 5B). The FNRAF enriched fractions had a brownish color suggesting the presence of a light absorbing cofactor associated with the protein (data not shown). The ultraviolet/visible spectrum of the protein, recorded under anoxic conditions (Figure 5C), showed the expected protein absorption maximum at 280 nm and a broad absorbance centered around 420 nm, consistent with a Fe-S cluster containing protein (Khoroshilova et al., 1995). In order to identify the type of Fe-S cluster coordinated by FNRAF, we measured the iron content of the purified FNRAF. We found approximately 4.5 mol iron per monomer of mol FNRAF, which is highly suggestive of a [4Fe-4S]2+ cluster per monomer of protein (Figure 5D).
FIGURE 5.
Biochemical validation and properties of FNRAF. (A) Western blot, antibodies prepared against FNREC were used to detect FNRAF, which was overproduced from plasmid pET100/D-TOPO fnrAF cloned into E. coli strain BL21. The gel shows the molecular mass marker (MM), crude cell free extract (approximately 10 μg of total protein) prepared prior to induction of fnrAF expression with IPTG (lane 1), 10 μg crude cell free extract 1 h after induction of fnrAF expression with IPTG (lane 2), and elution’s 1 to 4 of 10 μg purified FNRAF protein after elution from the nickel column (lanes 3–6). (B) Overproduction and purification of FNRAF from the pET11A fnrAF plasmid as shown by SDS-PAGE analysis. Gel lane 1 shows crude cell free extract (approximately 10 μg of total protein) prepared prior to induction of fnrAF expression with IPTG; lane 2, 10 μg of crude cell free extract 1 h after induction of fnrAF expression with IPTG; and lane 3, 10 μg FNRAF after the second cationic interchange chromatographic column. M, molecular mass marker (Mr are indicated). (C) Ultraviolet/visible absorption spectrum of a representative result of biological duplicates for purified FNRAF from anaerobically grown At. ferrooxidans with an inset of the ultraviolet/visible spectrum of anaerobically purified FNREC taken from Lazazzera et al. (1993). (D) Calculation of the FNRAF iron content by spectrophotometric assay.
FNRAF Reacts More Slowly With O2 in vitro Than FNREC
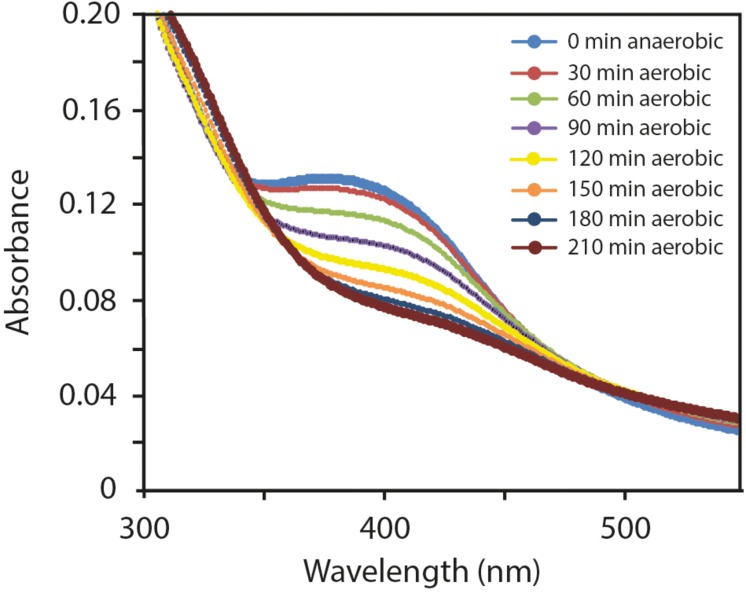
The ability to sense and adapt to changes in O2 concentration is critical for the regulatory function of FNR proteins. The ability of FNR to function as a transcription factor depends on the integrity of the [4Fe-4S]2+ cluster, which promotes a conformation amenable for dimerization, site-specific DNA binding, and transcriptional regulation [reviewed in (Crack and Le Brun, 2018; Mettert and Kiley, 2018)]. The O2 sensitivity of FNR is mediated by the [4Fe-4S]2+ cluster whereby in the presence of O2, the [4Fe-4S]2+ cluster is converted to [2Fe-2S]2+ both in vitro and in vivo. The [2Fe-2S]2+ form of FNR is monomeric in solution and is inactive for DNA binding and transcriptional regulation (Jordan et al., 1997; Khoroshilova et al., 1997; Popescu et al., 1998). To test whether FNRAF is O2 sensitive, the UV-visible spectrum of anaerobically purified FNRAF was recorded after it was exposed to air. A progressive decrease in absorbance, was observed consistent with the degradation of the [4Fe-4S]2+ by O2 and with complete degradation occurring by 210 min (Figure 6). The cluster decay was much slower than that observed for wild type FNREC (Crack et al., 2014) and other naturally O2-stable FNRs from Neisseria meningitidis (Edwards et al., 2010), Pseudomonas putida [FNR PP_3233; (Ibrahim et al., 2015)], and Paracoccus denitrificans (Crack et al., 2016). Furthermore, the appearance of a [2Fe-2S]2+ cluster product was not readily observed as found previously with E. coli FNR.
FIGURE 6.
Effect of O2 exposure on the ultraviolet/visible spectrum of anoxically purified FNRAF (10 μM). Changes are shown occurring in the 300–550 nm spectral region after 0, 30, 60, 90, 120, 150, 180, and 210 minutes exposure to O2.
Primary Amino Acid Sequence Differences Between FNRAF and FNREC Discussed in Light of the Three-Dimensional Crystal Structure of FNR From Aliivibrio fisheri
Note that in the following results, all amino acid locations in FNRAF are given based on the numbering system of FNREC in order to expedite comparisons in the text between the two sequences which have different lengths.
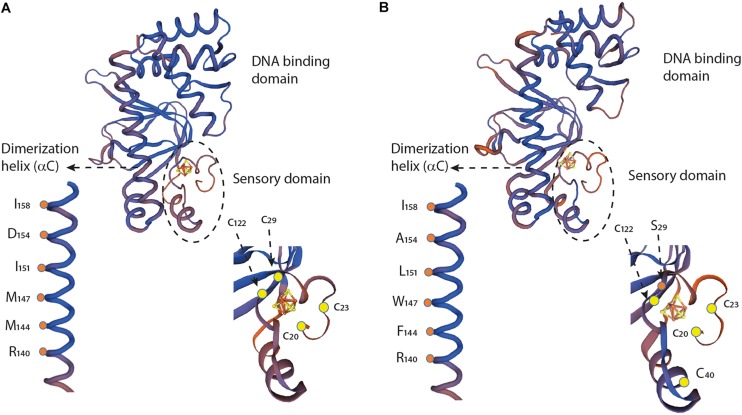
Despite the overall similarity of the primary amino acid sequences of FNRAF and FNREC, a number of important differences were observed. It is important to consider how these differences might affect the function of FNRAF and impact how FNRAF coordinates the [4Fe-4S] center and its increased resistance to O2. In order to address these issues, an alignment was carried out of the amino acid sequences of FNRAF, FNREC, and FNR from Aliivibrio fisheri (Figure 7). The primary amino acid sequences were then compared to three dimensional models of FNRAF and FNREC built using the crystal structure of FNR from A. fisheri (FNRAFi) as a template [PDB 5CVR (Volbeda et al., 2015)]. In agreement with the amino acid sequence evidence, the model shows that FNRAF shares similar global protein structure with important functional domains of FNREC, displaying a similar spatial distribution with an acceptable QMEAN score of −1.71 (Figure 8). These domains include the sensor domain that comprises a series of structural β-sheets with a [4Fe-4S]2+ coordination site, an α-helix promoting protein dimerization, and a DNA-binding domain composed of an HTH motif that allows recognition and binding to transcription factor binding sites (Myers et al., 2013).
FIGURE 7.

Comparison of the amino acid sequence of FNRAF (AFE_0270) with that of FNREC (WP_000916335). The sequence of FNR from Aliivibrio fisheri (FNRAFi; Q5E593) is included for comparison. The numbers correspond to the amino acid sequence of FNREC. In the sequence of FNREC, the four cysteines coordinating the [4Fe-4S]2+ center are highlighted in yellow. FNREC sequences corresponding to the dimerization helix and the DNA binding HTH motif are highlighted in gray and turquoise, respectively. Clear boxes indicate additional amino acids discussed in the text.
FIGURE 8.
Structural comparison and partial sequence comparison of (A) FNREC and (B) FNRAF. Models of the three-dimensional structures of the respective monomers, showing the sensory domains, dimerization helices, and DNA binding domains, were created using the crystal structure of FNRAFi (PDB 5e44) as a template. Cartoons of the sensory domains and dimerization helices are enlarged to illustrate specific amino acid locations discussed in the text. [4Fe-4S]2+ centers are illustrated with yellow and red Fe-S cage structures.
[4Fe-4S]2+ Center Coordination in FNRAF
A notable difference in amino acid sequence between FNRAF and FNREC is in the coordinating ligands of the [4Fe-4S]2+ center. In FNREC, coordination is carried out by four cysteines located at positions 20, 23, 29, and 122 (Figure 7). In FNRAF, cysteines are conserved at positions 20, 23, and 122 and, based on the 3D model, these are predicted to be located in positions that could potentially allow them to participate in coordinating the [4Fe-4S]2+ center (Figure 8). There is no corresponding cysteine at position 29 in FNRAF; there is, however, a Ser at this position that could potentially be the fourth coordinating ligand of the [4Fe-4S]2+ center. While mutational studies have shown that Ser can serve as a cluster ligand (Fujinaga et al., 1993; Vassiliev et al., 1995; Bentrop et al., 1996; Mansy et al., 2002) naturally occurring serine ligands are rare. Nevertheless, it is worth noting that the LipA enzyme contains an auxiliary [4Fe-4S] cluster that contains a 3Cys/Ser cluster ligation (Harmer et al., 2014).
An alternative hypothesis is that a Cys in FNRAF at position 40 could assume the function of the missing Cys29. Although this hypothesis cannot be rejected, we do not favor it. Inspection of the three-dimensional model of FNRAF suggests that the protein is unlikely to be able to fold to bring Cys40 into sufficient proximity to the [4Fe-4S]2+ center to facilitate the required coordination.
Amino Acid/Structural Changes That Could Help Explain the Observed Stability of FNRAF in Air
One of the major differences of FNRAF compared to FNREC is the increased stability of the Fe-S cluster in air. Here, we inspect the primary amino acid sequence and postulated 3D structure of FNRAF in order to propose hypotheses for explaining this unusual property.
Amino acid changes around the cluster ligand Cys23 have been shown to alter the O2 response of FNR in a number of organisms. In FNREC, replacement of Ser24, located immediately adjacent to the cluster ligand Cys23, by Pro results in increased aerobic FNR activity (Jervis et al., 2009). A natural variant of FNR from P. denitrificans has Pro in the position equivalent to Ser24 in FNREC and is at least six times less sensitive to O2 than FNREC (Crack et al., 2016). In P. putida a natural variant of FNR has an Arg in position 24 and is more stable to O2 than FNREC (Ibrahim et al., 2015). In At. ferrooxidans there is Leu in position 24 (Figure 7) and by analogy, this amino acid substitution could at least partially account for the lower O2 reactivity of FNRAF.
Amino acid changes at other positions next to the cluster-coordinating Cys residues are also known to influence the aerobic reactivity of FNREC. For example, substitution of Asp22 by Ala (Ibrahim et al., 2015) or by Gly (Kiley and Reznikoff, 1991) increased O2 activity of FNR. FNRAF has an Asp to His substitution at position 22. Interestingly, it has been found that juxtaposition of His to the cysteine-coordinated [4Fe-3S]2+ center of a subgroup of Ni hydrogenases provides stability in the presence of O2 (Frielingsdorf et al., 2014; Flanagan et al., 2018). Also, substitution of Leu28 by the positively charged His has been shown to stabilize the [4Fe-4S]2+ center in FNREC in the presence of O2 (Bates et al., 2000) perhaps by hindering conformational flexibility of the region (Volbeda et al., 2015). FNRAF has the bulky, polar, neutral amino acid Gln in this position and perhaps, like the L28F variant of FNREC (Jervis et al., 2009), this could hinder conformational flexibility by steric hindrance that results in greater activity in O2.
Changes in the dimerization helix may also alter the stability of the FNR dimer in O2. Two charged residues Arg140 and Asp130 have been reported to a play key role in the monomer-dimer equilibrium in FNREC (Moore and Kiley, 2001). An examination of the crystal structure of FNRAFi indicates that these residues could form a salt bridge between the a-C helix (Arg140) of one monomer with the opposite a-B helix (Asp130) of the other monomer, perhaps modulating monomer-dimer equilibrium in FNR (Volbeda et al., 2015). Both Arg140 and Asp130 are conserved in FNRAF implying conservation of the salt bridge and its role in monomer-dimer equilibrium in changing O2 environments (Figures 7, 8).
Of particular importance is the observation that when Asp154 in the dimer interface of FNREC is replaced with Ala154, FNREC exhibits increased activity under aerobic conditions (Moore et al., 2006). In FNRAF position 154 is occupied naturally by Ala, strongly suggesting that this change could, at least partially, explain its increased activity in O2 (Figures 7, 8).
Hydrophobic interactions have also been shown to be involved in dimer interaction and stabilization. These include Met144, Met147, Ile151, and Ile158 that lie on the dimer interface of FNREC as shown in the three-dimensional model (Figure 8; Moore and Kiley, 2001; Volbeda et al., 2015). Ile158 is conserved in FNRAF, but the other equivalently positioned residues in FNRAF are Phe144, Trp147, and Leu151, respectively. All are hydrophobic and potentially play a role in dimer stabilization. Of interest is the possibility that in FNRAF Trp147 of one monomer helix and Phe144 of the complementary monomer helix could interact through stacking of their respective aromatic rings, potentially providing additional stability to the interacting helices as has been observed in other proteins (McGaughey et al., 1998; Budyak et al., 2013; Madhusudan Makwana and Mahalakshmi, 2015).
Another difference is the presence of a truncated and divergent N terminal region of FNRAF compared to FNREC, in which FNRAF has only 7 amino acids just prior to the first Cys involved in [4Fe-4S]2+ center coordination instead of the 19 observed in FNREC (Figure 7). These amino acids form part of a flexible region with no predicted secondary structure. The truncation in FNRAF does not appear to be a result of sequence mis-annotation e.g., incorrect translation start site. Interestingly, deletion of N-terminal amino acid residues 2 to 16 and 2 to 17 FNREC, increased FNR activity under aerobic conditions (Yan and Kiley, 2008). These results suggest that the N-terminal region also contributes to the lability of the [4Fe-4S] cluster of FNR to O2 and that the removal of amino acids in this region may act to increase the stability of the cluster to O2. How these changes operate is not known.
Additional Discussion
Anaerobic culturing of At. ferrooxidans confirmed previous reports that it is capable of growth using Fe3+ as the final electron acceptor (Ohmura et al., 2002; Osorio et al., 2013) and that anaerobic growth is slower than with O2 as the electron acceptor (Osorio et al., 2013; Figure 1). The slower rate of growth for anaerobic cultures may be due to the greater amount of energy available from sulfur oxidation using O2 as terminal electron acceptor (−124 kcal/atom S0) compared with Fe3+ reduction (−75 Kcal/atom S0).
The ability to transition from aerobic reduction of O2 to utilizing ferric iron as a terminal electron acceptor suggests that At. ferrooxidans must regulate the expression of alternative electron transfer chains used in energy conservation. In this study, we provide evidence that At. ferrooxidans, a model organism for studying life at extremely low pH, contains a FNR-like protein (FNRAF) that is a member of the CRP FNR superfamily of regulators. FNRAF exhibits sequence (Figure 2) and structural similarity (Figure 8) with the archetypal FNR from E. coli (FNREC) that was recently deduced from the crystal structure of FNR from Al. fisheri (Mettert and Kiley, 2018). FNRAF reacts with antibodies prepared against FNREC (Figure 5A) and is able to drive expression from the FNR-responsive E. coli PnarG promoter, suggesting that it is functionally active as an FNR-like protein at least in the surrogate host E. coli.
Despite high levels of structural and protein sequence similarity, FNRAF exhibits several properties that differ from FNREC. First, RNA encoded by fnrAF, although detected in aerobic cultures, increases in amount in the stationary phase of anaerobically grown cultures (Figure 4), suggesting that depletion of O2, and/or culture age upregulate fnr expression or modifies post-transcriptional processing of fnr RNA. Upregulation of fnr in anaerobic conditions has also been observed in B. subtilis (Cruz Ramos et al., 1995). In contrast, although there is negative auto-regulation of FNREC expression (Mettert and Kiley, 2007), in E. coli it has been demonstrated that FNR activity is predominantly regulated at the protein level where FNR appears to cycle between active [4Fe-4S]2+, inactive [2Fe-2S], and apo forms, with the level of O2 determining which form predominates and therefore, the extent to which FNR is transcriptionally active. Such a mechanism requires that the levels of FNR in the cell are tightly controlled (Spiro and Guest, 1987; Sutton et al., 2004; Mettert and Kiley, 2007; Jervis et al., 2009). The suggestion that expression of fnrAF is regulated opens up opportunities to investigate the underlying mechanism(s) involved.
Another important consideration is the significantly increased stability of FNRAF compared to FNREC. What could be the evolutionary advantage of this strategy? We hypothesize that it allows the control of genes in its network over a wide range of O2 concentrations without the need to resort to recycling between active and inactive forms of FNR, as in E. coli, or to differential transcriptional regulation of FNR as exhibited by B. subtilis, P. putida, H. seropedicae, B. cenocepacia, and R. eutropha. These mechanisms are energetically costly and time consuming. Speed of response to increased environmental concentrations of O2 might be particularly critical for At. ferrooxidans as it needs to transition rapidly from anaerobic to highly oxidizing Fe-rich environments at very low pH such as found in bioheaps for industrial copper recovery (Jerez, 2008) and in biofilms in naturally occurring acidic environments (Wilmes et al., 2009; Liljeqvist et al., 2015) and these responses need to be made in an organism with a relatively slow growth rate and whose energy budget allocation is restricted by living at the thermodynamic edge of life.
Data Availability
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/genome/1014?genome_assembly_id=300479.
Author Contributions
DH, EJ, and PK designed the study. HO and EM carried out the experiments. All authors analyzed the data. DH and MD drafted the manuscript and all authors agreed on the final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
HO thanks the doctoral program Mecesup UAB0802 for supporting a stay in the laboratory of PK. The authors wish to thank a reviewer for drawing our attention to additional literature on the replacement of Cys by Ser as one of the potential coordinating ligands of the Fe-S cluster.
Funding. This work was supported by the Programa de Apoyo a Centros con Financiamiento Basal AFB 17004 to Fundación Ciencia & Vida and Fondecyt 1181717 to DH. The work in the Kiley lab was supported by the NIH grants R01-GM115894 and R01-GM045844 to PK.
References
- Baker-Austin C., Potrykus J., Wexler M., Bond P. L., Dopson M. (2010). Biofilm development in the extremely acidophilic archaeon ‘Ferroplasma acidarmanus’ Fer1. Extremophiles 14 485–491. 10.1007/s00792-010-0328-1 [DOI] [PubMed] [Google Scholar]
- Bates D. M., Popescu C. V., Khoroshilova N., Vogt K., Beinert H., Munck E., et al. (2000). Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S]2+ cluster to oxygen. J. Biol. Chem. 275 6234–6240. 10.1074/jbc.275.9.6234 [DOI] [PubMed] [Google Scholar]
- Bentrop D., Bertini I., Capozzi F., Dikiy A., Eltis L., Luchinat C. (1996). Three-dimensional structure of the reduced C77S mutant of the Chromatium vinosum high-potential iron-sulfur protein through nuclear magnetic resonance: comparison with the solution structure of the wild-type protein. Biochemistry 35 5928–5936. 10.1021/bi9528513 [DOI] [PubMed] [Google Scholar]
- Bonnefoy V., Holmes D. S. (2012). Genomic insights into microbial oxidation and iron homeostasis in extremely acidic environments. Environ. Microbiol. 14 1597–1611. 10.1111/j.1462-2920.2011.02626.x [DOI] [PubMed] [Google Scholar]
- Brierley C. L., Brierley J. A. (2013). Progress in bioleaching: part B: applications of microbial processes by the minerals industries. Appl. Microbiol. Biotechnol. 97 7543–7552. 10.1007/s00253-013-5095-3 [DOI] [PubMed] [Google Scholar]
- Budyak I. L., Zhuravleva A., Gierasch L. M. (2013). The role of aromatic-aromatic interactions in strand-strand stabilization of β-sheets. J. Mol. Biol. 425 3522–3535. 10.1016/j.jmb.2013.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno E., Mesa S., Bedmar E. J., Richardson D. J., Delgado M. J. (2012). Bacterial adaptation of respiration from oxic to microoxic and anoxic conditions: redox control. Antioxid. Redox Signal. 16 819–852. 10.1089/ars.2011.4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinidou C., Hobman J. L., Griffiths L., Patel M. D., Penn C. W., Cole J. A., et al. (2006). A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 281 4802–4815. 10.1074/jbc.m512312200 [DOI] [PubMed] [Google Scholar]
- Crack J. C., Green J., Thomson A. J., Brun N. E. L. (2014). Iron–sulfur clusters as biological sensors: the chemistry of reactions with molecular oxygen and nitric oxide. Accounts Chem. Res. 47 3196–3205. 10.1021/ar5002507 [DOI] [PubMed] [Google Scholar]
- Crack J. C., Hutchings M. I., Thomson A. J., Le Brun N. E. (2016). Biochemical properties of Paracoccus denitrificans FnrP: reactions with molecular oxygen and nitric oxide. J. Biol. Inorg. Chem. 21 71–82. 10.1007/s00775-015-1326-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crack J. C., Le Brun N. E. (2018). Redox-sensing iron-sulfur cluster regulators. Antioxid. Redox Signal. 29 1809–1829. 10.1089/ars.2017.7361 [DOI] [PubMed] [Google Scholar]
- Cruz Ramos H., Boursier L., Moszer I., Kunst F., Danchin A., Glaser P. (1995). Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 14 5984–5994. 10.1002/j.1460-2075.1995.tb00287.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demergasso C. S., Galleguillos P. P. A., Escudero G. L. V., Zepeda A. V. J., Castillo D., Casamayor E. O. (2005). Molecular characterization of microbial populations in a low-grade copper ore bioleaching test heap. Hydrometallurgy 80 241–253. 10.1016/j.hydromet.2005.07.013 [DOI] [Google Scholar]
- Dopson M. (2016). “Physiological and phylogenetic diversity of acidophilic bacteria,” in Acidophiles: Life in Extremely Acidic Environments, eds Quatrini R., Johnson D. B. (Poole: Caister Academic Press; ), 79–92. 10.21775/9781910190333.05 [DOI] [Google Scholar]
- Dopson M., Holmes D. S. (2014). Metal resistance in acidophilic microorganisms and its significance for biotechnologies. Appl. Microbiol. Biotechnol. 98:8133. 10.1007/s00253-014-5982-2 [DOI] [PubMed] [Google Scholar]
- Dopson M., Johnson D. B. (2012). Biodiversity, metabolism and applications of acidophilic sulfur- metabolizing micro-organisms. Environ. Microbiol. 14 2620–2631. 10.1111/j.1462-2920.2012.02749.x [DOI] [PubMed] [Google Scholar]
- Dopson M., Lövgren L., Boström D. (2009). Silicate mineral dissolution in the presence of acidophilic microorganisms: implications for heap bioleaching. Hydrometallurgy 96 288–293. 10.1016/j.hydromet.2008.11.004 [DOI] [Google Scholar]
- Edwards J., Cole L. J., Green J. B., Thomson M. J., Wood A. J., Whittingham J. L., et al. (2010). Binding to DNA protects Neisseria meningitidis fumarate and nitrate reductase regulator (FNR) from oxygen. J. Biol. Chem. 285 1105–1112. 10.1074/jbc.M109.057810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N., Eramian D., Webb B., Shen M. Y., Sali A. (2008). Protein structure modeling with MODELLER. Methods Mol. Biol. 426 145–159. 10.1007/978-1-60327-058-8_8 [DOI] [PubMed] [Google Scholar]
- Flanagan L. A., Chidwick H. S., Walton J., Moir J. W. B., Parkin A. (2018). Conserved histidine adjacent to the proximal cluster tunes the anaerobic reductive activation of Escherichia coli membrane-bound [NiFe] hydrogenase-1. ChemElectroChem 5 855–860. 10.1002/celc.201800047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischhacker A. S., Kiley P. J. (2011). Iron-containing transcription factors and their roles as sensors. Curr. Opin. Chem. Biol. 15 335–341. 10.1016/j.cbpa.2011.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frielingsdorf S., Fritsch J., Schmidt A., Hammer M., Lowenstein J., Siebert E., et al. (2014). Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase. Nat. Chem. Biol. 10 378–385. 10.1038/nchembio.1500 [DOI] [PubMed] [Google Scholar]
- Fujinaga J., Gaillard J., Meyer J. (1993). Mutated forms of a [2Fe-2S] ferredoxin with serine ligands to the iron-sulfur cluster. Biochem. Biophys. Res. Commun. 194 104–111. 10.1006/bbrc.1993.1791 [DOI] [PubMed] [Google Scholar]
- Green J., Crack J. C., Thomson A. J., Lebrun N. E. (2009). Bacterial sensors of oxygen. Curr. Opin. Microbiol. 12 145–151. 10.1016/j.mib.2009.01.008 [DOI] [PubMed] [Google Scholar]
- Gumulya Y., Boxall N., Khaleque H., Santala V., Carlson R., Kaksonen A. (2018). In a quest for engineering acidophiles for biomining applications: challenges and opportunities. Genes 9:16. 10.3390/genes9020116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halinen A.-K., Beecroft N. J., Määttä K., Nurmi P., Laukkanen K., Kaksonen A. H., et al. (2012). Microbial community dynamics during a demonstration-scale bioheap leaching operation. Hydrometallurgy 12 34–41. 10.1016/j.hydromet.2012.05.001 [DOI] [Google Scholar]
- Harmer J. E., Hiscox M. J., Dinis P. C., Fox S. J., Iliopoulos A., Hussey J. E., et al. (2014). Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions. Biochem. J. 464 123–133. 10.1042/BJ20140895 [DOI] [PubMed] [Google Scholar]
- Hedrich S., Johnson D. B. (2013). Aerobic and anaerobic oxidation of hydrogen by acidophilic bacteria. FEMS Microbiol. Lett. 349 40–45. 10.1111/1574-6968.12290 [DOI] [PubMed] [Google Scholar]
- Ibrahim S. A., Crack J. C., Rolfe M. D., Borrero-De Acuna J. M., Thomson A. J., Le Brun N. E., et al. (2015). Three Pseudomonas putida FNR family proteins with different sensitivities to O2. J. Biol. Chem. 290 16812–16823. 10.1074/jbc.M115.654079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba Y., Banerjee I., Kernan T., Banta S. (2018). Transposase-mediated chromosomal integration of exogenous genes in Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 84:e01381-18. 10.1128/aem.01381-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerez C. A. (2008). The use of genomics, proteomics and other OMICS technologies for the global understanding of biomining microorganisms. Hydrometallurgy 94 162–169. 10.1016/j.hydromet.2008.05.032 [DOI] [Google Scholar]
- Jervis A. J., Crack J. C., White G., Artymiuk P. J., Cheesman M. R., Thomson A. J., et al. (2009). The O2 sensitivity of the transcription factor FNR is controlled by Ser24 modulating the kinetics of [4Fe-4S] to [2Fe-2S] conversion. Proc. Natl. Acad. Sci. U.S.A. 106 4659–4664. 10.1073/pnas.0804943106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. A., Thomson A. J., Ralph E. T., Guest J. R., Green J. (1997). FNR is a direct oxygen sensor having a biphasic response curve. FEBS Lett. 416 349–352. 10.1016/s0014-5793(97)01219-2 [DOI] [PubMed] [Google Scholar]
- Khoroshilova N., Beinert H., Kiley P. J. (1995). Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA binding. Proc. Nat. Acad. Sci. U.S.A. 92 2499–2503. 10.1073/pnas.92.7.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoroshilova N., Popescu C., Munck E., Beinert H., Kiley P. J. (1997). Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc. Natl. Acad. Sci. U.S.A. 94 6087–6092. 10.1073/pnas.94.12.6087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiley P. J., Reznikoff W. S. (1991). Fnr mutants that activate gene expression in the presence of oxygen. J. Bacteriol. 173 16–22. 10.1128/jb.173.1.16-22.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera B. A., Bates D. M., Kiley P. J. (1993). The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 7 1993–2005. 10.1101/gad.7.10.1993 [DOI] [PubMed] [Google Scholar]
- Liljeqvist M., Ossandon F. J., González C., Rajan S., Stell A., Valdes J., et al. (2015). Metagenomic analysis reveals adaptations to a cold-adapted lifestyle in a low-temperature acid mine drainage stream. FEMS Microb. Ecol. 91:fiv011. 10.1093/femsec/fiv1011 [DOI] [PubMed] [Google Scholar]
- Madhusudan Makwana K., Mahalakshmi R. (2015). Implications of aromatic-aromatic interactions: From protein structures to peptide models. Protein Sci. 24 1920–1933. 10.1002/pro.2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansy S. S., Xiong Y., Hemann C., Hille R., Sundaralingam M., Cowan J. A. (2002). Crystal structure and stability studies of C77S HiPIP: a serine ligated [4Fe-4S] cluster. Biochemistry 41 1195–1201. 10.1021/bi011811y [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Zheng C., Chitsaz F., Derbyshire M. K., Geer L. Y., Geer R. C., et al. (2013). CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41 D348–D352. 10.1093/nar/gks1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. H. (2009). Handbook of Biological Statistics. Baltimore, MD: Sparky House Publishing. [Google Scholar]
- McGaughey G. B., Gagne M., Rappe A. K. (1998). pi-Stacking interactions. Alive and well in proteins. J. Biol. Chem. 273 15458–15463. 10.1074/jbc.273.25.15458 [DOI] [PubMed] [Google Scholar]
- Mendez-Garcia C., Pelaez A. I., Mesa V., Sanchez J., Golyshina O. V., Ferrer M. (2015). Microbial diversity and metabolic networks in acid mine drainage habitats. Front. Microbiol. 6:475. 10.3389/fmicb.2015.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettert E. L., Kiley P. J. (2007). Contributions of [4Fe-4S]-FNR and integration host factor to fnr transcriptional regulation. J. Bacteriol. 189 3036–3043. 10.1128/jb.00052-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettert E. L., Kiley P. J. (2018). Reassessing the structure and function relationship of the O2 sensing transcription factor FNR. Antioxid. Redox Signal. 29 1830–1840. 10.1089/ars.2017.7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Moore L. J., Kiley P. J. (2001). Characterization of the dimerization domain in the FNR transcription factor. J. Biol. Chem. 276 45744–45750. 10.1074/jbc.m106569200 [DOI] [PubMed] [Google Scholar]
- Moore L. J., Mettert E. L., Kiley P. J. (2006). Regulation of FNR dimerization by subunit charge repulsion. J. Biol. Chem. 281 33268–33275. 10.1074/jbc.m608331200 [DOI] [PubMed] [Google Scholar]
- Myers K. S., Yan H., Ong I. M., Chung D., Liang K., Tran F., et al. (2013). Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet. 9:e1003565. 10.1371/journal.pgen.1003565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nancucheo I., Johnson D. B. (2010). Production of glycolic acid by chemolithotrophic iron- and sulfur-oxidizing bacteria and its role in delineating and sustaining acidophilic sulfide mineral-oxidizing consortia. Appl. Environ. Microbiol. 76 461–467. 10.1128/AEM.01832-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto P. A., Covarrubias P. C., Jedlicki E., Holmes D. S., Quatrini R. (2009). Selection and evaluation of reference genes for improved interrogation of microbial transcriptomes: case study with the extremophile Acidithiobacillus ferrooxidans. BMC Mol. Biol. 10:63. 10.1186/1471-2199-10-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmura N., Sasaki K., Matsumoto N., Saiki H. (2002). Anaerobic respiration using Fe3+, S0, and H2 in the chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans. J. Bacteriol. 184 2081–2087. 10.1128/jb.184.8.2081-2087.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio H., Cardenas J. P., Valdes J., Holmes D. S. (2009). Prediction of FNR regulated genes and metabolic pathways potentially involved in anaerobic growth of Acidithiobacillus ferrooxidans. Adv. Mat. Res. 7 195–198. 10.4028/www.scientific.net/amr.71-73.195 [DOI] [Google Scholar]
- Osorio H., Mangold S., Denis Y., Nancucheo I., Johnson D. B., Bonnefoy V., et al. (2013). Anaerobic sulfur metabolism coupled to dissimilatory iron reduction in the extremophile Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 79 2172–2181. 10.1128/AEM.03057-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio H., Martinez V., Nieto P. A., Holmes D. S., Quatrini R. (2008a). Microbial iron management mechanisms in extremely acidic environments: comparative genomics evidence for diversity and versatility. BMC Microbiol. 8:203. 10.1186/1471-2180-1188-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio H., Martinez V., Veloso F. A., Pedroso I., Valdes J., Jedlicki E., et al. (2008b). Iron homeostasis strategies in acidophilic iron oxidizers: Studies in Acidithiobacillus and Leptospirillum. Hydrometallurgy 94 175–179. 10.1111/j.1462-2920.2011.02626.x [DOI] [PubMed] [Google Scholar]
- Petersen J. (2010). Determination of oxygen gas–liquid mass transfer rates in heap bioleach reactors. Min. Engin. 23 504–510. 10.1016/j.mineng.2010.01.006 [DOI] [Google Scholar]
- Popescu C. V., Bates D. M., Beinert H., Munck E., Kiley P. J. (1998). Mossbauer spectroscopy as a tool for the study of activation/inactivation of the transcription regulator FNR in whole cells of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 95 13431–13435. 10.1073/pnas.95.23.13431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk J. T., Liem K., Bos P., Keunen J. G. (1991). Energy transduction by anaerobic ferric iron respiration in Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 57 2063–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatrini R., Appia-Ayme C., Denis Y., Jedlicki E., Holmes D., Bonnefoy V. (2009). Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10:394. 10.1186/1471-2164-10-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remonsellez F., Galleguillos F., Moreno-Paz M., Parro V., Acosta M., Demergasso C. (2009). Dynamic of active microorganisms inhabiting a bioleaching industrial heap of low-grade copper sulfide ore monitored by real-time PCR and oligonucleotide prokaryotic acidophile microarray. Microb. Biotechnol. 2 613–624. 10.1111/j.1751-7915.2009.00112.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. S., Prisant M. G., Richardson D. C. (2013). Crystallographic model validation: from diagnosis to healing. Curr. Opin. Struct. Biol. 23 707–714. 10.1016/j.sbi.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekkola-Vanhanen M. (2013). Talvivaara mining company – From a project to a mine. Min. Engin. 48 2–9. 10.1016/j.mineng.2013.04.018 [DOI] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor. [Google Scholar]
- Sievers F., Higgins D. G. (2018). Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27 135–145. 10.1002/pro.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M. P., Lundgren D. G. (1959). Studies on the chemoautotrophic iron bacterium Ferrobacillus ferrooxidans I. An improved medium and a harvesting procedure for securing high cell yields. J. Bacteriol. 77 642–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. (1987). Regulation and over-expression of the fnr gene of Escherichia coli. J. Gen. Microbiol. 133 3279–3288. 10.1099/00221287-133-12-3279 [DOI] [PubMed] [Google Scholar]
- Sutton V. R., Mettert E. L., Beinert H., Kiley P. J. (2004). Kinetic analysis of the oxidative conversion of the [4Fe-4S]2+ cluster of FNR to a [2Fe-2S]2+ cluster. J. Bacteriol. 186 8018–8025. 10.1128/jb.186.23.8018-8025.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng W., Kuang J., Luo Z., Shu W. (2017). Microbial diversity and community assembly across environmental gradients in acid mine drainage. Minerals 7:106 10.3390/min7060106 [DOI] [Google Scholar]
- Tupikina O. V., Minnaar S. H., Van Hille R. P., Van Wyk N., Rautenbach G. F., Dew D., et al. (2013). Determining the effect of acid stress on the persistence and growth of thermophilic microbial species after mesophilic colonisation of low grade ore in a heap leach environment. Min. Engin. 53 152–159. 10.1016/j.mineng.2013.07.015 [DOI] [Google Scholar]
- Valdés J., Cárdenas J. P., Quatrini R., Esparza M., Osorio H., Duarte F., et al. (2010). Comparative genomics begins to unravel the ecophysiology of bioleaching. Hydrometallurgy 104 471–476. 10.1016/j.hydromet.2010.03.028 [DOI] [Google Scholar]
- Vassiliev I. R., Jung Y. S., Smart L. B., Schulz R., Mcintosh L., Golbeck J. H. (1995). A mixed-ligand iron-sulfur cluster (C556SPaB or C565SPsaB) in the Fx-binding site leads to a decreased quantum efficiency of electron transfer in photosystem I. Biophys. J. 69 1544–1553. 10.1016/s0006-3495(95)80026-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera M., Schippers A., Sand W. (2013). Progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation–part A. Appl. Microbiol. Biotechnol. 97 7529–7541. 10.1007/s00253-013-4954-2 [DOI] [PubMed] [Google Scholar]
- Volbeda A., Darnault C., Renoux O., Nicolet Y., Fontecilla-Camps J. C. (2015). The crystal structure of the global anaerobic transcriptional regulator FNR explains its extremely fine-tuned monomer-dimer equilibrium. Sci. Adv. 1:e1501086. 10.1126/sciadv.1501086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46 W296–W303. 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widder S., Allen R. J., Pfeiffer T., Curtis T. P., Wiuf C., Sloan W. T., et al. (2016). Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J. 10 2557–2568. 10.1038/ismej.2016.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes P., Remis J. P., Hwang M., Auer M., Thelen M. P., Banfield J. F. (2009). Natural acidophilic biofilm communities reflect distinct organismal and functional organization. ISME J. 3 266–270. 10.1038/ismej.2008.90 [DOI] [PubMed] [Google Scholar]
- Yan A., Kiley P. J. (2008). Dissecting the role of the N-terminal region of the Escherichia coli global transcription factor FNR. J. Bacteriol. 190 8230–8233. 10.1128/JB.01242-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan A., Kiley P. J. (2009). “Techniques to Isolate O2-Sensitive Proteins: [4Fe–4S]-FNR as an Example,” in Methods in Enzymology, eds Richard R. B., Murray P. D. (Cambridge: Academic Press; ), 787–805. 10.1016/s0076-6879(09)63042-1 [DOI] [PubMed] [Google Scholar]
- Yin S.-H., Wu A.-X., Li X.-W., Wang Y.-M. (2011). Mathematical model for coupled reactive flow and solute transport during heap bioleaching of copper sulfide. J. Cent. South Univ. Technol. 18:1434 10.1007/s11771-011-0858-4 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/genome/1014?genome_assembly_id=300479.