Abstract
The mechanistic target of rapamycin (mTOR) exerts both rapamycin-sensitive and rapamycin-insensitive signaling events, and the rapamycin-sensitive components of mTOR signaling have been widely implicated in the pathway through which resistance exercise induces skeletal muscle hypertrophy. This review explores the hypothesis that rapamycin-insensitive components of mTOR signaling also contribute to this highly important process.
Keywords: Exercise, Hypertrophy, Protein Synthesis, mTOR, mTORC1, mTORC2, Rapamycin
Summary:
This review examines whether both rapamycin-sensitive, and rapamycin-insensitive, components of mTOR signaling contribute to the hypertrophic effects of resistance exercise.
INTRODUCTION
It is well known that resistance exercise induces an increase in skeletal muscle protein synthesis and hypertrophy (1). Signaling by the mechanistic target of rapamycin (mTOR), which exists in at least 2 protein complexes (mTORC1 and mTORC2), is activated by resistance exercise, and it has been widely assumed that rapamycin-sensitive/mTORC1-dependent signaling is necessary for the increase in protein synthesis and hypertrophy that occurs in response to resistance exercise. The basis for this assumption largely stems from previous studies which have shown that models of chronic mechanical overload (e.g., synergist ablation) induce hypertrophy through a fully rapamycin-sensitive and mTORC1-dependent process (2–4). However, using a rodent model of resistance exercise, we recently discovered that rapamycin only partially inhibits the resistance exercise-induced changes in protein synthesis and hypertrophy (5). Yet, pharmacological inhibition of all mTOR kinase activity was able to completely block the resistance exercise-induced increase in protein synthesis (6). These observations lead to our hypothesis that both rapamycin-sensitive and rapamycin-insensitive components of mTOR signaling contribute to the hypertrophic effects of resistance exercise. In this review, we will: i) summarize the history behind the studies which led to the assumption that resistance exercise-induced changes in protein synthesis and hypertrophy are mediated by rapamycin-sensitive/mTORC1-dependent signaling events, ii) inform the reader about the important differences that exists between mTORC1 versus mTORC2 and rapamycin-sensitive versus rapamycin-insensitive components of mTOR signaling, iii) summarize how rapamycin-insensitive components of mTOR signaling could contribute to the hypertrophic effects of resistance exercise, and iv) highlight some of the primary outstanding questions in this field.
RAPAMYCIN AND ITS MECHANISM OF ACTION
In 1965, an antifungal-antibiotic compound produced by microbes was discovered in the soil on the island of Rapa Nui (Easter Island) and subsequently named rapamycin (7, 8). Several years later, it was determined that rapamycin can form a complex with the FK506 binding protein 12 (FKBP12), and that this complex enables rapamycin to function as a potent immunosuppressant (8, 9). Moreover, it was shown that rapamycin can prevent the ability of growth factors to induce an increase in the phosphorylation of the 70 kDa ribosomal protein S6 kinase (p70S6K), suggesting that rapamycin targets a kinase (10). In 1994, the kinase responsible for rapamycin’s effect was discovered by several laboratories and given the names: rapamycin target 1 (RAPT1); FKBP-rapamycin associated protein (FRAP); and rapamycin and FKBP target 1 (RAFT1). Over time these names became standardized to the mammalian target of rapamycin. However, in 2009, the gene name was officially changed by the HUGO Gene Nomenclature Committee to the “mechanistic target of rapamycin” abbreviated as “mTOR” (11).
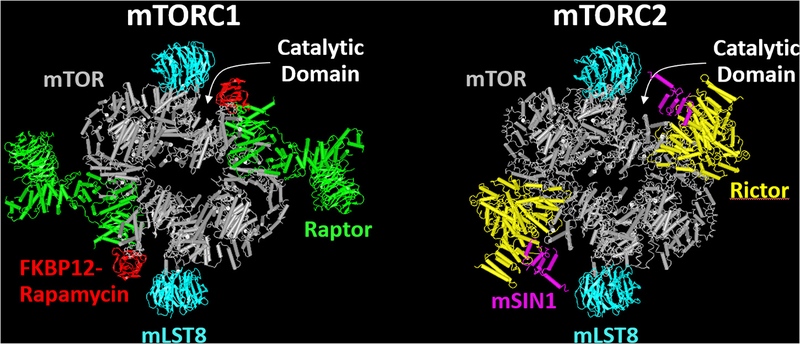
Structurally, mTOR is a large protein consisting of 2549 amino acids, with a predicted molecular mass of 289 kDa (12). However, size exclusion chromatography revealed that mTOR had an apparent molecular mass of ~1–2 MDa, suggesting that mTOR exists within large multi-protein complexes (13). Indeed, further biochemical and genetic analyses revealed that mTOR exists within two functionally distinct protein complexes called mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). As illustrated in Figure 1, the core components of mTORC1 include the regulatory-associated protein of TOR (raptor), and the mammalian lethal with sec-13 protein 8 (mLST8), whereas the core components of mTORC2 include the rapamycin-insensitive companion of mTOR (rictor), the mammalian stress-activated protein kinase interacting protein 1 (mSIN1) and mLST8 (14, 15). Importantly, although both complexes contain mTOR, only signaling by mTORC1 is sensitive to the acute inhibitory effects of rapamycin (15, 16).
Figure 1.
The web-based version of iCn3D was used to visualize and compare the structures of mechanistic target of rapamycin complex 1 (mTORC1) and mTORC2 that were reported by Aylett et al., 2016 (17) and Chen et al., 2018 (20), respectively. Both mTORC1 and mTORC2 are composed of two mTOR (gray) and two mLST8 (blue) molecules. The defining feature of mTORC1 is the presence of two raptor molecules (green) which deliver mTORC1-specific substrates to the catalytic domain of mTOR. The FKBP12-rapamycin complexes (red) bind to mTOR in the region that lies between raptor and mTOR’s catalytic domain, blocking access of some, but not all, of mTORC1’s substrates to the catalytic domain. The defining feature of mTORC2 is the presence of two rictor (yellow) and two mSIN1 (purple) molecules which sterically inhibit the ability of FKBP12-rapamycin complexes to bind to mTOR.
During the last few years a number of studies have been aimed at elucidating the structure of the mTOR complexes (17–20). Based on these studies it has been concluded that mTORC1 is a lozenge shaped protein complex whose core components consist of two mTOR, two raptor and two mLST8 molecules (Figure 1). It has also been shown that raptor recognizes and delivers mTORC1-specific substrates to the catalytic domain of mTOR (19). Furthermore, these studies have demonstrated that the FKBP12-rapamycin complex binds to mTOR in a region that lies between raptor and mTOR’s catalytic domain. Combined, these observations have helped to establish that the FKBP12-rapamycin complex does not directly inhibit mTOR catalytic activity. Instead, it inhibits mTORC1 signaling by hindering the ability of substrates to gain access to mTOR’s catalytic domain (18). However, it has also been concluded that, in the presence of the FKBP12-rapamycin complex, ample space for substrate association with the catalytic domain still exists (18) (See Figure 1). This is a significant point because, as addressed later in the review, it potentially explains why rapamycin inhibits some, but not all, mTORC1-dependent signaling events. As illustrated in Figure 1, mTORC2 also forms a dimeric structure, and the general shape of this structure is quite similar to that of mTORC1 (20). Importantly, however, the combined presence of rictor and mSIN1 sterically inhibits the ability of the FKBP12-rapamycin complex to bind to mTOR (20). This is another noteworthy point because it helps to account for why rapamycin does not exert an acute inhibitory effect on signaling by mTORC2 (15, 16).
DEFINING mTOR-DEPENDENT SIGNALING EVENTS: COMMON MISCONCEPTIONS
Shortly after the discoveries of mTORC1 and mTORC2, it became widely assumed that rapamycin-sensitive signaling events are mediated by mTORC1, and that if a signaling event was rapamycin-insensitive, then that meant that it was mediated by an mTORC1-independent process. However, studies over the last decade have revealed that these assumptions are not always correct. For instance, it is now known that rapamycin can potently inhibit some, but not all, mTORC1-dependent signaling events (21). As a case in point, the phosphorylation of the T389 residue on p70S6K and the T37/46 residues on eIF4E binding protein 1 (4E-BP1) are two of the most commonly used readouts of mTORC1 signaling, and numerous studies have shown that rapamycin potently inhibits p70S6K1 T389 phosphorylation while only exerting a mild, and often undetectable, inhibition of 4E-BP1 T37/46 phosphorylation (5, 21). Evidence that rapamycin does not inhibit all mTORC1-dependent signaling events has also come from studies that employed inhibitors of mTOR catalytic activity (e.g., PP242, Torin1, AZD8055). For example, in cells that lack ricto/mTORC2 (i.e., cells in which mTOR would presumably only be found in mTORC1), it has been shown that Torin1 can inhibit protein synthesis to a greater extent than rapamycin (22). Thus, when defining mTOR-dependent events, it is critical to appreciate that rapamycin-insensitive does not necessarily mean mTORC1-independent.
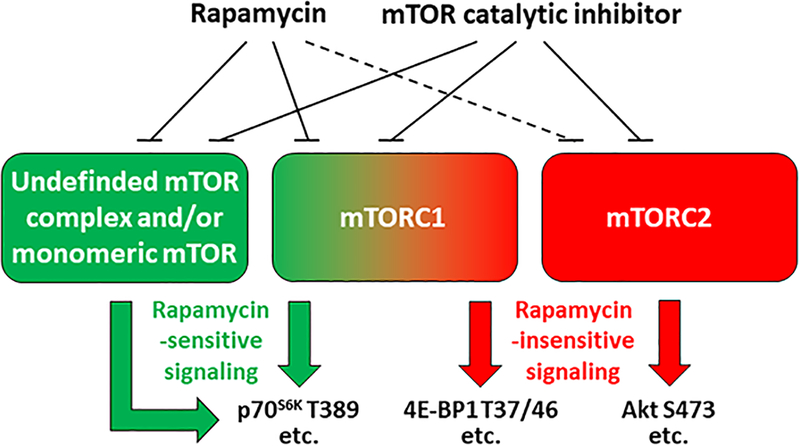
Another common misconception is that if a signaling event is rapamycin-sensitive, then that means that it is mediated by mTORC1. However, several studies have challenged the validity of this assumption. For instance, it has been reported that both rapamycin, and the knock down of mTOR, can potently inhibit the translation of 5’TOP mRNAs; yet, knocking out raptor/mTORC1 only slightly impairs the translation of these mRNAs (23). Previous studies have also revealed that myogenesis proceeds through a rapamycin-sensitive mechanism, and that mTOR is the rapamycin-sensitive element that confers this event, but it does not require raptor/mTORC1 (24). Finally, it has been shown that, even in the absence of raptor, mTOR can still induce changes in the phosphorylation of p70S6K1 T389 through a rapamycin-sensitive process (25, 26). Interestingly, the mechanism(s) behind these rapamycin-sensitive but raptor/mTORC1-independent effects have not been defined, but one possibility is that mTOR is capable of functioning in a monomeric state that remains rapamycin-sensitive. Another noteworthy point is that high-dose/long-term administration of rapamycin can lead to the disassembly and subsequent inhibition of mTORC2, thus once again illustrating that rapamycin-sensitive does not exclusively imply mTORC1-dependent (14). A summary of the key points from this section are shown in Figure 2.
Figure 2.
Overview of the signaling events that are mediated by the different mechanistic target of rapamycin (mTOR) complexes, along with the sensitivity of these events to rapamycin and inhibitors of mTOR catalytic activity. Dashed line indicates an indirect affect.
THE ROLE OF mTOR IN CHRONIC MECHANICAL OVERLOAD-INDUCED HYPERTROPHY
In 1999, Baar and Esser published a study which demonstrated that a bout of resistance exercise-like contractions in rats could induce an increase p70S6K phosphorylation, and that the magnitude of the increase in p70S6K phosphorylation was highly correlated with the induction of hypertrophy (27). In 1999, it was already well known that changes in p70S6K phosphorylation are largely mediated through a rapamycin-sensitive mechanism, and hence, the study of Baar and Esser effectively became the first to suggest that rapamycin-sensitive signaling might play a role in resistance exercise-induced hypertrophy (27). In 2001, Bodine et al. extended this concept by demonstrating that systemic administration of rapamycin (1.5 mg/kg/d) could prevent the increase p70S6k phosphorylation that occurs in response to the synergist ablation model of chronic mechanical overload (28). More importantly, this study also demonstrated that rapamycin could prevent synergist ablation from inducing a hypertrophic response. These observations were quickly confirmed by independent groups, and paralleled by additional studies which indicated that the activation of rapamycin-sensitive signaling was not only necessary, but also sufficient, for the induction of hypertrophy (29, 30).
A key question that remained after these early studies was whether the anti-hypertrophic effects of rapamycin were due to the inhibition of mTOR within the skeletal muscle cells or the inhibition of mTOR in other cell types (e.g., immune cells, satellite cells). This was an important question because rapamycin is a potent immunosuppressant, and several studies had indicated that immune cells might be required for chronic mechanical overload-induced hypertrophy (31). Thus, to address this question, we utilized a line of transgenic mice that expressed a rapamycin-resistant mutant of mTOR exclusively within the skeletal muscle cells (RR-mTOR) (2). Similar to the results of Bodine et al. (28), we found that rapamycin (0.6, 1,0, and 3.0 mg/kg/d but not 0.3 mg/kg/d) could abolish the hypertrophic effects of synergist ablation in muscles from wild-type mice, but it did not prevent the induction of hypertrophy in the muscles of RR-mTOR mice. Accordingly, it was concluded that signaling through a rapamycin-sensitive component of mTOR, within the skeletal muscle fibers themselves, is necessary for chronic mechanical overload-induced hypertrophy.
As detailed above, it has been widely assumed that rapamycin-sensitive means mTORC1 dependent. To test this assumption, Bentzinger et al. (2013) subjected constitutive skeletal muscle specific raptor knockout mice (RAmKO) to synergist ablation and concluded that raptor/mTORC1 was necessary for the induction of hypertrophy (3). However, the interpretation of these results from this study are confounded by a number of traits that are inherent to the RAmKO mice. For instance, the muscles of RAmKO mice have a significant reduction in mitochondrial content along with numerous signs of myopathy, including a decrease in mass and fiber size (32). Moreover, the daily voluntary activity of RAmKO mice is dramatically reduced when compared to control mice (32). The reduction in voluntary activity is particularly problematic because, in the synergist ablation model, the amount of mechanical overload that is placed on the muscles is directly proportional to the animals’ level of activity. Hence, the lack of a hypertrophic response in the RAmKO mice might have simply resulted from an insufficient amount of mechanical overload being placed on the muscles. Therefore, to overcome this limitation, we generated skeletal muscle-specific and tamoxifen-inducible raptor knockout mice (4). These mice did not present with the confounding traits of the RAmKO mice, yet consistent with the findings of Bentzinger et al. (3), chronic mechanical overload-induced hypertrophy was completely abolished by the loss of mTORC1 (4). Thus, in models of chronic mechanical overload, it appears that rapamycin-sensitive mTOR/mTORC1 is fully required for the induction of a hypertrophic response. In the next section, we will address whether the same point holds true in models of resistance exercise.
THE ROLE OF mTOR IN RESISTANCE EXERCISE-INDUCED HYPERTROPHY
The results from studies that have employed models of chronic mechanical overload helped to forge the concept that resistance exercise-induced hypertrophy is also mediated through a rapamycin-sensitive mTOR/mTORC1-dependent process. In support of this notion, numerous human studies have reported that resistance exercise robustly increases p70S6K phosphorylation, and that the extent of the increase in p70S6K phosphorylation is strongly correlated with the hypertrophy that occurs after repeated bouts of training (33). Accordingly, it seemed safe to assume that just like chronic mechanical overload-induced hypertrophy, resistance exercise-induced hypertrophy is also mediated through a rapamycin-sensitive mTOR/mTORC1-dependent process. Indeed, this concept has been engrained in the literature since the early 2000’s: yet, it wasn’t directly tested until 2016. Specifically, in 2016, we employed a rodent model of resistance exercise and, much to our surprise, we found that rapamycin (1.5 mg/kg, 3 d/week) only partially (~50%) blocked the increase in fiber cross-sectional area that occurred after 12 bouts of training (5). In other words, unlike the results that have been obtained in models of chronic mechanical overload, the outcomes of our study suggested that signaling by rapamycin-sensitive mTOR/mTORC1 is only partially necessary for the hypertrophic effects of resistance exercise.
RESISTANCE EXERCISE-INDUCED HYPERTROPHY: THINKING BEYOND THE CLASSIC RAPAMYCIN-SENSITIVE mTOR/mTORC1-DEPENDENT MECHANISMS
Protein Synthesis
Skeletal muscle mass is regulated by the balance between the rates of protein synthesis and protein degradation (34). Previous studies have shown that resistance exercise increases the rate of both protein synthesis and degradation, with the extent of the increase in protein synthesis exceeding that of degradation (35). Thus, the increase in protein synthesis that occurs in response to resistance exercise is considered to be a critical part of the process through which resistance exercise induces hypertrophy.
As mentioned in the introduction, rapamycin-sensitive mTOR/mTORC1-dependent signaling events have been widely implicated in the pathway through which resistance exercise induces an increase in protein synthesis (36). In support of this point, Drummond et al. (2009) reported that, in humans, the increase in protein synthesis that occurs during the early recovery period (~2 h) after resistance exercise is completely inhibited by rapamycin (approximately 0.15 mg/kg) (37). On the other hand, West et al. (2016) used a rat model of resistance exercise to conclude that the increase in protein synthesis that occurs 18 h after the bout of resistance exercise is largely rapamycin-insensitive (rapamycin dose was 1.5 mg/kg) (38). Using a different rat model, we have also found that the increase in protein synthesis that occurs at later time points following a bout of resistance exercise is rapamycin-insensitive (rapamycin dose was 1.5 mg/kg) (5, 6). With this model, we have also used AZD8055 (an inhibitor of mTOR catalytic activity) to determine whether the rapamycin-insensitive increase in protein synthesis is regulated by rapamycin-insensitive mTOR. Importantly, the outcomes revealed that the increase in protein synthesis was completely abolished by AZD8055 (6). Thus, it appears that resistance exercise can induce an increase in protein synthesis through an mTOR-dependent process that involves both rapamycin-sensitive and rapamycin-insensitive signaling events (6).
Currently, the rapamycin-insensitive components of mTOR signaling that contribute to the resistance exercise-induced increase in protein synthesis are not known. However, one possibility is that these components involve rapamycin-insensitive mTORC1-dependent signaling events. For instance, the regulation of 4E-BP1 phosphorylation by mTORC1 is well known for its ability to control protein synthesis and, as mentioned at the beginning of this article, many of the phosphorylation sites on 4E-BP1 are regulated through a rapamycin-insensitive but mTORC1-dependent process (6). Another potential rapamycin-insensitive component of mTOR involves the regulation of eEF2 T56 phosphorylation. Specifically, eEF2 T56 phosphorylation can inhibit the elongation phase of protein synthesis, and we have determined that resistance exercise induces a decrease in eEF2 T56 phosphorylation through a mechanism that is rapamycin-insensitive but completely abolished by AZD8055 (6). Accordingly, we suspect that an mTORC1-independent decrease in eEF2 T56 phosphorylation might be one of the rapamycin-insensitive components of mTOR signaling that regulates the resistance exercise-induced increase in protein synthesis; however, this hypothesis requires further investigation.
The possibility that protein synthesis can be regulated through an mTORC1-independent mechanism is further highlighted by recent work from our lab which demonstrated that both intermittent passive stretch, as well as chronic mechanical overload, can induce an increase in protein synthesis despite the tamoxifen-induced KO of skeletal muscle raptor and, therefore, the loss of mTORC1 (4). Although it’s possible that a residual amount of raptor (and thus mTORC1) may have still been present in these muscles, and that this residual raptor was sufficient to elicit a robust increase in protein synthesis, this possibility seems unlikely. Instead, it is our conviction that mTORC1-independent mechanisms contributed to the increase in protein synthesis.
Finally, recent studies have suggested that mTORC2 might regulate protein synthesis via a mechanism that involves the transcription factor c-Myc (39), and thus, a potential role for mTORC2 in the resistance exercise-induced increase in protein synthesis should also be considered.
Protein Degradation
In addition to its widely appreciated role in the regulation of protein synthesis, signaling by mTOR can also play a prominent role in the regulation of protein degradation. Indeed, previous non-muscle studies have reported that the mTORC1 can have both positive and negative effects on protein degradation (40, 41). In skeletal muscle, the sustained long-term activation of mTORC1 induced by the deletion of the mTORC1 inhibitor, tuberous sclerosis complex (TSC), activates the ubiquitin-proteasome system (UPS) (3) and accelerates denervation-induced atrophy (42). Although these muscle studies suggest that signaling by mTOR activates the UPS, this effect may be secondary to other deleterious effects of sustained long-term mTOR activation, a condition that would not occur in response to resistance exercise training. Nonetheless, it is currently unknown if mTOR signaling regulates the UPS in response to acute or chronic resistance exercise.
Another cellular process involved in protein degradation is autophagy and it is well known that mTORC1 can inhibit autophagy through a mechanism that involves phosphorylation of the S757 residue on ULK1 (43). Interestingly, work from our group has shown that resistance exercise induces an increase in ULK1 S757 phosphorylation through a mechanism that is rapamycin-insensitive, yet completely abolished by AZD8055 (5,6). Thus, a rapamycin-insensitive component of mTOR signaling might help to repress the induction of autophagy that occurs after a bout of resistance-exercise (44).
Recent studies have also indicated that mTORC2 can regulate protein degradation via control of the UPS. For example, it is known that members of the forkhead box O family of transcription factors (FoxO) play a prominent role in the regulation the UPS, and that the transcriptional activity of FoxO’s (e.g., nuclear localization) is potently regulated by protein kinase B (Akt) (45). Furthermore, it has been shown that the S473 phosphorylation/activity of Akt can be regulated by mTORC2, and that the knockdown of rictor/mTORC2 leads to nuclear accumulation of FoxO3 in skeletal muscle (46). Thus, a potential role for mTORC2 in the regulation of UPS must be acknowledged. Moreover, recent studies have indicated that rapamycin-sensitive components of mTOR signaling can also regulate the activity of the UPS (47). Based on these points, it’s surprising that no studies have directly examined the role that mTOR plays in the regulation of protein degradation following resistance-exercise, and this will certainly be an area that is worthy of further investigation.
Other Metabolic Pathways
When considering the potential role that mTOR plays in regulation of resistance exercise-induced hypertrophy, one should also remain cognisant of the effect that mTOR signaling can have on a variety of different metabolic pathways. For instance, signaling through rapamycin-sensitive mTOR/mTORC1 has been shown to regulate the synthesis of the lipids that would be required for the expansion of the cell membrane. Furthermore, the synthesis of the nucleotides that would be required for the formation of new ribosomes as well as the transcription of both mitochondrial and nuclear DNA are also regulated by rapamycin-sensitive mTOR/mTORC1 (48).
In addition to rapamycin-sensitive mTOR/mTORC1, signaling by mTORC2 can also play a prominent role in the regulation of metabolism. For instance, it has been shown that a single injection of AZD8055, but not rapamycin, can transiently lower the respiratory exchange ratio and induce insulin resistance (49). The same study also demonstrated that the incubation of isolated skeletal muscle strips with AZD8055, but not rapamycin, blocked insulin-stimulated Akt S473 phosphorylation and reduced insulin-stimulated glucose uptake (49). Consistent with the effects of AZD8055, mice with skeletal muscle specific knockout of rictor/mTORC2 display increased whole-body fat oxidation, increased intramuscular TG storage, and a decreased ability to stimulate muscle glucose uptake in vivo (49, 50). Thus, when considering previous studies which have shown that: i) the activity of mTORC2 can be inhibited by AZD8055; ii) the S473 phosphorylation/activity of Akt can be regulated by mTORC2; and iii) the activity of Akt mediates many of the actions of insulin and is required for insulin-stimulated glucose uptake in skeletal muscle, it would appear that inhibition of mTORC2 -> Akt signaling could explain the effects that AZD8055 exerts on metabolism. However, the skeletal muscle specific knockout of rictor/mTORC2 only reduces insulin-stimulated glucose uptake at a submaximal, but not maximal insulin, concentrations (51). In contrast, AZD8055 can inhibit maximal insulin-stimulated glucose uptake (49), which suggests that AZD8055 might alter substrate metabolism through mechanisms that extend beyond the inhibition of the mTORC2->Akt signaling. Regardless of this possibility, it is apparent that rapamycin-insensitive components of mTOR signaling (e.g., signaling by mTORC2) can play an important role in the regulation of metabolism.
Another important aspect of an mTORC2 signaling relates to its ability to control the activity of c-Myc (39). In fact, we find the link between mTORC2 and c-Myc to be particularly intriguing because c-Myc is often described as a “master regulator” of the metabolic processes that support cellular growth such as nucleic acid, lipid and protein synthesis (52). Moreover, previous studies have shown that the expression of c-Myc is increased by resistance exercise in both humans and rodents, and its induction can be blocked by long-term, but not acute, rapamycin injections (5). As noted in section III, long-term administration of rapamycin can inhibit mTORC2 signaling (14), and thus, it’s tempting to suggest that the resistance exercise induced increase in c-Myc might be driven by an mTORC2-dependent mechanism. Finally, a recent study that compared the phosphoproteome of wild type and skeletal muscle specific rictor knockout mice following a bout of endurance exercise revealed the mTORC2 likely controls the phosphorylation of a large number of previously unrecognized substrates (53). Considering this point, along with other studies which have implicated mTORC2 in the regulation of metabolism and overall cellular growth (54), it is easy to envision that mTORC2 might play an important role in resistance exercise-induced hypertrophy.
CONCLUSION
In contrast to the current dogma, recent studies have indicated that both rapamycin-sensitive and rapamycin-insensitive components of mTOR signaling contribute to the hypertrophic effects of resistance exercise. Additional studies will be needed to address this novel concept.
Key Points:
Resistance exercise induces hypertrophy through a mechanism that is, at least in part, dependent on an increase in the rate of protein synthesis.
A long-standing contention in the field is that the rapamycin-sensitive components of mTOR signaling are necessary for a resistance exercise-induced increase in the rate of protein synthesis. However, a number of recent studies have demonstrated that this is not always the case. In fact, an emerging body of evidence suggests that rapamycin-insensitive components of mTOR signaling play a key role in this process.
In this review we will summarize the evidence which indicates that rapamycin-insensitive components of mTOR signaling could play a key role in the pathway through which resistance exercise induces an increase in protein synthesis and the concomitant hypertrophic response.
Funding:
The work in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (NIH) under award number AR057347 to T.A.H, a Novo Nordisk Foundation Excellence project grant to T.E.J (15182), as well as JSPS KAKENHI Grant No. 26702028 and 17KK0192 to R.O.
Footnotes
Conflicts of Interest: None
REFERENCES
- 1.Brook MS, Wilkinson DJ, Mitchell WK et al. Skeletal muscle hypertrophy adaptations predominate in the early stages of resistance exercise training, matching deuterium oxide-derived measures of muscle protein synthesis and mechanistic target of rapamycin complex 1 signaling. FASEB J. 2015;29(11):4485–96. [DOI] [PubMed] [Google Scholar]
- 2.Goodman CA, Frey JW, Mabrey DM et al. The role of skeletal muscle mTOR in the regulation of mechanical load-induced growth. J Physiol. 2011;589(Pt 22):5485–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentzinger CF, Lin S, Romanino K et al. Differential response of skeletal muscles to mTORC1 signaling during atrophy and hypertrophy. Skeletal muscle. 2013;3(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.You JS, McNally RM, Jacobs BL et al. The role of raptor in the mechanical load-induced regulation of mTOR signaling, protein synthesis, and skeletal muscle hypertrophy. FASEB J. 2018:fj201801653RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogasawara R, Fujita S, Hornberger TA et al. The role of mTOR signalling in the regulation of skeletal muscle mass in a rodent model of resistance exercise. Sci Rep. 2016;6:31142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogasawara R, Suginohara T. Rapamycin-insensitive mechanistic target of rapamycin regulates basal and resistance exercise-induced muscle protein synthesis. FASEB J. 2018:fj201701422R. [DOI] [PubMed] [Google Scholar]
- 7.Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo). 1975;28(10):721–6. [DOI] [PubMed] [Google Scholar]
- 8.Sehgal SN. Sirolimus: its discovery, biological properties, and mechanism of action. Transplant Proc. 2003;35(3 Suppl):7S–14S. [DOI] [PubMed] [Google Scholar]
- 9.Harding MW, Galat A, Uehling DE, Schreiber SL. A receptor for the immuno-suppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature. 1989;341(6244):758–60. [DOI] [PubMed] [Google Scholar]
- 10.Price DJ, Grove, Calvo V, Avruch J, Bierer BE. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science (New York, N.Y 1992;257(5072):973–7. [DOI] [PubMed] [Google Scholar]
- 11.Hall MN. On mTOR nomenclature. Biochemical Society transactions. 2013;41(4):887–8. [DOI] [PubMed] [Google Scholar]
- 12.Sabers CJ, Martin MM, Brunn GJ et al. Isolation of a Protein Target of the FKBP12-Rapamycin Complex in Mammalian Cells. The Journal of biological chemistry. 1995;270(2):815–22. [DOI] [PubMed] [Google Scholar]
- 13.Kim D-H, Sarbassov DD, Ali SM et al. mTOR Interacts with Raptor to Form a Nutrient-Sensitive Complex that Signals to the Cell Growth Machinery. Cell. 2002;110(2):163–75. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19(3):373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou H, Huang S. The complexes of mammalian target of rapamycin. Current protein & peptide science. 2010;11(6):409–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aylett CH, Sauer E, Imseng S et al. Architecture of human mTOR complex 1. Science. 2016;351(6268):48–52. [DOI] [PubMed] [Google Scholar]
- 18.Yang H, Wang J, Liu M et al. 4.4 A Resolution Cryo-EM structure of human mTOR Complex 1. Protein Cell. 2016;7(12):878–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H, Jiang X, Li B et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552(7685):368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Liu M, Tian Y et al. Cryo-EM structure of human mTOR complex 2. Cell Res. 2018;28(5):518–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang SA, Pacold ME, Cervantes CL et al. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341(6144):1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thoreen CC, Kang SA, Chang JW et al. An ATP-competitive Mammalian Target of Rapamycin Inhibitor Reveals Rapamycin-resistant Functions of mTORC1. The Journal of biological chemistry. 2009;284(12):8023–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patursky-Polischuk I, Stolovich-Rain M, Hausner-Hanochi M et al. The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol Cell Biol. 2009;29(3):640–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge Y, Chen J. Mammalian target of rapamycin (mTOR) signaling network in skeletal myogenesis. J Biol Chem. 2012;287(52):43928–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You JS, Lincoln HC, Kim CR et al. The role of diacylglycerol kinase zeta and phosphatidic acid in the mechanical activation of mammalian target of rapamycin (mTOR) signaling and skeletal muscle hypertrophy. J Biol Chem. 2014;289(3):1551–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell. 2010;38(5):768–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baar K, Esser K. Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. The American journal of physiology. 1999;276(1 Pt 1):C120–7. [DOI] [PubMed] [Google Scholar]
- 28.Bodine SC, Stitt TN, Gonzalez M et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3(11):1014–9. [DOI] [PubMed] [Google Scholar]
- 29.Hornberger TA, McLoughlin TJ, Leszczynski JK et al. Selenoprotein-deficient transgenic mice exhibit enhanced exercise-induced muscle growth. J Nutr. 2003;133(10):3091–7. [DOI] [PubMed] [Google Scholar]
- 30.Pallafacchina G, Calabria E, Serrano AL, Kalhovde JM, Schiaffino S. A protein kinase B-dependent and rapamycin-sensitive pathway controls skeletal muscle growth but not fiber type specification. Proc Natl Acad Sci U S A. 2002;99(14):9213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiPasquale DM, Cheng M, Billich W et al. Urokinase-type plasminogen activator and macrophages are required for skeletal muscle hypertrophy in mice. Am J Physiol Cell Physiol. 2007;293(4):C1278–85. [DOI] [PubMed] [Google Scholar]
- 32.Bentzinger CF, Romanino K, Cloetta D et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8(5):411–24. [DOI] [PubMed] [Google Scholar]
- 33.Terzis G, Georgiadis G, Stratakos G et al. Resistance exercise-induced increase in muscle mass correlates with p70S6 kinase phosphorylation in human subjects. Eur J Appl Physiol. 2008;102(2):145–52. [DOI] [PubMed] [Google Scholar]
- 34.Atherton PJ, Phillips BE, Wilkinson DJ. Exercise and Regulation of Protein Metabolism. Prog Mol Biol Transl Sci. 2015;135:75–98. [DOI] [PubMed] [Google Scholar]
- 35.Phillips SM, Tipton KD, Aarsland A, Wolf SE, Wolfe RR. Mixed muscle protein synthesis and breakdown after resistance exercise in humans. The American journal of physiology. 1997;273(1 Pt 1):E99–107. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez AM, Hoffman JR, Stout JR, Fukuda DH, Willoughby DS. Intramuscular Anabolic Signaling and Endocrine Response Following Resistance Exercise: Implications for Muscle Hypertrophy. Sports Med. 2016;46(5):671–85. [DOI] [PubMed] [Google Scholar]
- 37.Drummond MJ, Fry CS, Glynn EL et al. Rapamycin administration in humans blocks the contraction-induced increase in skeletal muscle protein synthesis. J Physiol. 2009;587(Pt 7):1535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.West DW, Baehr LM, Marcotte GR et al. Acute resistance exercise activates rapamycin-sensitive and -insensitive mechanisms that control translational activity and capacity in skeletal muscle. J Physiol. 2016;594(2):453–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masui K, Tanaka K, Akhavan D et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18(5):726–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, Goldberg AL. Coordinate regulation of autophagy and the ubiquitin proteasome system by MTOR. Autophagy. 2016:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Nicholatos J, Dreier JR et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513(7518):440–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang H, Inoki K, Lee M et al. mTORC1 promotes denervation-induced muscle atrophy through a mechanism involving the activation of FoxO and E3 ubiquitin ligases. Science signaling. 2014;7(314):ra18. [DOI] [PubMed] [Google Scholar]
- 43.Su KH, Dai C. mTORC1 senses stresses: Coupling stress to proteostasis. BioEssays : news and reviews in molecular, cellular and developmental biology. 2017;39(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ulbricht A, Gehlert S, Leciejewski B, Schiffer T, Bloch W, Hohfeld J. Induction and adaptation of chaperone-assisted selective autophagy CASA in response to resistance exercise in human skeletal muscle. Autophagy. 2015;11(3):538–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends in biochemical sciences. 2014;39(4):159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mammucari C, Milan G, Romanello V et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–71. [DOI] [PubMed] [Google Scholar]
- 47.Zhao J, Zhai B, Gygi SP, Goldberg AL. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci U S A. 2015;112(52):15790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;169(2):361–71. [DOI] [PubMed] [Google Scholar]
- 49.Kleinert M, Sylow L, Fazakerley DJ et al. Acute mTOR inhibition induces insulin resistance and alters substrate utilization in vivo. Mol Metab. 2014;3(6):630–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleinert M, Parker BL, Chaudhuri R et al. mTORC2 and AMPK differentially regulate muscle triglyceride content via Perilipin 3. Mol Metab. 2016;5(8):646–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleinert M, Sylow L, Jensen TE, Oxboll A-J, Richter E. Muscle-specific deletion of mTORC2 (Rictor) blocks insulin stimulated Akt Ser 473 phosphorylation and impairs submaximal but not maximal insulin induced glucose uptake. The FASEB Journal. 2013;27(1_supplement):1109.10–.10. [Google Scholar]
- 52.Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18(20):5546–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kleinert M, Parker BL, Fritzen AM et al. Mammalian target of rapamycin complex 2 regulates muscle glucose uptake during exercise in mice. J Physiol. 2017;595(14):4845–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuo Y, Huang H, Cai T, Wang T. Target of Rapamycin Complex 2 regulates cell growth via Myc in Drosophila. Sci Rep. 2015;5:10339. [DOI] [PMC free article] [PubMed] [Google Scholar]