Abstract
Expression of DAZ-like (DAZL) is a hallmark of vertebrate germ cells, and is essential for embryonic germ cell development and differentiation, yet the gametogenic function of DAZL has not been fully characterized and most of its in vivo direct targets remain unknown. We showed that postnatal stage-specific deletion of Dazl in mouse germ cells did not affect female fertility, but caused complete male sterility with gradual loss of spermatogonial stem cells, meiotic arrest and spermatid arrest. Using the genome-wide high-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation and mass spectrometry approach, we found that DAZL bound to a large number of testicular mRNA transcripts (at least 3008) at the 3′-untranslated region and interacted with translation proteins including poly(A) binding protein. In the absence of DAZL, polysome-associated target transcripts, but not their total transcripts, were significantly decreased, resulting in a drastic reduction of an array of spermatogenic proteins and thus developmental arrest. Thus, DAZL is a master translational regulator essential for spermatogenesis.
Keywords: RNA binding proteins, CLIPs, infertility, translational regulation, DAZ
INTRODUCTION
Germ cells are considered immortal as they are the only cells that pass from one generation to the next, while somatic cells die within a single generation [1]. It has long been thought that germ cells utilize a unique set of tools and mechanisms to achieve their distinct functions [1,2]. One such tool is conserved germ cell-specific expression of RNA binding proteins among animals [1,3]. Deleted in Azoospermia-like (DAZL) is one of those germ cell-specific RNA binding proteins and its expression is a hallmark of vertebrate germ cells [4–6]. DAZL belongs to a human fertility protein family, the Deleted in AZoospermia (DAZ) family, which consists of DAZ, DAZL and BOULE [7,8]. DAZ and BOULE appear to be required only for male fertility [9,10], but DAZL is required for both male and female fertility [11]. Genetic and epigenetic variations affecting human DAZL protein are associated with human infertility [12–15]. In addition, DAZL promotes in vitro differentiation of human embryonic stem cells (ESCs) towards haploid gametes [16]. Hence, better understanding of the molecular function of DAZL could provide insights into not only fundamental features of germ cells, but also into human infertility and the development of in vitro gamete production technology.
Mouse Dazl was shown to be critically important in early primordial germ cells (PGCs), and for both male and female fertility [11,17,18] in the mixed background of Dazl knockout mice. Analysis of mutant mice in a C57/B6 pure background revealed essential roles for development and sexual differentiation of PGCs [19–21]. Such a PGC requirement also appears to be conserved in other vertebrates [4]. In contrast, the role of Dazl in gametogenesis is less clear, partly due to the extensive loss of embryonic germ cells in Dazl knockout mice. Studies of the remaining spermatogenic cells in Dazl mutant testes suggested defects in spermatogonial transition and a final block at the leptotene stage of meiosis [17,18]. Stage-specific examination of Dazl function bypassing the early PGC requirement is hence needed to provide a full picture of Dazl’s function in gametogenesis.
Dazl has been proposed to be important for mouse oocyte maturation and the oocyte–zygote transition based on Dazl knockdown via RNA interference [22]. This is consistent with studies implicating Xenopus Dazl protein's roles in the translational control of oocyte maturation [23–25]. Surprisingly, conditional knockout of Dazl by oocyte-specific GDNF9-Cre produced a normal number of pups [26], although the contribution of Dazl at or before primordial follicles has not be excluded.
The first clue regarding the DAZ family proteins’ molecular function came from a study on the Drosophila boule homolog, which was shown to be required for posttranscriptional control of the CDC25 homolog twine [27]. While twine was not shown to be a direct target of Boule, a series of studies on mammalian DAZL protein revealed potential binding motifs and targets. DAZL bound to the 5′UTR of Cdc25c by (GUn)n using Systematic Evolution of Ligands by Exponential Enrichment (SELEX) and a tri-hybrid system [28]. Another in vitro experiment (Specific Nucleic Acids Associated with Proteins technique (SNAAP)) revealed that mDAZL protein could specifically bind to at least nine distinct mRNAs, including the 3′UTR of Tpx-1 [29]. The crystal structures of the RNA Recognition Motif (RRM) from murine DAZL were characterized and verified to be able to bind the motif GUU [30]. Reynolds et al. performed DAZL testis immunoprecipitation and microarray hybridization, and identified at least 11 DAZL targets from the testes directly. Using a similar RNA Immunoprecipitation (RIP) approach, DAZL targets from mouse PGCs and human fetal ovaries were also identified [21,31]. However, most of the in vivo direct targets of DAZL remain unknown. Systematic identification of those direct targets in their physiological context is key if we are to understand how DAZL regulates those targets during spermatogenesis and why DAZ family proteins are critical in sperm development.
While posttranscriptional regulation has been established for DAZ proteins, increasing evidence points to a major role of DAZL in translational control. Tsui et al. reported that DAZL could bind to poly(A) RNAs, implicating DAZL in translation control [32]. Using a tethering translation assay in Xenopus laevis oocytes, Collier et al. demonstrated nicely that DAZL promotes target translation by recruiting poly(A) binding protein (PABP) [24]. Among 11 testicular mRNA targets, Reynolds et al. showed that the translation of two of them (Vasa and Sycp3) could be enhanced by DAZL based on a few surviving germ cells in Dazl knockout mouse testes [33,34]. Polysome profiling of mouse oocytes at different stages revealed extensive translational regulation, and DAZL was proposed to be one of the key translational regulators working synergistically with Cytoplasmic Polyadenylation Element Binding protein (CPEB) [22,35]. However, the translational role of DAZL appears to be context-dependent, as both translational promotion and repression were reported for mammalian DAZL [21,36]. Despite progress studying DAZL protein, our understanding of DAZL’s functions in spermatogenesis remains incomplete. A major limitation is a lack of systemic knowledge of its binding targets and interacting proteins in their native context. Thus, comprehensive identification of DAZL targets in gametogenesis is necessary to understand their roles and why they are so critical in human fertility. Therefore, we determine the stage-specific requirements of Dazl in postnatal gametogenesis, then identified comprehensive direct targets and protein partners of DAZL in the mouse testis in vivo via both transcriptome-wide high-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation (HITS-CLIP) and immunoprecipitation (IP) mass spectrometry, and finally interrogate the impact of stage-specific deletion of DAZL on the identified DAZL targets, to unravel the molecular circuitry of DAZL-mediated posttranscriptional regulation.
RESULTS
Dynamic and continuous Dazl expression from spermatogonial stem cells to the round spermatid stage
Although DAZL was previously found to be highly expressed in various stages of germ cell development using different antibodies and a DAZL-GFP reporter [8,11,37], the extent of DAZL expression, especially in specific stages such as spermatogonial stem cells (SSCs) or round spermatids, was not clearly defined. We decided to perform a systematic analysis of DAZL expression at both the RNA and protein levels, using our validated DAZL antibody to gain precise and detailed expression and localization patterns from ESCs through postnatal gametogenesis, supported by western blot analysis of STA-PUT method-purified spermatogenic cells at different stages and immunofluorescent co-localization with stage-specific markers. DAZL protein expression in SSCs and round spermatids was clearly established by co-localization with stage-specific markers, and western analysis of purified SSCs and round spermatids (see Supplementary Figs S1C and F). We confirmed DAZL expression in the embryonic gonads, and throughout postnatal male germ cell development and differentiation (See Supplementary Fig. S1), validating its expression as a bona fide germ cell marker, and suggesting a global and central role during spermatogenesis.
Removal of Dazl in gonocytes, spermatogonia and spermatocytes led to infertility with distinct spermatogenic phenotypes
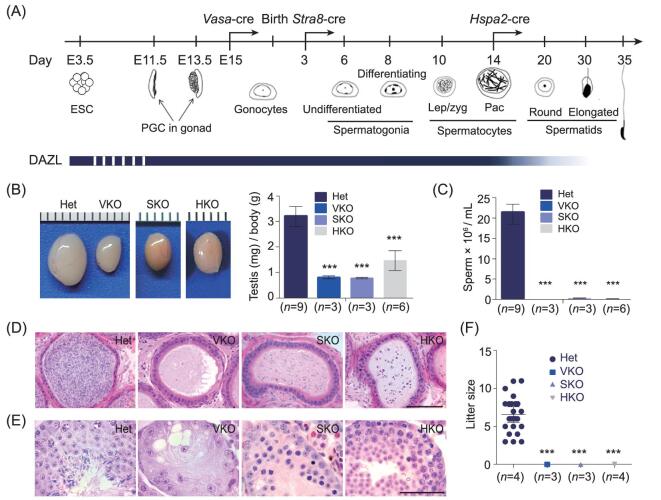
In order to investigate Dazl function throughout spermatogenesis, we constructed a conditional Dazl knockout mouse and confirmed its nature as a loss of function mutation after the deletion of exons 4, 5 and 6, in comparison to the Dazl whole-body knockout mouse (see Supplementary Fig. S2) [11]. The availability of a conditional Dazl knockout in combination with germ cell-specific Cre active at different time points of germline development allowed us to determine the requirement of Dazl systematically in postnatal gametogenesis. Remarkably, deletion of Dazl at the gonocyte (Vasa-Cre; Dazlf/−, VKO), spermatogonia (Stra8-Cre; Dazlf/−, SKO) and spermatocyte stages (Hspa2-Cre; Dazlf/−, HKO) [38–40] all led to complete sterility, with reduced testis size and absence of sperm in the epididymis (see Fig. 1A–D), revealing a persistent critical requirement in at least three stages of spermatogenesis. Heterozygotes appeared indistinguishable from those of wild type mice and hence were used as littermate controls in the experiments. Examination of adult testis sections showed an absence of germ cells in VKO, an arrest at the zygotene stage in SKO, and an arrest at the round spermatid stage in HKO mice (see Fig. 1E). Hence, Dazl is critically required in SSCs, meiosis and spermiogenesis.
Figure 1.
Removal of Dazl in gonocytes, spermatogonia and spermatocyte respectively led to infertility with distinct spermatogenic phenotype. (A) Strategy for conditional removal of Dazl in gonocytes, spermatogonia and spermatocyte via three germ cell-Cre. The onset expression of Cre was indicated by an arrow on developmental time line. DAZL protein expression was shown as solid bar in blue color. Dotted line indicated that DAZL expression is unclear during this period. (B) Adult testes size was reduced to different extent in three conditional knockout mice. (C) No sperm in adult VKO and barely any sperm in SKO and HKO were observed in epididymis. (D) H&E stained epididymis sections showed no or little sperm in conditional knockout (cKO) mouse testes. Scale bar: 100 μm. (E) H&E stained testis sections from heterozygotes (Het), VKO, SKO and HKO showing distinct spermatogenic defects inside adult seminiferous tubules. Scale bar: 50 μm. (F) All Dazl cKO mouse were sterile. Error bars indicate SD. *P < 0.05, **P < 0.01, ***P < 0.005.
DAZL is also highly expressed in ovaries and in oocytes of all stages (see Supplementary Fig. S3A) [22,35]. Full-body Dazl knockout led to an ovary without any follicles, similar to the previously reported knockout (see Supplementary Fig. S3B) [11]. Surprisingly, both Dazl VKO (see Supplementary Figs S3B, E and F) and Gdnf9-Cre; Dazlf/− (data not shown) [26] were fertile, with the number of progeny and of pups per litter similar to those of wild type. We confirmed the absence of Dazl wild type transcripts and protein in conditional Knockout (cKO) oocytes (see Supplementary Fig. S3C and D). To exclude any compensatory effects from the Dazl paralog Boule [41], we also determined the RNA level of Boule transcripts in Dazl VKO ovaries. Boule transcripts remained undetectable in the absence of Dazl (see Supplementary Fig. S3C). Furthermore, we constructed a Vasa-Cre; Dazlf/−; Boule−/− double knockout mouse; those Dazl and Boule double knockout females remained fertile with no significant difference from wild type or VKO mice (see Supplementary Figs S3E and F). We hence conclude that neither DAZL nor BOULE is required for postnatal female gametogenesis.
DAZL globally binds mRNAs associated with spermatogenesis at the 3′UTR via the motif UGUU
To shed light on how DAZL functions during spermatogenesis, we performed high-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation (HITS-CLIP) to identify DAZL binding targets [42,43]. DAZL antibody was used to pull-down RNAs bound by DAZL from a pool of 16 testes from eight 25-d postpartum (dpp) mice. Pulled down RNAs were detected at the predicted DAZL band and at a slightly higher band in the UV cross-linked samples, but not in the non-UV cross-linked samples (see Fig. 2A and B). The RNAs excised from both the main bands and the slightly higher bands from two independent pull-downs were recovered to construct three cDNA libraries (see Fig. 2C and Supplementary Table S1). The cDNA library constructed from the higher bands overlapped extensively with the cDNA libraries from the predicted main DAZL band, suggesting that the higher bands also contain DAZL targets. Therefore, we used all three libraries for DAZL target analysis. RNA sequencing (RNAseq) of 25-dpp testes was used as background. The CLIP peaks of at least twofold enrichment when compared with the background were considered significant. DAZL binding sites appeared to distribute throughout the genome, with the majority of sites (51%) located on the 3′UTR of mRNA, consistent with its role in posttranscriptional regulation (see Fig. 2D). Thus, we focused our analysis on mRNA targets with DAZL binding sites at the 3′UTR and predominant binding sites for mRNAs were located on the 3′UTR. Out of 1373 shared mRNA targets among all three libraries, 1235 targets contained binding sites at the 3′UTR (see Supplementary Figs S4A and S2E). For mRNAs shared by at least two libraries, 3008 out of 3470 targets contained binding sites at the 3′UTR. Those targets (1235 or 3008 targets) were significantly enriched for pathways involved in RNA metabolism, such as mRNA processing, RNA splicing and translational regulation. The cell cycle and spermatogenesis pathways were also highly enriched (see Fig. 2F). To determine the validity of the DAZL target mRNAs identified by HITS-CLIP, we randomly picked 8 targets from among the 1235 3′UTR targets shared by all three libraries and 7 targets from among the less stringent 3008 3′UTR targets (shared by at least two libraries), and found that they were all significantly enriched in DAZL IP (see Fig. 2G). In contrast, Sertoli cell transcripts Wt1, Gata4 and other non-targets such as Nanos2, Neurog3 (Ngn3), Tnp1 and Tnp2 were not enriched (see Fig. 2G and Supplementary Fig. S4B). This suggested that the 3008 shared targets were reliable DAZL targets. We further established the quality and reproducibility of the three libraries by comparing the binding sites of the targets. We observed a consistent peak distribution for the same target transcripts among the three libraries (see Supplementary Fig. S4C).
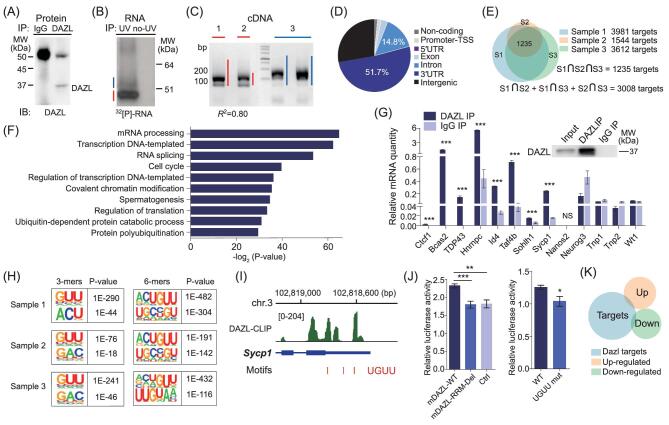
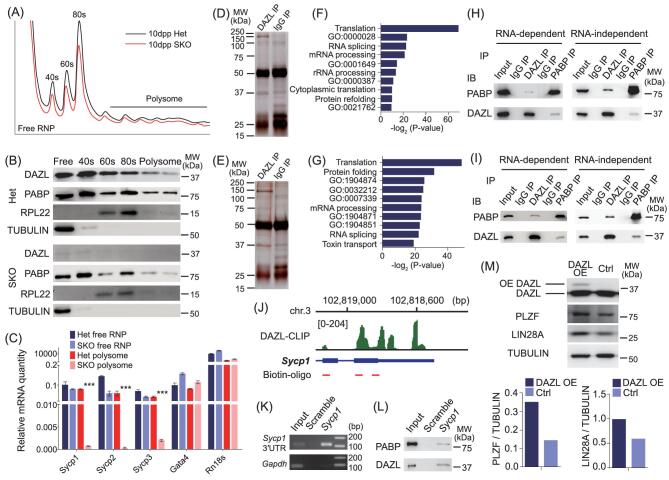
Figure 2.
Identification of DAZL targets in mouse testes by HITS-CLIP. (A) Immunoblot (IB) analysis of DAZL immunoprecipitation (IP). (B) Autoradiogram of 32P-labeled RNA crosslinked to DAZL pulled down by IP. The RNA marked by blue and red bars was excised for library construction. (C) RNA excised from (B) was ligated to linker (36nt) and amplified by PCR. The amplified cDNA product marked by blue and red bars was derived from the band of the same color in (B). (D) Distribution of all DAZL binding regions based on their genomic locations. (E) Venn diagram of DAZL targets bound at 3′UTR from three cDNA libraries, including two main bands (sample 1 and 2) and a mixture of mRNA combined from the two higher bands (sample 3). (F) Gene ontology analysis result of DAZL targets revealed by DAVID. (G) Validation of HITS-CLIP result by RNA immunoprecipitation [49] of 8 target genes. (H) Consensus motifs within DAZL clusters from 3 samples identified by HITS-CLIP using the HOMER algorithm. (I) Consensus motif distribution at the 3′UTR of a DAZL target Sycp1. (J) DAZL bound to the 3′UTR of Sycp1 around the motif ‘UGUU’ and enhanced luciferase expression. Error bars indicate SD. *P < 0.05, **P < 0.01, ***P < 0.005. (K) DAZL target genes showed little overlap with up or down regulated genes in 10dpp Dazl SKO testes.
To identify the consensus sequence of DAZL binding sites, we used the motif discovery algorithm Hypergeometric Optimization of Motif EnRichment (HOMER) [44] to search for the mRNA motifs bound by DAZL. Of the most enriched 3-mer motifs, the GUU triplet ranked highest in all three samples with much higher statistical significance than other motifs, consistent with the binding motif determined by crystal structure and binding affinity analysis of the DAZL RRM-RNA complex [30] (see Fig. 2H). Among the most enriched 6-mer motifs, VVUGUU ranked highest. To test the UGUU binding motif, we evaluated the 3′UTR of the DAZL target Sycp1 using a dual luciferase assay (see Fig. 2I). When we mutated UGUU to ACAA, relative luciferase activity decreased significantly, confirming the consensus binding motif of DAZL (see Fig. 2J).
DAZL does not significantly impact the stability of its target mRNAs
We next examined the transcript abundance of the identified DAZL targets in the absence of DAZL. To our surprise, there was little overlap between the DAZL targets identified by HITS-CLIP (see Fig. 2K) and genes whose transcript levels significantly changed in the 10-dpp Dazl SKO testes. We chose 10-dpp testes because knockout testes were comparable to those of controls in terms of cell composition. A lack of transcript level change in the knockout testes among DAZL targets could not be attributed to the different time points of testes used, as the majority (2896 out of 3008) of DAZL targets were detectable (Fragments Per Kilobase Million (FPKM) >30) in 10-dpp testes. This result suggested that loss of DAZL did not significantly impact target transcript levels, leading us to propose that mouse DAZL regulates translation rather than transcript stability. Furthermore, changes in transcript levels of non-target genes in Dazl mutant testes compared to wild type testes were likely a secondary effect of Dazl loss.
Protein expression of DAZL SSC-associated targets is essential for the maintenance of SSCs
Since spermatogenic genes were highly enriched among DAZL targets, we mapped all the DAZL binding sites of spermatogenic targets. Remarkably, all the spermatogenic target mRNAs contained binding sites exclusively in their 3′UTR (124 out of 124, classified by the Database for Annotation, Visualization and Integrated Discovery (DAVID), see Supplementary Table S2), consistent with DAZL being a translational regulator [45]. We also found that DAZL target transcripts were present throughout spermatogenesis, with enrichment for genes associated with key steps such as SSC self-renewal, meiosis and spermatid differentiation. Such stage enrichment corresponded nicely with the stages when Dazl is shown to be required from our genetic analysis above, leading us to hypothesize that DAZL may promote spermatogenesis by regulating the protein expression of many target transcripts critical for each stage of spermatogenesis.
Among the spermatogenic mRNAs bound by DAZL, there was a remarkable enrichment of genes linked to SSCs. Out of 13 genes shown to be important for maintenance of the spermatogonial progenitor pool [46], the mRNAs for eight of these genes were found to be DAZL targets (see Fig. 3A), suggesting a previously unknown function of DAZL in the regulation of SSCs. Careful examination of binding peaks among those eight genes revealed distinct peaks on their 3′UTRs (see Fig. 3B). We then determined DAZL expression in SSCs, and found that both DAZL mRNA and protein were detectable in neonatal testes and in purified primary SSCs, and that DAZL co-expressed with PLZF+ and LIN28A+ cells (see Supplementary Figs S1A–C and F), further supporting a role for DAZL in SSCs [47].
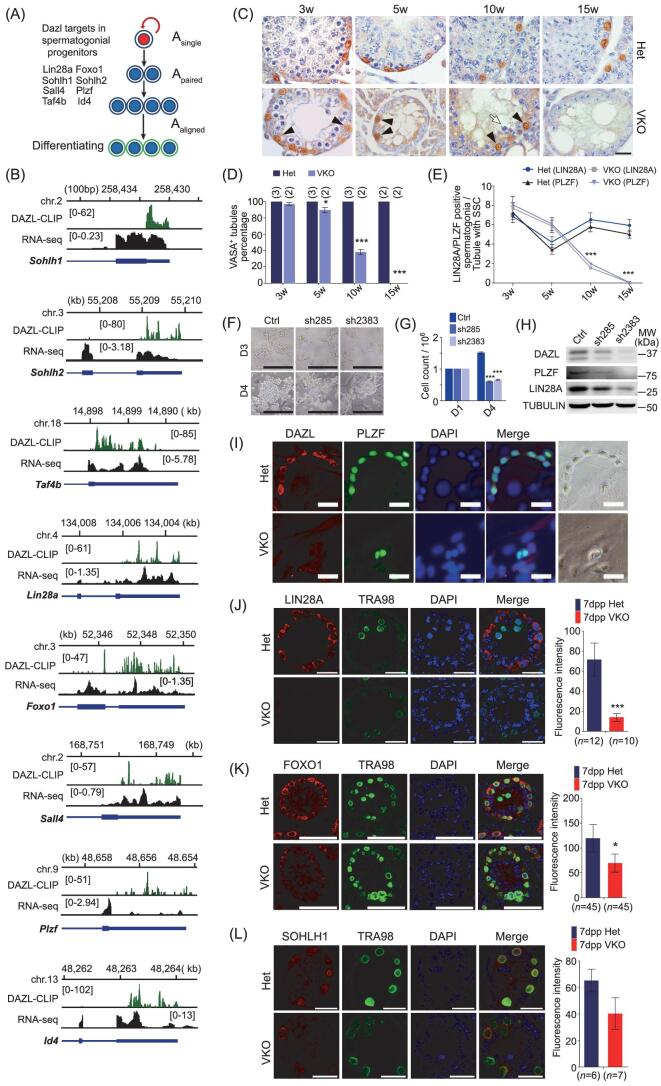
Figure 3.
DAZL targets are enriched for SSC-associated genes and loss of DAZL leads to a defect in spermatogonial progenitor maintenance. (A) Schematic illustration of SSC-associated genes bound by DAZL. (B) Genome browser tracks showing binding peak distributions (DAZL-CLIP) and transcript levels (RNA-seq) of SSC-associated target genes. (C) Immunohistochemical staining of the SSC marker PLZF in Dazl VKO testes. Scale bar: 20 μm. (D) Gradual germ cell loss in testes of Dazl VKO mice. Testes sections from 3, 5, 10 and 15-week-old mice were stained for VASA expression to detect the number of VASA-positive tubules. A seminiferous tubule containing at least one VASA-positive cell was classified as a VASA-positive tubule. The number of mice at each time point is shown in parentheses. More than 100 cross-sections were scored for samples taken from random slides. (E) Comparison of the number of LIN28A+ and PLZF+ SSCs in cross-sections of testes from Dazl heterozygotes (Het) and Dazl VKO mice at different ages. The number of mice is the same as in (D). A seminiferous tubule containing at least one SSC was classified as tubule with SSCs. Data are mean ± SEM for 20 round seminiferous tubules of both genotypes at each age. *P < 0.05, **P < 0.01, ***P < 0.005. (F) Dazl knockdown led to a significant reduction in the size of SSC clones at day 4 following shRNA lentiviral transduction. Scale bar: 200 μm. (G) Cell count of SSCs at day 4 following lentiviral transduction. (H) SSC markers (LIN28A, PLZF) were downregulated at day 4 following lentiviral transduction. (I) Established SSC clones immunostained for DAZL and PLZF after 4 days of culture of Thy1+ cells from 7dpp Dazl heterozygotes and Dazl VKO testes. Scale bar: 50 μm. (J-L) Fluorescence intensity of DAZL targets was compared between Dazl heterozygotes and Dazl VKO testes at 7dpp by immunofluorescent staining in TRA98-positive cells. Scale bar: 20 μm. Error bars indicate SEM. *P < 0.05, **P < 0.01, ***P < 0.005.
We then investigated the effect of removing Dazl from SSCs by generating Dazl VKO mice; these mice were expected to produce no functional DAZL in postnatal germ cells, including SSCs [38]. In testes from 3-week-old Dazl VKO mice, despite a significant reduction in germ cells, the number of both LIN28A+ and PLZF+ cells were comparable to controls, suggesting that the initial pool of SSCs was not different between heterozygotes and knockout mice. However, by 5 and 10 weeks of age, LIN28A+ and PLZF+ cells had decreased gradually and had completely disappeared by 15 weeks of age (see Fig. 3C–E, and Supplementary Fig. S5A and B), resembling the reported phenotypes of SSC maintenance-defective mutant mice [48]. Short hairpin RNA (shRNA) knockdown of Dazl in established SSC cultures led to significantly fewer SSC clones with reduced size and cell number (see Fig. 3F and G). PLZF and LIN28A proteins were also significantly reduced in the Dazl knocked-down SSCs, establishing a role for DAZL in SSC self-renewal and development by promoting the protein expression of SSC-associated target genes (see Fig. 3H).
To examine the self-renewal ability of Dazl knockout SSCs, we isolated THY1+ cells from Dazl VKO testes to generate in vitro SSC clones without DAZL protein. The Dazl knockout SSC clones were established in vitro and had similar morphology to heterozygotes SSCs on the first and second days of culture (data not shown). However, the knockout SSC clones failed to grow further, forming only chains or clusters of two-to-three cells; in contrast, the heterozygote SSCs formed long chains and large clusters (see Fig. 3I and Supplementary Fig. S5C). These data demonstrate an essential role of DAZL in the maintenance of the spermatogonial progenitor pool.
Furthermore, we examined protein expression of DAZL targets in Dazl knockout germ cells. Based on fluorescence intensity relative to the germ cell marker TRA98, the expression of SSC-associated DAZL target proteins was decreased in 7-dpp Dazl VKO mouse testes, with LIN28A and FOXO1 showing a statistically significant decrease compared to controls (see Fig. 3J–L). This finding further supports the notion that DAZL-mediated SSC-associated protein expression is critical for SSC maintenance.
DAZL-mediated protein expression of meiotic genes is essential for synaptonemal complex assembly and DNA repair during meiosis
Other spermatogenic genes significantly enriched in the DAZL targets were associated with the meiotic cell cycle (see Fig. 4A and B). The list of DAZL-targeted meiosis-associated genes included the previously reported Sycp3 as a direct target [34], but contained a number of other meiotic genes such as major synaptonemal complex (SC) genes (Sycp1 and Sycp2) and DNA repair pathway genes (Rad51, Msh4, etc). We investigated the effect of loss of Dazl on the expression of these DAZL targets, as well as on key meiotic events. To bypass the embryonic and SSC reqiurement for Dazl we utilized Stra8-Cre, which drives Cre expression starting in the 3-dpp testis [39]. We found that the resulting Dazl SKO mice are sterile and have significantly smaller testes compared to heterozygotes (see Fig. 1B and C). Spermatogenesis was arrested at the transition from zygotene to pachytene stage in adult testes, and in the first wave of spermatogenesis of both Dazl VKO and Dazl SKO animals (see Supplementary Figs S5A and S6A), confirming a meiotic block in the absence of Dazl [18]. Expression of the SSC marker PLZF, and early meiotic markers such as STRA8 and SPO11, was not affected in 9-dpp Dazl SKO mice, suggesting that those knockout germ cells were able to enter meiosis (see Fig. 4C). Indeed, meiotic chromosome spread revealed a similar proportion of leptotene cells in the Dazl SKO mice compared to wild type but significantly more zygotene and pachytene-like cells, suggesting an arrest at the transition from zygotene to pachytene (see Supplementary Fig. S6B).
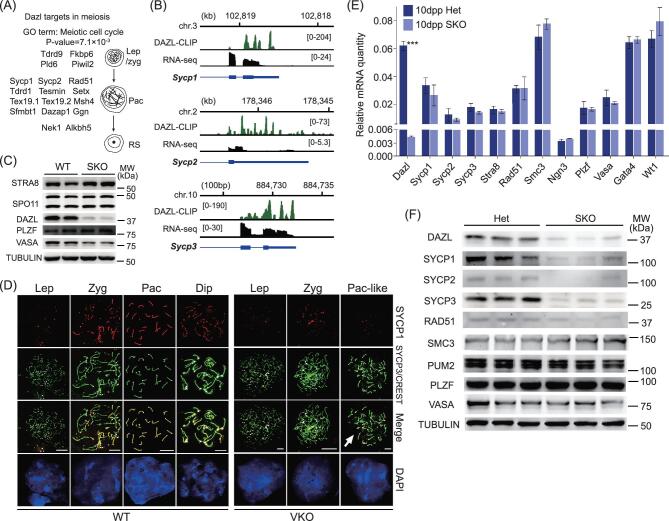
Figure 4.
Synapsis defect in Dazl knockout spermatocytes results from reduced protein expression of DAZL targets Sycp1, Sycp2 and Sycp3. (A) Schematic illustration of meiotic genes bound by DAZL. (B) Genome browser tracks showing binding peak distributions and transcript levels of meiotic target genes of the synaptonemal complex. (C) Expression of proteins involved in meiotic initiation and SSC-associated proteins were not significantly changed in 9dpp Dazl SKO testes. (D) Meiotic spread of 21dpp wild type and Dazl VKO spermatocytes co-stained with SYCP1, SYCP3 and CREST antibodies. Completion of synapsis is determined by a single CREST signal (white points) for a given pair of chromosomes. (E) Transcript levels of meiotic target genes were not changed significantly in 10dpp Dazl SKO testes. (F) Protein level of meiotic target genes was significantly reduced in 10dpp Dazl SKO testes.
In DAZL-deficient leptotene nuclei, short stretches of condensed SYCP3 signals were observed, indicating that chromosome core-associated SYCP3 protein was loaded normally during early meiotic prophase. However, loss of Dazl resulted in an increase in the unpaired SC in zygotene-stage spermatocytes (see Fig. 4D). Despite the presence of SYCP3-positive axial elements, nearly all of the Dazl VKO zygotene spermatocytes had weak SYCP1 staining, suggesting a partial loss of transverse filaments of the SC (white arrow, see Fig. 4D). Defects in synapsis formation were also observed in Dazl VKO pachytene spermatocytes. Typical pachytene nuclei with 20 synapsed chromosomes were never observed in Dazl VKO cells. Instead, most nuclei contained 40 unsynapsed chromosomes (n = 222) with <5% exhibiting partial synapsis. The vast majority of axial elements in Dazl VKO spermatocytes failed to align and instead remained separated from each other as univalent chromosomes. The presence of about 40 short SCs suggested that the chromosomes were condensed and that the spermatocytes were at a stage corresponding to early pachytene; we refer to these as ‘pachytene-like’. The same abnormal pachytene-like cells were found in Dazl SKO testes (see Supplementary Fig. S6C). These results suggest that the loss of Dazl disrupted the assembly of the SC and likely led to meiotic arrest at early pachytene.
In addition to defective synapsis and disruption of the SC, we also observed increased DNA damage in Dazl VKO spermatocytes. Phosphorylated H2AX (called γ-H2AX or H2AFX) is a marker of double strand breaks during leptotene and zygotene, and of the XY body during pachytene [49,50]. While Dazl VKO leptotene spermatocytes showed similar staining for H2AFX, the signal did not decrease in zygotene, and instead increased in both zygotene and pachytene-like cells. The single XY body observed in wild type pachytene spermatocytes was not present in the Dazl VKO pachytene spermatocytes. A number of dispersed H2AFX domains were retained along each chromosome (see Supplementary Fig. S6D). A similar dispersed H2AFX staining pattern was seen in Dazl SKO spermatocytes, implicating DNA repair defects in the absence of Dazl (see Supplementary Fig. S6C).
DAZL meiotic target mRNA expression was not affected in the SKO testes but their protein level was significantly reduced (see Figs 2K, and 4E and F), consistent with a defect in the SC, synapsis and DNA repair in the mutant testes. These findings further support a role for DAZL in translational regulation rather than mRNA stability in the regulation of spermatogenesis.
DAZL is required for translation of its target mRNAs and interacts with PABP in mouse testes
Given the requirement of DAZL throughout spermatogenesis, we next examined the mechanism of DAZL action at the molecular level, specifically, how it controls protein expression of its targets. Several previous reports have suggested that DAZL plays important roles as an enhancer of mRNA translation in germ cell development [33–35]. Because meiotic arrest was the prominent phenotype in Dazl conditional knockout testes and genes encoding components of the SC were identified as DAZL targets, we investigated how expression of Sycp1, Sycp2 and Sycp3 was affected by Dazl knockout at the translational level via polysome analysis. RNA was isolated from the fractions of free ribonucleoprotein and polysomes (see Fig. 5A and B). Sycp1, Sycp2 and Sycp3 transcripts were not significantly decreased in total transcripts but were dramatically decreased in polysome fractions (see Figs 4E and 5C). Hence, reduction of protein expression of those DAZL targets resulted from reduced protein translation rather than reduced mRNA levels, further supporting a role for DAZL in translational regulation.
Figure 5.
DAZL recruits PABP to regulate translation of its target. (A) Sucrose density gradient fractionation of 10dpp mouse testes with and without DAZL. (B) Immunoblot analysis of different fractions. (C) Target mRNAs from polysome fractions decreased dramatically with Dazl KO. (D) Silver staining of SDS-PAGE gel of DAZL and IgG immunoprecipitation (IP) in 8dpp mouse testes. Proteins were identified by mass spectrometry from excised bands (the whole lane except heavy and light chain). (E) Silver staining of SDS-PAGE gel of DAZL and IgG IP in 25dpp mouse testes. Proteins were identified by mass spectrometry from excised bands (the whole lane except heavy and light chain). (F) Gene ontology analysis results of proteins identified by mass spectrometry from (D). GO:0000028: ribosomal small subunit assembly; GO:0001649: osteoblast differentiation; GO:0000387: spliceosomal snRNP assembly; GO:0021762: substantial nigra development. (G) Gene ontology analysis results of proteins identified by mass spectrometry from (E). GO:1904874: positive regulation of telomerase RNA localization to Cajal body; GO:0032212: positive regulation of telomere maintenance via telomerase; GO:1904871: positive regulation of protein localization to Cajal body; GO:1904851: positive regulation of establishment of protein localization to telomere. (H) Co-IP of DAZL and PABP in 8dpp mouse testes. (I) Co-IP of DAZL and PABP in 25dpp mouse testes. (J) Biotin-oligo was designed to reverse complement the 3′UTR of Sycp1 mRNA. (K) 3′UTR of Sycp1 mRNA was enriched in Sycp1 pull-downed samples. (L) DAZL and PABP proteins were enriched in Sycp1 pull-downed samples. (M) Overexpression (OE) of DAZL in SSCs enhanced the expression of PLZF and LIN28A. Relative expression of PLZF and LIN28A in different samples was quantified using ImageJ.
Next, we performed mass spectrometry to identify proteins that interact with DAZL protein via IP (see Fig. 5D and E). We identified >100 proteins that potentially interact with DAZL (see Supplementary Table S3). Among them, proteins involved in translation were significantly enriched: specifically, a series of ribosomal proteins and PABP, which stimulates translation when bound to a poly(A) tract (see Fig. 5F and G, and Supplementary Fig. S7A and B). PABP was previously found to interact with DAZL in frog oocytes [24]. To determine if DAZL directly interacts with mouse PABP in the testes, we performed co-IP of DAZL and PABP in 8-dpp and 25-dpp mouse testis extract. DAZL and PABP reciprocally pulled down each other independent of RNA, establishing a direct interaction between these two proteins in vivo (see Fig. 5H and I). We then asked if we could identify a DAZL-PABP complex bound to a DAZL mRNA target directly by pulling down the 3′UTR of DAZL’s target. Using three biotin-labeled oligo probes complementary to the 3′UTR of a meiotic target of DAZL, Sycp1, we could pull-down Sycp1 3′UTR from the testis extract (see Fig. 5J and K). Remarkably, we could also pull-down DAZL and PABP proteins at the same time (see Fig. 5L, demonstrating DAZL-PABP interaction directly on a DAZL target. To confirm the nature of the translational regulation of DAZL, we next overexpressed DAZL in SSCs in vitro. The protein levels of both DAZL targets Plzf and Lin28a were enhanced with DAZL overexpression (see Fig. 5M). Thus, DAZL functions by recruiting PABP to mRNA targets of DAZL to promote their translation. Since DAZL bound to the 3′UTR of mRNA targets associated with SSCs, meiosis and round spermatids, we propose that DAZL facilitates translation by recruiting PABP to those mRNA targets to form active translational circles (see Fig. 6). Without DAZL, the translation of key proteins central for SSC maintenance, the SC, DNA repair and spermatid differentiation is severely impaired, resulting in a progressive loss of germ cells, failure of synapsis, arrest in meiosis and round spermatids. Indeed, we found that DAZL and PABP are concomitantly expressed from the SSC to round spermatid stages during spermatogenesis (see Supplementary Fig. S7C), and co-localize in spermatogenic cells in 10-dpp and 25-dpp mouse testes (white arrow, see Supplementary Fig. S7D). These findings establish a central and global role of DAZL-mediated translational control throughout spermatogenesis (see Fig. 6).
Figure 6.
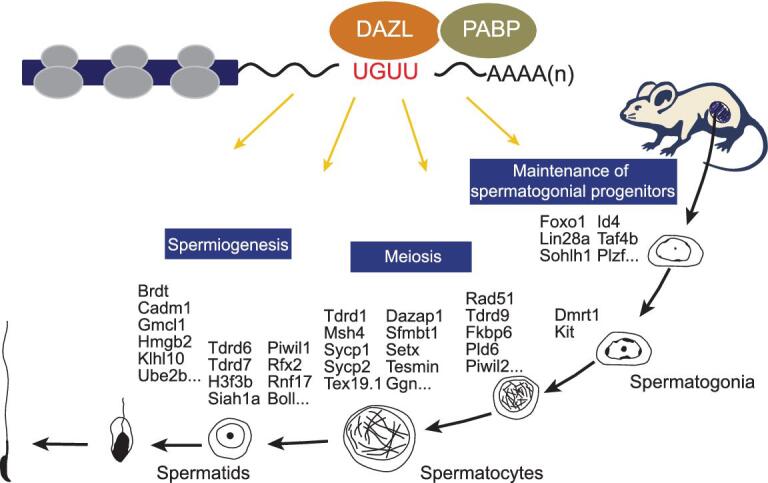
Model for DAZL-mediated master translational control in at least three key steps of spermatogenesis.
DISCUSSION
Our finding suggests that DAZL expression is a hallmark of mouse germ cells, and is also critically required throughout male gametogenesis but not in female gametogenesis. While earlier studies have demonstrated a requirement for DAZL in fetal germ cell development and differentiation [20], our findings extend DAZL’s requirement into postnatal male gametogenesis and argue for a central regulatory role of DAZL throughout germ cell development, from embryonic to postnatal sperm development. Our genome-wide identification of DAZL targets and its protein partners, in combination with systemic interrogation of the requirement for DAZL, has led us to propose that DAZL-mediated translational regulation is critical throughout spermatogenesis, and that DAZL acts as a master translational regulator to ensure the proper translation of key spermatogenic factors and thus fertility.
Ample genetic evidence regarding the human DAZ family (DAZ, DAZL and BOULE) genes and their homologs among animals supports their essential roles in human and animal fertility [8–11,51,52]. However, molecular mechanisms by which DAZ proteins regulate sperm development remain elusive, with their direct in vivo targets staying largely unknown. The identification of more than 3000 direct targets from our transcriptome-wide HITS-CLIP experiments hence provides a key mechanistic clue regarding DAZL function. A caveat of the HITS-CLIP approach is that the signal/noise ratio may depend on the specificity of the antibody used, relative RNA abundance and bioinformatics strategies. We have validated the specificity of the DAZL antibody with knockout mice (Figs 3I and S2D) and normalized HITS-CLIP data against RNAseq data from the testes of mice of the same age (Fig. 3B). We only considered mRNA targets ≥ twofold higher relative to the RNAseq data to be DAZL targets and successfully validated a number of spermatogenic targets from the target list. Hence, false positives are likely to be minimal in our DAZL target list. We showed that DAZL protein directly regulates the translation of their target mRNAs via binding to their 3′UTRs, without apparent an effect on mRNA levels, supporting a conserved translational function in mammalian germ cells [22–24,33,34,53]. Among a number of protein partners of DAZL identified through mass spectrometry, we showed that mouse DAZL interacts with PABP in the testes to promote the translation of spermatogenic targets, extending the seminal finding on DAZL interacting with PABP to activate translation in frog oocytes to the mouse testis [23,24]. Our work further supported a general and conserved function of DAZL in translation. Bioinformatics analysis of target sequences revealed 3mer GUU and 4mer UGUU to be consensus binding motifs, consistent with the results from the crystal structure analysis of DAZL RRM [30]. Spermatogenic transcripts are highly enriched among DAZL targets and pathway analysis revealed potential roles in SSC development, meiosis and spermatid differentiation, among which only two transcripts were previously reported to be regulated [33,34]
Previously, Dazl knockout germ cells exhibited a defect in spermatogonial transition from Aalign- to A1-type spermatogonia [17]. We showed a clear requirement for DAZL in SSC maintenance. Conditional deletion of Dazl by either Vasa-cre or Stra8-cre led to a gradual loss of germ cells and SSCs marked by PLZF+ and LIN28A+ cells, with no apparent increase in KIT+ spermatogonia (unpublished). This finding established a role for DAZL in SSC maintenance but not in spermatogonia differentiation [54]. The meiotic roles of DAZL were suggested as Dazl knockout germ cells failed to enter meiosis, or were arrested at or before leptotene during the first wave of spermatogenesis [18,55]; we found that spermatogenic cells lacking DAZL could apparently differentiate and enter meiosis but were arrested at the zygotene/pachytene transition stage, supporting the notion of a critical meiotic requirement for DAZL [18]. Differences in the specific arrest stage may reflect the unique properties of knockout germ cells from the whole-body DAZL knockout or the limitations of the very few germ cells available for analysis [18,55].
DAZL’s role in SSCs revealed a novel layer of translational regulation in SSC maintenance and differentiation. SSC regulation involves a hierarchy of transcriptional regulators and signaling pathways [46,56]. The only posttranscriptional regulator that is required in SSC maintenance is Nanos2, another RNA binding protein [57], although the molecular mechanism by which NANOS2 regulates SSCs has not been determined. Interestingly, NANOS2 repressed DAZL expression in sexually differentiating germ cells at embryonic stages [58], raising the possibility that NANOS2 might exert its influence in SSCs via DAZL. Indeed Nanos2 overexpression resulted in a reduction of SSC chains and an increase of As, similar to our Dazl knockdown phenotype (see Fig. 3I and Supplementary Fig. S5C) and the effects of Dazl conditional knockout on the testes [57]. Hence, NANOS2 may regulate levels of DAZL expression to balance SSC maintenance and differentiation. Future experiments are needed to determine the contribution of NANOS2 and DAZL to SSC maintenance, and their interaction in maintaining the homeostasis of spermatogenesis.
Master developmental regulators are the few key regulators in developmental pathways that regulate a large number of downstream targets in critical developmental steps. DAZL binds to many spermatogenic target transcripts and appears to regulate their translation to ensure sufficient protein expression for key steps of spermatogenesis. Hence, DAZL may represent a master translational regulator that facilitates efficient sperm production from the embryonic germ cell stage to postnatal spermatogenesis. Firstly, DAZL is most extensively expressed from the embryonic germ cell throughout postnatal spermatogenesis, validating its expression as a hallmark of germ cells. Secondly, DAZL is required persistently throughout germ line development including most stages of spermatogenesis, without which male germ cell development is arrested thereafter [11,19,20] (this work). Thirdly, DAZL binds to a large number of spermatogenic transcripts that are key to the progression of spermatogenesis and recruits PABP to the 3′UTR of those target transcripts. Without DAZL, translation of those key spermatogenic regulators is disrupted, resulting in an arrest in subsequent steps. Hence, we propose that DAZL functions as a master translational regulator during spermatogenesis (see Fig. 6). The insights and mechanistic frameworks established by this work could advance our understanding not only of the fundamental mechanisms that distinguish germ cells from somatic cells, but also of human infertility involving the DAZ family of proteins.
An additional note; during the preparation of this manuscript, another manuscript reported their CLIP analysis on DAZL and validation using FACS-sorted germ cells from whole-body Dazl knockout testes [59]. Two thirds of our targets overlapped with their targets, confirming the validity of our targets. However, Zagore et al. found that a portion of the target transcripts were downregulated in the Dazl knockout spermatogenic cells, leading to their conclusion that DAZL regulates the stability of a subset of its target transcripts. In contrast, our analysis showed that DAZL knockout does not affect the transcript levels of DAZL targets but mainly affect their translation. Such a difference may result from the different animal models used for the RNAseq studies, as the Dazl knockout mice used by Zagore et al. were full-body knockouts while our model was a stage-specific knockout. Despite this difference, a large portion of DAZL target transcript levels were still not changed in the absence of DAZL in the paper by Zagore et al. These unchanged target transcripts in Dazl knockout germ cells may correspond to the transcripts under DAZL translational regulation in our study.
METHODS
The detailed methods and materials are available as Supplementary data at NSR online.
Supplementary Material
Acknowledgements
The Dazlf/f ESCs used for this research project were generated by the trans-National Institutes of Health Knock-Out Mouse Project (KOMP) and obtained from the KOMP Repository (www.komp.org) at the University of California, Davis and Children's Hospital Oakland Research Institute (CHORI) (U42RR024244). We would like to thank Drs Ke Zheng and Xin Wu for advice and assistance on HITS-CLIP and SSC experiments, respectively, as well as discussion throughout the project; Jieli Chen, Guihua Du, Fan Yang, Xinrui Wang and Jiachen Sun for technical assistance; Dr Renee Reijo Pera Kehkooi Kee and Youqiang Su for comments on the manuscript; Genergy Biotech, Shanghai, China for bioinformatics analysis; Dr P. Jeremy Wang for the SYCP2 antibody; Dr Stephen Kistler for the TNP2 antibody; and Dr Aleksandar Rajkovic for the SOHLH1 antibody.
Author contributions
E.Y.X. conceptualized the research; E.Y.X. and H. L. designed the study; H.L. performed most of the experiments; Z.L., J. Y., D.W., H.W., and E. Y. X. performed and interpreted experiments; B.G. performed bioinformatics analyses for DAZL-CLIP; E.Y.X. supervised the study and wrote the paper with H.L.
FUNDING
This work was supported by the National Basic Research Program of China (2015CB943002 and 2013CB945201), the National Natural Science Foundation of China (31771652, 81270737 and 81401256) and the Natural Science Foundation of Jiangsu Province (BK2012838).
Conflict of interest statement. None declared.
REFERENCES
- 1. Cinalli RM, Rangan P, Lehmann R. Germ cells are forever. Cell 2008; 132: 559–62. [DOI] [PubMed] [Google Scholar]
- 2. Seydoux G, Braun RE. Pathway to Totipotency: lessons from germ cells. Cell 2006; 127: 891–904. [DOI] [PubMed] [Google Scholar]
- 3. Zhang C, Gao L, Xu EY. LncRNA, a new component of expanding RNA-protein regulatory network important for animal sperm development. Semin Cell Dev Biol 2016; 59: 110–7. [DOI] [PubMed] [Google Scholar]
- 4. Houston DW, King ML. A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development 2000; 127: 447–56.10631166 [Google Scholar]
- 5. Johnson AD, Bachvarova RF, Drum Met al.. Expression of axolotl DAZL RNA, a marker of germ plasm: widespread maternal RNA and onset of expression in germ cells approaching the gonad. Dev Biol 2001; 234: 402–15. [DOI] [PubMed] [Google Scholar]
- 6. Lee HC, Choi HJ, Lee HGet al.. DAZL Expression explains origin and central formation of primordial germ cells in chickens. Stem Cells Dev 2016; 25: 68–79. [DOI] [PubMed] [Google Scholar]
- 7. Shah C, Vangompel MJ, Naeem Vet al.. Widespread presence of human BOULE homologs among animals and conservation of their ancient reproductive function. PLoS Genet 2010; 6: e1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu EY, Moore FL, Pera RA. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc Natl Acad Sci USA 2001; 98: 7414–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reijo R, Lee TY, Salo Pet al.. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet. 1995; 10: 383–93. [DOI] [PubMed] [Google Scholar]
- 10. VanGompel MJ, Xu EY. A novel requirement in mammalian spermatid differentiation for the DAZ-family protein Boule. Hum Mol Genet 2010; 19: 2360–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ruggiu M, Speed R, Taggart Met al.. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 1997; 389: 73–7. [DOI] [PubMed] [Google Scholar]
- 12. Tung JY, Rosen MP, Nelson LMet al.. Variants in Deleted in AZoospermia-Like (DAZL) are correlated with reproductive parameters in men and women. Hum Genet 2006; 118: 730–40. [DOI] [PubMed] [Google Scholar]
- 13. Tung JY, Rosen MP, Nelson LMet al.. Novel missense mutations of the Deleted-in-AZoospermia-Like (DAZL) gene in infertile women and men. Reprod Biol Endocrinol 2006; 4: 40. doi:10.1186/1477-7827-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Navarro-Costa P, Nogueira P, Carvalho Met al.. Incorrect DNA methylation of the DAZL promoter CpG island associates with defective human sperm. Hum Reprod 2010; 25: 2647–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang C, Xue P, Gao Let al.. Highly conserved epigenetic regulation of BOULE and DAZL is associated with human fertility. FASEB J 2016; 2016: 3424–40. [DOI] [PubMed] [Google Scholar]
- 16. Kee K, Angeles VT, Flores Met al.. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature 2009; 462: 222–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schrans-Stassen BH, Saunders PT, Cooke HJet al.. Nature of the spermatogenic arrest in Dazl -/- mice. Biol Reprod 2001; 65: 771–6. [DOI] [PubMed] [Google Scholar]
- 18. Saunders PT, Turner JM, Ruggiu Met al.. Absence of mDazl produces a final block on germ cell development at meiosis. Reproduction 2003; 126: 589–97. [DOI] [PubMed] [Google Scholar]
- 19. Lin Y, Page DC. DAZL deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Dev Biol 2005; 288: 309–16. [DOI] [PubMed] [Google Scholar]
- 20. Gill ME, Hu YC, Lin Yet al.. Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc Natl Acad Sci USA 2011; 108: 7443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen HH, Welling M, Bloch DBet al.. DAZL limits pluripotency, differentiation, and apoptosis in developing primordial germ cells. Stem Cell Rep 2014; 3: 892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen J, Melton C, Suh Net al.. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev 2011; 25: 755–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brook M, Smith JW, Gray NK. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction 2009; 137: 595–617. [DOI] [PubMed] [Google Scholar]
- 24. Collier B, Gorgoni B, Loveridge Cet al.. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J 2005; 24: 2656–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Padmanabhan K, Richter JD. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev 2006; 20: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fukuda K, Masuda A, Naka Tet al.. Requirement of the 3′-UTR-dependent suppression of DAZL in oocytes for pre-implantation mouse development. PLoS Genet 2018; 14: e1007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maines JZ, Wasserman SA. Post-transcriptional regulation of the meiotic Cdc25 protein twine by the DAZL Orthologue boule. Nat Cell Biol 1999; 1: 171–4. [DOI] [PubMed] [Google Scholar]
- 28. Venables JP, Ruggiu M, Cooke HJ. The RNA-binding specificity of the mouse DAZL protein. Nucleic Acids Res 2001; 29: 2479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiao X, Trifillis P, Kiledjian M. Identification of target messenger RNA substrates for the murine deleted in Azoospermia-Like RNA-Binding protein. Biol Reprod 2002; 66: 475–85. [DOI] [PubMed] [Google Scholar]
- 30. Jenkins HT, Malkova B, Edwards TA. Kinked beta-strands mediate high-affinity recognition of mRNA targets by the germ-cell regulator DAZL. Proc Natl Acad Sci USA 2011; 108: 18266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rosario R, Smith RW, Adams IRet al.. RNA immunoprecipitation identifies novel targets of DAZL in human foetal ovary. Mol Hum Reprod 2017; 23: 177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsui S, Dai T, Warren STet al.. Association of the mouse infertility factor DAZL1 with actively translating polyribosomes. Biol Reprod 2000; 62: 1655–60. [DOI] [PubMed] [Google Scholar]
- 33. Reynolds N, Collier B, Maratou Ket al.. Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum Mol Genet 2005; 14: 3899–909. [DOI] [PubMed] [Google Scholar]
- 34. Reynolds N, Collier B, Bingham Vet al.. Translation of the synaptonemal complex component Sycp3 is enhanced in vivo by the germ cell specific regulator Dazl. RNA 2007; 13: 974–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martins JP, Liu X, Oke Aet al.. DAZL and CPEB1 regulate mRNA translation synergistically during oocyte maturation. J Cell Sci 2016; 129: 1271–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu X, Tan X, Lin Qet al.. Mouse Dazl and its novel splice variant functions in translational repression of target mRNAs in embryonic stem cells. Biochim Biophys Acta. 2013; 1829: 425–35. [DOI] [PubMed] [Google Scholar]
- 37. Nicholas CR, Xu EY, Banani SFet al.. Characterization of a Dazl-GFP germ cell-specific reporter. Genesis 2009; 47: 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gallardo T, Shirley L, John GBet al.. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 2007; 45: 413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sadate-Ngatchou PI, Payne CJ, Dearth ATet al.. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 2008; 46: 738–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Inselman AL, Nakamura N, Brown PRet al.. Heat shock protein 2 promoter drives Cre expression in spermatocytes of transgenic mice. Genesis 2010; 48: 114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He J, Stewart K, Kinnell HLet al.. A developmental stage-specific switch from DAZL to BOLL occurs during fetal oogenesis in humans, but not mice. PLoS ONE 2013; 8: e73996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ule J, Jensen K, Mele Aet al.. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 2005; 37: 376–86. [DOI] [PubMed] [Google Scholar]
- 43. Vourekas A, Zheng K, Fu Qet al.. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev 2015; 29: 617–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Heinz S, Benner C, Spann Net al.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 46. Chan F, Oatley MJ, Kaucher AVet al.. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev 2014; 28: 1351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu X, Schmidt JA, Avarbock MRet al.. Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. Proc Natl Acad Sci USA 2009; 106: 21672–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buaas FW, Kirsh AL, Sharma Met al.. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 2004; 36: 647–52. [DOI] [PubMed] [Google Scholar]
- 49. Gray S, Cohen PE. Control of meiotic crossovers: from double-strand break formation to designation. Annu Rev Genet 2016; 50: 175–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luo M, Yang F, Leu NAet al.. MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat Commun 2013; 4: 2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Eberhart CG, Maines JZ, Wasserman SA. Meiotic cell cycle requirement for a fly homologue of human deleted in Azoospermia. Nature 1996; 381: 783–5. [DOI] [PubMed] [Google Scholar]
- 52. VanGompel MJ, Xu EY. The roles of the DAZ family in spermatogenesis–more than just translation? Spermatogenesis 2011; 1: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Welling M, Chen HH, Munoz Jet al.. DAZL regulates Tet1 translation in murine embryonic stem cells. EMBO Rep 2015; 16: 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oatley JM, Avarbock MR, Telaranta AIet al.. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci USA 2006; 103: 9524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Koubova J, Hu YC, Bhattacharyya Tet al.. Retinoic Acid activates two pathways required for meiosis in mice. PLoS Genet 2014; 10: e1004541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Spradling A, Fuller MT, Braun REet al.. Germline stem cells. Cold Spring Harb Perspect Biol 2011; 3: a002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sada A, Suzuki A, Suzuki Het al.. The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science 2009; 325: 1394–8. [DOI] [PubMed] [Google Scholar]
- 58. Kato Y, Katsuki T, Kokubo Het al.. Dazl is a target RNA suppressed by mammalian NANOS2 in sexually differentiating male germ cells. Nat Commun 2016; 7: 11272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zagore LL, Sweet TJ, Hannigan MMet al.. DAZL regulates germ cell survival through a network of PolyA-Proximal mRNA interactions. Cell Rep 2018; 25: 1225–40.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.