Abstract
CRISPR-Cas systems provide microbes with adaptive immunity to infectious nucleic acids and are widely employed as genome editing tools. These tools utilize RNA-guided Cas proteins whose large size (950—1400 amino acids) has been considered essential to their specific DNA- or RNA-targeting activities. Here we present a set of CRISPR-Cas systems from uncultivated archaea that contain Cas14, a family of exceptionally compact RNA-guided nucleases (400—700 amino acids). Despite their small size, Cas14 proteins are capable of targeted single-stranded DNA (ssDNA) cleavage without restrictive sequence requirements. Moreover, target recognition by Cas14 triggers non-specific cutting of ssDNA molecules, an activity that enables high-fidelity SNP genotyping (Cas14-DETECTR). Metagenomic data show that multiple CRISPR-Cas14 systems evolved independently and suggest a potential evolutionary origin of single-effector CRISPR-based adaptive immunity.
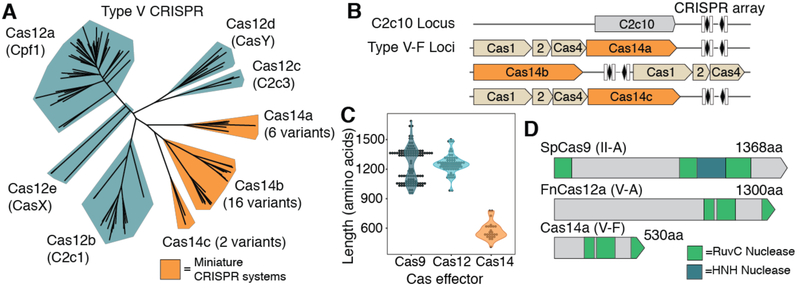
Competition between microbes and viruses stimulated the evolution of CRISPR-based adaptive immunity to provide protection against infectious agents (1, 2). In class 2 CRISPR-Cas systems, a single 100–200 kilodalton (kDa) CRISPR-associated (Cas) protein with multiple functional domains carries out RNA-guided binding and cutting of DNA or RNA substrates (3, 4). To determine whether simpler, smaller RNA-guided proteins occur in nature, we queried terabase-scale metagenomic datasets (5-9) for uncharacterized genes proximal to both a CRISPR array and cas1, the gene that encodes the universal CRISPR integrase (10, 11). This analysis identified a diverse family of CRISPR-Cas systems that contain cas1, cas2, cas4, and a new gene, cas14, encoding a 40–70 kDa polypeptide (Fig. 1A). We initially identified 24 different cas14 gene variants that cluster into three subgroups (Cas14a–c) based on comparative sequence analysis (Fig. 1A-B, fig. S1, S2). Cas14 proteins are ~400–700 amino acids (aa), about half the size of previously known class 2 CRISPR RNA-guided enzymes (950—1400 aa) (Fig. 1C-D). While the identified Cas14 proteins exhibit considerable sequence diversity, all are united by the presence of a predicted RuvC nuclease domain, whose organization is characteristic of Type V CRISPR-Cas DNA-targeting enzymes (Fig. 1D) (3, 12, 13).
Fig. 1. Architecture and phylogeny of CRISPR-Cas14 genomic loci.
(A) Phylogenetic tree of Type V CRISPR systems. Newly identified miniature CRISPR systems are highlighted in orange. (B) Representative loci architectures for C2c10 and CRISPR-Cas14 systems. (C) Length distribution of Cas14a–c systems compared to Cas12a-e and Cas9. (D) Domain organization of Cas14a compared to Cas9 and Cas12a with the nuclease domains (RuvC and HNH) indicated. Protein lengths are drawn to scale.
The Cas14 proteins we identified occur almost exclusively within DPANN, a super-phylum of symbiotic archaea characterized by small cell and genome sizes (14, 15). Phylogenetic comparisons showed that Cas14 proteins are widely diverse with similarities to C2c10 and C2c9, families of bacterial RuvC-domain-containing proteins that are sometimes found near a CRISPR array but not together with other cas genes (Fig. 1B, fig. S1) (3). This observation and the small size of c2c10, c2c9 and cas14 genes made it improbable that these systems could function as standalone CRISPR effectors (3).
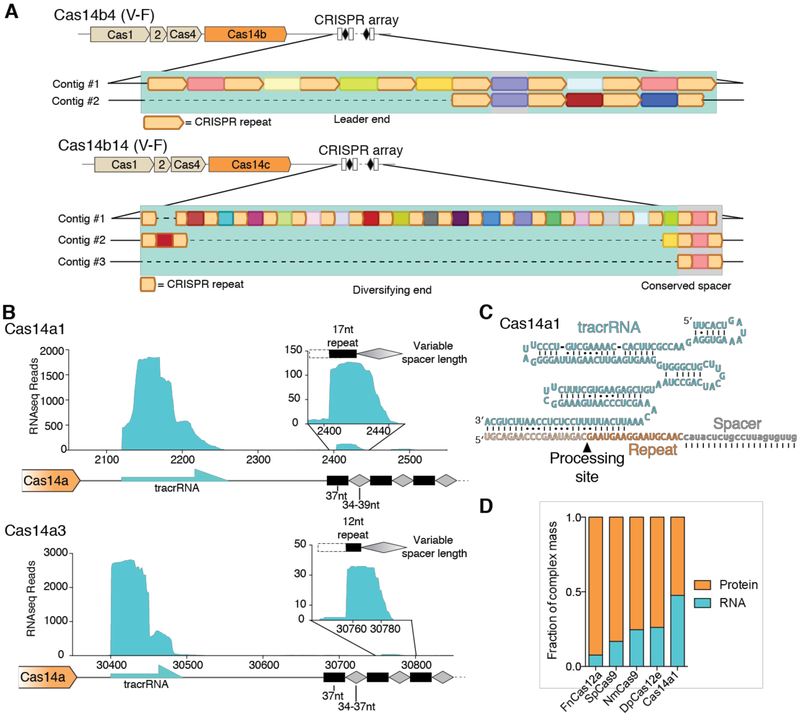
Based on their proximity to conserved genes responsible for creating genetic memory of infection (cas1, cas2, cas4) (fig. S3A), we explored whether CRISPR-Cas14 systems can actively acquire DNA sequences into their CRISPR arrays. Assembled metagenomic contiguous DNA sequences (contigs) for multiple CRISPR-Cas14 loci revealed that otherwise identical CRISPR systems showed diversity in their CRISPR arrays. These results are consistent with active adaptation to new infections although without longitudinal sampling this data could also be explained by alternative biological mechanisms (Fig. 2A, fig. S3B) (13). The evidence suggesting acquisition of new DNA sequences led us to hypothesize that these CRISPR-Cas14 loci encode functional enzymes with nucleic acid targeting activity despite their small size. To test this possibility, we first investigated whether RNA components are produced from CRISPR-Cas14 loci. Environmental metatranscriptomic sequencing data were analyzed for the presence of RNA from the native archaeal host that contains CRISPR-Cas14a (Fig. 2B, fig. S4A). In addition to CRISPR RNAs (crRNAs), a highly abundant non-coding RNA was mapped to a ~130-base pair sequence located between cas14a and the adjacent CRISPR array. Notably, the 3′ end of this transcript was mostly complementary to the repeat segment of the crRNA (Fig. 2C, fig. S4B), as observed for trans-activating CRISPR RNAs (tracrRNAs) found in association with Cas9, Cas12b and Cas12e CRISPR systems (12, 13, 16). In these previously studied systems, the double-stranded-RNA-cutting enzyme Ribonuclease III (RNase III) generates mature tracrRNAs and crRNAs, but no genes encoding RNase III were present in cas14-containing reconstructed genomes (fig. S5A), nor did Cas14a cleave its own pre-crRNA when tested biochemically (fig. S5B). These observations imply that an alternative mechanism for CRISPR-associated RNA processing exists in these hosts.
Fig. 2. CRISPR-Cas14a actively adapts and encodes a tracrRNA.
(A) Spacer diversity for Cas14b4 and Cas14b14 with CRISPR repeats diagramed in tan and unique spacers shown in different colors. (B) Metatranscriptomics reads mapped to Cas14a1 and Cas14a3. Inset shows expansion of most abundant repeat and spacer sequence. (C) In silico predicted structure of Cas14a1 crRNA and tracrRNA. Notably, RNase III orthologs were not identified in host genomes (fig. S5A). (D) Fraction of various CRISPR complexes mass made up of by RNA and protein.
To test whether the Cas14a proteins and associated RNA components can assemble together in a heterologous organism, we introduced a plasmid into E. coli containing a minimal CRISPR-Cas14a locus that includes the Cas14 gene, the CRISPR array and intergenic regions containing the putative tracrRNA. Affinity purification of the Cas14a protein from cell lysate and sequencing of co-purifying RNA revealed a highly abundant mature crRNA as well as the putative tracrRNA, albeit in lower relative abundance than environmental metatranscriptomics, suggesting that Cas14 associates with both crRNA and tracrRNA (fig. S5B). The calculated mass of the assembled Cas14a protein-tracrRNA-crRNA particle is 48% RNA by weight compared to just 17% for S. pyogenes Cas9 (SpCas9) and 8% for F. novicida Cas12a (FnCas12a) (Fig. 2D), hinting at a central role of the RNA in the architecture of the Cas14a complex. Known class 2 CRISPR systems require a short sequence called a protospacer adjacent motif (PAM) to target double-stranded DNA (dsDNA) (17). To test whether Cas14a requires a PAM and can conduct dsDNA interference, we transformed E. coli expressing a minimal Cas14a locus with a dsDNA plasmid containing a randomized PAM region next to a sequence matching the target-encoding sequence (spacer) in the Cas14 array. Notably, no depletion of a PAM sequence was detected among E. coli transformants, suggesting that the CRISPR-Cas14a system is either unable to target dsDNA, can do so without requiring a PAM, or is inactive in this heterologous host (fig. S6A, B).
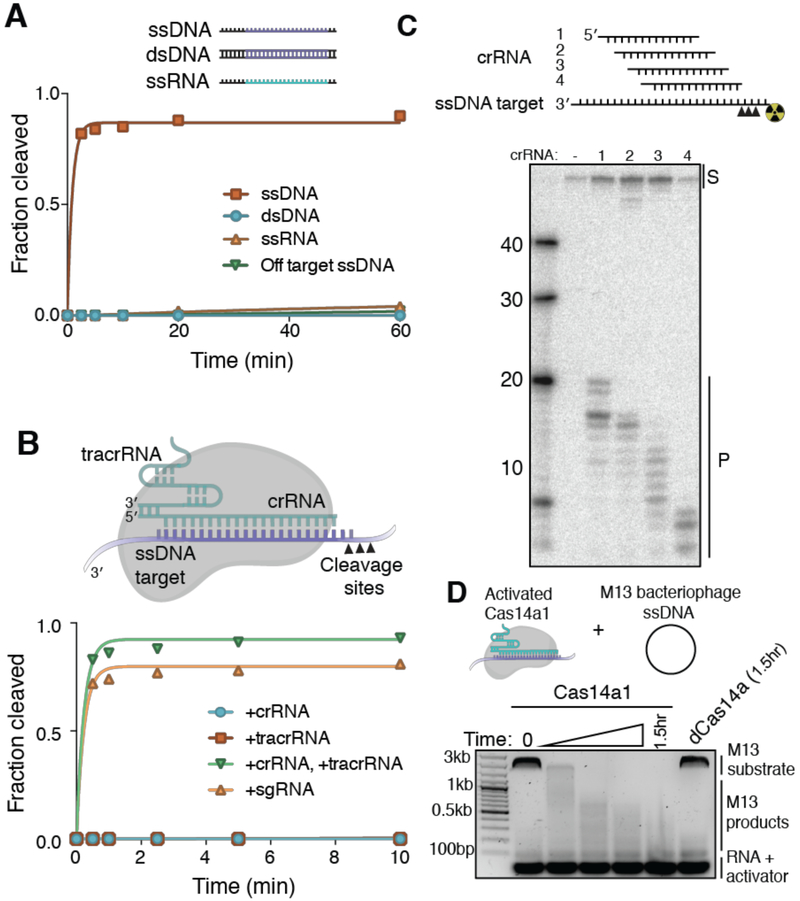
We next tested whether purified Cas14a-tracrRNA-crRNA complexes are capable of RNA-guided nucleic acid cleavage in vitro. All currently reconstituted DNA-targeting class 2 interference complexes are able to recognize both dsDNA and ssDNA substrates (18-20). We incubated purified Cas14a-tracrRNA-crRNA complexes with radiolabeled target oligonucleotides (ssDNA, dsDNA, and ssRNA) bearing 20-nucleotide sequence complementary to the crRNA guide sequence, or a non-complementary ssDNA, and we analyzed these substrates for Cas14a-mediated cleavage. Only in the presence of a complementary ssDNA substrate was any cleavage product detected (Fig. 3A, fig. S7A-C), and cleavage was dependent on the presence of both tracrRNA and crRNA, which could also be combined into a single-guide RNA (sgRNA) (Fig. 3B, fig. S8). The lack of detectable dsDNA cleavage suggests that Cas14a targets ssDNA selectively, although it is possible that some other host factor or sequence requirement could enable dsDNA recognition in the native host. Mutation of the conserved active site residues in the Cas14a RuvC domain eliminated cleavage activity (fig. S7D-E), implicating RuvC as the domain responsible for DNA cutting. Moreover, Cas14a DNA cleavage was sensitive to truncation of the RNA components to lengths shorter than the naturally produced sequences (fig. S9A-D). These results establish Cas14a as the smallest class 2 CRISPR effector demonstrated to conduct programmable RNA-guided DNA cleavage thus far.
Fig. 3. CRISPR-Cas14a is an RNA-guided DNA-endonuclease.
(A) Cleavage kinetics of Cas14a1 targeting ssDNA, dsDNA, ssRNA and off-target ssDNA. (B) Diagram of Cas14a RNP bound to target ssDNA and Cas14a1 cleavage kinetics of radiolabeled ssDNA in the presence of various RNA components. (C) Tiling of a ssDNA substrate by Cas14a1 guide sequences. (D) Cleavage of the ssDNA viral M13 genome with activated Cas14a1.
Although we were unable to identify a dsDNA PAM in vivo, we tested whether Cas14a requires a PAM for ssDNA cleavage in vitro by tiling Cas14a guides across a ssDNA substrate (Fig. 3C). Despite sequence variation adjacent to the targets of these different guides, we observed cleavage for all four sequences. Notably, the cleavage sites occur beyond the guide-complementary region of the ssDNA and shift in response to guide binding position (Fig. 3C). These data demonstrate Cas14a is a ssDNA-targeting CRISPR endonuclease that does not require a PAM for activation.
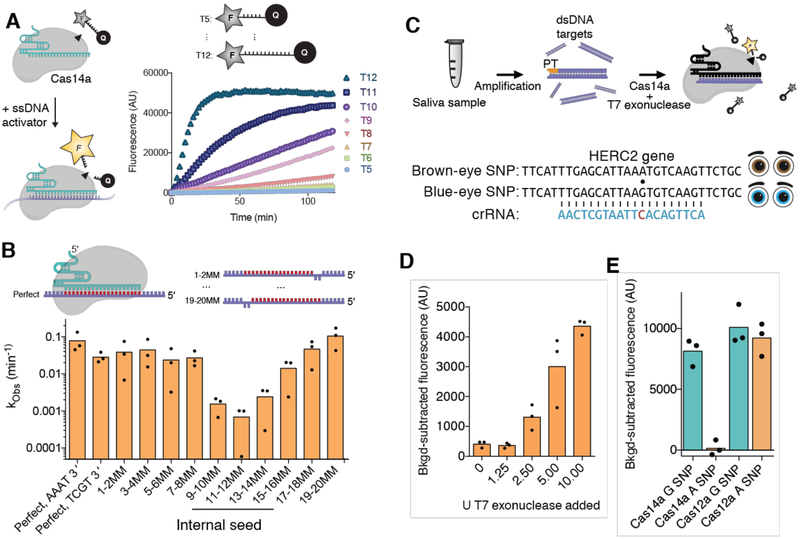
Based on the observation that Cas14a cuts outside of the crRNA/DNA targeting heteroduplex, we hypothesized that Cas14a might possess target-activated non-specific ssDNA cleavage activity, similar to the RuvC-containing enzyme Cas12a (20, 21). To test this possibility, we incubated Cas14a-tracrRNA-crRNA with a complementary activator DNA and an aliquot of M13 bacteriophage ssDNA bearing no sequence complementarity to the Cas14a crRNA or activator (Fig. 3D). The M13 ssDNA was rapidly degraded to small fragments, an activity that was eliminated by mutation of the conserved Cas14a RuvC active site, suggesting that activation of Cas14a results in non-specific ssDNA degradation. However, we were unable to observe Cas14a-mediated interference against the ssDNA bacteriophage ΦX174 when we expressed Cas14a heterologously in E. coli (Fig. S10A-C), possibly due to the dissimilarity between E. coli and Cas14a’s native archaeal host. To investigate the specificity of target-dependent non-specific DNA cutting activity by Cas14a, we adapted a fluorophore-quencher (FQ) assay in which cleavage of dye-labeled ssDNA generates a fluorescent signal (Fig. 4A) (22). When Cas14a was incubated with various guide RNA-target ssDNA pairs, a fluorescent signal was observed only in the presence of the cognate target and showed strong preference for longer FQ-containing substrates (fig. S10D, Fig. 4A). We next tested Cas14a mismatch tolerance by tiling 2-nt mismatches across the targeted region in various ssDNA substrates. Surprisingly, mismatches near the middle of the ssDNA target strongly inhibited Cas14a activity, revealing an internal seed sequence that is distinct from the PAM-proximal seed region observed for dsDNA-targeting CRISPR-Cas systems (Fig. 4B, fig. S11A-D). Moreover, DNA substrates containing strong secondary structure resulted in reduced activation of Cas14a (fig. S11E). Truncation of ssDNA substrates also resulted in reduced or undetectable trans cleavage (fig. S11F). Together, these results suggest a mechanism of fidelity distinct from dsDNA-targeting class 2 CRISPR systems, possibly utilizing a mechanism similar to the ssRNA-targeting Cas13a enzymes (23-25).
Fig. 4. High fidelity ssDNA SNP detection by CRISPR-Cas14a.
(A) Fluorophore-quencher (FQ) assay for detection of ssDNA by Cas14a1 and the cleavage kinetics for various length FQ substrates. (B) Cleavage kinetics for Cas14a1 with mismatches tiled across the substrate (individual points represent replicate measurements). (C) Diagram of Cas14-DETECTR strategy and HERC2 eye color SNP. (D) Titration of T7 exonuclease and impact on Cas14a-DETECTR. (E) SNP detection using Cas14a-DETECTR with a blue-eye targeting guide for a blue-eyed and brown-eyed saliva sample compared to ssDNA detection using Cas12a.
The target-dependent, non-specific DNase activity of Cas12a serves as a DNA detection platform (DNA endonuclease-targeted CRISPR trans reporter; DETECTR) for diagnostic uses (20, 26). While Cas12a exhibits low fidelity in discriminating against ssDNA substrates (20), Cas14a requires complementarity in the seed region for ssDNA substrate recognition. This improved specificity raised the possibility of using Cas14a for high-fidelity detection of DNA single nucleotide polymorphisms (SNPs) without the constraint of a PAM sequence. To test this idea, DNA substrates were amplified using a phosphorothioate (PT)-containing primer to protect one strand from degradation by exonucleases. Upon addition of T7 exonuclease, the unmodified strand was degraded, leaving ssDNA substrates that can be detected by Cas14a (Fig. 4C-D). As a proof of principle, we aimed to detect the human HERC2 gene, which contains a SNP responsible for eye color (27). We amplified the HERC2 gene from DNA in human saliva from both blue-eyed and brown-eyed individuals, using the PT amplification approach described above. When programmed with a guide RNA targeting the blue-eyed SNP, Cas12a failed to discriminate between the two ssDNA targets, exhibiting robust trans activity in both cases, while Cas14a exhibited strong activation in recognition of the blue-eyed SNP with near-background signal for the brown-eyed sample (Fig. 4E). The development of Cas14-DETECTR now allows for CRISPR-based detection of medically and ecologically important ssDNA pathogens as well as high-fidelity detection of SNPs without the constraint of a PAM sequence.
Further investigation of compact Type V systems in metagenomic data revealed a large diversity of systems that, like Cas14a–c, include a gene encoding a short RuvC-containing protein adjacent to acquisition-associated cas genes and a CRISPR array. We found 20 additional such systems in various uncultivated microbes that cluster into five main families (Cas14d–h). Excluding cas14g, which is related to cas12b, the cas14-like genes form separate clades on the type V effector phylogeny (fig. S12A, B), suggesting these families evolved from independent domestication events of TnpB, the transposase-associated protein implicated as the evolutionary ancestor of type V CRISPR effectors (28). Phylogenetic reconstruction of their associated cas1 genes indicated that they too have different origins for the cas14 subtypes (fig. S2A). Altogether we identified 38 CRISPR-Cas14 systems belonging to eight families (Cas14a–h) and eight additional systems that could not be clustered with our analysis (termed Cas14u, data S1).
The small size of the Cas14 proteins described here and their resemblance to type V effector proteins suggest that RNA-guided ssDNA cleavage may have existed as an ancestral class 2 CRISPR system (29, 30). In this scenario, a small, domesticated TnpB-like ssDNA interference complex may have gained additional domains over time, gradually improving dsDNA recognition and cleavage. Related to this hypothesis, smaller Cas9 orthologs exhibit weaker dsDNA-targeting activity than their larger counterparts but retain the ability to robustly cleave ssDNA (19). Aside from the evolutionary implications, the ability of Cas14 to specifically target ssDNA suggests a role in defense against ssDNA viruses or mobile genetic elements (MGEs) that propagate through ssDNA intermediates (31). A ssDNA-targeting CRISPR system would be particularly advantageous in certain ecosystems where ssDNA viruses comprise the vast majority of viral abundance (32). The unexpected finding that these miniature CRISPR proteins can conduct targeted DNA cleavage highlights the diversity of CRISPR systems hidden in uncultivated organisms. Ongoing exploration of these underrepresented microbial lineages will likely continue to reveal new, unexpected insights into this microscopic arms race and lead to continued development of valuable CRISPR-based technologies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Nan Ma and Kaihong Zhou for technical assistance. We thank Parker Harrington for graphic design assistance. D.B. was supported by a grant from the Paul Allen Frontiers Group, L.B.H., J.C.C. and J.S.C. by US National Science Foundation Graduate Research Fellowships. J.A.D. is an Investigator of the Howard Hughes Medical Institute. This work was supported in part by a Frontiers Science award from the Paul Allen Institute to J.A.D. and J.F.B., a grant from the National Science Foundation (MCB-1244557 to J.A.D.), the Lawrence Berkeley National Laboratory’s Sustainable Systems Scientific Focus Area funded by the U.S. Department of Energy, (DE-AC02-05CH11231 to J.F.B.) and the Office of Science of the US Department of Energy under Contract No. DE-AC02–05CH11231.
Footnotes
COMPETING INTERESTS
UC Regents have filed patents related to this work on which D.B., J.F.B., L.B.H., D.P.E., J.S.C. and J.A.D are inventors. L.B.H. and J.S.C. are co-founders of Mammoth Biosciences. I.P.W. is a consultant for Mammoth Biosciences. J.F.B. is a founder of Metagenomi. J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics, and Mammoth Biosciences. J.A.D. is a scientific advisory board member of Caribous Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Synthego, Metagenomi, Mammoth Biosciences and Inari. J.A.D is a member of the board of directors at Driver and Johnson & Johnson and has sponsored research projects by Roche Biopharma and Biogen.
DATA AVAILABILITY
Data S1 specifies the accession numbers, coordinates and samples of origin for all CRISPR-Cas14 systems described in this study.
REFERENCES AND NOTES
- 1.Barrangou R et al. , CRISPR provides acquired resistance against viruses in prokaryotes. Science. 315, 1709–12 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Jackson SA et al. , CRISPR-Cas: Adapting to change. Science (80-.). 356 (2017), pp. 1–9. [DOI] [PubMed] [Google Scholar]
- 3.Shmakov S et al. , Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol 15, 169–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen JS, Doudna JA, The chemistry of Cas9 and its CRISPR colleagues. Nat. Rev. Chem 1,0078 (2017). [Google Scholar]
- 5.Brown CT et al. , Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 523, 208–211 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Anantharaman K et al. , Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun 7, 13219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markowitz VM et al. , IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 42, 568–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Probst AJ et al. , Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2concentrations. Environ. Microbiol 19, 459–474 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Chen IMA et al. , IMG/M: Integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45, D507–D516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yosef I, Goren MG, Qimron U, Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nuñez JK, Lee ASY, Engelman A, Doudna J. a., Integrase-mediated spacer acquisition during CRISPR–Cas adaptive immunity. Nature. 519, 193–198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shmakov S et al. , Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 60, 385–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burstein D et al. , New CRISPR-Cas systems from uncultivated microbes. Nature. 542, 237–241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rinke C et al. , Insights into the phylogeny and coding potential of microbial dark matter. Nature. 499, 431–437 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Castelle CJ et al. , Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr. Biol 25, 690–701 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Deltcheva E et al. , CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 471,602–607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C, Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 155, 733–740 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Rajan R, Seifert HS, Mondragón A, Sontheimer EJ, DNase H Activity of Neisseria meningitidis Cas9. Mol. Cell 60, 242–255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma E, Harrington LB, O’Connell MR, Zhou K, Doudna JA, Single-Stranded DNA Cleavage by Divergent CRISPR-Cas9 Enzymes. Mol. Cell 60, 398–407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen JS et al. , CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 360, 436–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zetsche B et al. , Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.East-Seletsky A et al. , Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 538, 270–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L et al. , The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell. 170, 714–726.e10 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Abudayyeh OO et al. , C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science (80-.). 353, 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knott GJ et al. , Guide-bound structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme. Nat. Struct. Mol. Biol 24, 825–833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li SY et al. , CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 28, 491–493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eiberg H et al. , Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum. Genet 123, 177–187 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Shmakov S et al. , Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol 15, 169–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.V Koonin E, Makarova KS, Zhang F, Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol 37, 67–78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makarova KS et al. , An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol, 1–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barabas O et al. , Mechanism of IS 200 / IS 605 Family DNA Transposases : Activation and Transposon- Directed Target Site Selection, 208–220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida M et al. , Quantitative viral community DNA analysis reveals the dominance of single-stranded DNA viruses in offshore upper bathyal sediment from Tohoku, Japan. Front. Microbiol 9, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown CT et al. , Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 523, 208–211 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Anantharaman K et al. , Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun 7, 13219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Probst AJ et al. , Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2concentrations. Environ. Microbiol 19, 459–474 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Markowitz VM et al. , IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 42, 568–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markowitz VM et al. , IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40, 115–122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen IMA et al. , IMG/M: Integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45, D507–D516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finn RD, Clements J, Eddy SR, HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burstein D et al. , New CRISPR-Cas systems from uncultivated microbes. Nature. 542, 237–241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grissa I, Vergnaud G, Pourcel C, CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biswas A, Staals RHJ, Morales SE, Fineran PC, Brown CM, CRISPRDetect: A flexible algorithm to define CRISPR arrays. BMC Genomics. 17, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamatakis A, RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Letunic I, Bork P, Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat Methods. 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogata H et al. , KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrington LB et al. , A thermostable Cas9 with increased lifetime in human plasma. Nat. Commun 8, 1–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crooks G, Hon G, Chandonia J, Brenner S, NCBI GenBank FTP Site\nWebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrington LB et al. , A Broad-Spectrum Inhibitor of CRISPR-Cas9. Cell. 170, 1224–1233.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen JS et al. , CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 360, 436–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.