Abstract
Purpose: Breast cancer (BC) is a common malignancy in women, but the survival rate for BC is not very encouraging. Especially for triple-negative breast cancer (TNBC), a kind of breast cancer that does not have any of the receptors that are commonly found in BC. We investigated the impact of microRNA-206 (miR-206) on transmembrane 4 L6 family member 1 (TM4SF1) in TNBC for therapeutic purpose.
Patients and methods: Twenty BC tissues from diagnosed BC patients were analyzed via real-time PCR and Western blotting for expression of TM4SF1 and miR-206. The expression of TM4SF1 was studied in relationship with miR-206 in MDA-MB-231 cells. The biological impact of TM4SF1 and miR-206 on MDA-MB-231 cells and BALB/c nude mice model was studied using proliferation, transwell migration, and invasion assays both in vitro and in vivo.
Results: The expression of TM4SF1 in BC tissues was significantly higher than that in adjacent normal breast tissues. In contrast, miR-206 showed a decreased expression level in BC tissues, especially for subtype basal like. Overexpression of miR-206 in MDA-MB-231 cells by transfecting miR-206 resulted in downregulation of TM4SF1. In contrast, knockdown miR-206 expression reversed miR-206-mediated phenotype in MDA-MB-231 cells. Expression level of TM4SF1 in MDA-MB-231 cells was associated with cell migration and invasion capabilities in vitro. Breast tumor burden was correlated with the expression level of TM4SF1 in vivo.
Conclusion: Taken together, our results showed the involvement of TM4SF1 in TNBC migration and invasion. miR-206 negatively regulated gene expression of TM4SF1. These findings indicate that miR-206 could be used as a potential therapeutic agent for TNBC.
Keywords: miR-206, TM4SF1, TNBC, migration, invasion
Introduction
Breast cancer (BC) is a disease in which cell growth in the breast is out of control. BC can occur in both men and women, but it is much more common in women. Around 18% of women who are diagnosed with malignancies is BC. The 5-year survival rate for BC is about 50–60%.1 Treatments are available for BC including surgery, hormonal therapy, radiation therapy, chemotherapy, biological therapy and specific medicine for different BC subtypes.2,3 Although these methods can control and suppress tumor growth at varying degrees, it is common to observe tumor invasion and metastasis from clinical studies.4,5 Thus, it is an urgent need to understand the mechanisms of tumor invasion and metastasis so that new oncogenes can be identified and used as therapeutic targets for BC treatment.
Transmembrane 4 L-six family member 1 (TM4SF1) is a small plasma membrane glycoprotein that belongs to the transmembrane 4 superfamily, also known as the tetraspanin family.6 TM4SF1 plays important roles in the regulation of cell growth, development, activation and motility through modulating cell signaling transduction.7 TM4SF1 was first identified as an oncogene and was shown to correlate with tumor cell migration and invasion during metastasis in 1986. Normally, the expression of TM4SF1 is maintained at a low level in healthy tissues.8,9 However, TM4SF1 is found to be overexpressed in different tumors and is involved in regulating cell growth, proliferation, invasion and migration in many tumors such as colon cancer, ovarian cancer and BC.10–14 Therefore, TM4SF1 is an attractive target gene for BC therapy, and it is noteworthy to investigate whether the expression level of TM4SF1 will have an impact on BC metastasis. One approach to regulate gene expression is taking advantage of microRNA. MicroRNAs are a class of regulatory noncoding RNAs that bind to the 3′-UTR of target mRNAs to repress gene expression.15 MicroRNAs have been shown to participate in multiple cellular events including cell survival, proliferation, and apoptosis in many cancer cell types.16 Among these microRNAs, miR-206 has been identified as a tumor suppressor in many human malignancies.17–20 For instance, miRNA-206 suppresses PGE2-induced colorectal cancer cell proliferation, migration, and invasion by targeting TM4SF1.21 Given the specific interaction between TM4SF1 and miR-206, we hypothesis that miRNA-206 could have potential to suppress TM4SF1 to regulate BC cell proliferation, migration, and invasion. However, the relationships between BC, TM4SF1, and miRNA-206 have not been well characterized.
In this study, we investigated the impact of TM4SF1 expression on BC cell migration and invasion. We showed that TM4SF1 was overexpressed in BC tissues. miR-206 negatively regulated gene expression of TM4SF1 in MDA-MB-231 cells. The expression level of TM4SF1 was correlated with migration and invasion capabilities of BC cells both in vitro and in vivo. We propose a therapeutic approach by using miR-206 to restrict BC migration and invasion.
Materials and methods
Clinical BC specimens
From a total of 20 BC patients, who underwent surgical excision at our hospital in 2017, BC tissues were frozen in liquid nitrogen and stored at −80°C until further study. As shown in Table 1, patient age and tumor histologic type were obtained from medical records. All clinical specimens used in this study were approved with patients’ written informed consent and were done in accordance with the World Medical Association Declaration of Helsinki.
Table 1.
Clinical pathologic characteristics of the 20 breast cancer patients
| Variables | Number, n (%) |
|---|---|
| Age (years) | |
| <65 | 15 (75.0) |
| ≥65 | 5 (25.0) |
| Type | |
| Luminal A | 5 (25.0) |
| Luminal B | 5 (25.0) |
| Her+ | 5 (25.0) |
| Basal like | 5 (25.0) |
Cell culture and transfection
The human triple-negative BC cell line (MDA-MB-231) was ordered from the Institute of Cell Biology of the Chinese Academy of Sciences (Shanghai, China). MDA-MB-231 cells were grown in DMEM (Gibco, Grand Island, NY,USA) supplemented with 10% heat-inactivated (HI) FBS (Gibco) and 1% streptomycin at 37°C in a humidified chamber containing 5% CO2. Cells were transfected with recombinant lentiviruses containing miR-206 mimics, anti-miR-206, or a negative control (NC) using FECT™ CP Transfection kit (Ribo, Guangzhou, China) following the manufacturer’s instructions.
Generation of TM4SF1-knockout MDA-MB-231 cell line using CRISPR/Cas9
The single guide (sgRNA) was designed against exons 2 of the TM4SF1 gene (NM_014220.2) using the CRISPR sgRNA design tool (http://cripr.dbcls.jp/). The genomic DNA sequence as target: cr5 (5′-GCATTTGTCTTCATTGGGCT-3′). Guide sequences were subcloned into lenti-CRISPR v2-derived vector. Plasmids were digested by DraI, purified, and transcribed using Hiscribe T7 high yield transcription kit (OMEGA, Doraville, GA, USA). The resulting products were purified using EZNA microelute RNA cleanup kit (OMEGA) following the manufacturer’s instructions. Positive transfected cells were selected with medium containing puromycin (5 μg/mL).
Reverse transcription and real-time PCR
Total RNA was extracted from either frozen tissues or cells using TRIzol (Invitrogen, Carlsbad, CA, USA) and 1 μg of RNA was used to generate cDNA using the ReverTra Ace qPCR RT Kit (TOYOBO, Tokyo, Japan) as described by the manufacturer. Quantitative real-time PCR was carried out by using the SYBR Green PCR Master Mix (TOYOBO). Primers used include TM4SF1 forward 5′-CAGCCCTTGGCTTAGCAGA-3′, TM4SF1 reverse 5′-CCACAATGCTTGGGTTCA-3′ and GAPDH forward 5′-GAGTCAACGGATTTGGTCGT-3′, GAPDH reverse 5′-GACAAGCTTCCCGTTCTCAG-3′. The relative amount of miR-206 was detected using the miRcute miRNA qPCR Detection Kit (SYBR Green) with primers: miR-206 forward 5′-CGCTGGAATGTAAGGAAGTGTGTGG-3′ and U6 forward 5′-GCTTCGGCAGCACATATACTAAAAT-3′ as described by the manufacturer. Relative gene or miRNA expression was determined using the 2−ΔΔCt method.
Western blotting
Total protein was isolated from cells or tissues using cell lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, and 1% Protease Inhibitor Cocktail). Lysates were analyzed by SDS-PAGE and transferred to PVDF membranes (0.45 μM, Millipore, Billerica, MA, USA). Membranes were blocked with 5% skimmed milk and then immunoblotted at 4°C using the following primary antibodies: anti-TM4SF1 (1:1,000, Abcam, Cambridge, MA, USA), anti-β-actin (1:500, Abcam). Membranes were probed at room temperature for 1 hr with horseradish peroxidase-conjugated goat anti-rabbit antibody (Santa Cruz, CA, USA). Blots were detected using the Dyne ECL STAR Western Blot Detection kit (Millipore). All experiments were performed in triplicate.
Cell viability and cell cycle assays
1×104 cells per well were seeded into a 96-well plate and allowed to attach overnight. MTT cell viability assays were performed using parental MDA-MB-231 cells or TM4SF1-/- MDA-MB-231 cells for over 7 days according to the manufacturer’s instructions. The cell cycle was determined by propidium iodide staining and flow cytometry. To fix the cells, 70% cold ethanol was added and incubated overnight at −20°C. Cells were washed and incubated in RNase A at 37°C for 30 mins. After incubation, propidium iodide was added, and cells were incubated in the dark for 30 mins. Cell cycle status was analyzed using flow cytometer.
Invasion assays
The invasion assay was performed using a specialized invasion chamber (Chemicon, Temecula, CA, USA). The inserts contained 8 μm pore size polycarbonate membranes with a precoated thin layer of basement membrane matrix (ECMatrix). In brief, DMEM supplemented with 10% FBS was pre-coated into the lower chamber as a hemoattractant. MDA-MB-231 cells were trypsinized and resuspended in DMEM. 3 × 105 cells were seeded into the upper chambers. After 24 hrs of incubation at 37°C, noninvasive cells were removed from the upper surface of the membrane using a moist cotton-tipped swab. Invasive cells on the lower surface of the membrane, which had invaded the ECMatrix and had migrated through the polycarbonate membrane, were stained with the staining solution for 20 mins and rinsed with distilled water several times. Invasiveness was quantitated by selecting 5 different views and counting the number of invasion cells.
Immunofluorescence staining
Cells were fixed with methanol and washed with PBS. After blocking with 1% BSA, cells were incubated with anti-TM4SF1 primary antibody (1:200, Abcam) overnight at 4°C. The next day, cells were washed and incubated with FITC-labeled goat anti-rabbit secondary antibody (1:1,000, Abcam). DAPI was included to stain for nuclei (Sigma, St. Louis, MO, USA).
Mouse mammary pad injection
Six-week-old, 18 g female BALB/c nude mice were purchased from Vitallihua (Beijing, China). The BC cells were trypsinized and resuspended in HBSS/Matrigel (1:1 volume) to a final concentration of 107 cell/mL. Xenografts were generated by injecting 0.2 mL cell suspension into the area of the mammary fat pad. Weigh the tumors after the end of the experiment. This study was approved by the Institutional Animal Care and Use Committee (IACUC) of Jilin University. All animal experiments were performed in an animal biosafety level 2 plus facility in accordance with the Jilin University IACUC. We followed the guidelines of GB14925-2010 in China for the welfare of the animals.
H&E
After aseptically shaving the hair around the gland, the tumors were collected for histological examination. The tumors were fixed in 10% neutral buffered formalin for 24 hrs, processed on an automatic tissue processor, and embedded in paraffin wax. Sections were cut at 4 μm thickness at three levels and stained by the H&E stain.
Immunohistochemistry (IHC)
Sections were deparaffinized and rehydrated with xylene and graded alcohols. Antigen retrieval was carried out in 5 mM citrate buffer. After the inactivation of endogenous peroxidase with 3% H2O2, the sections were blocked with goat serum and incubated with TM4SF1 antibody (1:100, Abcam) overnight at 4°C. The sections were first rinsed in PBS, incubated with biotinylated secondary antibody at 37°C for 20 mins, and then washed with PBS three times. Diaminobenzidine was used as a chromogen substrate. Finally, the sections were counterstained with hematoxylin.
Statistical analysis
All statistical analyses and visualizations in this study were performed using GraphPad Prism software. All data were expressed as mean ± SD unless otherwise stated. An independent sample t-test was used to test for significance between groups, P<0.05 was considered statistically significant.
Results
TM4SF1 was overexpressed in TNBC and inversely correlated with miR-206
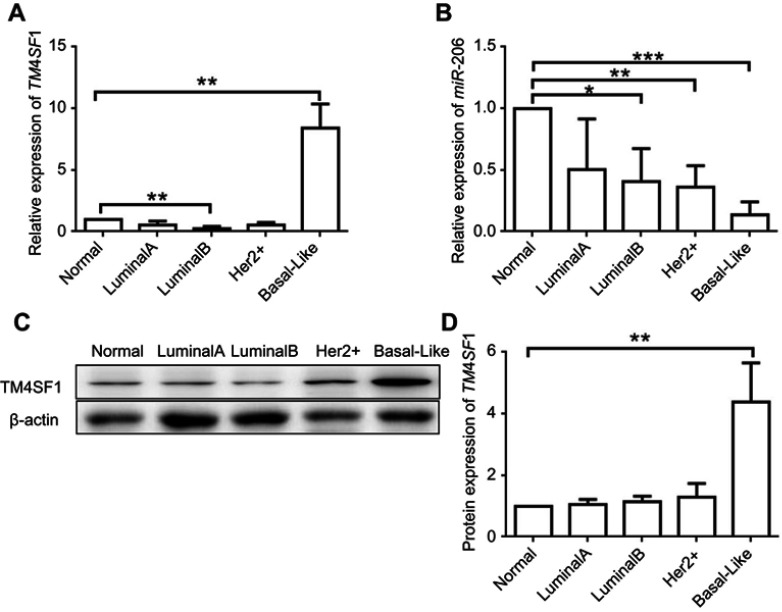
To investigate the expression level of TM4SF1 in BC tissues, we examined four molecular subtypes of BCs. Subtype luminal A, luminal B, and Her 2+ showed similar TM4SF1 expression level compared to normal breast tissue. Importantly, in subtype basal-Like (a TNBC subtype), TM4SF1 expression was 10-fold higher than normal breast tissue at mRNA level (Figure 1A) and was 4-fold higher at protein level (Figure 1C and D). We further analyzed the correlation between TM4SF1 and miR-206. TM4SF1 expression levels were inversely correlated with miR-206 in all four subtypes of BCs. It was worth noting that miR-206 showed the lowest expression level in subtype basal-Like (Figure 1B). These results indicated that TM4SF1 was overexpressed in BC tissues and was inversely correlated with the expression level of miR-206, especially for a TNBC subtype basal-Like.
Figure 1.
Upregulation of TM4SF1 and downregulation of miR-206 in clinical breast cancer tissues compared with the levels in adjacent normal breast tissues.
Notes: The mRNA expression of TM4SF1 (A) and miR-206 (B) was detected by real-time PCR in four subtypes of clinical breast cancer (Luminal A, Luminal B, Her2+, and basal like. N=5 for each subtype). (C) Western blotting analysis of TM4SF1 expression in four subtypes of clinical breast cancer. β-Actin served as the internal control. (D) The relative fold change of TM4SF1/β-actin in (C) was detected by imageJ software. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: TM4SF1, transmembrane 4 L6 family member 1; miR-206, microRNA-206.
miR-206 negatively regulated gene expression of TM4SF1 in MDA-MB-231 cells
We then investigated the impact of miR-206 on gene expression level of TM4SF1 in MDA-MB-231 cells. MDA-MB-231 cells were transfected with miR-206, anti-miR-206, scramble, or mock controls. As shown in Figure 2A, RT-PCR results showed that the overexpression of miR-206 led to a comparable reduction of TM4SF1 expression compared to scramble or mock controls. Furthermore, MDA-MB-231 cells that were transfected with anti-miR-206 reversed miR-206-mediated phenotype. Western blotting results further confirmed the RT-PCR results (Figure 2B and C). These observations suggest that miR-206 negatively regulated gene expression of TM4SF1 in MDA-MB-231 cells.
Figure 2.
miR-206 negatively regulated gene expression of TM4SF1 in MDA-MB-231 cells.
Notes: (A) MDA-MB-231 cells were transfected with 100 nM miR-206, anti-miR-206, or NC for 24 hrs. The mRNA expression of TM4SF1 was detected by real-time PCR. (B) Western blotting analysis of TM4SF1 expression in (A). β-Actin served as the internal control. (C) The relative fold change of TM4SF1/β-actin in (B) was detected by imageJ software. *P<0.05; **P<0.01.
Abbreviations: TM4SF1, transmembrane 4 L6 family member 1; NC, negative control; miR-206, microRNA-206.
Establishment of TM4SF1-knockout MDA-MB-231 cell line
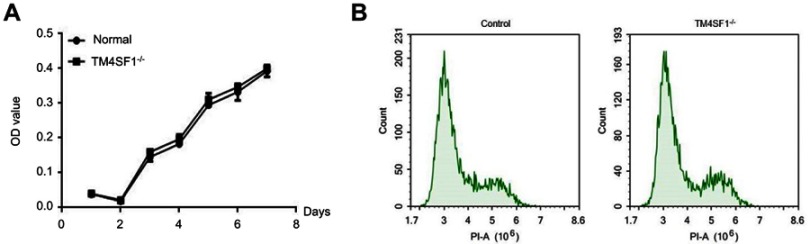
To further investigate the functional importance of TM4SF1 in TNBC, we knocked out the TM4SF1 gene in a TNBC cell line MDA-MB-231 using CRISPR/Cas9 technology. We successfully generated a TM4SF1-KO MDA-MB-231 cell line (Figure 3A), in which production of TM4SF1 was dramatically disrupted at both mRNA level (Figure 3B) and protein level (Figure 3C). We confirmed the TM4SF1 knockout by immunostaining. As shown in Figure 3D, TM4SF1 signal was only detected in the parental MDA-MB-231 cells but not the TM4SF1-KO MDA-MB-231 cells. We further analyzed the proliferation of these two cell lines by MTT assay. There was no evidence of cell growth defects in TM4SF1-KO MDA-MB-231 cells (Figure 4A). We next investigated the role of TM4SF1 in self-renewal-related cell cycle regulation. As shown in Figure 4B, no significant phase difference was observed in TM4SF1-KO MDA-MB-231 cells by DNA content analyses with PI staining. These results indicated the successful generation of TM4SF1-KO MDA-MB-231 cells.
Figure 3.
Generation of TM4SF1-knockout MDA-MB-231 cells.
Notes: (A) Sanger sequencing of TM4SF1 gene in the knockout MDA-MB-231 cells. Two bases of nucleic acid were deleted of coding sequence resulting in frameshift mutation. (B) The mRNA expression of TM4SF1 was detected by real-time PCR in the knockout MDA-MB-231 cells. (C) Western blotting analysis of TM4SF1 expression in (B). β-actin served as the internal control. (D) Immunostaining of TM4SF1 in the knockout MDA-MB-231 cells. DAPI was used to stain nuclei. ***P<0.001.
Abbreviations: TM4SF1, transmembrane 4 L6 family member 1; KO, knockout; DAPI, 4′,6-diamidino-2-phenylindole.
Figure 4.
TM4SF1 knockout had no effect on cell viability and cycle in MDA-MB-231 cells.
Notes: (A) MTT cell viability assays were performed using parental MDA-MB-231 cells or TM4SF1 KO MDA-MB-231 cells at the indicated time points. (B) Cell cycle was determined by propidium iodide staining and detected by flow cytometry.
Abbreviations: TM4SF1, transmembrane 4 L6 family member 1; -/-, TM4SF1-knockout MDA-MB-231 cells; PI, propidium iodide.
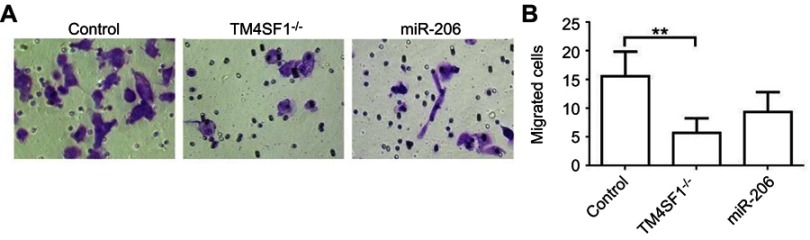
Suppression of TM4SF1 in MDA-MB-231 cells inhibited cell migration and invasion capabilities in vitro
We next analyzed the effect of TM4SF1 on the promotion of migration and invasion in MDA-MB-231 cells. In this experiment, three different MDA-MB-231 cell lines were used: the parental MDA-MB-231 cells, TM4SF1-KO MDA-MB-231 cells, and miR-206 transfected MDA-MB-231 cells. As shown in Figure 5A and B, both TM4SF1-KO MDA-MB-231 cells and miR-206 transfected MDA-MB-231 cells significantly decreased cell migration and invasion capabilities compared to the parental MDA-MB-231 cells. These results indicated that TM4SF1 was an important factor for the migration and invasion capabilities of MDA-MB-231 cells. More importantly, miR-206 was a potential negative regulator to control the migration and invasion capabilities of MDA-MB-231 cells.
Figure 5.
TM4SF1 KO MDA-MB-231 cells displayed reduced cell migration and invasion capabilities in vitro.
Notes: (A) Migration activity of TM4SF1 KO MDA-MB-231 cells or KO MDA-MB-231 cells transfected with miR-206 was measured by transwell assay. (B) Relative transmitted cell numbers in the transwell assay in (A). **P<0.01.
Abbreviations: TM4SF1, transmembrane 4 L6 family member 1; miR-206, microRNA-206; -/-, TM4SF1-knockout MDA-MB-231 cells.
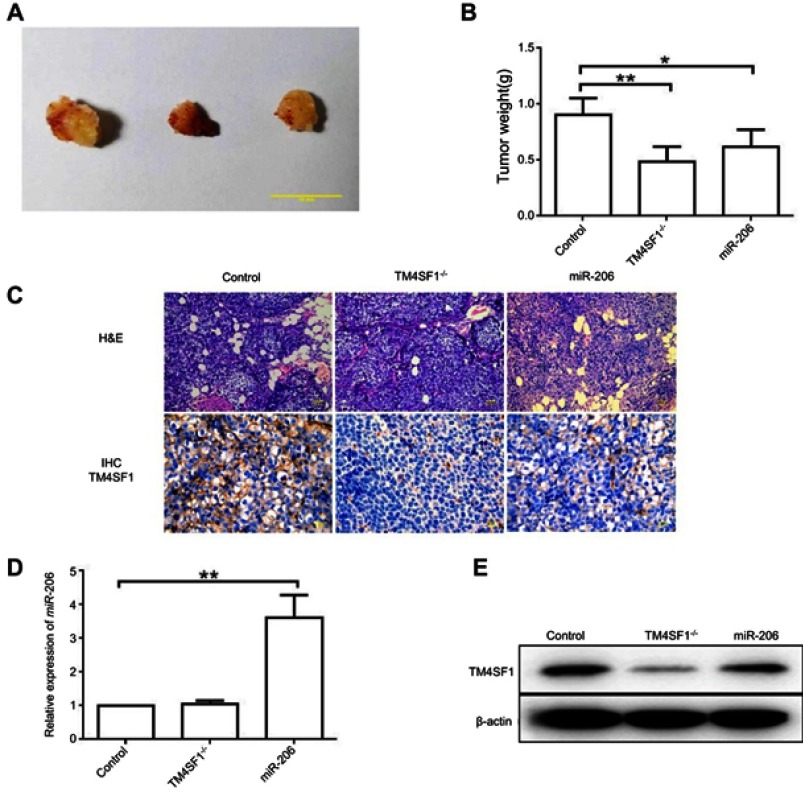
TM4SF1-KO MDA-MB-231 cells displayed a reduced tumor burden in BALB/c nude mice
Based on the above finding that TM4SF1 was an important factor for the migration and invasion capabilities of MDA-MB-231 cells, we hypothesized that BALB/c nude mice that were implanted with TM4SF1-KO MDA-MB-231 cells would display a reduced tumor burden. To test this, BALB/c nude mice were implanted with MDA-MB-231 or TM4SF1-KO MDA-MB-231 cells. MDA-MB-231 group mice were injected with either PBS (N=6) or 2.5 nmol of miR-206 (N=6) for every 2 days via tail vein injections. Figure 6A shows representative pictures of excised tumors from each experimental group. Both TM4SF1-KO- and miR-206-treated tumors were smaller than the control PBS-treated tumors, but the effect of miR-206 was less dramatic than that seen with the TM4SF1-KO tumor. This observation was consistent with the results shown in Figure 6B, where the average tumor weight for TM4SF1-KO group and the group treated with miR-206 was significantly lower than the PBS group. In addition, to verify the delivery of miR-206 to tumors, we examined the miR-206 levels in these three groups by RT-PCR. As demonstrated in Figure 6D, the miR-206 levels in miR-206-treated tumors were significantly higher than those in TM4SF1-KO- or PBS-treated tumors. Moreover, we examined the expression level of TM4SF1 in tumors from these three groups. Only limited TM4SF1 signal was detected in the TM4SF1-KO- and miR-206-treated tumors when compared to the PBS-treated tumors as shown in the Immunohistochemistry staining (Figure 6C) and Western blotting (Figure 6E) results. Taken together, our results suggested that TM4SF1 played a positive role in TNBC growth, and miR-206 had a potential to be used as a therapeutic agent to reduce the tumor burden in BALB/c nude mice model.
Figure 6.
TM4SF1-KO MDA-MB-231 cells displayed a reduced tumor burden in BALB/c nude mice.
Notes: (A) Representative images of breast tumors in control group, TM4SF1-/- group, and miR-206 group (N=6). (B) Average tumor weight of control group, TM4SF1-/- group, and miR-206 group (N=6). (C) Representative pictures of H&E staining and immunohistochemistry of TM4SF1 for breast tumors in the control group, TM4SF1-/- group and miR-206 group (N=6). (D) The miR-206 amount in the tumors of control group, TM4SF1-/- group and miR-206 group (N=6) was detected by real-time PCR. (E) Western blotting analysis of TM4SF1 expression in the tumors of the control group, TM4SF1-/- group, and miR-206 group (N=6). *P<0.05; **P<0.01.
Abbreviations: TM4SF1, transmembrane 4 L6 family member 1; miR-206, microRNA-206; -/-, TM4SF1-knockout MDA-MB-231 cells.
Discussion
BC is a complex disease with no simple solution. It is important to understand the basic biology of BC, from gene expression pattern in BC cells to the environment that is favored for BC cells to grow. Multiple genes have been identified as critical regulators of the metastasis of BC. There are a growing number of studies addressing that miRNAs are functional as tumor suppressor by targeting the regulators of the metastasis of BC in different types of tumors. Several targets of miR-206 have been identified thus far. For instance, miR-206 has been shown to repress tumor proliferation and invasion in BC by targeting Cx43.22 miRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation in HeLa cells.23 MET is targeted by miR-206 in papillary thyroid carcinoma.24 In this study, we found that miR-206 expression levels were downregulated and were inversely correlated with TM4SF1, a member of the transmembrane 4 superfamily in all four subtypes of BCs, especially for the TNBC.
Recent study has been shown that the expression level of TM4SF1 is correlated with miR-206. Bioinformatics analysis revealed that miR-206 directly targeted the 3ʹ-UTR of TM4SF1, and TM4SF1 expression was reduced by miR-206 overexpression which led to the suppression of colorectal cancer cell proliferation, migration, and invasion.21 Therefore, we investigated the impact of miR-206 on gene expression level of TM4SF1 in MDA-MB-231 cells. We found that miR-206 negatively regulated gene expression of TM4SF1.
Recently, numerous studies have shown that TM4SF1 plays an important role in migration and invasion of several malignant cancers, including liver and BCs.25–27 TM4SF1 promotes proliferation, invasion, and metastasis in human liver cancer cells.25 Knockdown of TM4SF1 decreased pancreatic tumor growth and increased responsiveness to treatments using gemcitabine in pancreas tumor models.28 These studies suggest that TM4SF1 may have an essential role in the tumorigenesis and progression in TNBC. We then studied the biological importance of miR-206 and TM4SF1 to the migration and invasion rate of MDA-MB-231 cells. We found that TM4SF1 was an important factor for the migration and invasion capabilities of MDA-MB-231 cells. Moreover, miR-206 was a potential negative regulator to control the migration and invasion capabilities of MDA-MB-231 cells.
We then performed in vivo experiments to verify our in vitro observation. BALB/c nude mice were implanted with MDA-MB-231 or TM4SF1-KO MDA-MB-231 cells. MDA-MB-231 group mice were injected with either PBS or 2.5 nmol of miR-206 for every 2 days via tail vein injections. We found that BALB/c nude mice that were implanted with TM4SF1-KO MDA-MB-231 cells displayed a reduced tumor burden. Moreover, MDA-MB-231 group mice that were treated with miR-206 also showed a reduced tumor burden.
Conclusion
Our results demonstrated that TM4SF1 played an important role in the progression and metastasis of TNBC. miR-206 negatively regulated gene expression of TM4SF1. Overexpression of miR-206 reduced proliferation, migration, and invasion of MDA-MB-231 cells. Our results indicated that miR-206 could be used as a potential therapeutic agent for TNBC.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. [DOI] [PubMed] [Google Scholar]
- 2.Harder H, Langridge C, Solis-Trapala I, et al. Post-operative exercises after breast cancer surgery: results of a RCT evaluating standard care versus standard care plus additional yoga exercise. Eur J Integr Med. 2015;7(3):202–210. doi: 10.1016/j.eujim.2015.02.002 [DOI] [Google Scholar]
- 3.Courneya KS, Segal RJ, Gelmon K, et al. Predictors of adherence to different types and doses of supervised exercise during breast cancer chemotherapy. Int J Behav Nutr Phys Act. 2014;11:85. doi: 10.1186/s12966-014-0085-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aceto N, Bardia A, Miyamoto DT, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F, Drabsch Y, Dekker TJ, et al. Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-β signalling. Nature Commun. 2014;5:3388. doi: 10.1038/ncomms4388 [DOI] [PubMed] [Google Scholar]
- 6.Wright MD, Ni J, Rudy GB. The L6 membrane proteins – a new four-transmembrane superfamily. Protein Sci. 2000;9:1594–1600. doi: 10.1110/ps.9.8.1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zukauskas A, Merley A, Li D, et al. TM4SF1: a tetraspanin-like protein necessary for nanopodia formation and endothelial cell migration. Angiogenesis. 2011;14:345–354. doi: 10.1007/s10456-011-9218-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Donnell RT, DeNardo SJ, Shi XB, et al. L6 monoclonal antibody binds prostate cancer. Prostate. 1998;37(2):91–97. doi: [DOI] [PubMed] [Google Scholar]
- 9.Marken JS, Schieven GL, Hellstrom I, Hellstrom KE, Aruffo A. Cloning and expression of the tumor-associated antigen L6. Proc Natl Acad Sci U S A. 1992;89(8):3503–3507. doi: 10.1073/pnas.89.8.3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S, Kim TY, Kwak TK, et al. Transmembrane 4 L six family member 5 (TM4SF5) enhances migration and invasion of hepatocytes for effective metastasis. J Cell Biochem. 2010;111:59–66. doi: 10.1002/jcb.22870 [DOI] [PubMed] [Google Scholar]
- 11.Shih SC, Zukauskas A, Li D, et al. The L6 protein TM4SF1 is critical for endothelial cell function and tumor angiogenesis. Cancer Res. 2009;69:3272–3277. doi: 10.1158/0008-5472.CAN-08-3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn S, Koch M, Nübel T, et al. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res. 2007;5:553–567. doi: 10.1158/1541-7786.MCR-06-0384 [DOI] [PubMed] [Google Scholar]
- 13.Giordano TJ, Shedden KA, Schwartz DR, et al. Organ-specific molecular classification of primary lung, colon, and ovarian adenocarcinomas using gene expression profiles. Am J Pathol. 2001;159:1231–1238. doi: 10.1016/S0002-9440(10)62509-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y, Xu Y, Xu J, Lu D, Wang J. Role of TM4SF1 in regulating breast cancer cell migration and apoptosis through PI3K/AKT/mTOR pathway. Int J Clin Exp Pathol. 2015;8(8):9081–9088. [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 16.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb1826 [DOI] [PubMed] [Google Scholar]
- 17.Vickers MM, Bar J, Gorn-Hondermann I, et al. Stage-dependent differential expression of microRNAs in colorectal cancer: potential role as markers of metastatic disease. Clin Exp Metastasis. 2012;29:123–132. doi: 10.1007/s10585-011-9435-3 [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Ling C, Bai Y, Zhao J. MicroRNA-206 is associated with invasion and metastasis of lung cancer. Anat Rec (Hoboken). 2011;294:88–92. doi: 10.1002/ar.21287 [DOI] [PubMed] [Google Scholar]
- 19.Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68:5004–5008. doi: 10.1158/0008-5472.CAN-08-0180 [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Yan Q, Li S, et al. Expression of the tumor suppressor miR-206 is associated with cellular proliferative inhibition and impairs invasion in ERalpha-positive endometrioid adenocarcinoma. Cancer Lett. 2012;314:41–53. doi: 10.1016/j.canlet.2011.09.014 [DOI] [PubMed] [Google Scholar]
- 21.Park YR, Seo SY, Kim SL, et al. MicroRNA-206 suppresses PGE2-induced colorectal cancer cell proliferation, migration, and invasion by targeting TM4SF1. Biosci Rep. 2018;38:BSR20180664. doi: 10.1042/BSR20180664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y, Zhao ZM, He QZ, Jiang BQ, Wu Y, Zhuang ZG. Hsa-miR-206 represses the proliferation and invasion of breast cancer cells by targeting Cx43. Eur Rev Med Pharmacol Sci. 2015;19(11):2091–2104. [PubMed] [Google Scholar]
- 23.Song G, Zhang Y, Wang L. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J Biol Chem. 2009;284:31921–31927. doi: 10.1074/jbc.M109.046862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, He M, Hou Y, et al. Expression profiles of microRNAs and their target genes in papillary thyroid carcinoma. Oncol Rep. 2013;29:1215–1220. [DOI] [PubMed] [Google Scholar]
- 25.Huang YK, Fan XG, Qiu F. TM4SF1 promotes proliferation, invasion, and metastasis in human liver cancer cells. Int J Mol Sci. 2016;17:661. doi: 10.3390/ijms17050661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao J, Yang JC, Ramachandran V, et al. TM4SF1 regulates pancreatic cancer migration and invasion in vitro and in vivo. Cell Physiol Biochem. 2016;38:740–750. doi: 10.1159/000445664 [DOI] [PubMed] [Google Scholar]
- 27.Yang JC, Zhang Y, He SJ, et al. TM4SF1 promotes metastasis of pancreatic cancer via regulating the expression of DDR1. Sci Rep. 2017;7:4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]