Abstract
Desmoplakin (Dsp) is a unique and critical desmosomal protein, that is integral to epidermal development. However, it is unclear whether this protein is required specifically for epidermal morphogenesis. Using morpholinos or Crispr/Cas9 mutagenesis we decreased the function of Dsp in frog embryos to better understand its role during epidermal development. Dsp morphant and mutant embryos had developmental defects such as epidermal fragility that mimicked what has been reported in mammals. Most importantly, we also uncovered a novel function for Dsp in the morphogenesis of the epidermis in X. laevis. In particular, Dsp is required during the process of radial intercalation where basally located cells move into the outer epidermal layer. Once inserted these newly intercalated cells expand their apical surface and then they differentiate into specific epidermal cell types. Decreased levels of Dsp resulted in the failure of the radially intercalating cells to expand their apical surface, thereby reducing the number of differentiated multiciliated and secretory cells. Such defects correlate with changes in E-cadherin levels and actin and microtubule localization which could explain the defects in apical expansion. A mutated form of Dsp that maintains cell-cell adhesion but eliminates the connections to the cytoskeleton results in the same epidermal morphogenesis defect. These results suggest a specific role for Dsp in the apical expansion of cells during radial intercalation. We have developed a novel system, in the frog, to demonstrate for the first time that desmosomes not only protect against mechanical stress but are also critical for epidermal morphogenesis.
Keywords: Desmosome, Desmoplakin, Xenopus laevis, Epidermal development, Radial intercalation, multiciliated cells
INTRODUCTION
Desmosomes together with tight and adherens junctions make up the main complexes that provide adhesion between cells. These cell-cell junctions also integrate signals which can regulate a range of cellular processes such as differentiation and cell migration (reviewed in (Celentano et al., 2017; Delva et al., 2009; Dusek et al., 2007; Garrod and Chidgey, 2008; Green and Gaudry, 2000; Green and Simpson, 2007; Johnson et al., 2014)). But surprisingly, much less is known about the desmosome relative to its other junctional counterparts during embryonic development. The importance of desmosomes to the embryo though is clear since birth defects caused by mutations to various desmosomal proteins can be devastating and are often lethal in humans (Celentano et al., 2017; Johnson et al., 2014). Further, mice with null mutations in desmosomal proteins die early in development and both Xenopus and zebrafish embryos with deficient desmosomal proteins show major developmental defects that would be incompatible with life (Berika and Garrod, 2014; Cheng et al., 2005; DeMarais and Moon, 1992; Gallicano et al., 2001; Gallicano et al., 1998; Goonesinghe et al., 2012; Kofron et al., 2002; Kofron et al., 1997; Munoz et al., 2012). From these studies, it is clear that desmosomes are essential to maintain the integrity and provide resilience to the embryo, however, it is unknown whether they play specific roles in morphogenesis.
The desmosome, like the adherens junction, is a modular structure that links adjacent cells to each other and to the intracellular cytoskeleton (see reviews such as (Garrod and Chidgey, 2008; Green and Gaudry, 2000; Green and Simpson, 2007)). The transmembrane desmosomal cadherins consist of desmogleins and desmocollins and bridge the extracellular space by homophilic binding to complement cadherins on the adjacent cell. These cadherins are anchored in the cytoplasm by the armadillo proteins, plakoglobin, and plakophilin. Such armadillo proteins in turn bind to desmoplakin which then forms the critical link to the intermediate filaments such as keratins and microtubules. Desmoplakin (Dsp) is composed of a central rod region that facilitates dimerization, a globular head plakin domain that is required for protein-protein interactions and a C-terminal tail that has three plakin repeat domains, the last of which interacts with cytoskeletal proteins including intermediate filaments (Delva et al., 2009; Garrod and Chidgey, 2008). Since Dsp serves as the connection between the transmembrane and cytoskeletal components it could be essential for integrating signals necessary for morphogenesis. Therefore, we have utilized the frog, Xenopus laevis, as a tractable vertebrate model to better define the developmental roles of Dsp in the embryo.
The requirement of Dsp has been examined in mammals using several approaches. First, complete loss of dsp in mice was shown to be embryonic lethal at very early stages due to defects in the extraembryonic tissue (Gallicano et al., 1998). In another approach, where tetraploid aggregation was used to rescue dsp expression in the trophectoderm, the requirement of this protein for embryonic development could be evaluated (Gallicano et al., 2001). These rescued dsp mutant embryos had major developmental defects in the heart, brain, and epidermal formation. Further, decreased dsp expression resulted in significantly smaller embryos than the wildtype siblings. A third study used a conditional loss of function approach to deplete dsp specifically in the epidermis before the simple epidermis stratifies (Vasioukhin et al., 2001). These embryos had skin fragility, abnormal cell morphologies and disrupted cytoskeletal architecture (Vasioukhin et al., 2001). However, it was difficult to determine whether the abnormal epidermis was due to problems associated with coordinated cell movements or organization during epidermal stratification in this system.
The Xenopus embryonic epidermis presents an attractive system in which to study epidermal morphogenesis. It forms as a simple bi-layered epidermis that undergoes tractable cell movements and differentiation that has been exceptionally well studied. Specifically, the epidermis is comprised of an outer or superficial layer and inner or sensorial layer (Billett and Gould, 1971; Drysdale and Elinson, 1992). The outer epidermal layer contains specific differentiated cells which include goblet cells, small secretory cells (SSCs), ionocytes and multiciliated cells (MCCs). Together these cells protect the embryo from toxins and bacteria by secreting substances and moving fluids (Dubaissi et al., 2014). The inner epidermal layer contains cells with stem cell properties, that is, they continually divide and supply new cells to the outer layer. These inner cells are specified and then move into the outer epidermal layer through a process called radial intercalation (Drysdale and Elinson, 1992; Dubaissi and Papalopulu, 2011; Stubbs et al., 2006; Walck-Shannon and Hardin, 2014). During this process, an inner epidermal cell must first move apically and insert between outer epidermal cells at the vertex of 3–4 cells. Then the apical surface of the inner cell expands as it joins the outer layer (Sedzinski et al., 2016). Radial intercalation in the epidermis not only provides specific cell types to the outer epidermis but also allows the skin to rapidly expand as the embryo develops.
Several of the players regulating epidermal radial intercalation has been defined in Xenopus especially concerning the MCCs (reviewed in (Walck-Shannon and Hardin, 2014)). These cells are first specified by the Delta-Notch activation of lateral inhibition in the inner cell layer (Stubbs et al., 2006). Several transcription factors such as multicilin, Foxj1, and RFX2 are then responsible for the activation of a multitude of genes that are necessary for MCC specification and differentiation (Chung et al., 2012; Stubbs et al., 2008; Stubbs et al., 2012). PCP signals together with microtubule and associated proteins are important for polarizing and inserting the inner cells into the outer epidermal layer (Kim et al., 2012; Kim et al., 2018; Mitchell et al., 2009; Ossipova et al., 2015; Werner et al., 2014). Finally, apical expansion of the intercalating MCC cell is regulated by RhoA signals combined with formins and actin (Sedzinski et al., 2016, 2017). Whether desmosomes have a role in radial intercalation of the epidermis has never been explored.
In this study, we demonstrate that the frog desmosomal protein, Dsp, has shared functions with mammals, such as providing mechanical integrity and is important for the size of the embryo. Importantly, we have developed a novel system, in Xenopus, to demonstrate that Dsp is critical for epidermal morphogenesis.
METHODS
X. laevis Adults and Embryos
Xenopus laevis adults were created in our breeding colony and purchased from Nasco. All procedures were approved by the VCU Institutional Animal Care and Use Committee (IACUC protocol number 5AD20261). Embryos were collected using standard procedures (Sive et al., 2000) and were staged according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1967). Embryos were cultured in 0.1× MBS (Modified Barth’s Serum), refreshed daily, and housed in a 23°C or 15 °C incubator (Torrey Pines Scientific, Cat. No. IN3 0). After the experiments were completed all embryos were given a lethal dose of anesthetic (10% tricaine for 1 hour).
Bioinformatics analysis of Dsp
Full-length (Desmoplakin) Dsp protein and mRNA sequences for Homo sapiens Desmoplakin I (NP_004406), Mus musculus Desmoplakin (NP_076331) and Danio rerio Desmoplakin isoform X1 (XP_001919901) X. laevis Dsp.L (XB-GENE-866134, Genome Build 9.1, http://www.xenbase.org), were aligned using the LALIGN tool (EMBL-EBI) and the EMBOSS Water tool (Smith-Waterman algorithm) (EMBL-EBI) was used to determine similarity. Protein domains were based on those identified in the human Dsp protein (Al-Jassar et al., 2011; Green et al., 1990; Virata et al., 1992).
TEM to examine desmosome ultrastructure and localization in the epidermis
Embryos were processed for EM at the VCU microscopy core using standard protocols. Briefly, embryos were fixed with 2% glutaraldehyde (MP Biomedicals, 198595) in 0.1M sodiμm cacodylate buffer (Electron Microscopy Services, 12300) overnight (4°C) and then refixed in 2% osmiμm tetroxide in 0.1M cacodylate buffer (one hour). They were dehydrated in ethanol and then infiltrated with propylene oxide (EMS, 20401) and Poly/Bed 812 resin mix (50:50, Polyscienes, 08792–1) overnight. Finally, embryos were incubated in EMbed 812 resin (EMS, 14120, overnight), placed in molds and heated 60°C (2 days). 700–900Å thick sections were created on a Leica EM UC6i Ultramicrotome (Leica Microsystems) and stained with 5% Uranyl acetate (EMS, 22400) and Reynold’s Lead Citrate (Lead Nitrate (EMS, 17900) and Sodium Citrate (EMS, 21140)). A JEOL JEM-1230 TEM (JEOL USA, Inc.) with the Gatan Orius SC1000 digital camera (Gatan Inc., Pleasanton, CA) was used to image the ultrastructure of the epidermis.
Dsp Morpholinos and RT-PCR tests for splicing defects
Functional analysis was performed utilizing splice blocking antisense Morpholinos (Genetools). DspMO1 (5’-ACAGTTACTACTTACTCTATGCTGC-3’) targets the donor site of exon 4 and DspMO2 (5’–TTGATGCAGAGCAAAGTTCAAACCT-3’) targets the donor site of exon 14. The standard control morpholino (CMO) was used in all experiments at 34–68 ng/embryo. Morpholinos were labeled with fluorescein to gauge injection success and for blastomere targeting. A FemtoJet microinjector (Eppendorf) and a SteREO Discovery.V8 (Zeiss) stereoscope were used for microinjections. Embryos injected at the 1 cell stage were injected with 17–35 ng while those injected at the 16 cell stage were injected with 1–3 ng of Dsp morpholinos.
RT-PCR was performed as described previously (Dickinson and Sive, 2009) to test for splicing defects. RNA was extracted with TRIzol (Invitrogen) followed by lithium chloride precipitation. cDNA was prepared using the High-capacity cDNA Reverse Transcription kit (Applied Biosystems). Dsp primers flanking the targeted exons (suppl. Table 1) were used together with the Apex™ Hot Start Taq DNA Polymerase Master Mix.
Dsp CRISPR/Cas9 mutational analysis
Injectable gRNA was created as described previously (Shah et al., 2016). CHOPCHOP (http://chopchop.cbu.uib.no/) was used to design two CRISPR gRNA sequences with no mismatches and high-efficiency targeting exons 8 (dspCrispr1) and 19 (dspCrispr2) of dsp.L (these correspond to exons 8 and 17 in dsp.S, respectively). The gRNA sequences were GGTGCTGGTTCATGATAAGCTGG (DspCrispr1) GGTCGCATCTGACAGTTTGATGG (DspCrispr2). A T7 primer sequence and loop-specific sequence (5’-AATTAATACGACTCACTATA-(N)20-GTTTTAGAGCTAGAAATAGC-3’) were added to create a DspCrispr Oligo (ordered from IDT). Double-stranded DNA template was produced combining the customs designed oligo with a generic Scaffold Oligo (5’GATCCGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCT AGCTCTAAAAC-3’; IDT) and PCR performed with the following parameters; 98°C (30s), 98°C (10s), 61°C (10s), 72°C (15s) × 45 cycles, followed by a 72°C (5 min) extension. DNA was then processed through the Clean & Concentrator Kit (Zymo Research, D4014) to purify and concentrate the template. The Ambion MEGAscript T7 kit (Thermo Fisher Scientific, AM1333) was used for in vitro transcription following the manufacturer’s instructions with long incubation (16hrs-24hrs). To create F0 mosaic mutants, embryos were co-injected with 1ng gRNA and 1.5ng Cas9 protein (1mg/ml PNA-Bio, CP01) at the one-cell stage. Wild-type embryos were used as controls.
The T7 endonuclease I assay was used to detect mutations induced by CRISPR/Cas9 as described previously (Mashal et al., 1995). Embryos were incubated in a lysis buffer consisting of 25 mM NaOH (Fisher Scientific, BP359) and 0.2 mM Na2+-EDTA (OmniPur, 4050)) at 95°C for 40 minutes. After the samples cooled an equal volume of neutralization buffer (40 mM Tris-HCl (Sigma-Aldrich, T3253)) was added. PCR was performed with 2× Phusion master mix and primers flanking the predicted mutation site (primer sequences provided in suppl. Table 2), using the following parameters 98°C (30s), 98°C (5s), 61°C (10s), 72°C (2 0s) for 36 cycles with a 72°C (2min) extension. PCR product s were purified with the DNA Clean and Concentrator kit (Zymo Research, D4014), eluted into nuclease-free water, and quantified using the NanoDrop Lite spectrophotometer (Thermo Fisher Scientific). Purified PCR product (200ng) was used in the following protocol: 95°C (5 min.), 95°C-85°C (−2°C/s), 85°C −25°C (−0.1°C/s). Then, 1 μl of T7 endonuclease I (NEB, M0302) was added and the solution incubated at 37°C for 15 min. Samples were run on a 3% agarose gel to identify the presence of mutations.
Mechanical stress assays
Dropping Assay:
Embryos were vertically dropped from a height of 15 cm using a transfer pipette (Fisher Scientific, 13-711-7M) onto a 150 × 15mm petri dish lined on the bottom with 5mm of 2% agarose (Bioline, 41025). 0.1×MBS was added to the dish to submerge the embryo and then it was visualized and photographed. Images were taken on both left and right lateral sides of the embryos before and after the assay was performed. This procedure was done blindly to avoid any handling bias while pipetting embryos.
Rotational Assay:
Embryos were placed in 50ml plastic polypropylene tubes (USA Scientific, 1500–1811) with 15 ml 0.1 × MBS (Modified Barth’s Saline). The tubes were then rotated using an RKVSD vertical rotating mixer (ATR Biotech) at 55 rpm for a total of 25 rotations.
Quantification and Statistical Analysis:
Embryos were scored as either damaged or undamaged based on whether the epidermis was visually intact or not. Chi-Squared tests were performed in Excel to determine statistical relationships between treatment and control groups. Error bars representing standard error were also calculated in Excel.
Histology
Histology was performed as described (Dickinson and Sive, 2006; Houssin et al., 2017) with some modifications. Briefly, embryos were fixed in 2% PFA and 2% glutaraldehyde in PBT buffer for 24 hr and then embedded in plastic resin (JB-4 Plus) and sectioned at 5–7 μm using a tungsten carbide knife. Sections were stained with Giemsa at 1:20 for 1 hr followed by 10 sec 0.05% acetic acid differentiation wash. Slides were dried and covered with Permount and imaged on a Nikon compound microscope fitted with a digital camera (VCU Biology microscopy core).
Immunohistochemistry, Phalloidin, Peanut Agglutinin
Embryos were fixed in 4% Paraformaldehyde (PFA) or Dent’s fixative (80% methanol:20% DMSO) and then labeled whole or after vibratome sectioning. For sectioning, embryos were embedded in 5% low-melt agarose (SeaPlaque GTG Cambrex) and sectioned using a 5000 Series Vibratome into 150–200μm sections. Primary antibodies included; mouse anti-desmoplakin I+II (Abcam, ab16434, diluted 1:75), mouse anti α-tubulin (Developmental Studies Hybridoma Bank (DSHB), AA4.3, 1:50), mouse anti-cytokeratin type II (DSHB, 1h5, 1:25), anti-β-catenin (Invitrogen, 71–2700, 1:500), and mouse anti-E-cadherin extracellular domain (DSHB, 5D3, 1:25). Dsp was detected using tyramide amplification (Alexa Fluor 488 Tyramide Superboost, goat anti-mouse, Invitrogen b40941). All other antibodies were detected with anti-mouse or anti-rabbit Alexa Fluor’s used at 1:500 (Invitrogen). Fluorescently labeled substrates included Lectin Peanut Agglutinin (PNA) (Alexa Fluor 488, Invitrogen, L21409, 1:1000) and phalloidin (Rhodamine, Life Technologies, R415, 1:50).
Relative keratin intensity profiles
Keratin intensity profiles were generated from images acquired from a C2 Nikon confocal microscope. In the accompanying Nikon Elements software Intensity profile lines (white lines with an arrow) were added to the image so that they crossed beta-catenin labeling at cell junctions and extended into the cytoplasm of each cell. The pixel intensity of beta-catenin and keratin labeling along the intensity profile line was plotted in Elements software.
Biotin labeling
Epidermal surface labeling was achieved by incubating embryos for 1 hour in EZ-Link Sulfo-NHS-LC-Biotin (10mg/ml; Thermo Fisher Scientific, 21335) dissolved in 0.1× MBS. This was followed by a 10μm glycine wash for 10 minutes and then a washout period of 4 hours in 0.1× MBS. Embryos were fixed in 4% PFA overnight at 4°C, blocked with 1% BSA and then labeled with Streptavidin conjugated to Alexa Fluor 568 (Life Technologies, S11226, 1:500). Biotin labeling was imaged with a C2 Nikon confocal.
Quantification of surface areas and numbers of cells
In all quantification experiments, images were taken of the lateral trunk midway between the head and tail tip using the 20× objective on a C2 Nikon confocal.
The relative surface areas of MCC cells labeled with anti-tubulin and phalloidin were measured using the magic wand tool in Photoshop. The wand was drawn along the outside of the cortical actin labeling to estimate cell surface area. Ten cells were measured in each confocal image field. A total of 10 embryos were sampled from three different experiments. The surface areas of the biotin negative cells were similarly measured using the magic wand tool in Photoshop by outlining the border of the cell at the junction of where labeled met unlabeled. All cells in each confocal image field were measured. Statistical Analyses: Sigma Stat software was utilized to determine differences between Dsp morphants and control groups. Normality tests were first performed using the Shapiro-Wilks test. In the Biotin surface analysis the normality test failed and therefore a Mann-Whitney Rank Sum Test was performed. In the analysis of surface areas of tubulin-positive cells, the normality test passed and a student t-test was performed. Sigma Stat or Sigma Plot was used to create box and whisker plots where the 5th and 95th interval are shown and median and mean lines provided. Histograms were created with equal binning and automated scaling adjusted for equivalent comparisons.
The number of tubulin-positive cells, PNA positive regions or cells unlabeled with biotin were quantified by counting all cells (including unlabeled cells) in each image field to achieve relative quantities. Image fields taken from 10–11 embryos (from at least 2 biological replicates) were quantified. Statistical Analyses: SigmaStat software or Excel was utilized to determine differences between Dsp morphants and control groups as well as to create box and whisker plots or bar graphs. Normality tests passed in all analyses and therefore student t-tests were utilized.
Confocal imaging and Photoshop processing
Embryos were imaged in 90–100% glycerol in PBT using a Nikon C2 confocal microscope located in the Biology Department Microscopy Core. Z-stacks were created with a step size of 0.3–0.5 μm, for a total thickness between 2–15 μm. Using NIS-Elements AR 4.50.00 software, Z-stacks were converted to maximum projection images depending on the experiment. Adobe Photoshop was used to further process images, which include increasing the brightness, cropping, and labeling. Changes to the channel color were performed to ensure consistency and accommodate those with color blindness. All images for each experiment were processed in the same way.
DP-NTP expression in embryos
DP-NTP, created by Dr. Kathleen Green, was cloned into the pCS2+ plasmid by Dr. Daniel Conway (Department of Biomedical Engineering, VCU) and provided as a kind gift. To create mRNA, the plasmid was digested with HpaI and then was transcribed using SP6 mMessage mMachine kit (Ambion). 1 ng was injected into embryos at the one cell stage.
RESULTS
1. Desmosomes and desmoplakin exist in the developing epidermis
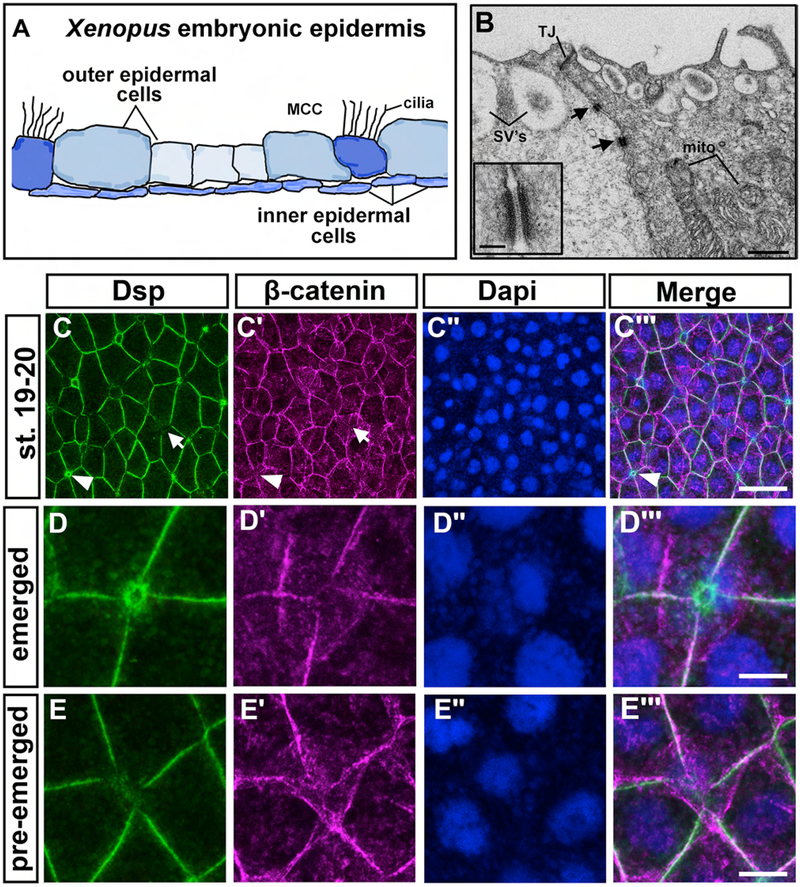
Our first goal was to characterize desmosomes in the developing epidermis of X. laevis. In this species, this epidermal tissue is composed of two layers; an inner and an outer layer (Fig. 1 A). We utilized a combination of transmission electron microscopy (TEM) and immunofluorescence to examine desmosomes and desmoplakin (Dsp) the inner and outer epidermal layers during development.
Figure 1:
Desmosomes and desmoplakin in the epidermis of X. laevis embryos. A) Schematic showing a transverse section of the X. laevis embryonic epidermis based on actual histological sections. B) TEM images where black arrows indicate desmosomes located between outer epidermal cells. Scale bar=1.8μm. Inset shows higher magnification of a desmosome. Scale bar= 135nm. C-D‴) Immunofluorescence of Dsp and beta-catenin in the epidermis. Dapi labeling of the nuclei was used as a counter stain. C-C‴) st. 19–20, in H arrows indicate regions where there is a less Dsp labeling and arrowheads indicate where there is enrichment of Dsp labeling. The same cells are indicated in panel C’. Scale bar=27μm. D-D‴) High magnification of a cell that appears to be apically emerging with high levels of Dsp and lower beta-catenin. Scale bar = 6μm. D-D‴) High magnification of a cell that appears to be beginning to intercalate and has low levels of Dsp. Scale bar=6μm. Abbreviations= MCC= multiciliated cell, SVs=secretory vesicles, mito=mitochondria, TJ=tight junction, oe=outer epidermis, ie=inner epidermis.
1a). TEM reveals that desmosomes are present in the epidermis during embryonic development of X. laevis
The location of desmosomes in the epidermal layers was determined using TEM. Desmosomes were observed frequently at junctions between outer epidermal cells at all stages examined ranging from stage 10 (9hpf) to stage 45 (96hpf) (Fig. 1B and Fig. S1). Desmosomes were always observed below a tight junction and could be observed between different cell types (Fig. 1B, shows two different cell types). At higher magnification, the ultrastructure of X. laevis embryonic desmosomes resembled desmosomes described in other vertebrates and a subset contained a classic dense intercellular midline (Borysenko and Revel, 1973; Garrod et al., 2005) (Fig. 1B inset). We also observed desmosomes to have connections to a filamentous network consistent with intermediate filaments (Fig. S1A). These complexes were also infrequently detected between outer and inner epidermal cells (Fig. S1B–C, black arrowheads) as well as between the inner epidermal cells (Fig. S1B–C, white arrowhead). Thus, desmosome ultrastructure, position in the cell and the connections they make in the developing frog embryo are remarkably similar to what has been reported in mammals.
1b). Desmoplakin is a conserved protein that is expressed in the epidermis of the developing X. laevis embryo
Desmoplakin (Dsp) is unique to the desmosome and the primary connection to both the cytoskeleton and the desmosomal cadherins. Therefore, this protein serves as a good proxy to understand the development and function of desmosomes in the X. laevis embryo. To support this idea, dsp is expressed throughout development as assessed by RT-PCR (Fig. S2 A) which is consistent with published RNAseq data (Session et al., 2016). Sequence alignments revealed that the X. laevis Dsp mRNA and protein are also highly similar to human and mouse (Table 1). For example, the X. laevis Dsp protein is 82% similar to the human Dsp protein. Additionally, the frog Dsp protein has all the same functional domains as the human Dsp protein unlike zebrafish (Table 1). Taken together, this data supports the idea that X. laevis Dsp is a useful protein to examine and perturb to study the role of desmosomes during epidermal development.
Table 1.
Comparative analysis of X. laevis Dsp.L desmoplakin mRNA and protein sequences with human, mouse, and zebrafish. % Similarity is provided. Sequences not present or available is indicated by an asterisk. Xenbase was used to access genome builds for X. laevis.
| Species | Source | Protein Domains | Whole Protein | mRNA | ||||
|---|---|---|---|---|---|---|---|---|
| PLAKIN | ROD | A | B | C | ||||
| H. sapiens | NP_004406 | 80.5 | 81.8 | 86.9 | 93.0 | 91.4 | 82.2 | 65.6 |
| M. musculus | NP_076331 | 80.0 | 81.9 | 87.4 | 93.4 | 91.4 | 82.1 | 63.7 |
| D. rerio | XP_001919901 | 70.1 | 61.4 | * | 87.3 | 81.1 | 59.7 | 50.8 |
Immunofluorescence revealed Dsp protein in the outer ectoderm and epidermis, from gastrulation to early tadpole stages (Fig. 1 C–E‴, Fig. S2B–E‴). Beta-catenin and Dapi were used as a counterstain in our imaging of Dsp to better visualize the developing epidermis. During gastrulation, Dsp protein was localized in a discontinuous pattern at the cell periphery (Fig. S2 B–B‴). At stages 19–20 (20–22 hpf), when morphogenesis and differentiation of the epidermis begins, we noted that Dsp was not equal in intensity in all outer epidermal cells (Fig. 1C–C‴). For example, some cells with small apical surface areas appeared to be enriched with Dsp (Fig. 1 C–C‴, arrowheads, Fig. 1 D–D‴, suppl. movie 1). Such cells with small apical surfaces are consistent with newly emerged intercalated cells. At the vertices of 4–5 cells, where cells were in the process of inserting in the outer ectoderm, Dsp levels appeared lower (Fig. 1 C–C‴, arrows, Fig. 1 E–E‴ and suppl. movie 2). Later, at stages 24–25 (24–27 hpf) and 32–33 (40–45 hpf), Dsp appeared to be more evenly distributed in the outer epidermal cells (Fig. S2 D–E‴) and could be visualized in a punctuate pattern in the inner epidermal cells at lower intensities (Fig. S2 F–G‴, arrows).
These results demonstrate that Dsp was present in cells of the developing epidermis and differences in localization are evident in cells undergoing radial intercalation.
2. Morpholinos and Crispr/Cas9 mediated depletion of Dsp reveal its importance in X. laevis embryonic development
We demonstrated that desmosomes and the desmosomal component, desmoplakin (Dsp), are present in the epidermis of the embryo. To better determine the specific functions of desmosomes and Dsp during epidermal development and morphogenesis morpholino (MO) and Crispr/Cas9 technologies were utilized to deplete Dsp.
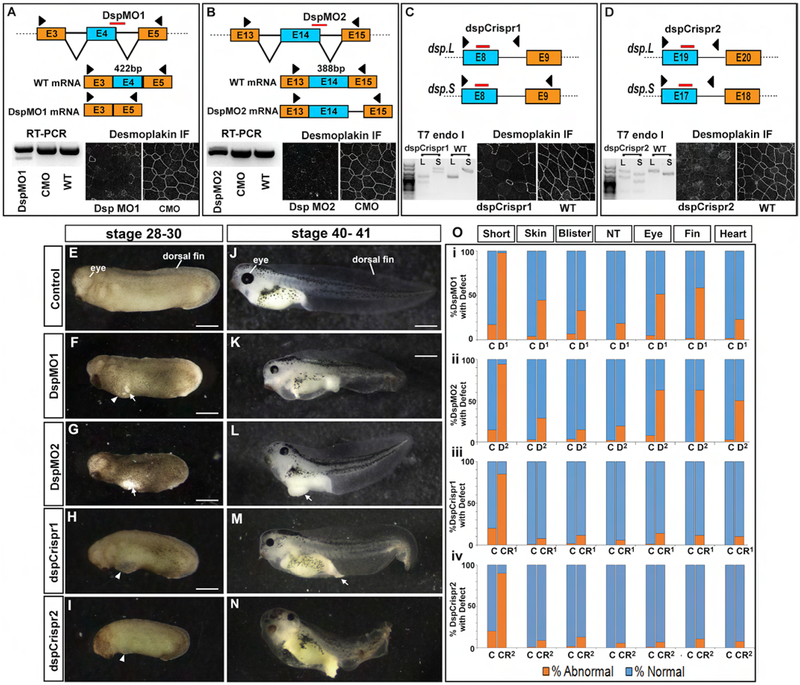
2a). Morpholinos and Crispr/Cas9 technologies result in Dsp reduction.
Since MOs are titratable, these can be utilized to examine the effects of decreased dosage of Dsp in the embryo. This is especially useful when a complete knockout might be lethal as Dsp is in the mouse (Gallicano et al., 1998). Two antisense splice-blocking MOs targeted to the splice donor sites of exon 4 (DspMO1) and exon 14 (DspMO2) (Fig. 2A, B) were utilized. Various amounts of MO were tested and we determined that 34ng/embryo DspMO1 and 17ng/embryo of DspMO2 caused similar morphant phenotypes. To determine the efficacy of these concentrations of morpholinos, a subset of the morphants were analyzed for splicing defects using RT-PCR as well as Dsp protein reduction using immunofluorescence. Results revealed that indeed DspMO1 and DspMO2 morphants had Dsp splicing defects (100%, n=10, 2 experiments, Fig. 2 A, B) and a reduction in Dsp protein (100%, n=10, 2 experiments, Fig. 2 A, B). To further explore whether decreased Dsp resulted in changes in desmosomes we examined the epidermis of representative DspMO1 morphants by TEM. In these experiments, embryos were injected at the 1-cell stage and examined at stages ranging from 26–28 (30–35 hpf). TEM demonstrated that desmosomes were missing in the Dsp morphants (observations made from 50 different cell-cell contacts taken from 2 embryos, Fig. S3 A–D). Missing desmosomes were rarely observed in the control embryos at this stage (2 embryos, 50 images of cell-cell contacts, Fig S3 A–D).
Figure 2:
Decreased function of Dsp in the embryo.(A) Schematic showing where the DspMO1 (red bar) is predicted to bind to dsp.L mRNA as well as the predicted spliced products in Dsp morphants and controls. Black arrow heads indicate the region of primer sequences used for RT-PCR analysis. RT-PCR of dsp showing there is an alternative splice product in the DspMO1 morphants. Immunofluorescence of desmoplakin (white) in DspMO1 and CMO morphants. (B) Schematic showing where the DspMO2 (red bar) is predicted to bind to dsp.L mRNA as well as the predicted spliced products in Dsp morphants and controls. Black arrow heads indicate the region of primer sequences used for RT-PCR analysis. RT-PCR of dsp showing there is an alternative splice product in the DspMO1 morphants. Immunofluorescence of desmoplakin (white) in DspMO2 and CMO morphants. (C) Schematic showing where the dspCrispr1 (red bars) is predicted to target the dsp.L and dsp.S genes. Black arrow heads indicate the region of primer sequences used for mutation analysis. Results of a T7 endonuclease assay are shown of dspCrispr mutants showing there is an alternative product indicative of mutation. Immunofluorescence of desmoplakin (white) in dspCrispr mutants and controls. (D) Schematic showing where the dspCrispr2 (red bars) is predicted to target the dsp.L and dsp.S genes. Black arrow heads indicate the region of primer sequences used for mutation analysis. Results of a T7 endonuclease assay are shown of dspCrispr mutants showing there is an alternative product indicative of mutation. Immunofluorescence of desmoplakin (white) in dspCrispr mutants and controls. E-I) Lateral view of control, Dsp morphants and mutant embryos at stage 28–30. Anterior is to the left. Arrows indicate regions where the epidermis is broken and arrowheads point to blister like structures. Scale bars=450μm. J-N) Later view of control, Dsp morphants and mutant embryos at stage 40–41. Anterior is to the left. Arrows indicate regions where the epidermis is broken and arrowheads point to blister like structures. Scale bars=450μm. O) Analysis of phenotypic frequency in i) DspMO1 morphants (D1), ii) DspMO2 morphants (D2), iii) dspCrispr1 mutants (Cr1), dspCrispr2 mutants (Cr2) compared to controls (C). Orange indicates percentage with the defect indicated at the top. Abbreviations: WT=wildtype uninjected sibling embryos, E=exon, IF=immunofluorescence.
While morpholinos are an effective method to test the role of gene function in X. laevis, non-specific effects are possible (Blum et al., 2015; Eisen and Smith, 2008; Heasman, 2002). Attempts to create a rescue construct failed and therefore, to ensure that the phenotypes produced by the Dsp MOs are specific to Dsp reduction, we also utilized the CRISPR/Cas9 system to mutate dsp (Bhattacharya et al., 2015; Nakayama et al., 2013; Wang et al., 2015). Two different mosaic F0 dsp CRISPR mutants were created where some cells contained the mutation while others did not. To do this, gRNAs targeting exons of the dsp gene (dspCrispr1, Fig. 2C, and dspCrispr2, Fig. 2D) were injected with Cas9 protein into embryos. To determine the efficacy of this technique, a subset of injected embryos (displaying a phenotype) were analyzed for mutations using T7 endonuclease assay. Results revealed that indeed prospective dspCrispr1 and dspCrispr2 mutant embryos were positive for regions of non-perfectly matched DNA as predicted (100%, n=12, 2 experiments, Fig. 2 C, D). To test whether these mutations resulted in a reduction in Dsp protein, immunofluorescence was utilized. dspCrispr1 and dspCrispr2 mutant embryos had regions where cells displayed a clear reduction in Dsp protein (100%, n=12, 2 experiments, Fig. 2C, D).
In summary, these results together demonstrate that morpholino and Crispr/Cas9 methods caused splicing defects and mutations respectively, which in turn effectively reduced Dsp protein in developing embryos.
2c). Decreased Dsp using morpholinos or Crisprs results in identical developmental abnormalities in whole embryos.
The defects observed in the DspMO morphants and the dspCrispr F0 mutants were remarkably similar (Fig. 2E–N). Various phenotypes were analyzed in a subset of injected embryos. At tailbud stages (st. 24–28) both the Dsp morphant and mutant embryos were smaller and had epidermal tears, hyperpigmentation, ventral blisters and neural tube closure defects (Fig. 2 E–I, O, suppl. table 3,4). Later, at larval stages (st. 40–41), many morphants and some mutant embryos were more likely to have craniofacial defects, malformed eye as well as heart edema and ruffled tail fins (Fig. 2 J–N, O, suppl. table 3,4). The percentage of embryos, (at both stages) with these phenotypes was lower in the dspCrispr injected embryos (Fig. 2O). It is possible that the mosaic F0 dspCripsr mutants have such reduced defects because the phenotypes are caused by a large region of epidermal depletion of Dsp in the morphants. The interspersion of normal cells in dspCrispr mutants may prevent the epidermal tearing for example.
In summary, these results suggest that depletion of desmoplakin is required for embryonic growth, epidermal development and for the formation of the face, heart, and fin. Importantly, the striking similarities in the Dsp morphants and dspCrispr mutants suggest that these abnormal phenotypes are specifically caused by decreased in Dsp.
2d). Dsp is specifically required for the formation of the embryonic mouth, eye, and fin.
Ectodermally derived structures that showed abnormalities in our morphant and mutant analysis included the embryonic mouth, eye/lens, and fin. Therefore, we next sought to determine whether Dsp is specifically required in the morphogenesis of these structures. We targeted Dsp morpholinos to these structures using blastomere injections to rule out secondary effects from gastrulation or heart defects. DspMO1 was injected into a dorsal blastomere (D1.2; Xenbase) at the 16 cell stage that fate maps to the face as well as the eye (Fig. S4 A). In these embryos, the injected side of the embryo showed a failure of the embryonic mouth to form in 100% of the embryos (n=20, 2 experiments; Fig. S4 B–C”). In addition, eye defects including a protruding lens were also observed in 90% of the DspMO1 morphants (n=20, 2 experiments; Fig S4 D–F”). In embryos where a ventral blastomere (V.2.2, Xenbase) at the 16 cell stage was targeted, the fin was labeled (Fig. S4 G). In these partial morphants the fin was ruffled, lacking the smooth appearance of the controls (n=23, 2 experiments; Fig. S4 H–I”). These results demonstrate the requirement of Dsp for mouth, eye and fin development. Importantly, these results also confirm the specificity of the phenotypes.
3. Dsp is specifically required for epidermal integrity and resilience.
Since desmosomes are well known for their role in providing mechanical resilience to the epidermis in mammals we next tested whether this was also true in X. laevis embryos. Quantitative assays were developed to test whether the epidermis is more susceptible to mechanical insults when depleted of Dsp. Further, the effect of specific loss of Dsp in the epidermis was assessed.
3a). Dsp morphants are more susceptible to mechanical stress
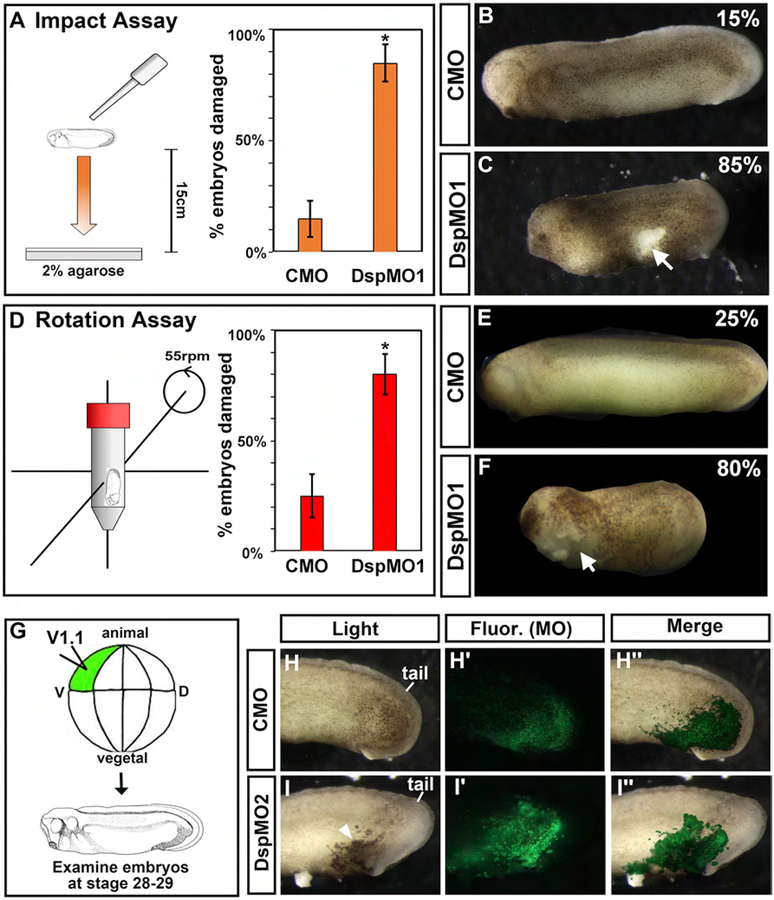
One explanation for increased epidermal tears in the Dsp morphant and mutant embryos is that they are more fragile and thus less resistant to mechanical perturbations. Here, we developed two methods to quantify the effects of mechanical insults to the epidermis of the Dsp morphants.
In the “Dropping Assay”, embryos were dropped from a height of 15 cm into a dish lined with agarose (Fig. 3A and suppl. movie 3). We expected that this assay could cause shear and impact stresses as the embryo fell and contacted the agarose. Results demonstrated that indeed significantly more DspMO1 morphants (85%) had damage to the epidermis compared to controls (15%) (Chi-squared test, p=9.55E-06; Fig. 3 A–C).
Figure 3:
Decreased Dsp results in less mechanical resilience and is specific to the epidermis. A) Schematic of experimental design for the Dropping Assay. Bar graphs summarizing quantification of the proportion of CMO and DspMO1 morphants with a ruptured epidermis after undergoing the Dropping Assay.*=statistical significance using Chi-Squared test. B-C) Lateral views of a representative control and Dsp morphant at stage 28–30. Anterior is to the left. Arrow indicates regions where the epidermis is broken and arrowheads point to blister like structures. Scale bars=450μm. D) Schematic of experimental design for the Rotation Assay. Bar graphs summarizing quantification of the proportion of CMO and DspMO1 morphants with a ruptured epidermis after undergoing the Rotation Assay. *=statistical significance using Chi-Squared test. E-F) Lateral views of a representative control and Dsp morphant at stage 28–30. Anterior is to the left. Arrow indicates regions where the epidermis is broken and arrowheads point to blister like structures. Scale bars=450μm. G) Schematic of epidermal target injections. H-I‴) Representative lateral views of the posterior of embryos as in G showing the embryo under light (Light), fluorescence (Fluor. MO) to visualize the morpholino alone and merged (Merge). Scale bars=450μm. H-H”) Control MO (CMO) I-I”) DspMO1 morphants.
In the “Rotational Assay”, embryos were placed in a buffer-containing 50 ml conical tube that underwent rotations in a vertical rotating mixer (Fig. 3D and suppl. movie 4). The embryos likely experienced multiple mechanical stresses as they contacted the air/media interface as well as the sides of the tube. Results demonstrated that again significantly more DspMO1 morphants (80%) had damage to the epidermis compared to controls (25%) (Chi-squared test, p=0.000478; Fig. 3 D–F).
These assays demonstrate that the epidermis of DspMO1 morphants has quantitatively decreased resistance to mechanical stress such as impact, shear and/or tensional forces.
3b). Dsp targeted to the trunk epidermis reveals a specific requirement for integrity in this tissue.
We next tested whether the epidermal integrity defects caused by the Dsp MO were due to a specific requirement for Dsp in the epidermis. It is possible that the epidermal integrity defects observed in Dsp morphants and mutants are due to secondary effects generated from abnormal gastrulation or smaller size of the embryo. To circumvent such non-specific possibilities, targeted injections of fluorescently-tagged DspMO1 were performed. This allowed us to investigate the effects of depleting a subset of epidermal cells of the Dsp protein in the context of a relatively normal embryo. DspMO1 or CMO was injected into a ventrally located blastomere fated to become the trunk epidermis (D1.1; Xenbase, Fig 3G). Results revealed that epidermal regions containing DspMO1 formed tears or ruptures in 100% of the embryos. Such tears were never observed in CMO injected embryos (n=30, 3 experiments, Fig 3H–I”). These results reveal that indeed epidermal integrity is due to Dsp depletion in the epidermis and not due to defects in other tissues.
4. Epidermal morphogenesis is perturbed in Dsp morphants.
We next asked whether the lack of Dsp in the epidermis specifically affected the morphogenesis and differentiation of this tissue. The X. laevis outer epidermal layer is comprised of several differentiated cell types including Multiciliated Cells (MCCs) and Small Secretory Cells (SSCs). These cells are born in the inner epidermis and then move to their final location in the outer epidermis by radial intercalation. During this process, the cell inserts itself at the vertex of 3–5 cells and then the apical surface expands. A combination of histology, immunofluorescence and cell tracking was utilized to assess the development of the epidermis including radially intercalating cells.
4a). Decreased Dsp results in abnormal morphology of epidermal cells.
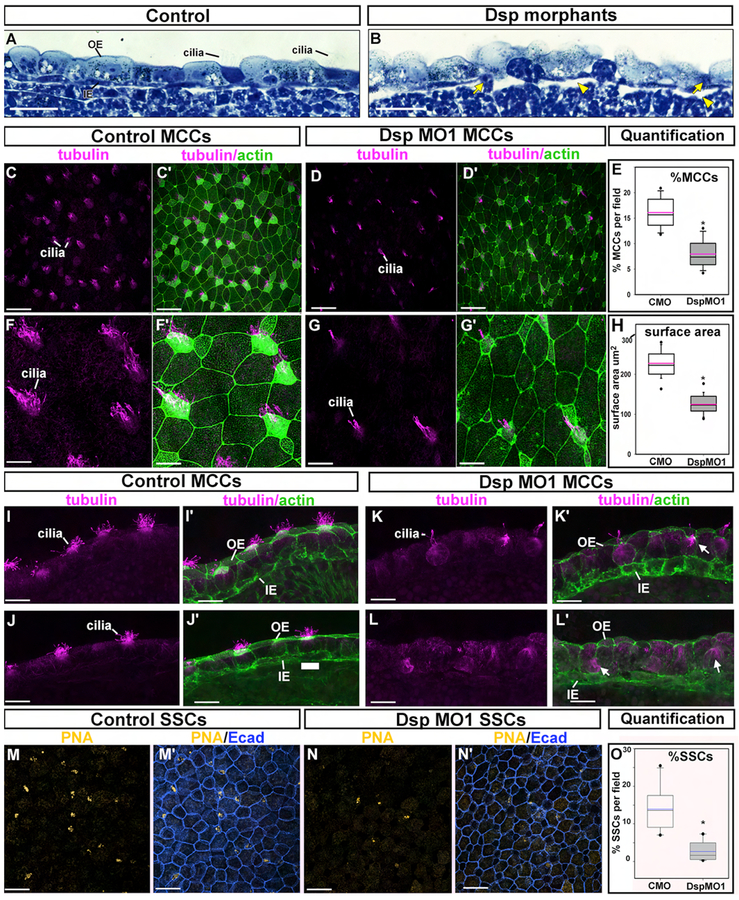
General epidermal morphology was initially assessed in histological sections at stage 26–28 (hpf) when MCC cells are present in the outer epidermis. In the control embryos, the epidermis was comprised of an outer layer of cuboidal-like cells showing various morphologies. For example, darker stained multiciliated cells (possibly MCCs) were interspersed between the larger lighter stained cells (Fig. 4 A, B). The inner epidermis was comprised of flat, oblong cells that were stained dark blue (Fig. 4 A, white arrow). A reduction of Dsp resulted in regions where the outer epidermal cells were less organized (Fig. 4 B). For example, the large lighter stained cells were more globular in nature than the controls. The darker stained cells appeared “sunken” compared to the surrounding lighter stained cells (Fig. 4 B, yellow arrows). Furthermore, multiple tufts of cilia were rarely observed on the surfaces of any cell. Also in Dsp morphants there appeared to be regions where there was more extracellular space underlying the inner epidermal cell layer (Fig. 4 B, arrowheads). These observations demonstrate that a reduction in Dsp results in morphological perturbations in the embryonic epidermis.
Figure 4:
Decreased Desmoplakin results in abnormal epidermal morphology, MCC and SSC development. A-B) Histological sections of the epidermis in control (A) and DspMO1 morphants (B). Yellow arrowheads indicate space underlying the epidermis. Scale bars= 45μm. C-C’) Representative control trunk epidermis labeled with tubulin (C), phalloidin and tubulin (C’). Scale bars = 50μm. D-D’) Representative DspMO1 trunk epidermis labeled with tubulin (D), phalloidin and tubulin (D’). Scale bars = 50μm. E) Quantification of the percentage of tubulin positive cells in control compared to DspMO1 morphants. *=statistical significance. F-F’) Representative control trunk epidermis labeled with tubulin (F), phalloidin and tubulin (F’). 2× magnified region of C and C’. Scale bars =11μm. G-G’) Representative DspMO1 trunk epidermis labeled with tubulin (G), phalloidin and tubulin (G’). 2× magnified region of D and D’. Scale bar =11μm. H) Quantification of the surface area of tubulin positive cells in control compared to DspMO1 morphants. *=statistical significance. I-J’) Sections of two representative control trunk epidermis labeled with tubulin (I,J), phalloidin and tubulin (I’,J’). Scale bars = 22μm. K-L’) Sections of two representative DspMO1 trunk epidermis labeled with tubulin (J), phalloidin and tubulin (G’). Scale bar = 22μm. M-M’) Representative control trunk epidermis labeled with PNA (M), PNA and E-cadherin (M’). Scale bar =50μm. N-N’) Representative DspMO1 trunk epidermis labeled with PNA (N), PNA and E-cadherin (N). Scale bar =50μm. O) Quantification of the percentage of PNA positive cells in control compared to DspMO1 morphants. Blue line represents the mean. *=statistical significance. Abbreviations; PNA-peanut agglutinin, MCC=multiciliated cell, SSC=small secretory cell, OE=outer ectoderm, IE=inner ectoderm.
4b). Decreased Dsp results in defects in MCC number and morphology.
Our histological observations revealed epidermal defects that specifically included MCCs. These cells form multiple cilia on their apical surface that are identified here using alpha-tubulin labeling. Results demonstrated a 2.8 fold decrease in the relative number of alpha-tubulin positive patches on the surface of the epidermis of Dsp MO1 compared to controls (n=30, 3 experiments, p<0.0001, Fig. 4 C–E).
In addition to a reduction in ciliated cells, it was also noted that Dsp MO1 morphants had a reduced MCC cell surface often accompanied by fewer cilia (Fig. 4 F–G’). Therefore, the surface area of a subset of the cells was measured using the phalloidin counterstain that marked the intense F-actin at the cell perimeter. Results indicated that indeed the DspMO1 morphants had an average 1.8 fold smaller MCC surface area when compared to the same cells in the controls (n=20, p<0.0001, Fig. 4 F–H).
We next characterized the MCCs further in transverse sections of the epidermis labeled with alpha-tubulin and phalloidin. In control embryos, tubulin positive cilia were evident at the apical surfaces of cells of the outer ectoderm (Fig. 4 I–J’). In the DspMO1 morphants where ciliated cells were observed, there were fewer cilia and the apical surfaces of these cells seemed to be sunken below surrounding cells (Fig. 4 K,K’, arrows). In epidermal regions that seemed more severely affected, there was a less organized appearance of the inner and outer ectoderm (Fig. 4 L,L’). Tubulin enriched within the cell the body consistent with radially intercalating cells were observed at the basal region of the outer ectoderm (Fig. 4 L,L’ arrow). These observations are consistent with the histological observations. Such results suggest that intercalating cells are present but fail to fully integrate with the outer epidermis.
To determine if the decreased numbers of MCCs was specific to the reduction in Dsp in the epidermis or whether it could be a more general non-specific effect, targeted injections to the trunk epidermis were performed as described above (Fig. S5 A). In control embryos, a uniform pattern of tubulin positive MCCs was apparent (Fig. S5 B). However, tubulin-positive cells were decreased in the DspMO1 positive regions (Fig. S5 C, D) but not in adjacent regions with little or no morpholino present (Fig. S4C, D). These results suggest that indeed the loss of Dsp in the epidermis itself explains why there are defects in ciliated epidermal cells. In addition, in these targeted injected embryos the epidermis was examined at stage 32 (40 hpf) and therefore any reduction we observed at earlier stages is not likely due to a delay in MCC development.
To determine whether other cells in the outer epidermis were also affected in DspMO morphants we next examined Small Secretory Cells (SSCs). These cells are also born in the inner epidermal layer and migrate to the outer epidermis by radial intercalation similar to the MCCs. SSCs were examined utilizing the marker peanut agglutinin (PNA) which is enriched in the secretory vesicles in these cells. Results revealed a 5.79 fold fewer cells with clear PNA positive accumulations in the epidermis of DspMO1 morphants compared to controls (n=20, p<0.0001, Fig. 5 M–O). Together, these results suggest that Dsp is required for the development of at least two differentiated cell types in the epidermis.
Figure 5:
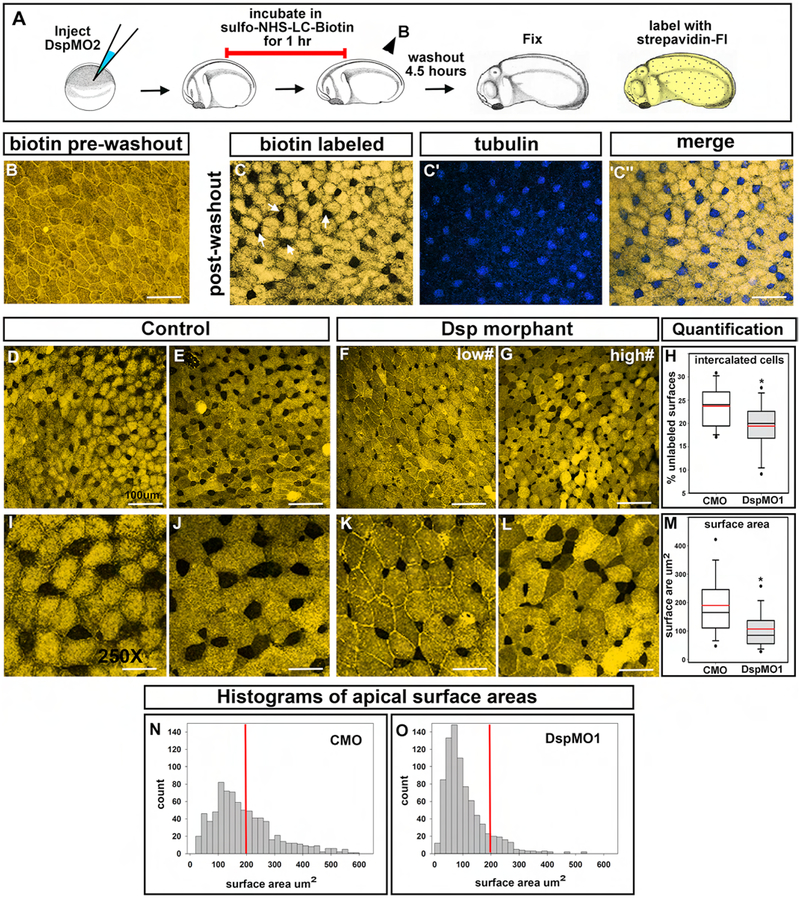
Tracking the epidermis. A) Schematic of outer epidermis labeling experiment. B) Representative image of trunk epidermis labeled with biotin directly after labeling without a washout period. Scale bar= 40μm. C-C”) Representative image of trunk epidermis double labeled with biotin (C) and tubulin (blue, C’) and merge (C”). Scale bar= 40μm. D-E) Two representative images of trunk epidermis from a control embryos after labeling with biotin. Scale bars=45μm. F-G) Two representative images of trunk epidermis from a DspMO1 morphant embryos after labeling with biotin. Scale bars =45μm. H) Quantification of the percentage of unlabeled cells in DspMO1 morphants compared to controls. Red line represents the mean. *= statistical significance. I-J) Two representative images of trunk epidermis from a control embryos after labeling with biotin at higher magnification. Scale bars=20μm. K-L) Two representative images of trunk epidermis from a DspMO1 morphant embryos after labeling with biotin at high magnification. Scale bars =20μm. M) Quantification of the relative change in surface area of unlabeled cells in DspMO1 morphants compared to controls. Red line represents the mean. *= statistical significance. N-O) Histogram analysis of surface areas of unlabeled cells in control (N) and DspMO1 (O). The red line indicates the surface area of 200μm2 for comparisons.
4c). Decreased Dsp results in defects in radial intercalation in the epidermis.
Since both MCCs and SCCs develop by a process of radial intercalation we next tested whether this process was perturbed in the Dsp morphants. To track radial intercalation, the epidermis was pulse labeled with biotin that adheres to the membranes of the outer ectodermal cells. Such labeling was performed from st.19 (21hpf) until st.20 (22hpf) during the first period of radial intercalation of primarily MCCs (Fig. 5 A). Embryos that were fixed and labeled directly after this treatment demonstrated complete and relatively uniform biotin labeling of the epidermis (Fig. 5 B). Biotin incubation was then followed by a 4 hour washout to allow unlabeled inner epidermal cells to emerge into the outer layer. Control embryos fixed after this washout had many biotin-negative regions (Fig. 5 C, arrows) that were interspersed among biotin positive cells (Fig 5 C, yellow labeling). The biotin-negative regions represented the cells that radially intercalated during the washout period. In the control embryos, the majority of such unlabeled biotin cells were also positive for alpha-tubulin, confirming that these cells indeed originated in the inner epidermal layer (Fig. 5 C’–C”). Surprisingly, the DspMO1 morphants had only a small reduction in the average number of unlabeled cells that was marginally significant (p=0.0475, n= 11, 3 experiments; Fig. 5 D–H). The Box plot indicates a large amount of variability, where a subset of morphant embryos had the same or even more unlabeled cells than a subset of the controls (Fig. 5 H). However, we also noted that in these embryos, especially those with high numbers of unlabeled cells, the size of the apical surfaces of such cells appeared smaller (Fig 5 I–L). Thus, the surface area of the biotin-negative cells was quantified in the same images. Results revealed that indeed there was a statistically significant reduction in the average surface area of biotin-negative cells in the DspMO1 morphants when compared to controls (Mann Whitney, p=<0.0001, n=11, 3 experiments; Fig. 5 M). A histogram analysis revealed that in controls the unlabeled surface areas followed a binomial distribution with a large proportion of cells greater than 100 μm2 (Fig 5N, see yellow line). On the other hand, in the Dsp MO1 morphants, a larger proportion of the unlabeled surface areas were less than 100 μm2 (Fig. 5 O, see yellow line). These results reveal that DspMO1 morphants had more intercalating cells that had smaller apical surfaces. This result is consistent with the smaller apical surfaces of MCCs quantified in the previous section.
Taken together, our results reveal that Dsp is required for the process of apical expansion.
5. Decreased Dsp alters the cytoskeleton and the adherens junction in the outer epidermis.
Integrity and morphogenesis of the epidermis of the Xenopus embryo depends on a functioning cytoskeleton and adherens junctions. To begin to understand possible mechanisms underlying the embryonic defects in embryos with reduced Dsp we characterized type II keratins, alpha-tubulin, F-actin and E-cadherin. Immunofluorescence was used to examine the distribution of these cellular components in Dsp morphants. In each experiment at least 20 embryos were examined in two biological replicates.
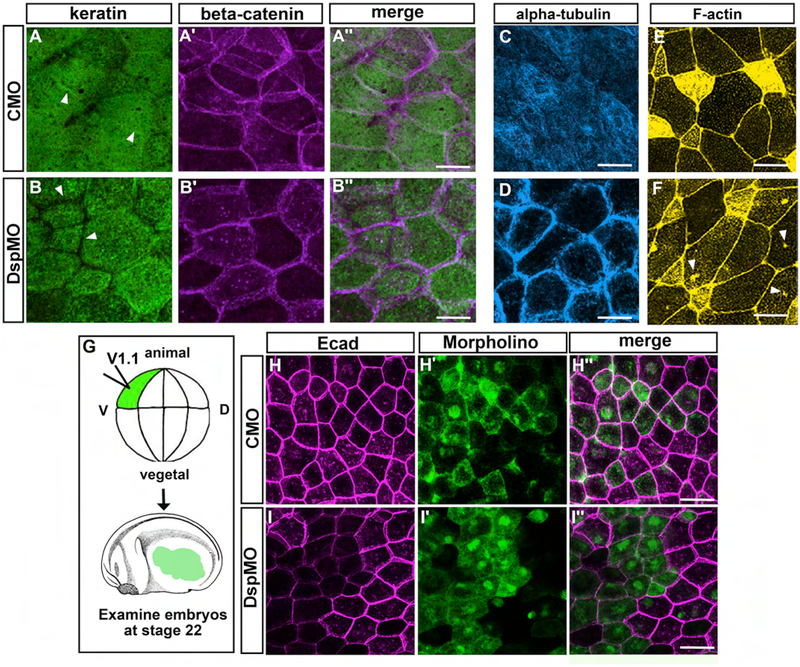
In control embryos (st. 30–31, 72–76 hpf), epidermal keratin appeared as a uniform network that extended to the periphery where junctional beta-catenin labeling was observed (Fig. 6A–A”). However, in Dsp morphants, the keratin labeling did not extend to the periphery (Fig. 6B–B”, arrowheads). This defect in keratin localization was also observed in the Crispr mutants and in DspMO2 morphants (Fig. S6).
Figure 6:
A-B”) Representative control and DspMO1 morphant trunk epidermis labeled with keratin (A,B; green), beta-catenin (A’,B’; pink) and merge (A”,B”). Scale bars = 25μm. Beta-catenin labeling (pink) was used to provide a marker of the cortical region of the cell. C,D) alpha-tubulin labeling of cells from control (C) and DspMO1 morphants (D). Scale bars =25 μm. E,F) Phalloidin labeling of F-actin from control (C) and DspMo1 morphants (D). Scale bars =25 μm. G) Schematic of the experimental design for H-I”. H-H”) Representative control trunk epidermis labeled with E-cadherin (J), showing the fluorescein labeled MO (H’) and merge (H”). Scale bar =25μm. K-K”) Representative control trunk epidermis labeled with E-cadherin (I), showing the Fluorescein labeled MO (I’) and merge (I”). Scale bar =25μm. Abbreviations= Ecad=E-cadherin.
Our next goal was to examine cytoskeletal microtubules, specifically alpha-tubulin. Since this protein is enriched in cilia and its labeling can obscure imaging of the cell body we examined alpha-tubulin at st. 19–20, prior to MCC development. Similar to keratin, alpha-tubulin also appeared as a uniform network in the outer epidermal cells at this stage in controls (CMO, Fig. 6 D). However, in the DspMO1 morphants, the alpha-tubulin appeared concentrated at the cell periphery (Fig. 6 D).
F-actin appeared to be concentrated at the cortex of the all epidermal cells of both the control and DspMO1 morphants at st. 30–31 (72–76 hpf) (Fig. 6 E, F). In addition, F-actin was enriched at the apical surface of MCCs (Fig. 6 E, F) consistent with previous reports (Park et al., 2006; Park et al., 2008; Sedzinski et al., 2016, 2017). Such an enrichment appears decreased in the DspMO1 morphants (Fig. 6 F). There is also a punctate appearance of F-actin in other cells of the epidermis of both controls and morphants (Fig 6 E, F). Further, in the morphants, there were additional and larger puncta of F-actin within the outer epidermal cells (Fig. 6 F, arrowheads) that was not present in the controls (CMO).
Since epidermal integrity was so severely affected in Dsp morphants we also asked whether other junctions could also be defective. While the EM images revealed intact tight junctions in Dsp morphants, it was more difficult to assess the adherens junctions. Therefore, a major adherens junction component, E-cadherin, was examined in Dsp morphants at st. 24 (25–26hpf). However, we could not discern major differences in the localization of E-cadherin labeling when DspMO1 was injected into whole embryos. We did note however that the labeling of E-cadherin appeared to be decreased in the morphants (not shown). To better show this we performed a mosaic analysis where we injected morpholinos into only one blastomere fated to become trunk epidermis (V1.1, 16 cell stage, Fig. 6 G). In these experiments, morphant and normal cells could be directly compared side by side in the same embryo. Control morpholinos targeted in this way did not result in any noticeable change in E-cadherin labeling when control cells were compared to MO-positive cells (Fig 6H–H”). However, a clear reduction in E-cadherin was observed in targeted Dsp morphant cells compared to their morpholino-free counterparts in the same embryo (Fig. 6 I–I”).
Together these results suggest that the cytoskeletal elements and adherens junctions were altered in outer epidermal cells of embryos lacking adequate amounts of Dsp. Such changes could have profound effects on epidermal integrity and morphogenesis.
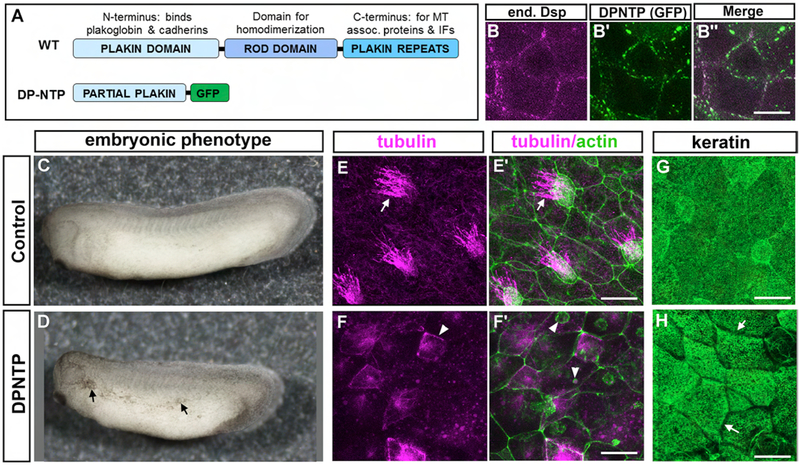
5. Expression of a mutant Dsp in X. laevis results in decreased MCCs and cytoskeletal defects
The localization of both microtubules and keratin were perturbed in embryos deficient in Dsp. Dsp interacts directly with both these cytoskeletal elements at its C-terminus. To test whether these interactions specifically were integral to the phenotypes we observed we next employed a mutant form of the human Dsp (DP-NTP). This construct is missing the C terminal domains of Dsp which impairs interactions with microtubules and keratin. But, the N-terminus is present in DP-NTP and therefore desmosomal cadherin associations and cell-cell adhesions are presumably maintained (Fig. 7A). In X. laevis outer epidermis, we confirmed that DP-NTP displayed membrane association and partially replaced endogenous Dsp (Fig. 7 B–B”). Embryos expressing DP-NTP were smaller in size and displayed hyperpigmentation similar to Dsp morphants and mutants (Fig. 7 C, D). These results corroborate our other loss of function techniques suggesting the phenotypes caused by the morpholinos effects and Crispr induced mutations are specific. DP-NTP expressing embryos also displayed a reduction in MCCs (Fig. 7E, F’, arrows indicate MCC cilia), had cells with increased cortically localized alpha-tubulin (Fig 7 F arrowhead) and large abnormal puncta of F-actin (fig 7 F’, arrowheads). In addition, keratin was reduced in the cortical regions of cells expressing DP-NTP similar to the Dsp morphants and mutants (Fig. 7G, H, arrows). These results mimicked our cellular observations of the DspMO1 morphants and dsp mutants again confirming specificity. Importantly, these results suggest that the loss of Dsp’s effect on the cellular changes and MCC development and could primarily be due to its effect on the changes to the cytoskeleton.
Figure 7:
DP-NTP expression in X. laevis embryos. A) Schematic of the major domains of desmoplakin in wildtype (WT) and DP-NTP contruct. B) Desmoplakin labeled with an antibody B’) DP-NTP-GFP expression B”) Merge of B and B’. C,D) Lateral views of representative control (C) and DP-NTP expressing embryos. E-E’) Trunk epidermis of a representative control embryo, labeled with tubulin (pink, E) and phalloidin (green) merged with tubulin (E’). F-F’) Trunk epidermis of a representative DP-NTP expressing embryo, labeled with tubulin (pink, F) and phalloidin (green) merged with tubulin (F’). G,H). Keratin localization in the trunk epidermis of a representative control (G) DP-NTP expressing embryo (H). Scale bars =25 μm.
Discussion
The functions of desmoplakin are conserved during X. laevis development
Desmosomes are especially distinct by TEM imaging, where they appear as electron-dense structures consisting of two identical cytoplasmic plaques and a central core that spans the intercellular space (reviewed in (Berika and Garrod, 2014; Garrod and Chidgey, 2008)). This ultrastructure is observed in all vertebrates examined to date including X. laevis embryos (Borysenko and Revel, 1973; Farquhar and Palade, 1965; Fleming et al., 1991; Garrod and Chidgey, 2008; Goonesinghe et al., 2012; Holbrook and Odland, 1975). Desmoplakin, a unique and critical protein member of the desmosome, is also highly similar in both sequence and composition of the functional domains in vertebrates. X. laevis desmoplakin is no exception. These structural similarities suggest the strong possibility that the desmosome and, desmoplakin, in particular, have well-conserved roles across vertebrates including X. laevis embryos. Certainly, our experiments demonstrate that desmoplakin does indeed have some conserved functions in X. laevis embryos. For example, one of the most well-known roles for desmosomes in mammals is to protect the epidermis from mechanical stress. Humans and mice with mutations in Dsp and other desmosomal genes have skin fragility (Smith et al., 2012; Whittock et al., 1999; Whittock et al., 2002). Similarly, in X. laevis embryos, a reduction in Dsp resulted in skin fragility and reduced resistance to mechanical stresses. The epidermis became damaged upon impact and shear forces more readily than controls. Thus skin fragility, possibly due to decreased cell adhesion between cells of the epidermis, is a common defect in vertebrates (including X. laevis embryos) lacking Dsp.
Another common feature of humans with desmosomal diseases is skin blistering. One possible reason for this problem is that the epidermal layers do not adhere to each other and this results in fluid-filled spaces between such layers. In agreement with this idea, separations between skin layers, specifically the basal and spinous layers were reported in mice with dsp mutations (Gallicano et al., 2001; Vasioukhin et al., 2001). In X. laevis, we also observed similar spaces between embryonic tissue layers and a percentage of Dsp morphants and mutants also had blister-like formations. Together, these data demonstrate a role for desmosomes in maintaining overall epidermal connectivity not just within a skin layer but also between different layers in vertebrates, X. laevis embryos included.
Decreased Dsp also resulted in overall similar developmental defects in Xenopus when compared to other vertebrates. For example, Xenopus Dsp morphants and mutants were shorter and had smaller heads consistent with convergent extension defects during gastrulation and early development. Consistently, zebrafish injected with translation blocking morpholinos targeting desmosomal cadherins have convergent extension defects (Goonesinghe et al., 2012). Dsp knockout mice did proceed through gastrulation but were extremely small with both head and neural defects characteristic of convergent extension problems (Gallicano et al., 2001). Combining splice-blocking morpholinos with targeted blastomere injections has allowed us to investigate later roles of Dsp without large scale interruption of gastrulation. In doing so we were able to observe defects in ectodermally derived structures such as the mouth, eye, and fin that develop much later in development.
The current study shows that the fundamental roles of Dsp and the developmental effects when this protein is depleted are similar in X. laevis and other vertebrates. Therefore, X. laevis could prove to be a valuable tool to begin to uncover the mechanisms by which Dsp is involved in morphogenesis which in turn could help explain why particular defects occur in humans with desmosomal defects.
Dsp is required for the apical expansion of radially intercalating epidermal cells
A major morphogenetic process in the epidermis of X. laevis is radial intercalation. During this process, the inner cell layer migrates to a position in the outer cell layer and then expands its apical surface. Here we have uncovered a requirement for Dsp in the apical expansion of radially intercalating multiciliated cells (MCCs). First, newly emerged cells appear to be enriched in Dsp apically suggesting it is required during apical expansion. Furthermore, pulse labeling the outer epidermis of Dsp morphants just as MCCs are intercalating resulted in emerging cells with smaller apical surface areas. Correlated to the decrease in apical surface size and a reduction in intercalating cells, was a decrease in the number of MCCs. The MCCs that did differentiate had smaller surface areas and appeared to have fewer cilia. These results suggest that Dsp is predominantly required for the apical expansion aspect of MCC radial intercalation. Since another intercalating cell type, SSCs, are also decreased in Dsp morphants, Dsp may be more generally required in the apical expansion of radially intercalating epidermal cells.
It is possible that Dsp does not directly regulate the apical expansion of intercalating cells and rather it is the lack of adhesion between cells that causes the defects in this process. However, the fact that MCCs are decreased in embryos expressing DP-NTP suggests that Dsp may have a more direct role. DP-NTP has been shown to maintain the cell-cell adhesion function of desmoplakin but lacks association with the cytoskeleton and associated intercellular proteins. Therefore, apical expansion of MCCs and other epidermal cells could more likely depend upon the Dsp interaction with intercellular structural components such as keratin, microtubules, actin, and cadherin.
Dsp is required for cytoskeletal organization during radial intercalation.
In the epidermis, Dsp can connect to the cytoskeletal network through direct interaction with keratins. As a result, mutations in the dsp gene is associated with reduced keratin attachment to the desmosome (Favre et al., 2018). In mice, where DSP has been specifically deleted in the epidermis, the keratin network does not extend to the periphery of the cells (Vasioukhin et al., 2001). A similar change in the distribution of keratin was also observed in desmosomal deficient cells in vitro (Vasioukhin et al., 2001). We too observed a reduction of keratin at the epidermal cell-cell boundaries in X. laevis Dsp morphant embryos. This alteration of keratin distribution could be expected to have dramatic effects during development especially since keratins are important for cell morphology and migration (Nishimura et al., 2018; Seltmann et al., 2013). For example, loss of keratin in the embryo results in misdirected lamellipodial protrusions and disorganized actin networks that translate into defects in tissue level collective migration (Sonavane et al., 2017). Additionally, keratins convey stiffness to cells important for cell shape changes (Ramms et al., 2013). Thus, it is possible that keratins can integrate the cell mechanics necessary for morphogenetic movements and cell shape changes (Klymkowsky et al., 1992) necessary during radial intercalation.
In addition to keratins, Dsp can also interact with microtubule associated proteins such as ninein, Lis1, Ndel1 and CLIP170. These proteins are recruited to the desmosome and are required for changes in microtubule organization (Lechler and Fuchs, 2007; Sumigray et al., 2011; Sumigray and Lechler, 2011). In the absence of Dsp in both our system and in mice results in mislocalized microtubules (Lechler and Fuchs, 2007). Microtubule rearrangements are also essential for radial intercalation of the epidermis. The microtubule associated protein CLAMP facilitates rearrangements of microtubules and the localization of polarity protein which together are necessary for PCP directed movement of inner epidermal cells (Kim et al., 2018; Werner et al., 2014). Therefore, we might speculate that Dsp facilitates microtubule organization during radial intercalation.
Loss of Dsp also affected other major components of the cell’s architecture in X. laevis, namely the adherens junctions and the actin cytoskeleton. Adherens junctions, like desmosomes, use cadherins (such as E- or N-cadherin) to bridge the extracellular space and connect cells to each other. The intercellular domain of cadherins bind to catenins which, together with other accessory proteins, link the junction to actin filaments (reviewed in (Yonemura, 2011)). In Dsp morphants, there is reduced E-cadherin associated with the adherens junctions in the developing epidermis. Furthermore, we also noted a decrease in the apical accumulation of actin in multiciliated epidermal cells. This is consistent with reports in mice where conditional knockout of Dsp in the epidermis also resulted in a decrease in adherens junctions in some skin layers as well as a disorganized actin cytoskeleton (Vasioukhin et al., 2001). In addition, mouse epithelium deficient in desmoplakin had decreased actin stability and altered microvillar structure (Sumigray and Lechler, 2012). Together these results indicate possible crosstalk between desmosomes and the adherens junction and actin cytoskeleton in the developing embryo. Such crosstalk has certainly been shown in cultured cells. For example, adherens junction associated proteins, E-cadherin and actin are both required for desmosome assembly (Lowndes et al., 2014; Shafraz et al., 2018). Further, disruption of the desmosomal protein, Desmoglein 3, decreased E-cadherin at the junction and perturbed actin organization in cultured epithelial cells (Moftah et al., 2017). Therefore, our results and the work of others demonstrate that disruption of the desmosome can affect the adherens junctions and actin network.
Since adherens junction-actin complexes direct processes such as cell shape changes and migration (Harris and Tepass, 2010; Nishimura and Takeichi, 2009) then it is not surprising that loss of Dsp results in morphogenetic defects in the embryo. In particular, actin has a well-described role in the morphogenesis of the intercalating MCCs. Rho directed apical accumulation of formin-actin has been shown to be critical for the 2D pressure required for apical emergence of intercalating cells (Sedzinski et al., 2016, 2017). While E-cadherin was not correlated with such apical emergence in MCC, a decrease in this protein could disrupt the stability of the actin network and reduce the forces required for apical expansion.
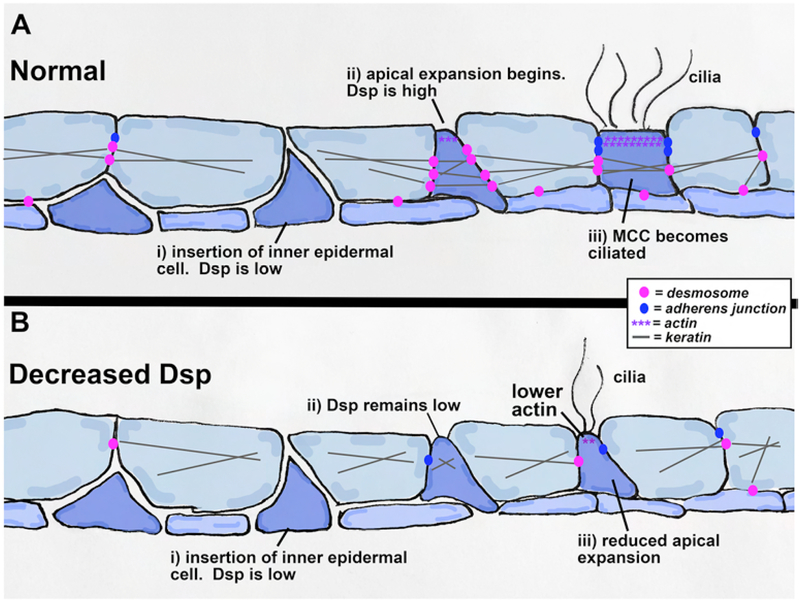
Model: Dsp is required for apical emergence during epidermal morphogenesis
Based on the data collected in this study we have developed a model of how Dsp regulates epidermal development. As a radially intercalating inner ectodermal cell inserts into the outer layer, Dsp is temporarily depleted from the vertex of 4–5 outer epidermal cells to allow for insertion (Fig. 8 Ai). Then Dsp becomes enriched in the apically emerging cell just prior to expansion (Fig. 8 Aii). At this time Dsp interacts with the cytoskeleton and adherens junctions to facilitate apical surface expansion. Cilia form on the apical surface of apically emergent cells (Fig. 8Aiii). While the precise role of Dsp in this process is unknown we predict that Dsp is necessary cell autonomously to ensure formation and/or maintenance of the actin cytoskeletal network that provides the forces needed for apical surface expansion. When the epidermis is lacking Dsp radially intercalating cells can begin to insert between outer ectodermal cells (Fig. 8Bi). However, the cells fail to expand their apical surface (Fig. 8Bii). This could be due to mislocalized keratin and microtubules as well as the reduction of are E-cadherin and actin all of which contribute to the inability to create the cell autonomous pressure needed for apical emergence.
Figure 8:
Model of how decreased Dsp affects radial intercalation of the epidermis of Xenopus. Dsp (pink) connects with keratins (grey) and is increased during apical expansion. With reduced Dsp, the intercalating cell does not have properly localized keratins or apical accumulation of actin and the apical surface does not expand sufficiently.
Supplementary Material
Acknowledgments
We would like to thank Daniel Conway (VCU, Department of Biomedical Engineering) for providing us with a modified DP-NTP construct (originally from Dr. K Green) and for his insightful contributions to the study. In addition, we would like to thank Jakub Sedzinski, Greg Walsh, James Lister, and Rita Shiang whose ideas and thoughts also helped shape this work. We thank the VCU Microscopy Facility, supported, in part, by funding from NIH-NCI Cancer Center Support Grant P30 CA016059 for helping with our TEM preparation, training, and imaging as well as the VCU Biology Microscopy Core for providing confocal use. We thank VCU undergraduate students Morgan Van Driest and Skyler Kuhn for their help with comparative bioinformatics analysis of Dsp. We especially thank Deborah Howton for helping with the frog care essential for this study. We apologize to those whose work could not be cited here due to space constraints.
Funding
This work was supported by the National Science Foundation (IOS-1349668) and National Institutes of Health (RAR065583A).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
No competing interests declared.
References
- Al-Jassar C, Knowles T, Jeeves M, Kami K, Behr E, Bikker H, Overduin M, Chidgey M, 2011. The nonlinear structure of the desmoplakin plakin domain and the effects of cardiomyopathy-linked mutations. J Mol Biol 411, 1049–1061. [DOI] [PubMed] [Google Scholar]
- Berika M, Garrod D, 2014. Desmosomal adhesion in vivo. Cell Commun Adhes 21, 65–75. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Marfo CA, Li D, Lane M, Khokha MK, 2015. CRISPR/Cas9: An inexpensive, efficient loss of function tool to screen human disease genes in Xenopus. Dev Biol 408, 196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billett FS, Gould RP, 1971. Fine structural changes in the differentiating epidermis of Xenopus laevis embryos. J Anat 108, 465–480. [PMC free article] [PubMed] [Google Scholar]
- Blum M, De Robertis EM, Wallingford JB, Niehrs C, 2015. Morpholinos: Antisense and Sensibility. Dev Cell 35, 145–149. [DOI] [PubMed] [Google Scholar]
- Borysenko JZ, Revel JP, 1973. Experimental manipulation of desmosome structure. Am J Anat 137, 403–421. [DOI] [PubMed] [Google Scholar]
- Celentano A, Mignogna MD, McCullough M, Cirillo N, 2017. Pathophysiology of the Desmo-Adhesome. J Cell Physiol 232, 496–505. [DOI] [PubMed] [Google Scholar]
- Cheng X, Den Z, Koch PJ, 2005. Desmosomal cell adhesion in mammalian development. Eur J Cell Biol 84, 215–223. [DOI] [PubMed] [Google Scholar]
- Chung MI, Peyrot SM, LeBoeuf S, Park TJ, McGary KL, Marcotte EM, Wallingford JB, 2012. RFX2 is broadly required for ciliogenesis during vertebrate development. Dev Biol 363, 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delva E, Tucker DK, Kowalczyk AP, 2009. The desmosome. Cold Spring Harb Perspect Biol 1, a002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarais AA, Moon RT, 1992. The armadillo homologs beta-catenin and plakoglobin are differentially expressed during early development of Xenopus laevis. Dev Biol 153, 337–346. [DOI] [PubMed] [Google Scholar]
- Dickinson AJ, Sive H, 2006. Development of the primary mouth in Xenopus laevis. Dev Biol 295, 700–713. [DOI] [PubMed] [Google Scholar]
- Dickinson AJ, Sive HL, 2009. The Wnt antagonists Frzb-1 and Crescent locally regulate basement membrane dissolution in the developing primary mouth. Development 136, 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale TA, Elinson RP, 1992. Cell Migration and Induction in the Development of the Surface Ectodermal Pattern of the Xenopus laevis Tadpole. Development, Growth & Differentiation 34, 51–59. [DOI] [PubMed] [Google Scholar]
- Dubaissi E, Papalopulu N, 2011. Embryonic frog epidermis: a model for the study of cell-cell interactions in the development of mucociliary disease. Dis Model Mech 4, 179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubaissi E, Rousseau K, Lea R, Soto X, Nardeosingh S, Schweickert A, Amaya E, Thornton DJ, Papalopulu N, 2014. A secretory cell type develops alongside multiciliated cells, ionocytes and goblet cells, and provides a protective, anti-infective function in the frog embryonic mucociliary epidermis. Development 141, 1514–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusek RL, Godsel LM, Green KJ, 2007. Discriminating roles of desmosomal cadherins: beyond desmosomal adhesion. J Dermatol Sci 45, 7–21. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Smith JC, 2008. Controlling morpholino experiments: don’t stop making antisense. Development 135, 1735–1743. [DOI] [PubMed] [Google Scholar]
- Farquhar MG, Palade GE, 1965. Cell junctions in amphibian skin. J Cell Biol 26, 263–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre B, Begre N, Borradori L, 2018. A recessive mutation in the DSP gene linked to cardiomyopathy, skin fragility and hair defects impairs the binding of desmoplakin to epidermal keratins and the muscle-specific intermediate filament desmin. Br J Dermatol 179, 797–799. [DOI] [PubMed] [Google Scholar]
- Fleming TP, Garrod DR, Elsmore AJ, 1991. Desmosome biogenesis in the mouse preimplantation embryo. Development 112, 527–539. [DOI] [PubMed] [Google Scholar]
- Gallicano GI, Bauer C, Fuchs E, 2001. Rescuing desmoplakin function in extra-embryonic ectoderm reveals the importance of this protein in embryonic heart, neuroepithelium, skin and vasculature. Development 128, 929–941. [DOI] [PubMed] [Google Scholar]
- Gallicano GI, Kouklis P, Bauer C, Yin M, Vasioukhin V, Degenstein L, Fuchs E, 1998. Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J Cell Biol 143, 2009–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod D, Chidgey M, 2008. Desmosome structure, composition and function. Biochim Biophys Acta 1778, 572–587. [DOI] [PubMed] [Google Scholar]
- Garrod DR, Berika MY, Bardsley WF, Holmes D, Tabernero L, 2005. Hyper-adhesion in desmosomes: its regulation in wound healing and possible relationship to cadherin crystal structure. J Cell Sci 118, 5743–5754. [DOI] [PubMed] [Google Scholar]
- Goonesinghe A, Luan XM, Hurlstone A, Garrod D, 2012. Desmosomal cadherins in zebrafish epiboly and gastrulation. BMC Dev Biol 12, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KJ, Gaudry CA, 2000. Are desmosomes more than tethers for intermediate filaments? Nat Rev Mol Cell Biol 1, 208–216. [DOI] [PubMed] [Google Scholar]
- Green KJ, Parry DA, Steinert PM, Virata ML, Wagner RM, Angst BD, Nilles LA, 1990. Structure of the human desmoplakins. Implications for function in the desmosomal plaque. J Biol Chem 265, 11406–11407. [PubMed] [Google Scholar]
- Green KJ, Simpson CL, 2007. Desmosomes: new perspectives on a classic. J Invest Dermatol 127, 2499–2515. [DOI] [PubMed] [Google Scholar]
- Harris TJ, Tepass U, 2010. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol 11, 502–514. [DOI] [PubMed] [Google Scholar]
- Heasman J, 2002. Morpholino oligos: making sense of antisense? Dev Biol 243, 209–214. [DOI] [PubMed] [Google Scholar]
- Holbrook KA, Odland GF, 1975. The fine structure of developing human epidermis: light, scanning, and transmission electron microscopy of the periderm. J Invest Dermatol 65, 16–38. [DOI] [PubMed] [Google Scholar]
- Houssin NS, Bharathan NK, Turner SD, Dickinson AJ, 2017. Role of JNK during buccopharyngeal membrane perforation, the last step of embryonic mouth formation. Dev Dyn 246, 100–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Najor NA, Green KJ, 2014. Desmosomes: regulators of cellular signaling and adhesion in epidermal health and disease. Cold Spring Harb Perspect Med 4, a015297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lake BB, Haremaki T, Weinstein DC, Sokol SY, 2012. Rab11 regulates planar polarity and migratory behavior of multiciliated cells in Xenopus embryonic epidermis. Dev Dyn 241, 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Zhang S, Werner ME, Brotslaw EJ, Mitchell JW, Altabbaa MM, Mitchell BJ, 2018. CLAMP/Spef1 regulates planar cell polarity signaling and asymmetric microtubule accumulation in the Xenopus ciliated epithelia. J Cell Biol 217, 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymkowsky MW, Shook DR, Maynell LA, 1992. Evidence that the deep keratin filament systems of the Xenopus embryo act to ensure normal gastrulation. Proc Natl Acad Sci U S A 89, 8736–8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofron M, Heasman J, Lang SA, Wylie CC, 2002. Plakoglobin is required for maintenance of the cortical actin skeleton in early Xenopus embryos and for cdc42-mediated wound healing. J Cell Biol 158, 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofron M, Spagnuolo A, Klymkowsky M, Wylie C, Heasman J, 1997. The roles of maternal alpha-catenin and plakoglobin in the early Xenopus embryo. Development 124, 1553–1560. [DOI] [PubMed] [Google Scholar]
- Lechler T, Fuchs E, 2007. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J Cell Biol 176, 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes M, Rakshit S, Shafraz O, Borghi N, Harmon RM, Green KJ, Sivasankar S, Nelson WJ, 2014. Different roles of cadherins in the assembly and structural integrity of the desmosome complex. J Cell Sci 127, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashal RD, Koontz J, Sklar J, 1995. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat Genet 9, 177–183. [DOI] [PubMed] [Google Scholar]
- Mitchell B, Stubbs JL, Huisman F, Taborek P, Yu C, Kintner C, 2009. The PCP pathway instructs the planar orientation of ciliated cells in the Xenopus larval skin. Curr Biol 19, 924–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moftah H, Dias K, Apu EH, Liu L, Uttagomol J, Bergmeier L, Kermorgant S, Wan H, 2017. Desmoglein 3 regulates membrane trafficking of cadherins, an implication in cell-cell adhesion. Cell adhesion & migration 11, 211–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz WA, Kloc M, Cho K, Lee M, Hofmann I, Sater A, Vleminckx K, McCrea PD, 2012. Plakophilin-3 is required for late embryonic amphibian development, exhibiting roles in ectodermal and neural tissues. PLoS One 7, e34342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM, 2013. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis 51, 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop P, Faber J, 1967. Normal table of Xenopus laevis (Daudin): a systematic and chronological survey of the development from the fertilized egg till the end of metamorphosis. North-Holland. [Google Scholar]
- Nishimura R, Kato K, Fujiwara S, Ohashi K, Mizuno K, 2018. Solo and Keratin Filaments Regulate Epithelial Tubule Morphology. Cell Struct Funct 43, 95–105. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Takeichi M, 2009. Remodeling of the adherens junctions during morphogenesis. Current topics in developmental biology 89, 33–54. [DOI] [PubMed] [Google Scholar]
- Ossipova O, Chu CW, Fillatre J, Brott BK, Itoh K, Sokol SY, 2015. The involvement of PCP proteins in radial cell intercalations during Xenopus embryonic development. Dev Biol 408, 316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Haigo SL, Wallingford JB, 2006. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat Genet 38, 303–311. [DOI] [PubMed] [Google Scholar]
- Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB, 2008. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet 40, 871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramms L, Fabris G, Windoffer R, Schwarz N, Springer R, Zhou C, Lazar J, Stiefel S, Hersch N, Schnakenberg U, Magin TM, Leube RE, Merkel R, Hoffmann B, 2013. Keratins as the main component for the mechanical integrity of keratinocytes. Proc Natl Acad Sci U S A 110, 18513–18518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedzinski J, Hannezo E, Tu F, Biro M, Wallingford JB, 2016. Emergence of an Apical Epithelial Cell Surface In Vivo. Dev Cell 36, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedzinski J, Hannezo E, Tu F, Biro M, Wallingford JB, 2017. RhoA regulates actin network dynamics during apical surface emergence in multiciliated epithelial cells. J Cell Sci 130, 420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltmann K, Fritsch AW, Kas JA, Magin TM, 2013. Keratins significantly contribute to cell stiffness and impact invasive behavior. Proc Natl Acad Sci U S A 110, 18507–18512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, van Heeringen SJ, Quigley I, Heinz S, Ogino H, Ochi H, Hellsten U, Lyons JB, Simakov O, Putnam N, Stites J, Kuroki Y, Tanaka T, Michiue T, Watanabe M, Bogdanovic O, Lister R, Georgiou G, Paranjpe SS, van Kruijsbergen I, Shu S, Carlson J, Kinoshita T, Ohta Y, Mawaribuchi S, Jenkins J, Grimwood J, Schmutz J, Mitros T, Mozaffari SV, Suzuki Y, Haramoto Y, Yamamoto TS, Takagi C, Heald R, Miller K, Haudenschild C, Kitzman J, Nakayama T, Izutsu Y, Robert J, Fortriede J, Burns K, Lotay V, Karimi K, Yasuoka Y, Dichmann DS, Flajnik MF, Houston DW, Shendure J, DuPasquier L, Vize PD, Zorn AM, Ito M, Marcotte EM, Wallingford JB, Ito Y, Asashima M, Ueno N, Matsuda Y, Veenstra GJ, Fujiyama A, Harland RM, Taira M, Rokhsar DS, 2016. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafraz O, Rubsam M, Stahley SN, Caldara AL, Kowalczyk AP, Niessen CM, Sivasankar S, 2018. E-cadherin binds to desmoglein to facilitate desmosome assembly. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AN, Moens CB, Miller AC, 2016. Targeted candidate gene screens using CRISPR/Cas9 technology. Methods Cell Biol 135, 89–106. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM, 2000. Early development of Xenopus laevis: a laboratory manual. [Google Scholar]
- Smith FJ, Wilson NJ, Moss C, Dopping-Hepenstal P, McGrath J, 2012. Compound heterozygous mutations in desmoplakin cause skin fragility and woolly hair. Br J Dermatol 166, 894–896. [DOI] [PubMed] [Google Scholar]
- Sonavane PR, Wang C, Dzamba B, Weber GF, Periasamy A, DeSimone DW, 2017. Mechanical and signaling roles for keratin intermediate filaments in the assembly and morphogenesis of Xenopus mesendoderm tissue at gastrulation. Development 144, 4363–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Davidson L, Keller R, Kintner C, 2006. Radial intercalation of ciliated cells during Xenopus skin development. Development 133, 2507–2515. [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C, 2008. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet 40, 1454–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Vladar EK, Axelrod JD, Kintner C, 2012. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol 14, 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumigray KD, Chen H, Lechler T, 2011. Lis1 is essential for cortical microtubule organization and desmosome stability in the epidermis. J Cell Biol 194, 631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumigray KD, Lechler T, 2011. Control of cortical microtubule organization and desmosome stability by centrosomal proteins. Bioarchitecture 1, 221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumigray KD, Lechler T, 2012. Desmoplakin controls microvilli length but not cell adhesion or keratin organization in the intestinal epithelium. Mol Biol Cell 23, 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Bowers E, Bauer C, Degenstein L, Fuchs E, 2001. Desmoplakin is essential in epidermal sheet formation. Nat Cell Biol 3, 1076–1085. [DOI] [PubMed] [Google Scholar]
- Virata ML, Wagner RM, Parry DA, Green KJ, 1992. Molecular structure of the human desmoplakin I and II amino terminus. Proc Natl Acad Sci U S A 89, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walck-Shannon E, Hardin J, 2014. Cell intercalation from top to bottom. Nat Rev Mol Cell Biol 15, 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Shi Z, Cui Y, Guo X, Shi YB, Chen Y, 2015. Targeted gene disruption in Xenopus laevis using CRISPR/Cas9. Cell Biosci 5, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Mitchell JW, Putzbach W, Bacon E, Kim SK, Mitchell BJ, 2014. Radial intercalation is regulated by the Par complex and the microtubule-stabilizing protein CLAMP/Spef1. J Cell Biol 206, 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittock NV, Ashton GH, Dopping-Hepenstal PJ, Gratian MJ, Keane FM, Eady RA, McGrath JA, 1999. Striate palmoplantar keratoderma resulting from desmoplakin haploinsufficiency. J Invest Dermatol 113, 940–946. [DOI] [PubMed] [Google Scholar]
- Whittock NV, Wan H, Morley SM, Garzon MC, Kristal L, Hyde P, McLean WH, Pulkkinen L, Uitto J, Christiano AM, Eady RA, McGrath JA, 2002. Compound heterozygosity for non-sense and mis-sense mutations in desmoplakin underlies skin fragility/woolly hair syndrome. J Invest Dermatol 118, 232–238. [DOI] [PubMed] [Google Scholar]
- Yonemura S, 2011. Cadherin-actin interactions at adherens junctions. Curr Opin Cell Biol 23, 515–522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.