Abstract
Plant viruses are responsible for losses in worldwide production of numerous economically important food and fuel crops. As obligate cellular parasites with very small genomes, viruses rely on their hosts for replication, assembly, intra- and intercellular movement, and attraction of vectors for dispersal. Chloroplasts are photosynthesis and are the site of replication for several viruses. When viruses replicate in chloroplasts, photosynthesis, an essential process in plant physiology, is inhibited. The mechanisms underlying molecular and biochemical changes during compatible and incompatible plants–virus interactions, are only beginning to be elucidated, including changes in proteomic profiles induced by virus infections. In this review, we highlight the importance of proteomic studies to understand plant–virus interactions, especially emphasizing the changes in photosynthesis-related protein accumulation. We focus on: (a) chloroplast proteins that differentially accumulate during viral infection; (b) the significance with respect to chloroplast-virus interaction; and (c) alterations in plant’s energetic metabolism and the subsequently the plant defense mechanisms to overcome viral infection.
Keywords: Plant–virus interactions, Virus replication in chloroplasts, Proteomics, Photosynthesis, Proteome
1. Introduction
Plants are under permanent attack by several pathogenic agents, such as fungi, oomycetes, nematodes and viruses. These pathogens are responsible for important agricultural losses that limit worldwide food production. Viral diseases can represent approximately 50% of the new emerging diseases affecting plants and new plant viruses are being discovered every day (Whitfield et al. 2015). Plant viruses are classified as DNA or RNA viruses according to Baltimore’s viral classification (Baltimore 1971). While both RNA and DNA plant viruses lead to lower crop production, RNA viruses are the most abundant (Laliberté and Sanfaçon 2010). All viruses are obligatory parasites that require a replication site within host cells upon infection. Once inside the cell, the viral genome serves as a template for the synthesis of several genomic copies that are transcribed and translated into viral proteins, which ultimately resulting in new viral particles (Nagy 2008; Nagy and Pogany 2012). To complete the replication cycle, viruses require cellular proteins and other resources, collectively known as host factors (Laliberté and Sanfaçon 2010; Nagy and Pogany 2012; Tajima et al. 2017).
In plants, virus infection induced numerous genetic, metabolic and physiological changes including changes in the photosynthetic apparatus in chloroplasts (Bhattacharyya and Chakraborty 2018; Zhao et al. 2016). These changes may prevent the establishment of plant defense mechanisms (Zhao et al. 2016) or promote virus susceptibility or simply occur as side effects of gene silencing (Garcia-Ruiz et al. 2016). Plants have evolved several resistance mechanisms that act in a complementary way to promote defense against virus infection. Antiviral RNA silencing is an essential component of antiviral immunity (Bologna and Voinnet 2014; Diaz-Pendon et al. 2004; Garcia-Ruiz et al. 2016). Briefly, dicer-like proteins cleave the double-stranded viral RNA (dsvRNA) into virus-derived small interference RNAs (vsiRNA), which are loaded into a RISC (RNA interference silencing complex); then AGO protein uses the vsiRNA to degrade viral RNA preventing viral replication (Garcia-Ruiz et al. 2016; Voinnet 2001). Other part of the process is called amplification of RNA silencing; this phase involves the replication of dsvRNAs by RNA-dependent RNA polymerase (RDRs), which result in a strongest local and systemic extend of RNA silencing antiviral immunity (Garcia- Ruiz et al. 2010, 2016).
Plant defense mechanisms, including RNA silencing, are highly expensive for the plant cell in energetic terms. Defense responses are usually associated with the consumption of high levels of reducing power, especially ATP and NADPH, as well as carbon skeletons provided by primary metabolism (Bolton 2009). In plant leaves, photosynthetic activity is the main mechanism for providing reducing power and metabolic intermediates to sustain plant defenses against infections (Bhattacharyya and Chakraborty 2018; de Torres Zabala et al. 2015; Zhao et al. 2016). Since the chloroplasts are the site of photosynthesis in plants, they are strategic organelles that viruses may target (Bhattacharyya and Chakraborty 2018; Zhao et al. 2016). Additionally, chloroplasts are capable of producing high amounts of reactive oxygen species (ROS) as a collateral effect of the redox reactions carried out in the photosynthetic electron transport chain, especially at PSII and PSI acceptor sides (Kale et al. 2017; Jimbo et al. 2018). Moreover, the photorespiratory pathway, which is initiated by the Rubisco oxygenase activity at chloroplasts, could generate high ROS amounts at peroxisomes and also represents an important source of photosynthesis-related-ROS in plants during viral infection (Bolton 2009). ROS play an important role in retrograde signaling pathways driven from organelles to nucleus, which could lead to increase expression of several nuclear genes involved in biotic stress defense. In addition, these highly reactive molecules are involved in several processes of cell degeneration, including programmed cell death (PCD). Consequently, ROS originated from photosynthesis-related processes, in parallel to NADPH oxidase sourced ROS, may also be related to hypersensitive response (HR) induction, acting directly in defensive mechanisms against viral infection (Bolton 2009; Caplan et al. 2008).
To establish infection, viruses have to suppress antiviral responses, including those mediated by photosynthesis-dependent defense components (Bhattacharyya and Chakraborty 2018). Several studies have reported effects of viral infection in chloroplasts (Prod’homme et al. 2003, 2001; Wei et al. 2013; Zhao et al. 2016; Souza et al. 2017), including accumulation of coating proteins at the PSII super-complex leading to decreased oxygen evolving activity and photoinhibition (Hodgson et al. 1989). Photosynthesis and photorespiration encompass an intricate network, cross-regulated, to optimize photosynthetic yield under the most variable environmental conditions, adding complexity to the mechanisms governing plant–virus interactions (Silveira and Carvalho 2016).
The use of integrative “omics” approaches has allowed the simultaneous analysis of several processes that plant cells employ in order to avoid or minimize the establishment of disease and has allowed a glimpse of the metabolic processes targeted by viruses. Proteomics is a powerful technique to identify massive changes in the cellular proteins and has been used to study the biochemical mechanisms involved in plant–virus interactions (Kundu et al. 2011, 2013; Mehta et al. 2008; Paiva et al. 2016; Varela et al. 2017; Zhong et al. 2017). Recent advances in mass spectrometry (MS), genomics and bioinformatics have also contributed to our understanding of plant–virus interactions. Other technologies such as gel-free and label-free approaches in tandem with MS analysis have provided critical information on the differential expression of thousands of proteins (Quirino et al. 2010). Combined, these approaches are a valuable tool in the identification of proteins differentially expressed during viral infection, enabling hypothesis-driven studies on plant–virus interactions (Beltran et al. 2017; Kundu et al. 2011, 2013; Lodha et al. 2013; Paiva et al. 2016; Quirino et al. 2010; Toby et al. 2016; Varela et al. 2017).
Thus, in the apogee of the “omics” era, this review addresses the most remarkable advances in the use of proteomic approaches to understand plant–virus inter-actions, with a special emphasis on the biochemical and physiological changes driven in photosynthesis-related processes. In literature several review papers have approached this issue in a fragmented way. In opposition, in this review an integrative view of different proteomic, physiological and molecular processes, is targeted. Therefore, we focused on (1) photosynthesis-related proteins commonly affected, (2) the physiological meaning of these effects, and (3) the causal nature of the reduction in photosynthesis-related proteins during infection.
2. Photosynthesis shut off and symptoms of virus infection
Photosynthesis is of great importance to plant antiviral defense mechanisms (Bolton 2009; Caplan et al. 2008). However, while viruses need to impair plant defense mechanisms dependent on photosynthesis-related processes, viruses also need the highly energetic expensive translational machinery of host plants to guarantee successful viral infection. Indeed, the development of several characteristic symptoms of viral infection, such mosaic, chlorosis and leaf distortion, are a consequence of widespread damage to the photosynthetic machinery. Symptoms are first associated with local infection, as results of viral replication at the initial sites of infection. Whole plant symptoms appear after the virus has established systemic infection through the entire plant (Fig. 1). After replication in the very first infected cell, viruses start the cell-to-cell movement to infect nearby cells, establishing then local infection (Fig. 1b, c). With the local infection spot established, virus continues cell-to-cell movement until reach veins and vascular tissues in the leave (Fig. 1d, white arrowheads). After colonize vascular tissue, virus can move to young healthy leaves to establish systemic infection (Fig. 1e) (Mochizuki et al. 2014; Rahoutei et al. 2000; Zhao et al. 2016).
Fig. 1.
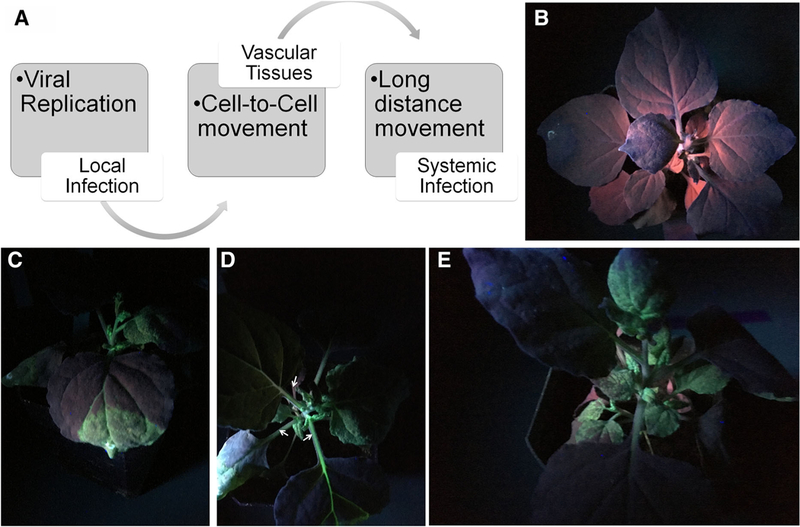
Establishment of virus infection through local cell-to-cell and systemic movement. Nicotiana benthamianaplants were agro-infiltrated with GPF-tagged Turnip mosaic virus (TuMV-GFP) or empty vector. Pictures were taken under UV light. a Model showing virus movement to establish local and systemic infection. b N. benthamiana plants infiltrated with empty. c N. benthamiana plants infiltrated with TuMV-GFP and showing local infection. d After cell-to-cell movement, TuMV reaches the veins and vascular tissues (white arrowheads). e Establishment of systemic infection after long-distance movement through the vascular system
Several metabolic and molecular changes in plant cells must occur simultaneously during infection development and a clear cause–effect relationship has not been established. Several reports suggest that changes in chloroplast components and functions may be the most important factor in development of the classic symptoms of chlorosis and mosaic formation (Liu et al. 2014; Rahoutei et al. 2000). During viral infection, chloroplasts suffer several structural changes induced by viruses such as appearance of atypical morphological structures (i.e., globules or membrane-bound extrusions), aggregation of unusual photosynthetic structures, dilatation of thylakoids and complete chloroplast degradation and grana disorganization, which in extreme cases can even lead to a decrease in total chloroplast number per leaf area (Laliberté and Sanfaçon 2010; Montasser and Al-Ajmy 2015; Zhao et al. 2016).
Interference with chlorophyll metabolism, or induction of chlorophyll degradation reduce photosynthesis yield, visible as yellow or white spots, or leaf mosaics (Liu et al. 2014; Shimura et al. 2011). Drastic reductions in chlorophyll levels during this process are associated with symptom development (Kundu et al. 2013; Liu et al. 2014; Shimura et al. 2011; Souza et al. 2017). In cowpea (Vigna unguiculata) plants susceptible to Cowpea severe mosaic virus (CPSMV, genus Comovirus) infection can induce decreases up to 32% and 40% in chlorophyll b and a, respectively (Souza et al. 2017). Moreover, in this study, the authors clearly suggest a similar trend of reduction in chlorophyll level and lower photosynthetic index in parallel with symptoms and disease establishment. Nevertheless, the mechanism by which viruses can interfere in chlorophyll metabolism remains elusive. Furthermore, during infection, viruses can negatively affect several important chloroplast functions such as electron transfer reactions, chlorophyll metabolism, autonomous chloroplast protein synthesis, CO2 fixation reactions and chlorophyll metabolism, all potentially involved in symptom development (Liu et al. 2014; Neilson et al. 2013; Souza et al. 2017; Varela et al. 2017; Zhao et al. 2016).
The establishment of characteristic symptoms in plants during viral infection is a complex process. An isolate of Cucumber mosaic virus carrying a Y-satRNA (CMV-YsatRNA) induces silencing of the plant mRNA encoding for magnesium chelatase subunit I (ChlI), which an important enzyme for chlorophyll biosynthesis (Shimura et al. 2011). In parallel, Liu et al. (2014) also reported that during plant infection by African cassava mosaic virus (ACMV genus Begomovirus) in Cassava (Manihot esculenta), several genes involved in chlorophyll degradation ware up regulated. A recent comparative genetic analysis showed that in a potyvirus and in an orthotospovirus, on one protein, the silencing suppressors HC-Pro or NSs, respectively, are needed for symptom development (Garcia-Ruiz et al. 2018). Collectively, these observations suggest a role for epigenetic regulation of chlorophyll degradation in symptom development by two possible mechanisms. Virus-encoded silencing suppressors that bind small RNAs may promote upregulation of genes involved in chlorophyll degradation or virus-derived siRNA may directly target and downregulate chlorophyll biosynthesis genes.
3. Variations in proteomic techniques
By definition proteomics is a technique applied to analyze a protein profile from a cell. This method can be performed by two ways (1) gel-based and (2) gel-free both coupled with mass spectrometer analysis (MS) (Fig. 2). The first is based in sodium dodecyl-sulfate polyacrylamide (SDS-PAGE) gel used in electrophoresis analysis. This approach allows the identification of many proteins. A variation of this technique called DIGE brought an upgrade to this type of analysis allowing the identification of thousands of proteins (Fig. 2) (Lodha et al. 2013; Quirino et al. 2010). However, limitations of this technique are related to high cost and to problems in identification of very large and very small proteins (Fig. 2). The second, the gel-free analysis is a great update in this type. In gel-free analysis proteins are applied directly in MS machine, in this case no gel analysis is needed. The main advantage of this technique is the possibility of identifies thousands of proteins (e.g. 3000, Paiva et al. 2016). The limitation of this analysis is the required high skill level to operate the MS machine and to analyze the data (Fig. 2) (Lodha et al. 2013; Di Carli et al. 2012; Quirino et al. 2010).
Fig. 2.
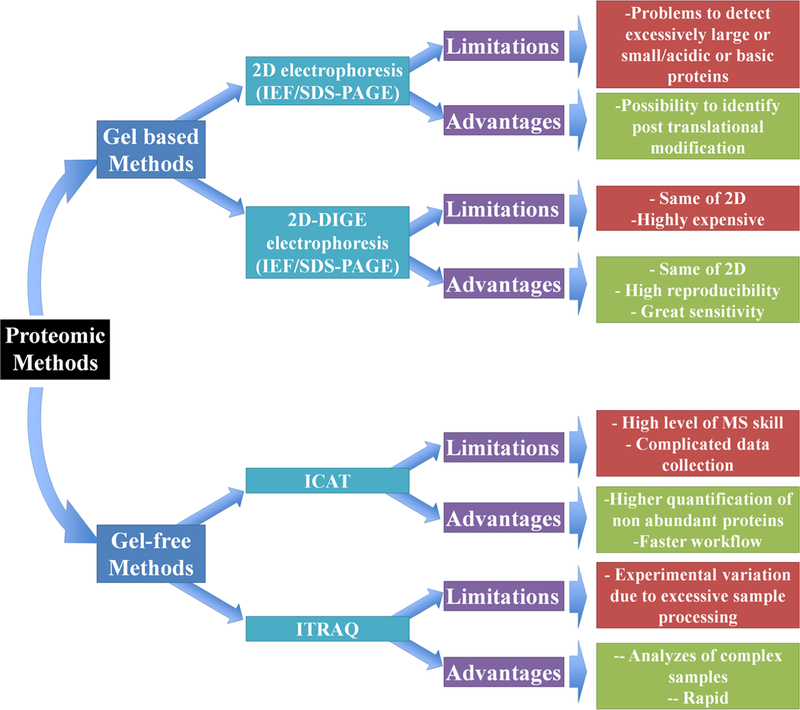
Advantages and limitations of gel-based and gel-free proteomics methods
Although, the field of proteomics has been evolving at an impressive rate, especially due to development of new methods to analyze simultaneously a large number of proteins, using either gel or gel-free techniques, in combination with MS analyses that now have higher sensitivity and precision (Fig. 2 and Table 1). Besides the great contribution of gel-free techniques, gel-based proteomic methods have also improved protein identification (Fig. 1 and Table 1). Pineda et al. (2010) identified only 55 proteins when analyzing N. benthamiana and Pepper mild mottle virus (PMMoV, genus Tobamovirus) interactions by 2DE-PAGE-MS analysis, whereas Serra-Soriano et al. (2015) identified 1046 differentially expressed protein species in plants infected with Cucumis melo and Melon necrotic spot virus (MNSV, genus Carmovirus) by 2DE-DIGE LC–MS analysis. Despite the high sensibility of 2DE-DIGE methodologies, it is used less often than gel-free approaches, mainly due to the high costs of dyes (Di Carli et al. 2012).
Table 1.
A summary of original published studies on plant–virus interaction using proteomic approach
| Plant species | Virus | Proteomic method | No. proteins identified |
References |
|---|---|---|---|---|
| Oryza sativa | Rice Yellow mottle virus | 2D-PAGE, MALDI, LC-MS/MS | 64 | Ventelon-Debout et al. (2004) |
| Nicotiana benthamiana | Pepper Mild Mottle Virus | 2D-PAGE/MS | 55 | Pineda et al. (2010) |
| Capsicum chinese | Pepper Mild Mottle Virus | 2D-PAGE, MALDI, LC–MS/MS | 17 | Elvira et al. (2008) |
| Vigna mungo | Mungbean Yellow Mosaic Virus | MALDI–TOF/TOF | 29 | Kundu et al. (2011) |
| Zea mays | Sugarcane Mosaic Virus | 2D-DIGE/MALDI–TOF–MS/MS | 93 | Wu et al. (2013) |
| Oryza sativa | Rice Black-Streaked Dwarf Virus | 2-DE-PAGE/MALDI-TOF/TOF–MS | 72 | Xu et al. (2013) |
| Nicotiana benthamiana | Oilseed rape mosaic virus | 1D-SDS-PAGE/LC–MS/MS | 2915 | Niehl et al. (2013) |
| Zea mays | Sugarcane Mosaic Virus | 2-D-PAGE/MALDI-TOF/MS–MS | 96 | Wu et al. (2013) |
| Cucumis melo | Melon Necotic Spot Virus | 2D-DIGE/LC–MS/MS | 1046 | Serra-Soriano et al. (2015) |
| Oryza sativa | Rice Stripe Virus | iTRAQ/LC–MS/MS | 681 | Wang et al. (2015) |
| Nicotiana tabacum | Tobacco Mosaic Virus | iTRAQ/LC–MS/MS | 2125 | Wang et al. (2016) |
| Solaum lycospercum | Tomato Yellow Curl Virus | 2D-DIGE/MALDI–TOF–MS/MS | 86 | Huang et al. (2016) |
| Solaum lycospercum | Potato Virus X | 2D-PAGE/MS | 85 | Cueto-Ginzo et al. (2016) |
| Nicotiana benthamiana | Tomato Yellow Leaf Curl China Virus | iTRAQ-Strong Cationic Chromatography/LC–MS | 1232 | Zhong et al. (2017) |
| Vigna unguiculata | Cowpea Severe Mosaic Virus | LC–MS/MS | 3000 | Paiva et al. (2016) |
| Solaum lycospercum | Tomato Chlorotic Mottle Virus | 2D-PAGE-nanoUPLC-HDMS | 534 | Carmo et al. (2017) |
| Pisum sativum | Pea Seed-Borne Mosaic Virus | LC–MS/MS | 2300 | Cerna et al. (2017) |
| Oryza sativa | Southern Rice Black Streaked Dwarf Virus | 1D-SDS-PAGE/LC–MS/MS | 2197 | Yang et al. (2017) |
| Vigna unguiculata | Cowpea Severe Mosaic Virus | LC–MS/MS | 2033 | Varela et al. (2017) |
4. Proteomics provides insights into plant–virus interactions
While photosynthetic shutdown during viral infection of plants is important, basic understanding of the molecular mechanisms underlying this phenomenon remains to be elucidated. The challenges are related to the complexity of chloroplast metabolism and to the complexity of its regulatory system, which includes signals originated from both nuclear and chloroplast genomes. A typical plastidial genome, such as in Arabidopsis thaliana, presents approximately 80 genes, which encode mostly for the photosynthetic electron transport chain (PETC) proteins and plastid translational machinery (Sato et al. 1999). On other hand, the majority of proteins involved in chloroplast metabolism, including those related to sugar, amino acid and lipid metabolisms are nuclear-encoded proteins. Therefore, to fully understand a complex biological phenomenon such as plant–irus interaction processes involving photosynthetic shutdown, an effective holistic approach able to uncover simultaneously several processes of plant cell metabolism is needed. Among several approaches which are able to fulfill this requirement, proteomics deserves a special attention.
Photosynthesis is involved in triggering and/or supporting plant defense responses to fight viral infection by providing energy to the plant cell and as a source of ROS. On the other hand, viruses activate mechanisms that break down important biochemical pathways in chloroplast. Consequently, these two different signaling waves triggered by virus infection might generate new metabolic dynamics in chloroplasts that ultimately involve extensive repercussions in the entire protein profile. Proteomics is a powerful tool to understand these changes. Despite several years of studies involving the use of proteomics approaches in the understanding of plant–virus interactions and a great amount of data collected, the physiological interpretation of the obtained results is a mayor landmark that has not been achieved. The central question is: What is the role of differentially expressed proteins during plant virus-interactions?
Virus infection triggers of drastic changes in the protein accumulation in the entire plant, including leaves. Leaves are a very interesting organ for plant–virus interaction studies, since the most important symptoms of viral infection developed in leaves. Several groups of proteins differentially accumulate in infected leaves, most of which are involved in photosynthesis-related processes (Table 2). Interestingly, changes in protein accumulation have been detected in incompatible plant–virus combinations (Di Carli et al. 2010; Kundu et al. 2013; Varela et al. 2017). These proteins may provide initial clues to the mechanisms of plant resistance to virus infection (Kundu et al. 2013; Varela et al. 2017). The class of differentially accumulated proteins may determine whether or not the plant can restrict viral infection, and consequently, whether or not it will develop disease signals (Kundu et al. 2013; Varela et al. 2017).
Table 2.
Photosynthesis-related proteins identified by proteomic studies that have accumulation affected by virus infection
| Plant species | Virus | Interaction | Protein species | References |
|---|---|---|---|---|
| Nicotiana benthamiana | Pepper Mild Mottle Virus | Compatible | Cyt. F; FNR (chain b); Cyt. F; ATPδ; ATPε; Lhca type III; RbcS; RbcL; GS; SBPase; PRK; Rca; PGK; PGK; SFBA; PsaD; Ribosomal protein L 14 | Pineda et al. (2010) |
| Nicotiana benthamiana | Pepper Mild Mottle Virus | Compatible | PsbP species A, B, C, and D | Pérez-Bueno et al. (2004) |
| Zea mays | Sugarcane Mosaic Virus | Incompatible | FNR; PS I subunit IV; PS I subunit I; OEE3; RuBisCO large subunit; RuBisCO small subunit; RuBisCo subunit binding-protein beta subunit | Wu et al. (2013) |
| Zea mays | Sugarcane Mosaic Virus | Compatible | FNR; PS I ycf4; Cab binding protein 6A; Cab binding protein 8; RuBisCO large subunit; RuBisCo subunit binding-protein beta subunit | Wu et al. (2013) |
| Vigna mungo | Mungbean Yellow Mosaic India Virus | Compatible | OEE1; RuBisCO activase; HSP70b; Chaperonin 60 alpha; PSI subunit II; RuBisCO large subunit; GS; PSI-D; PSI subunit IV B; PRK; RuBisCO small subunit; Triose-phosphate isomerise; Glycine hydroxymethyltransferase 2; Cab binding protein 4 b; OEE2; OEE3–2; GAPDH A; Plastoquinol-plastocyanin reductase; Rieske iron-sulfur protein precursor; SBPase; Glycine cleavage system protein H; Thioredoxin M; Chlorophyll magnesium chelatase; FNR; ATPε; Cytochrome c oxidase subunit 6b-1a; EF-Tu | Kundu et al. (2013) |
| Solanum lycopersicum | Cucumber Mosaic Virus | Incompatible/compatible | OEE2; OEE1; RuBisCO large subunit; RuBisCO activase; T-protein; Glycolate oxidase; AOAT2 | Di Carli et al. (2010) |
| Arabidopsis thaliana | Oilseed rape mosaic virus | Compatible | LHCB1.4 | Niehl et al. (2013) |
| Oryza sativa | Rice stripe virus | Compatible | Magnesium-chelatase; Uroporphyrinogen decarboxylase 1; Porphobilinogen deaminase; Magnesium-chelatase subunit D; Glutamate-1-semialdehyde 2,1-aminomutase; Uroporphyrinogen decarboxylase 2; Protoporphyrinogen oxidase; Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclise; Delta-aminolevulinic acid dehydratase Glutamyl-tRNA reductase; RuBisCO large subunit; OEE3; OEE2; PSI subunit XI; Cab binding protein; PSI subunit II; PsbQ domain protein family; LHCII type III; OEE3–2; PSI subunit N; Ribulose-phosphate 3-epimerase; FBPase; PsbP; FNR; FTR; SNT7 |
Wang et al. (2015) |
| Nicotiana benthamiana | Tobacco Mosaic Virus | Compatible | LHCa3; PsbL; PsbO; PetE; ATPβ; ATPδ; ATPβ’ | Wang et al. (2016) |
| Nicotiana benthamiana | Tobacco Mosaic Virus | Incompatible | LHCa3; LHCb1; LHCb4; LHCb5; LHCb6; PsbA; PsbL; PsbO; PsbQ; PsbS; PetB; PetD; PsaF; PsaG; PsaJ; PsaK; PetE; PetF; ATPβ; ATPγ; ATPδ; ATPβ’ | Wang et al. (2016) |
| Solanumlycopersicum | Tomato yellow leaf curl virus | Compatible/incompatible | Cab binding protein 4; Cab binding protein 8; OEE1; RuBisCO accumulation factor 1; RuBisCO activase 1; RuBisCO; RuBisCO large subunit; RuBisCO subunit beta; RuBisCO large chain OS; FNR; Protein TIC 62; EF-G; EF-TuB; GLDC | Huang et al. (2016) |
| Nicotiana benthamiana | Tomato yellow leaf curl China virus | Incompatible | PsbH; CP24; RuBisCO large subunit; NDH subunit K; Curvature thylakoid 1A; RuBisCO small subunit; PsbB; CP26 | Zhong et al. (2017) |
| Vigna unguiculata | Cowpea Severe Mosaic Virus | Compatible | CP26; thylakoid lumenal 17.9 kDa; Protein Ycf2 | Paiva et al. (2016) |
| Pisum sativum | Pea Seed-borne Mosaic Virus | Compatible/incompatible | ATPγ; OEE1; PsaK; Chaperone protein ClpC; Mg-protoporphyrin IX chelatase; PSI subunit III; Thylakoid lumenal 17.4 kDa protein; Cab binding protein complex I; Cab binding protein AB80; Cab binding protein 3; GAPDH | Cerna et al. (2017) |
| Oryza sativa | Rice Black-Streaked Dwarf Virus | Compatible | Cab binding protein; RuBisCO large subunit; FBA; RuBisCO small subunit; PGK; PRK; GAPDH A; ATPβ; FNR; GS; EF-Tu | Xu et al. (2013) |
| Vigna mungo | Mungbean Yellow Mosaic India Virus | Incompatible | Chaperonin 60 alpha; Glycine hydroxymethyltransferase; GAPDH A; OEE1; RuBisCO activase; PSI subunit D; PSI subunit IV B; Cab binding protein 4; OEE2; Rieske iron–sulfur protein precursor |
Kundu et al. (2011) |
Other generic groups of proteins also change during virus infection. Cytoskeleton and RNA binding proteins, photosynthesis, photorespiration and chlorophyll metabolism are commonly affected based on proteomics studies (Alexander and Cilia 2016; Paiva et al. 2016; Varela et al. 2017; Wang 2015). A precise prediction of specific proteins responding to each plant–virus combination is not currently available. However, the proteomic data obtained over the years could provide the best protein candidates to study at the mechanistic level and to pursue in plant breeding programs. Based on this proteomic data obtained with different plant species, we propose that the mechanism employed by viruses during infection could be divided into three main processes: (1) impairment in chlorophyll biosynthesis (2) decrease in the photochemical activity and (3) impairment of Calvin–Benson and photorespiratory reactions. These mechanisms are discussed in deeper details bellow.
5. Virus-induced changes in PETC proteins
Proteomic analyses comparing interactions between resistant and susceptible plants show that viruses can interfere in photochemical reactions by reducing the abundance of important proteins involved in the photosynthetic electron transport chain—PETC (Table 2) (Kundu et al. 2011, 2013; Wang 2015; Wu et al. 2013; Zhong et al. 2017). In addition, these studies have revealed at least four cellular targets of viruses: (1) the light harvesting complex proteins; (2) the PSII and PSI core subunits and the oxygen evolving extrinsic proteins and (3) the feredoxin NAD(P)+ reductase in the terminal part of PETC. These enzymes are all crucial for the process of energy trapping and conversion of photons quanta into reducing power inside plant chloroplasts. Therefore, the ability to control the accumulation of such proteins is critical for survival.
5.1. The light harvesting complex proteins
Among the proteins involved in light harvesting complexes, the most common proteins accumulated in response to viral interactions are generically named chlorophyll a/b binding proteins (ChlABB), because the great capacity of such proteins to bind chlorophyll, xanthophylls and other pigments (Table 2). Compatible interactions between N. benthamiana × PMMoV (Pineda et al. 2010), Arabidopsis thaliana × Oilseed rape mosaic virus (ORMV, genus Tobamovirus) (Niehl et al. 2013), N. benthamiana × Tobacco mosaic virus (TMV, genus Tobamovirus) —Wang et al. 2016), N. benthamiana × Tomato yellow leaf curl china virus (TYLCuCV, genus Begomovirus) (Zhong et al. 2017), V. unguiculata × CPSMV (Paiva et al. 2016) and Pisum sativum × Pea Seed-Borne Mosaic Virus (PSBMV, genus Potyvirus) (Cerna et al. 2017) were described previously (Table 2). All these works involving different viruses and different host plant species, shown clear evidence of decreased amounts of LHC proteins in the presence of compatible interactions. These concordant results demonstrate the importance of LHC during plant–virus interactions. Together these proteins are important in the initial steps of the photosynthesis, light absorption by LHC proteins, and chlorophyll excitation and decay in which light is converted into chemical energy (Shen 2015). Therefore, reduction of these proteins is very important to successful viral infection, as well as the reciprocal is also true.
The PSII and PSI antennas are a hetero-complexes composed by several different subunits. Summarizing, PSII antennas are subdivided into major and minor antennas, which are able to bind several different pigments, including chlorophylls and xanthophylls. The major antennas of PSII are constituted by two integral proteins, the CP43 and CP47 subunits. These proteins are closely related due to the excitation transference to the PSII reaction center (Van Ameron-gen and Croce 2013). In parallel, on the stromal face of thylakoid membranes, the minor antennas are another important complex involved in light capture. The PSII minor antennas encompass units of hetero-trimmer subunits (LHCb1, LHCb2 and LHCb3) and three monomers (LHCb4, LHCB5 and LHCb6). On the other hand, PSI antennas are simpler and are composed of LHC trimmers (LHCal, LHCa2, and LHCa3) and an LHCa4 monomer (Silveira and Carvalho 2016). Unfortunately, a great limitation in the interpretation of data obtained from proteomics studies regarding LHC proteins is the fact that several different names are currently employed to describe these proteins. For instance, the LHCb4 protein, also referred to CP29 in literature, is commonly identified in proteomics studies by the nomenclature chlorophyll a/b binding protein 29, or CB29 (Varela et al. 2018). These different names for the same protein represent important mistakes in literature and a strong limitation for the physiological interpretation of proteomics data obtained from different biological contexts, remarkably plant–virus interactions.
Indeed, the reduction in abundance of several chlorophyll a/b binding proteins is followed by low chlorophyll content and photosynthetic index, which lead to disease symptoms and establishment (Kundu et al. 2013; Souza et al. 2017). Kundu et al. (2013) reported a reduction in ChlABB proteins succeeded by the reduction of chlorophyll a and b content and consequent reduction in photosynthetic index in the interaction of susceptible V. mungo plants and Mung-bean yellow mosaic india virus (MYMIV, genus Begomovirus). Using iTRAQ-based quantitative proteomic analyses, Wang et al. (2016) reported a decrease in abundance in five different ChlABB protein species, which was associated with severe disease symptoms in susceptible O. sativa plants infected by Rice stripe virus (RSV, genus Tenuivirus). In addition, Liu et al. (2014) reported a direct correlation between reduction in ChlABB protein species and reduction in chlorophyll content associated with the formation of mosaic and yellow spots in leaves of Cassava infected with ACMV. Indeed, the interference by viruses on these plastidial proteins can compromise the entire photochemical reaction system, due to the impairment on the most primordial source for water oxidation in the oxygen evolving complex—OEC (Kundu et al. 2013; Souza et al. 2017).
Besides the importance of LHC proteins regarding the light energy capture, it is also very clear the occurrence of a high correlation between these proteins and non-photochemical quenching (NPQ) of chlorophyll a fluorescence (Elrad et al. 2002). NPQ is a multi-component mechanism triggered by plants in the presence of increasing light (Ruban 2017). The most important component of NPQ, called qE, is a photoprotective mechanism related to mitigation of excessive light energy captured by the pigments bound to PSII LHC proteins, dissipating the excess of energy as heat. Indeed, the qE component is believed to contribute up to 90% of total NPQ measured in illuminated leaves (Ruban et al. 2012). The molecular mechanism and action-site for qE have been intensely studied for the past 30 years, but there is still no consensus on this issue. There are supporting evidence that this mechanism is triggered by thylakoid lumen acidification induced by light (Ruban et al. 2003), which generates a signal perceived by the PsbS protein (Niyogi et al. 2005; Johnson and Ruban 2010) that is subsequently able to activate the violoxanthin de-epoxidase activity (Demmig-Adams 1990). However, the exact site for heat loss during qE triggering has remained elusive. The energy transfer by interactions between chlorophyll molecules (Horton et al. 1996), the quenching interactions between the lutein bound at the L1 site and chlorophylls a612–a611–a610 (Ruban et al. 2007), and xanthophyll–chlorophyll quenching interactions between zeaxanthin bound at the L2 site and chlorophylls A5 and B5 (Ahn et al. 2008) are possible candidates for the qE site in higher plants. Nevertheless, a hypothetical “all pigments”— quenching model has been recently proposed (Chmeliov et al. 2015). This model is based on the semi-empirical MNDOCAS-CI method and has successfully evaluated all the existing inter-pigment couplings and thus, generated a reasonable estimate of the quenching ability for various carotenoids binding to LHC antennas, simultaneously.
If NPQ mechanism is still poorly understood to date, the attempts to comprehend the involvement of this phenomenon in the plant–virus interaction is much more incipient. Pérez-Bueno et al. (2006) showed a specific increase in NPQ leaf areas exposed to PMMoV (Pérez-Bueno et al. 2006). In addition, Souza et al. (2017) reported that EMS-mutagenized resistant cowpea plants (V. unguiculata) to CPSMV had an increase of 51% in NPQ levels, whereas plants susceptible to CPSMV showed a decrease of 50.8% in NPQ levels. Based on these results, the idea emerged that NPQ plays a role in the defense of plants against viral infection. However, the precise underlying mechanisms regulating this process during plant–virus interaction is still unclear. Indeed, the increase of NPQ levels provides heat, which has been shown to lead to ROS accumulation in TMV-inoculated resistant tobacco leaves, probably by increasing the oxygen uptake (Chaerle et al. 1999). The main problem with this hypothesis is that qE induction is a competitive sink for energy in PETC, which is ultimately the source of ROS in chloroplasts, either in PSII (Kale et al. 2017) or at the PSI acceptor side level (Takagi et al. 2016; Huang et al. 2018). Thus, the increase in NPQ, in terms of energy balance, appears to be incompatible with the increase of thylakoidal ROS production.
Another interesting fact concerning NPQ induction during incompatible plant–virus interactions is the fact that susceptible plants also exhibit a decreased accumulation of antenna complex proteins, as discussed above. Therefore, the decrease in these protein amounts could be interpreted as an advanced virus mechanism to counteract NPQ defenses, by directly regulating its formation site. Consequently, an important question remains open: What is the benefit to resistant plants by presenting higher NPQ levels? Recently, some authors have proposed that changes in NPQ could be related with systemic signaling processes inducing a systemic acquired acclimation— SAA (Karpiński et al. 2013). Accordingly, during a stressful condition, NPQ wave-like changes, as well as electrical waves regarding changes in membrane potential and ROS signaling waves, can be induced as important players to achieve SAA (Karpiński et al. 2013).
5.2. PSII and PSI core subunits and the oxygen evolving extrinsic proteins
Additional important targets for virus-induced changes revealed by proteomic approaches are the core and low molecular mass proteins from PSII, as well as several subunits of the PSI complex (Table 2). The most notable consequence of changes in amount of PSII and PSI subunits is the potential decrease of electron transport rate in PETC, and consequently, the generation of reducing power should be limited. Indeed, viruses can interfere in photochemical activity by reducing accumulation of proteins involved in the oxygen-evolving complex (OEC), those involved in charge separation inside both PSII and PSI super-complexes, or by decreasing amount of proteins involved in electron transport in between PSII and PSI. Some of the most common differential proteins targeted by viruses during compatible interaction are: oxygen-evolving enhancer proteins (OEEP), the PSII core proteins D1 and D2, the PsaD subunit of PSI and ferredoxin-NADP reductase (FNR) (Table 2) (Kundu et al. 2013; Qiu et al. 2018; Wang 2015; Wang et al. 2016; Wu et al. 2013).
OEEP perform an important role in OEC activity carried out in the PSII core complex. The PSII core complex is composed by the plastidial-encoded proteins D1 and D2, the two major antennas CP43 and CP47 and the α and β subunits of cytochrome b559 (Bricker and Frankel 2011). By definition, these are “core” proteins because the oxygen evolving activity is not possible without any of these PSII subunits (Roose et al. 2016). On the other hand, the OEEP are nuclear-encoded proteins with molecular masses of 26.5 kDa (PsbO), 20.2 kDa (PsbP) and 16.5 kDa (PsbQ), also necessary for maximal rates of O2 evolution, but not crucial, because their absence only modestly affects PSII function and, therefore, are not classified as core proteins. The role of these extrinsic proteins is probably related to modulation of inorganic cofactors (manganese, calcium and chloride), which are requirements for maximum core activity (Roose et al. 2016). Thus, during viral compatible interactions with plant hosts, the capability to viruses interfere in the amount of OEEP might represent a very efficient strategy to down-regulate the photosynthetic activity, and subsequently the provision of energy for defense mechanisms, without promoting the entire cell energetic collapse.
Interestingly, OEEP have also been investigated for several other non-canonical functions. For example, evidence suggests that PsbO could present carbonic anhydrase (Shitov et al. 2009) and GTPase activity (Lundin et al. 2007). In parallel, PsbP and PsbQ subunits have been implicated as participants in grana stack formation (Yi et al. 2009). Furthermore, OEEP may also act as assembly/stability factors for photo-system II (Roose et al. 2016). Nevertheless, since these OEEP are highly abundant in leaves, the possibility that they act as reserves for C and N remobilization during stress conditions should not be ruled out (Silveira and Carvalho 2016). Therefore, viruses would benefit from decreased levels of OEEP for several reasons and, indeed, proteomic studies have shown that this is a frequent response in positive plant–virus interactions. The most interesting observation involving the participation of OEEP during virus compatible interactions was recently reported by Balasubramaniam et al. (2014), which described that PSII OEEP interacts specifically with the coat protein (CP) of Alfalfa mosaic virus (AMV, genus Alfamovirus) and this event could be related with inhibition of virus replication, thus demonstrating a direct role in antiviral defense. In concert with the functions discussed above, several proteomics approaches have been used to investigate incompatible plant–virus interactions in which the evidence indicates that resistant plants exhibit an accentuated increase in abundance of many isoforms of OEEP (Di Carli et al. 2010; Kundu et al. 2013, 2011; Varela et al. 2017; Wu etal. 2013).
In addition to a reduction in the abundance of OEEP, viral infection also commonly results in a lowered accumulation of D1 and D2 proteins in the PSII core (Table 2). The decrease in abundance of PsbA (D1) and PsbB (D2) proteins during viral infection indicates a decrease in the synthesis/degradation balance for these proteins. This lowered balance in the presence of virus infection leads to PSII photoinhibition (Aro et al. 1993) and consequently to a decrease in the reducing power available for carboxylation activity and photorespiration (Souza et al. 2017). In fact, Kundu et al. (2013), using a proteomic approach to the study of V. mungo plants infected with MYIMV found a great reduction in D1 and D2 proteins. Beyond that, by analyzing photochemical reactions via chlorophyll a fluorescence measurements, the authors reported that the very low levels of D1 and D2 proteins were associated with lower actual quantum efficiency of PSII in susceptible V. mungo plants, as expected. These results strongly suggest that MYMIV induces photoinhibition of PSII, which might limit energy conversion by light reactions.
During viral evolution, viruses acquired proteins that could perform multiple functions at the same time. Thus, while working in replication, some viral proteins also display activities that lead to the breakdown of plant defense mechanisms (Acosta-Leal et al. 2011; Huang et al. 2010; Hwang et al. 2015; Walsh and Mohr 2011; Wang 2015). Thus, by decreasing the amount of important PSII core proteins, viruses could affect the entire photosynthetic machinery in plant leaves, consequently, decreasing any potential defense capacity. Indeed, Hodgson et al. (1989) and Reinero and Beachy (1989) reported that accumulation of TMV CP in the PSII super-complexes (Fig. 3A) from leaves of spinach (Spinacia oleracea) and Tobacco (N. tabacum L.), respectively, was associated with reduction in the photosynthetic index. In addition, Pineda et al. (2010), Di Carli et al. (2010) and Wu et al. (2013), who employed proteomic analysis to infected susceptible plants, also reported a virus-induced CP accumulation in these compatible interactions.
Fig. 3.
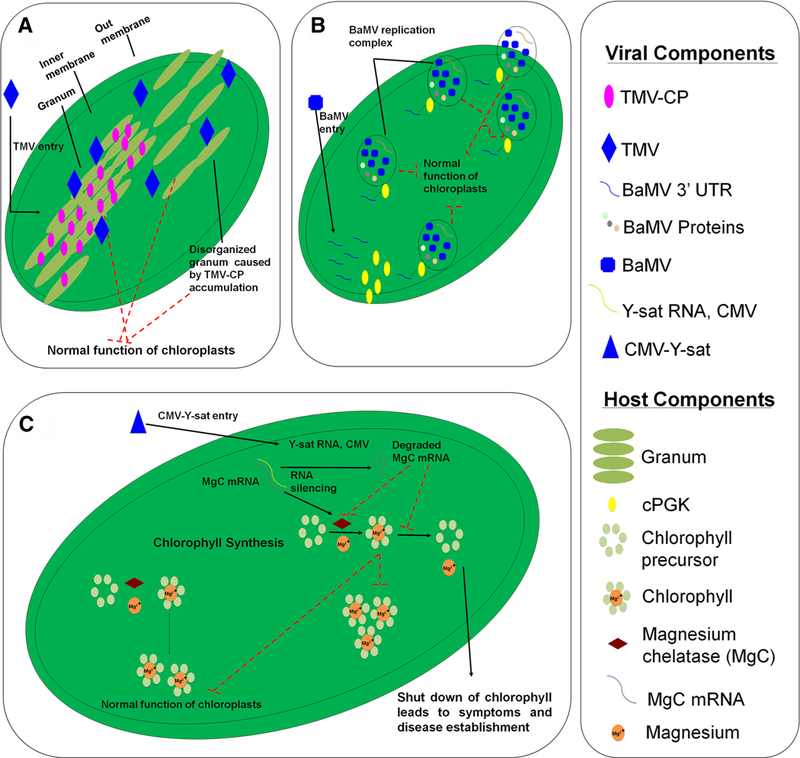
Model of virus-induced effects in chloroplast-defense related functions. a TMV accumulation in the granum leads to morphological disorders in chloroplasts. b cPGK is a host factor that participates in the formation of BaMV replication complexes in chloroplasts. c CMV-Y-sat replicates in the cytoplasm. Y-sat-derives siRNAs mediate silencing of resulting in depletion of chlorophyll content
Particularly, Pineda et al. (2010) performed a proteomic analysis of the enriched fraction of chloro-plasts obtained from tobacco leaves infected by PMMoV. Corroborating the previous data, in their study the CP accumulation at the PSII super-complex also led to inhibition of electron transport rate from PSII (ETRII), consequently leading to photoinhibition, disruption of grana organization and severe damage to chloroplasts (Fig. 2a) (Hodgson et al. 1989). Besides, several other proteomic studies have reported the reduction in D1 and D2 induced by viruses from different families and interacting with susceptible plants from different species (Wang et al. 2015, 2016; Wu et al. 2013; Zhong et al. 2017). Remarkably, the reciprocal is also true for incompatible interactions between plants and viruses. Wang et al. (2016) reported that tobacco (N. tabacum) plants, tolerant of TMV infection, were able to induce an increase in D1 and D2 proteins accumulation, whereas in TMV susceptible plants, these proteins decreased. Taken together, these studies provide strong evidence to support the idea that the control of D1 and D2 levels are a common strategic target disputed by different viruses and plants, in order to resist or establish the infection.
5.3. Ferredoxin NAD(P)+ reductase
By examining Table 2, it is also clear that ferredoxin-NADP reductase (FNR) accumulation is also commonly affected by several viral infections (Pineda et al. 2010; Qiu et al. 2018; Wang et al. 2016; Wu et al. 2013; Zhong et al. 2017). FNR is a central enzyme in PETC involved in the final electron transference to NADP+ producing NADPH. This is the primary reaction responsible for providing reducing power (NADPH) required for the reductive phase of the photosynthesis that leads to CO2 assimilation. In addition, the NADPH generated by FNR activity is also crucial for sustaining the activity of important antioxidative systems involved in the chloroplast redox protection, including NADPH-dependent thioredoxin reductase C (NTRC) system and glutathione reductase (GR) in the ascorbate–glutathione cycle (Kozuleva et al. 2016). Finally, FNR proteins are also involved in the cross-talking between chloroplasts and mitochondria, in terms of its importance for the chloroplastic NADPH exportation via the malate valve (Kozuleva et al. 2016). Thus, in addition with the changes in other proteins involved with PETC, the capacity to regulate negatively the accumulation of FNR during viral infection, should represent a strategic advantage to control the energetic machinery from plant cells, ensuring that the defense system will be minimized.
Taken together, proteomic is an approach to understand how viruses benefit from decreases of photochemical activity in the chloroplasts. However, an important question remains elusive: would the decrease in photochemical activity be a cause or consequence of compatible viral interaction? At this time, further experiments are needed in order to answer this question. By inhibiting the PETC functioning, virus-infected plants should present a subsequent ROS production by chloroplasts, leading to PCD events that interfere with viral infection (Kangasjärvi et al. 2012). Indeed, when PETC is over-reduced due to energetic imbalance between light input into antennas and lowered NADH consumption, the electron donation from P680 to oxidized plastoquinone is inhibited and charge recombination could lead to P680 triplet formation. In turn, the triplet form of chlorophyll P680 is possible to react with molecular oxygen, which will generate oxygen singlet, an important ROS involved in chloroplast retrograde signaling. Not so far, the excessive reducing power at PSI acceptor side, which is totally consistent with decreased FNR activity, could provide electrons to Mehler reaction, originating both radical ion superoxide and H2O2, also involved in chloroplast retrograde signaling, induction of defense mechanisms and even, PCD (Foyer 2018).
ROS accumulation is also an important factor regulating the synthesis of photochemical proteins, especially D1 turnover (Jimbo et al. 2018). Thus, by decreasing the activity of PETC proteins, viruses compatible interactions are able to decrease considerably the energy availability for the synthesis of defense proteins but, simultaneously they are prone to activate the entire wave of ROS retrograde signaling, which could lead to inducible expression of several plant defense genes. In this sense, the decrease of PETC proteins accumulation could be more beneficial than prejudicial to global plant defense against viruses. However, viruses able to establish compatible interactions are able to cheat all these factors through the precise regulation in synthesis/degradation of a single group of biomolecules: the chlorophylls. In the next section it will be discussed what proteomics studies have revealed about changes in chlorophyll metabolism in response to plant–virus interactions.
6. Virus-induced changes in chlorophyll metabolism
A recent proteomic study based on iTRAQ analyses of the interaction between O. sativa and RSV gave us important insights into viral interference on chlorophyll biosynthesis (Wang et al. 2015). In this study, the authors reported a decrease in abundance of proteins involved in chlorophyll biosynthesis, such as magnesium chelatase (Mg-chelatase) subunits I and D, protoporphyrinogen oxidase and magnesium-pro-toporphyrin IX monomethyl ester. All these proteins are important for Mg2+ insertion into a protoporphyrin IX molecule, which is an immediate precursor of chlorophyll biosynthesis (Walker and Weinstein 1991; Walker and Willows 1997). Reductions of Mg-chelatase in plants during infection by viruses had already been reported by Di Carli et al. (2010), however, the analysis reported by Wang et al. (2015) also suggested that other isoforms of Mg-chelatase were involved in the same response mechanism, providing new information on this issue (Fig. 3c). In addition, Shimura et al. (2011) and Liu et al. (2014) also reported the importance of Mg-chelatase shut off to the success of viral infection. These studies reported that viruses have different mechanisms to shut down this enzyme, generating a deep inactivation of plant defenses and allowing viral infection establishment (Fig. 3c).
Although there is a clear positive correlation between decreased Mg-chelatase levels and increases in disease signals development during viral infection, little is known about the viral mechanism that induces the shut-down of Mg-chelatase. Shimura et al. (2011) provided one interesting explanation while studying the interaction of CMV-Ysat and N. benthamiana. While CMV-Ysat does not replicate in chloroplasts, it does have an RNA sequence complementary to Mg-chelatase mRNA (Fig. 3c). During viral infection, CMV replicates in the cytoplasm and then moves to chloroplasts, inducing degradation of Mg-chelatase mRNA by plant cell RNA silencing machinery (Shimura et al. 2011). To date, this mechanism has only been described in CMV-Ysat infections of N. benthamiana. Indeed, other mechanism(s) for lowering Mg-chelatase levels may exist. Thus, further studies are warranted if we are going to fully understand how plant–virus interactions interfere with chlorophyll biosynthesis.
On the other hand, the lack of proteomics evidence suggesting negative changes in Mg-chelatase and other proteins related to chlorophyll metabolism during incompatible plant–virus interactions, reinforce the hypothesis that this is a primordial target for the disease establishment. In fact, pigments like chlorophyll are possible involved with NPQ formation, possibly via the quenching interactions between the lutein bound at the L1 site and chlorophylls a612–a611–a610 (Horton et al. 1996; Ruban et al. 2007). Moreover, if chlorophyll content is reduced the amount of light energy effectively absorbed will drop significantly and therefore viruses could be benefited with the decrease of ROS generation at both PSII and PSI levels and, ultimately, by the decrease of reducing power availability, crucial for the reduction phase of photosynthesis. In the next section, the importance of controlling the accumulation of proteins related to Calvin–Benson cycle during plant–virus interactions will be further discussed.
7. Virus effects on carbon assimilation reactions
Carbon assimilation reactions are extremely important to plants cope the viral infection. The trioses generated by the Calvin–Benson cycle are crucial to provide the primordial carbon skeletons that are employed in several biochemical pathways in plant cells, including for the production of important metabolites with roles related to defense against biotic stresses and hormones. As discussed previously, the virus ability to shut down the chlorophyll metabolism and PETC activity will ultimately reflect in the decrease of the carboxylation activity. However, during evolution, natural selection has favored virus strategies that ensure not only to shutting down proteins needed in photochemical reactions, but also the capability to regulate the accumulation of proteins related to the later reductive phase of photosynthesis. Indeed, numerous proteomic studies have reported decreased abundance of proteins involved in Calvin–Benson Cycle (Table 2). Abundance reduction has been more often reported for the RuBisCO small subunit, the RuBisCO large subunit, phosphoglycerate kinase (PGK), sedoheptulose-bisphosphatase (SBPase), fructose-bisphosphatase aldolase (SFBA), and rubisco activase (Table 2) (Huang et al. 2010; Kundu et al. 2013; Pineda et al. 2010; Wu et al. 2013; Zhong et al. 2017).
The observed decreases in abundance of proteins related to the Calvin–Benson cycle (CB-cycle) suggest a mechanism employed by some viruses to decrease CO2 assimilation in susceptible infected plants. Indeed, CO2 assimilation is involved with the higher demand for assimilates, required by plants to establish defense responses, including the carbon skeletons employed in the synthesis of proteins, nucleic acids and secondary metabolites (Neilson et al. 2013). For example, the accumulation of photosynthetic proteins in EMS-mutagenized cowpea plants challenged with CPSMV suggests that resistant plants evoke mechanisms to protect the photosynthetic machinery. Indeed, these results indicate that more than merely preserving it, these plants can improve the level of photosynthetic performance required for supporting the primary metabolism of plants and thereby, allowing these plants to display an effective mechanism to counterattack virus infection (data not published).
Among the proteins associated with the CB-cycle that have been shown to have differential accumulation in response to plant–virus-interactions, those of the RuBisCO large and small subunits deserve special attention. While RuBisCO is the most abundant enzyme in the planet, it is also the most inefficient. The holoenzyme is a complex comprised of four dimers of large subunits (RbcL) encoded by the chloroplast genome, and four dimers of small subunits (RbcS) encoded by the nuclear genome (Spreitzer and Salvucci 2002). However, despite changes in these proteins abundance have been very commonly reported in proteomic studies regarding plant–virus interactions, it is recommended an extra caution for interpreting this data. A special problem arises from the fact that these holoenzymes present a highly complex system of post-translational regulation (Carmo-Silva and Salvucci 2011). This regulatory system includes the carbamoylation and Mg2+ binding to Rubisco, which are crucial events that trigger the holoenzyme activation, but are also dependent on stromal alkaline pH. Moreover, RuBisCO is under the redox control by thioredoxin-dependent systems (García-Murria et al. 2018) and is also highly susceptible to inhibition by phosphate-sugars, such as 2-carboxyarabinitol-1-phosphate (CA1P), which together can ultimately limit in vivo RuBisCO activity (Andralojc et al. 2018).
RuBisCO activase is another important enzyme usually reported in the proteomic studies by its abundance changed during plant–virus interactions (Table 2). The biological activity exhibited by this enzyme is related to removal of phosphate-sugars from the RuBisCO catalytic site and, therefore, this protein is primordial for the final carboxylation activity (Carmo-Silva and Salvucci 2011). Thus, the fact that compatible plant–virus interactions generate negative changes in the abundance of both Rubisco and Rubisco activase might represent an additional evidence to support the hypothesis that viruses strategies to establish disease precludes down-regulation in the entire photosynthetic machinery, consequently allowing to shut-down plant defenses. Indeed, besides the changes in these two crucial enzymes abundance, several other proteins related to CB-cycle are commonly reported as differentially accumulated in plants undergoing virus infection.
Interestingly, phosphoglycerate kinase (PGK) should be focus of further study. Currently, proteomic data has not shown a consistent pattern in PGK levels. In some cases, PGK levels have increased regardless the resistance or susceptibility exhibited by plants, while in other cases they have simply decreased (Table 2). The fact that PGK proteins are commonly differently expressed during plant–virus interactions might indicate that this protein is anyway related to viral response. However, the understanding of the molecular mechanism in which this protein is involved during such interactions is much harder task to achieve. Possible, as previous noticed for OEEP, PGK proteins might be involved in other non-canonical functions, which are still poorly understood to date and probably could explain the complex pattern of response during exposure to different viruses. Indeed, PGK protein is an important host factor involved in the replication of several plant viruses. The 3′ UTR region of the Bamboo Mosaic Virus (BaMV genus Potexvirus) genomic RNA specifically interacts with PGK proteins, consequently driving the viral RNA into chloroplasts, and subsequently improving its replication (Fig. 3b). The interaction between BaMV 3′ UTR and PGK guides the formation of membrane-derived vesicles in chloroplasts, which is a supportative place for BaMV replication (Fig. 3b). The importance of this interaction was confirmed by using mutants that silenced to PGK protein, which lead to reduced BaMV RNA accumulation (Cheng et al. 2013; Lin et al. 2006). In addition, Arabidopsis plants that have a natural recessive resistance gene rwm1 against Watermelon Mosaic Virus (WMV, genus Potyvirus), encode a mutated version of PKG, which affects only WMV replication and infection without causing any harm to the hostplants (Lin et al. 2006; Ouibrahim et al. 2014).
Thus, the precise physiological interpretation of proteomic data regarding CB-cycle proteins differentially expressed in response to virus interactions is complex and requires cautions to avoid misinterpretations. The highly complexity of this mechanism, as well as its great importance to plant cell metabolism are consistent with this fact. In order to achieve more robust conclusions concerning the changes in abundance of these proteins is highly recommended to adopt an integrative approach, precluding the simultaneous analysis of the different CB-cycle proteins, in association with other enzymes crucial for the post-translational regulation of its activity, such as the identification of chloroplastic thioredoxins related to CB-cycle activation. Unfortunately, the identification of these proteins in proteomic studies is much less frequent and frequently neglected during data interpretation (Table 2). In addition, the use of simultaneous research/instrumentation approaches to estimate carboxylation activity, including in vitro techniques and/or gas exchange assays, are essential in order to avoid misinterpretation of proteomic data concerning the CB-cycle.
8. Changes in proteins related to photorespiratory metabolism
The photorespiratory pathway is an important meta-bolic route in C3 plants originated from the dual Rubisco affinity for both O2 and CO2. Since the oxygenation competes directly with the carboxylation activity, photorespiration has been long considered detrimental to plant productivity. However, a different role for this metabolic pathway has been arisen in the last decades, especially under excess energy conditions (Peterhansel et al. 2013). Under excess energy in PETC, photorespiration should act as an alternative electron sink, protecting the photosynthetic machinery against photoinhibition (Foyer et al. 2012). In addition, the photorespiratory N assimilatory metabolism probably might act as an important sink of carbon skeletons from chloroplasts, decreasing triose limitation of CB-cycle (Busch et al. 2018). Moreover, GO is also involved in a step of photorespiration in the peroxisome, which is responsible for the over-production of H2O2 (Corpas 2015). Thus, during compatible plant–virus interactions is crucial to viruses the capability to shut-down the photorespiratory pathway in parallel to photosynthesis in order to avoid ROS burst and, in parallel, decrease the functioning of N assimilatory machinery.
Indeed, the fact that Rubisco is the initial enzyme involved in two crucial biological processes, photosynthesis and photorespiration, could indicate that by changing the amounts of these enzyme, viruses are targeting the both routes simultaneously. However, besides the enzymes involved in the Calvin–Benson cycle, proteomic analyzes also indicate the decrease in amounts of glycolate oxidase (GO) and glutamine synthetase during compatible plant–virus interactions, important enzymes involved in photorespiration cycle. Photorespiration is the most important site of H2O2 generation in leaves (Corpas 2015), consequently the levels of this ROS should be also decreased as consequence of lower photorespiratory ratios during compatible interactions. Several studies have reported that H2O2 is an important signaling molecule involved in inducing PCD, which is crucial to prevent viral spreading. In addition, this ROS is involved in several routes of organelle retrograde signaling, which is capable to induce expression of numerous defense-related genes (Jones and Dangl 2006). Thus, the capability of virus to cause a precise and negative effect on proteins related to photorespiratory metabolism is an excellent strategy to over-come the most important plant defenses.
In this current review, a rigorous analysis of several different proteomic studies focused in plant–virus interactions and responses of photosynthesis-related metabolism, including the photorespiratory pathways, allowed us to depict a complex event regarding the regulation of proteins addressed to chloroplasts, notably those related to antenna complex, photosynthetic electron transport chain and CB-cycle. Moreover, similar trends for changes in proteins related to photorespiratory metabolism were observed, which is all these proteins are commonly strongly down-accumulated during compatible plant–virus interactions and the reciprocal is also true for incompatible relations. Indeed, in illuminated leaves it is estimated that the H2O2 production related to GO activity in peroxisomes might reach up to 10 μmol m−2 s−1, whereas in chloroplasts this ROS production should not overcome 5 μmol m−2 s−1 (Foyer and Noctor 2003). Despite similar studies involving the H2O2 rate production in different organelles during a compatible or incompatible plant–virus interaction is lacking in the literature, this simple comparison between chloroplasts and peroxisomes in terms of ROS generation could give us a nice view of the importance of photorespiratory metabolism during viral disease establishment. Moreover, beside the oxidative burst per se, it is important to highlight that any change in photorespiratory H2O2 production induced by viruses precludes alterations in the balance of reducing equivalents inside chloroplasts, the sink for carbon skeletons and triose-phosphate inhibition of CB-cycle and the ROS-dependent retrograde signaling pathways, all of which are important to eliminate plant defenses and support disease establishment (Bolton 2009). Thus, during evolution selective pressures might have allowed to virus the capability to interfere in photorespiratory metabolism as well as chlorophyll metabolism, LHC, PETC and CB-cycle enzymes, in order to complete viral life cycle.
However, despite several works have supported a decreased amount of proteins involved in photorespiration (Table 2), Kundu et al. (2013) working with V. mungo infected with MYMIV have reported for an increase of glycine hydroxymethyltransferase during a compatible plant–virus interaction. This enzyme is responsible for a crucial step in photorespiratory pathway and consequently, the comprehension of the relationship between virus infection and photorespiration remains elusive to date. Probably by example of what was noticed for PGK proteins, these transferases could exhibit other non-canonical function in plant cell during plant–virus interactions, which are not completely understood to date. If it is the case, only further studies will clarify this issue completely. Nevertheless, studies involving the photorespiratory pathway regulation and the susceptibility of plants against different viruses are promising in terms of select candidate genes to confer resistance to plants against viral infection.
9. Concluding remarks
Differential accumulation of important proteins in virus-infected plants is a complex phenomenon. Proteomic approaches are an important tool to understand plant–virus interactions. Proteins closely related with the photosynthetic metabolism, including the photorespiratory pathway, are differentially expressed in virus-infected plants. These proteins are more often related to the light harvest complex, the photosynthetic electron transport chain, the chlorophyll metabolism, the Calvin–Benson cycle and photorespirations. Two hypotheses may explain these observations (Fig. 4). Different biochemical pathways could act in sync to as an antiviral mechanism to provide resistance to virus infection. Identification of the proteins and genes involved in the interactions could be used to in plant breeding for resistance to viruses, increasing in this way the global crop productivity.
Fig. 4.
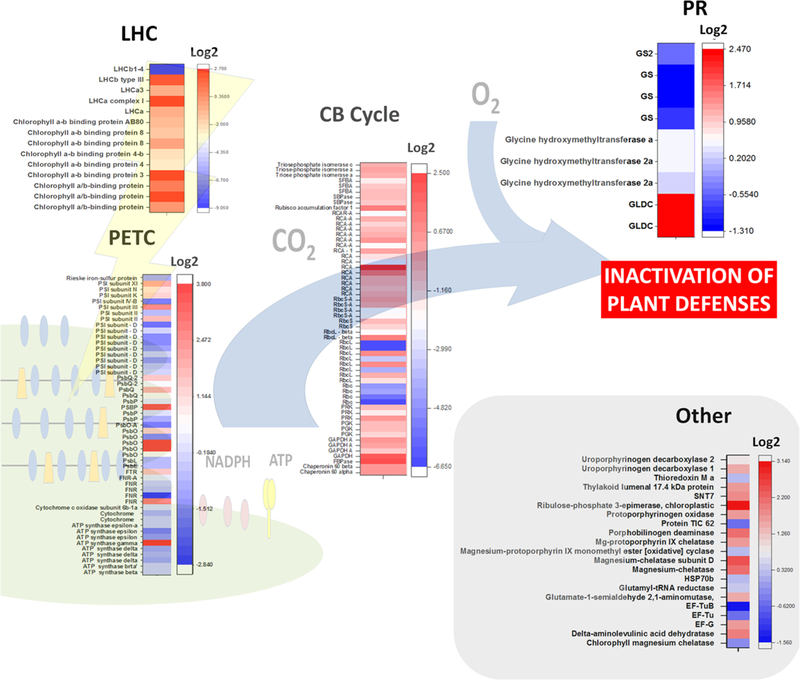
Heat map summarizing the changes in abundance of proteins related to photosynthesis and photorespiration in selected virus-infected plants. Proteomics data employed and plant species studied are indicated in Table 2. Changes in protein accumulation are expressed as log2. The heat maps were plotted using the OriginPro 2017 software (Origin Lab Corporation, Northampton, USA). CB Cycle Calvin–Benson cycle, EF-G chloroplastic enlogation factor G, EF-Tu chloro-plastic enlogation factor Tu, FNR ferredoxin-dependent NADP(H) oxireductase, FTR ferredoxin-dependent thioredoxin reductase, GAPDH glyceraldehyde 3-phosphate dehydrogenase, GLDC glycine dehydrogenase (decarboxylating), GS glutamine synthetase, HSP70b chloroplastic heat shock protein 70 b, LHC light harvesting complex, LHCa light harvesting complex from PSI, LHCb light harvesting complex from PSII, PETC photosynthetic electron transport chain, PGK phosphoglycerate kinase, PR photorespiration, PRK phosphoribulokinase, PsbO subunit O from PSII (OEE1), PsbP subunit P from PSII (OEE2), PsbQ subunit Q from PSII (OEE3), PSI photosystem I, PSII photosystem II, RbcL large subunit of Rubisco, RbcS small subunit of Rubisco, RCA Rubisco activase, SBPase sedoheptulose biphosphatase, SFBA sedoheptulose/frutose biphosphate aldolase, SNT7 serine/threonine-protein kinase
Acknowledgements
This study was supported by the following Brazilian institutions: CNPq (National Council for Scientific and Technological Development. Process Numbers: 308107/2013–6 and 306202/2017–4); CAPES (Coordination of Improvement of Higher Education. Toxinology Project, Process Number: 431511/2016–0) and Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico (FUNCAP). FELC is supported by FUNCAP/CAPES (Bolsista CAPES/BRASIL – Proc. 88887.162856/2018–00). Research at the Garcia-Ruiz lab is supported by NIH grant R01GM120108 to Hernan Garcia-Ruiz and by the Nebraska Agricultural Experiment Station with funding from the Hatch Act (Accession Number 1007272) through the USDA National Institute of Food and Agriculture.
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interest.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pedro F. N. Souza, Department of Plant Pathology, Nebraska Center for Virology, University of Nebraska-Lincoln, Lincoln, NE, USA Department of Biochemistry and Molecular Biology, Federal University of Ceará, Fortaleza, Ceará, Brazil.
Hernan Garcia-Ruiz, Department of Plant Pathology, Nebraska Center for Virology, University of Nebraska-Lincoln, Lincoln, NE, USA.
Fabricio E. L. Carvalho, Department of Biochemistry and Molecular Biology, Federal University of Ceará, Fortaleza, Ceará, Brazil
References
- Acosta-Leal R, Duffy S, Xiong Z, Hammond R, Elena SF (2011) Advances in plant virus evolution: translating evolutionary insights into better disease management. Phytopathology 101:1136–1148 [DOI] [PubMed] [Google Scholar]
- Ahn TK, Avenson TJ, Ballottari M, Cheng Y-C, Niyogi KK, Bassi R, Fleming GR (2008) Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320:794–797 [DOI] [PubMed] [Google Scholar]
- Alexander MM, Cilia M (2016) A molecular tug-of-war: global plant proteome changes during viral infection. Curr Plant Biol 5:13–24 [Google Scholar]
- Andralojc PJ, Carmo-Silva E, Degen GE, Parry MAJ (2018) Increasing metabolic potential: C-fixation. Essays Bio-chem 62:109–118 [DOI] [PubMed] [Google Scholar]
- Aro EM, McCaffery S, Anderson JM (1993) Photoinhibition and D1 protein degradation in peas acclimated to different growth irradiances. Plant Physiol 103:835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniam M, Kim B-S, Hutchens-Williams HM, Loesch-Fries LS (2014) The photosystem II oxygen-evolving complex protein PsbP interacts with the coat protein of Alfalfa mosaic virus and inhibits virus replication. Mol Plant Microbe Interact 27:1107–1118 [DOI] [PubMed] [Google Scholar]
- Baltimore D (1971) Expression of animal virus genomes. Bacteriol Rev 35:235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran PMJ, Federspiel JD, Sheng X, Cristea IM (2017) Proteomics and integrative omic approaches for understanding host–pathogen interactions and infectious diseases. Mol SystBiol 13:922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya D, Chakraborty S (2018) Chloroplast: the Trojan horse in plant–virus interaction. Mol Plant Pathol 19:504–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Voinnet O (2014) The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol 65:473–503 [DOI] [PubMed] [Google Scholar]
- Bolton MD (2009) Primary metabolism and plant defense—fuel for the fire. Mol Plant Microbe Interact 22:487–497 [DOI] [PubMed] [Google Scholar]
- Bricker TM, Frankel LK (2011) Auxiliary functions of the PsbO, PsbP and PsbQ proteins of higher plant photosystem II: a critical analysis. J Photochem Photobiol B Biol 104:165–178 [DOI] [PubMed] [Google Scholar]
- Busch FA, Sage RF, Farquhar GD (2018) Plants increase CO2 uptake by assimilating nitrogen via the photorespiratory pathway. Nat Plants 4:46–54 [DOI] [PubMed] [Google Scholar]
- Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar S (2008) Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132:449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo LS, Murad AM, Resende RO, Boiteux LS, Ribeiro SG, Jorrín-Novo JV, Mehta A (2017) Plant responses to tomato chlorotic mottle virus: proteomic view of the resistance mechanisms to bipartite begomovirus in tomato. J Proteom 151:284–292 [DOI] [PubMed] [Google Scholar]
- Carmo-Silva AE, Salvucci ME (2011) The activity of Rubisco’s molecular chaperone, Rubisco activase, in leaf extracts. Photosynth Res 108:143–155 [DOI] [PubMed] [Google Scholar]
- Cerna H, Černý M, Habánová H, Šafářová D, Abushamsiya K, Navrátil M, Brzobohatý B (2017) Proteomics offers insight to the mechanism behind Pisum sativum L. response to pea seed-borne mosaic virus (PSbMV). J Proteom 153:78–88 [DOI] [PubMed] [Google Scholar]
- Chaerle L, Van Caeneghem W, Messens E, Lambers H, Van Montagu M, Van Der Straeten D (1999) Presymptomatic visualization of plant–virus interactions by thermography. Nat Biotechnol 17:813–816 [DOI] [PubMed] [Google Scholar]
- Cheng S-F, Huang Y-P, Chen L-H, Hsu Y-H, Tsai C-H (2013) Chloroplast phosphoglycerate kinase is involved in the targeting of Bamboo mosaic virus to chloroplasts in Ni-cotiana benthamiana plants. Plant Physiol 163:1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmeliov J, Bricker WP, Lo C, Jouin E, Valkunas L, Ruban AV, Duffy CD (2015) An “all pigment” model of excitation quenching in LHCII. Phys Chem Chem Phys 17:15857–15867 [DOI] [PubMed] [Google Scholar]
- Corpas FJ (2015) What is the role of hydrogen peroxide in plant peroxisomes? Plant Biol 17:1099–1103 [DOI] [PubMed] [Google Scholar]
- Cueto-Ginzo AI, Serrano L, Bostock RM, Ferrio JP, Rodríguez R, Arcal L, Achon MÁ, Falcioni T, Luzuriaga WP, Medina V (2016) Salicylic acid mitigates physiological and proteomic changes induced by the SPCP1 strain of Potato virus X in tomato plants. Physiol Mol Plant Pathol 93:1–11 [Google Scholar]
- de Torres Zabala M, Littlejohn G, Jayaraman S, Studholme D, Bailey T, Lawson T, Tillich M, Licht D, Bölter B, Delfino L, Truman W, Mansfield J, Smirnoff N, Grant M (2015) Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat Plant 1:15074. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B (1990) Carotenoids and photoprotection in plants: a role for the xanthophyll zeaxanthin. BBA—Bioenergy 1020:1–24 [Google Scholar]
- Di Carli M, Villani ME, Bianco L, Lombardi R, Perrotta G, Benvenuto E, Donini M (2010) Proteomic analysis of the plant–virus interaction in Cucumber Mosaic Virus (CMV) resistant transgenic tomato. J Proteome Res 9:5684–5697 [DOI] [PubMed] [Google Scholar]
- Di Carli M, Benvenuto E, Donini M (2012) Recent insights into plant-virus interactions through proteomic analysis. J Proteome Res 11:4765–4780 [DOI] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Truniger V, Nieto C, Garcia-Mas J, Bendah-mane A, Aranda MA (2004) Advances in understanding recessive resistance to plant viruses. Mol Plant Pathol 5:223–233 [DOI] [PubMed] [Google Scholar]
- Elrad D, Niyogi KK, Grossman AR (2002) A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14:1801–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira M, Galdeano MM, Gilardi P, García-Luque I, Serra MT (2008) Proteomic analysis of pathogenesis-related proteins (PRs) indeuced by compatible and incompatible interactions of pepper mild mottle virus (PMMoV) in Capsicum Chinese L.3 plants. J Exp Bot 59:1253–1265 [DOI] [PubMed] [Google Scholar]
- Foyer CH (2018) Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ Exp Bot 154:134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2003) Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxi-somes and mitochondria. Physiol Plant 119:355–364 [Google Scholar]
- Foyer CH, Neukermans J, Queval G, Noctor G, Harbinson J (2012) Photosynthetic control of electron transport and the regulation of gene expression. J Exp Bot 63:1637–1661 [DOI] [PubMed] [Google Scholar]
- García-Murria MJ, Sudhani HPK, Marín-Navarro J, del Pino MMS, Moreno J (2018) Dissecting the individual contribution of conserved cysteines to the redox regulation of Rubisco. Photosynth Res 137:251–262 [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC (2010) Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during turnip mosaic virus infection. Plant Cell 22:481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Garcia Ruiz MT, Peralta G, Manuel S, Gabriel M, Betzabeth C, El-Mounadi K (2016) Mecanismos, aplicaciones y perspectivas del silenciamiento geínico de virus en plantas. Rev Mex Fitopatol 34:286–307 [Google Scholar]
- Garcia-Ruiz H, Peralta GSM, Harte-Maxwell PA (2018) Tomato spotted wilt virus NSs protein supports infection and systemic movement of a potyviruses and is a symptom determinant. Viruses 10:1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson RA, Beachy RN, Pakrasi HB (1989) Selective inhibition of photosystem II in spinach by tobacco mosaic virus: an effect of the viral coat protein. FEBS Lett 245:267–270 [DOI] [PubMed] [Google Scholar]
- Horton P, Ruban AV, Walters RG (1996) Regulation of light harvesting in green plants. Annu Rev Plant Physiol Plant Mol Biol 47:655–684 [DOI] [PubMed] [Google Scholar]
- Huang T-S, Wei T, Laliberte J-F, Wang A (2010) A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol 152:255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Ma HY, Huang W, Wang F, Xu ZS, Xiong AS (2016) Comparative proteomic analysis provides novel insight into the interaction between resistant vs susceptible tomato cultivars and TYLCV infection. BMC Plant Biol 16:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Tikkanen M, Zhang SB (2018) Photoinhibition of photosystem I in Nephrolepis falciformis depends on reactive oxygen species generated in the chloroplast stroma. Photosynth Res 137:129–140 [DOI] [PubMed] [Google Scholar]
- Hwang J, Lee S, Lee JH, Kang WH, Kang JH, Kang MY, Oh CS, Kang BC (2015) Plant translation elongation factor 1Bβ facilitates potato virus X (PVX) infection and interacts with PVX triple gene block protein 1. PLoS ONE 10:e0128014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimbo H, Yutthanasirikul R, Nagano T, Hisabori T, Hihara Y, Nishiyama Y (2018) Oxidation of translation factor EF-Tu inhibits the repair of photosystem II. Plant Physiol 176:2691–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MP, Ruban AV (2010) Arabidopsis plants lacking PsbS protein possess photoprotective energy dissipation. Plant J 61:283–289 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323. [DOI] [PubMed] [Google Scholar]
- Kale R, Hebert AE, Frankel LK, Sallans L, Bricker TM, Pospíšil P (2017) Amino acid oxidation of the D1 and D2 proteins by oxygen radicals during photoinhibition of photosystem II. Proc Natl Acad Sci 114:2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangasjärvi S, Neukermans J, Li S, Aro E-M, Noctor G (2012) Photosynthesis, photorespiration, and light signalling in defence responses. J Exp Bot 63:1619–1636 [DOI] [PubMed] [Google Scholar]
- Karpiński S, Szechyńska-Hebda M, Wituszyńska W, Burdiak P (2013) Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ 36:736–744 [DOI] [PubMed] [Google Scholar]
- Kozuleva M, Goss T, Twachtmann M, Rudi K, Trapka J, Selinski J, Ivanov B, Garapati P, Steinhoff HJ, Hase T, Scheibe R, Klare JP, Hanke GT (2016) Ferre-doxin:NADP(H) oxidoreductase abundance and location influences redox poise and stress tolerance. Plant Physiol 172:1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S, Chakraborty D, Pal A (2011) Proteomic analysis of salicylic acid induced resistance to Mungbean yellow mosaic india virus in Vigna mungo. J Proteom 74:337–349 [DOI] [PubMed] [Google Scholar]
- Kundu S, Chakraborty D, Kundu A, Pal A (2013) Proteomics approach combined with biochemical attributes to elucidate compatible and incompatible plant–virus interactions between Vigna mungo and Mungbean yellow mosaic india virus. Proteome Sci 11:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliberté J-F, Sanfaçon H (2010) Cellular remodeling during plant virus infection. Annu Rev Phytopathol 48:69–91 [DOI] [PubMed] [Google Scholar]
- Lin J-W, Ding M-P, Hsu Y-H, Tsai C-H (2006) Chloroplast phosphoglycerate kinase, a gluconeogenetic enzyme, is required for efficient accumulation of Bamboo mosaic virus. Nucl Acids Res 35:424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yang J, Bi H, Zhang P (2014) Why mosaic? Gene expression profiling of African Cassava Mosaic Virus-infected cassava reveals the effect of chlorophyll degradation on symptom development. J Integr Plant Biol 56:122–132 [DOI] [PubMed] [Google Scholar]
- Lodha TD, Hembram P, Nitile Tep JB (2013) Proteomics: a successful approach to understand the molecular mechanism of plant-pathogen interaction. Am J Plant Sci 4:1212–1226 [Google Scholar]
- Lundin B, Thuswaldner S, Shutova T, Eshaghi S, Samuelsson G, Barber J, Andersson B, Spetea C (2007) Subsequent events to GTP binding by the plant PsbO protein: structural changes, GTP hydrolysis and dissociation from the photosystem II complex. Biochim Biophys Acta 1767:500–508 [DOI] [PubMed] [Google Scholar]
- Mehta A, Brasileiro AC, Souza DS, Romano E, Campos MA, Grossi-de-Sá MF, Silva MS, Franco OL, Fragoso RR, Bevitori R, Rocha TL (2008) Plant–pathogen interactions: what is proteomics telling us? FEBS J 275:3731–3746 [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Ogata Y, Hirata Y, Ohki ST (2014) Quantitative transcriptional changes associated with chlorosis severity in mosaic leaves of tobacco plants infected with Cucumber mosaic virus. Mol Plant Pathol 15:242–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montasser M, Al-Ajmy A (2015) Histopathology for the influence of CMV infection on tomato cellular structures. FASEB J 29:887 [Google Scholar]
- Nagy PD (2008) Yeast as a model host to explore plant virus–host interactions. Annu Rev Phytopathol 46:217–242 [DOI] [PubMed] [Google Scholar]
- Nagy PD, Pogany J (2012) The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10:137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson EH, Goodger JQ, Woodrow IE, Møller BL (2013) Plant chemical defense: at what cost? Trends Plant Sci 18:250–258 [DOI] [PubMed] [Google Scholar]
- Niehl A, Zhang ZJ, Kuiper M, Peck SC, Heinlein M (2013) Label-free quantitative proteomic analysis of systemic responses to local wounding and virus infection in Ara-bidopsis thaliana. J Proteome Res 12:2491–2503 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Li XP, Rosenberg V, Jung HS (2005) Is PsbS the site of non-photochemical quenching in photosynthesis? J Exp Bot 56:375–382 [DOI] [PubMed] [Google Scholar]
- Ouibrahim L, Mazier M, Estevan J, Pagny G, Decroocq V, Desbiez C, Moretti A, Gallois JL, Caranta C (2014) Cloning of the Arabidopsis rwm1 gene for resistance to Watermelon mosaic virus points to a new function for natural virus resistance genes. Plant J 79:705–716 [DOI] [PubMed] [Google Scholar]
- Paiva AL, Oliveira JT, de Souza GA, Vasconcelos IM (2016) Label-free proteomic reveals that Cowpea Severe Mosaic Virus transiently suppresses the host leaf protein accumulation during the compatible interaction with cowpea (Vigna unguiculata [L.] Walp.). J Proteome Res 15:4208–4220 [DOI] [PubMed] [Google Scholar]
- Pérez-Bueno ML, Rahoutei J, Sajnani C, García-Luque I, Barón M (2004) Proteomic analysis of the oxygen-evolving complex of photosystem II under biotec stress: studies on Nicotiana benthamiana infected with tobamoviruses. Proteomics 4:418–425 [DOI] [PubMed] [Google Scholar]
- Perez-Bueno ML, Ciscato M, García-Luque I, Valcke R, Barón M (2006) Imaging viral infection: studies on Nicotiana benthamiana plants infected with the pepper mild mottle tobamovirus. Photosynth Res 90:111–123 [DOI] [PubMed] [Google Scholar]
- Peterhansel C, Krause K, Braun HP, Espie GS, Fernie AR, Hanson DT, Keech O, Maurino VG, Mielewczik M, Sage RF (2013) Engineering photorespiration: current state and future possibilities. Plant Biol 15:754–758 [DOI] [PubMed] [Google Scholar]
- Pineda M, Sajnani C, Baron M (2010) Changes induced by the Pepper mild mottle tobamovirus on the chloroplast proteome of Nicotiana benthamiana. Photosynth Res 103:31–45 [DOI] [PubMed] [Google Scholar]
- Prod’homme D, Le Panse S, Drugeon G, Jupin I (2001) Detection and subcellular localization of the turnip yellow mosaic virus 66 K replication protein in infected cells. Virology 281:88–101 [DOI] [PubMed] [Google Scholar]
- Prod’homme D, Jakubiec A, Tournier V, Drugeon G, Jupin I (2003) Targeting of the turnip yellow mosaic virus 66 K replication protein to the chloroplast envelope is mediated by the 140 K protein. J Virol 77:9124–9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Zhang Y, Wang C, Lei R, Wu Y, Li X, Zhu S (2018) Cucumber mosaic virus coat protein induces the development of chlorotic symptoms through interacting with the chloroplast ferredoxin I protein. Sci Rep 8:1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino B, Candido E, Campos P, Franco O, Krüger R (2010) Proteomic approaches to study plant–pathogen interactions. Phytochemistry 71:351–362 [DOI] [PubMed] [Google Scholar]
- Rahoutei J, García-Luque I, Barón M (2000) Inhibition of photosynthesis by viral infection: effect on PSII structure and function. Physiol Plant 110:286–292 [Google Scholar]
- Reinero A, Beachy RN (1989) Reduced photosystem II activity and accumulation of viral coat protein in chloroplasts of leaves infected with tobacco mosaic virus. Plant Physiol 89:111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose JL, Frankel LK, Mummadisetti MP, Bricker TM (2016) The extrinsic proteins of photosystem II: update. Planta 243:889–908 [DOI] [PubMed] [Google Scholar]
- Ruban AV (2017) Quantifying the efficiency of photoprotection. Philos Trans R Soc B Biol Sci 372:20160393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban AV, Wentworth M, Yakushevska AE, Andersson J, Lee PJ, Keegstra W, Dekker JP, Boekema EJ, Jansson S, Horton P (2003) Plants lacking the main light-harvesting complex retain photosystem II macro-organization. Nature 421:648–652 [DOI] [PubMed] [Google Scholar]
- Ruban AV, Berera R, Ilioaia C, van Stokkum IHM, Kennis JTM, Pascal AA, van Amerongen H, Robert B, Horton P, van Grondelle R (2007) Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450:575–578 [DOI] [PubMed] [Google Scholar]
- Ruban AV, Johnson MP, Duffy CDPP (2012) The photoprotective molecular switch in the photosystem II antenna. Biochim Biophys Acta 1817:167–181 [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu Tabata S (1999) Complete Structure of the chloroplast genome of Arabidopsis thaliana. DNA Res 290:283–290 [DOI] [PubMed] [Google Scholar]
- Serra-Soriano M, Navarro JA, Genoves A, Pallás V (2015) Comparative proteomic analysis of melon phloem exudates in response to viral infection. J Proteom 124:11–24 [DOI] [PubMed] [Google Scholar]
- Shen J-R (2015) The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu Rev Plant Biol 66:23–48 [DOI] [PubMed] [Google Scholar]
- Shimura H, Pantaleo V, Ishihara T, Myojo N, Inaba J-I, Sueda K, Burgyán J, Masuta C (2011) A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog 7:e1002021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitov AV, Pobeguts OV, Smolova TN, Allakhverdiev SI, Klimovet VV (2009) Manganese-dependent carboanhydrase activity of photosystem II proteins. Biochemistry 74:509–517 [DOI] [PubMed] [Google Scholar]
- Silveira JAG, Carvalho FEL (2016) Proteomics, photosynthesis and salt resistance in crops: an integrative view. J Proteom 143:24–35 [DOI] [PubMed] [Google Scholar]
- Souza PF, Silva FD, Carvalho FE, Silveira JA, Vasconcelos IM, Oliveira JT (2017) Photosynthetic and biochemical mechanisms of an EMS-mutagenized cowpea associated with its resistance to Cowpea severe mosaic virus. Plant Cell Rep 36:219–234 [DOI] [PubMed] [Google Scholar]
- Spreitzer RJ, Salvucci ME (2002) Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu Rev Plant Biol 53:449–475 [DOI] [PubMed] [Google Scholar]
- Tajima Y, Iwakawa H-O, Hyodo K, Kaido M, Mise K, Okuno T (2017) Requirement for eukaryotic translation initiation factors in cap-independent translation differs between bipartite genomic RNAs of red clover necrotic mosaic virus. Virology 509:152–158 [DOI] [PubMed] [Google Scholar]
- Takagi D, Takumi S, Hashiguchi M, Sejima T, Miyake C (2016) Superoxide and singlet oxygen produced within the thy-lakoid membranes both cause photosystem I photoinhibition. Plant Physiol 171:1626–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toby TK, Fornelli L, Kelleher NL (2016) Progress in top-down proteomics and the analysis of proteoforms. Annu Rev Anal Chem 9:499–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Amerongen H, Croce R (2013) Light harvesting in photo-system II. Photosynth Res 116:251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela ALN, Komatsu S, Wang X, Silva RGG, Souza PFN, Lobo AKM, Vasconcelos IM, Silveira JAG, Oliveira JTA (2017) Gel-free/label-free proteomic, photosynthetic, and biochemical analysis of cowpea (Vigna unguiculata [L.] Walp.) resistance against Cowpea severe mosaic virus (CPSMV). J Proteom 163:76–91 [DOI] [PubMed] [Google Scholar]
- Varela ALN, Oliveira JTA, Komatsu S et al. (2018) A resistant cowpea (Vigna unguiculata [L.] Walp.) genotype became susceptible to cowpea severe mosaic virus (CPSMV) after exposure to salt stress. J Proteom. 10.1016/j.jprot.2018.11.015 [DOI] [PubMed] [Google Scholar]
- Ventelon-Debout M, Delalande F, Brizard JP, Diemer H, Van Dorsselaer A, Brugidou C (2004) Proteome analysis of cultivar-specific deregulations of Oryza sativa indica and O. sativa japonica cellular suspensions undergoing Rice yellow mottle virus infection. Proteomics 4:216–225 [DOI] [PubMed] [Google Scholar]
- Voinnet O (2001) RNA silencing as a plant immune system against viruses. Trends Genet 17:449–459 [DOI] [PubMed] [Google Scholar]
- Walker CJ, Weinstein JD (1991) In vitro assay of the chlorophyll biosynthetic enzyme Mg-chelatase: resolution of the activity into soluble and membrane-bound fractions. Proc Natl Acad Sci 88:5789–5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC, Willows DR (1997) Mechanism and regulation of Mg-chelatase. Biochem J 327:321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I (2011) Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol 9:860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A (2015) Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu Rev Phytopathol 53:45–66 [DOI] [PubMed] [Google Scholar]
- Wang B, Ren Y, Lu C, Wang X (2015) iTRAQ-based quantitative proteomics analysis of rice leaves infected by Rice stripe virus reveals several proteins involved in symptom formation. Virol J 12:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang X-r, Zhou Q, Yang J-m, Guo H-x, Yang L-j, Liu W-q (2016) iTRAQ protein profile analysis provides integrated insight into mechanisms of tolerance to TMV in tobacco (Nicotiana tabacum). J Proteom 132:21–30 [DOI] [PubMed] [Google Scholar]
- Wei T, Zhang C, Hou X, Sanfacon H, Wang A (2013) The SNARE protein Syp71 is essential for turnip mosaic virus infection by mediating fusion of virus-induced vesicles with chloroplasts. PLoS Pathog 9:e1003378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield AE, Falk BW, Rotenberg D (2015) Insect vector-mediated transmission of plant viruses. Virology 480:278–289 [DOI] [PubMed] [Google Scholar]
- Wu L, Han Z, Wang S, Wang X, Sun A, Zu X, Chen Y (2013) Comparative proteomic analysis of the plant–virus interaction in resistant and susceptible ecotypes of maize infected with sugarcane mosaic virus. J Proteomics 89:124–140 [DOI] [PubMed] [Google Scholar]
- Xu Q, Ni H, Chen Q, Sun F, Zhou T, Lan Y, Zhou Y (2013) Comparative proteomic analysis reveals the cross-talk between the responses induced by H2O2 and by long-term Rice black-streaked dwarf virus infection in Rice. PLoS ONE 8:e81640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Yu L, Chen Z, Zhang S, Shi J, Zhao X, Yang Y, Hu D, Song B (2017) Label-free quantitative proteomic analysis of chitosan oligosaccharide-treated rice infected with southern rice black-streaked dwarf virus. Viruses 115:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Hargett SR, Frankel LK, Bricker TM (2009) The PsbP protein, but not the PsbQ protein, is required for normal thylakoid architecture in Arabidopsis thaliana. FEBS Lett 583:2142–2147 [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhang X, Hong Y, Liu Y (2016) Chloroplast in plant–virus interaction. Front Microbiol 7:1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Wang ZQ, Xiao R, Wang Y, Xie Y, Zhou X (2017) iTRAQ analysis of the tobacco leaf proteome reveals that RNA-directed DNA methylation (RdDM) has important roles in defense against geminivirus-betasatellite infection. J Proteom 152:88–101 [DOI] [PubMed] [Google Scholar]


