Abstract
LC/MS is an analytical technique that, due to its high sensitivity, has become increasingly popular for the generation of metabolic signatures in biological samples and for the building of metabolic data bases. However, to be able to create robust and interpretable (transparent) multivariate models for the comparison of many samples, the data must fulfil certain specific criteria: (i) that each sample is characterized by the same number of variables, (ii) that each of these variables is represented across all observations, and (iii) that a variable in one sample has the same biological meaning or represents the same metabolite in all other samples. In addition, the obtained models must have the ability to make predictions of, e.g. related and independent samples characterized accordingly to the model samples. This method involves the construction of a representative data set, including automatic peak detection, alignment, setting of retention time windows, summing in the chromatographic dimension and data compression by means of alternating regression, where the relevant metabolic variation is retained for further modelling using multivariate analysis. This approach has the advantage of allowing the comparison of large numbers of samples based on their LC/MS metabolic profiles, but also of creating a means for the interpretation of the investigated biological system. This includes finding relevant systematic patterns among samples, identifying influential variables, verifying the findings in the raw data, and finally using the models for predictions. The presented strategy was here applied to a population study using urine samples from two cohorts, Shanxi (People’s Republic of China) and Honolulu (USA). The results showed that the evaluation of the extracted information data using partial least square discriminant analysis (PLS-DA) provided a robust, predictive and transparent model for the metabolic differences between the two populations. The presented findings suggest that this is a general approach for data handling, analysis, and evaluation of large metabolic LC/MS data sets.
Introduction
Recent analytical advances within the area of system biology1–3 (e.g. DNA microarrays, NMR, GC/MS and LC/MS) have allowed the collection of large-scale multivariate data sets describing complex biological systems and events. These extremely information-rich data sets potentially provide great possibilities for the understanding of the development and treatment of human diseases, toxic responses and genetic modification. From a statistical modelling point of view such data sets pose a great challenge, given that organized and robust approaches will be crucial in order to handle the vast amounts of data generated as well as to extract the relevant information in the data by applying reliable and interpretable mathematical algorithms and models.
Within the areas of metabolomics and metabonomics, metabolite analysis in biofluids and tissues has been carried out using mainly NMR4–6 or GC/MS.7,8 However, due to recent instrumental advances, LC/MS has established itself as a powerful means for metabolite analysis,9–11 and should be regarded as an information-rich technique, complementary to NMR and GC/MS for research in the fields of metabolomics and metabonomics.
LC/MS is an analytical technique that, due to its high sensitivity, has become increasingly popular for the generation of metabolic signatures in biological samples and for the building of metabolic data bases. However, in order to characterize these multiparametric metabolic signatures and relate them to specific pathophysiological conditions or pathways, systematic interactions between different metabolites have to be considered and understood. Multivariate statistical projection methods (e.g. principal component analysis (PCA)12,13 and partial least squares (PLS)14,15) have proven to be valuable tools for the analysis of metabolic and related biological data in many applications.16,17 This is mainly due to the ability to handle many and correlated variables but also to the robustness and high interpretability (transparency) of the obtained models.
Critical issues when analysing LC/MS data are alignment and resolution. As a prerequisite for multivariate techniques to work, and hence to be able to compare samples in large data bases, a data table must be constructed where each sample is characterized by the same number of variables and each of these variables is represented across all observations. Additionally, it is vital that a variable in one sample is the same for all the other samples (i.e. has the same biological meaning or represents the same metabolite) otherwise the whole idea of comparing samples based on the systematic changes in metabolite patterns fails.
Another important issue when modelling complex biological data sets is the ability of an existing model to predict the outcome of new samples. This poses high demands both on the analytical procedures, in terms of robustness and reproducibility, and on the existing model, in terms of providing a good characterisation of the model samples, which is valid and robust over the whole variation space expected for future samples. For these purposes multivariate projection based methods are suitable provided that the sample characterisation (data presentation) is done uniformly over all samples in the model and that the variables used to describe new samples are consistent with the variables for the model samples. However, it is important to clarify that chemometric or multivariate tools should not be regarded as a replacement of the analytical chemists’ knowledge in interpreting the LC/MS data or the biologists/toxicologists/clinicians’ interpretation of the interactions and mechanisms occurring in the studied biological systems. Instead, these methods should be seen as the means for creating robust and highly interpretable multivariate models with the aim of finding and understanding patterns and trends among samples and variables in large and complex data sets, e.g. as generated by LC/MS of biofluids. These models will highlight important areas in the spectral and chromatographic data where further effort on interpretation and metabolite identification should be put in.
Unbiased extraction of information from LC/MS data prior to multivariate analysis can be done using different approaches. Examples of the most common approaches found in the literature are:
Peak detection: a peak detection algorithm detects all peaks in the data above some defined noise threshold. The area under the peak or the peak height is then used as a quantitative measure and the retention time and m/z are used for identification. Identical peaks from different samples should hence end up in the same column in the data table.9
Curve resolution or deconvolution: the spectral and the chromatographic profiles for each compound are resolved. The areas under the chromatographic profiles are used as a quantitative measure of the compound and the spectral profile and the retention time are used for identification. Identical compounds from different samples should hence end up in the same column in the data table.10
Summing of the data to obtain a total mass spectrum for each sample: this can be done by combining the whole chromatograms or by first dividing the chromatograms into segments and then combining all the segments prior to multivariate analysis.11
Although these approaches have shown very promising results, there is still a lot of work to be done before LC/MS can be considered a robust and efficient tool for metabolite analysis of biofluids.
Here we present a strategy for screening of large-scale LC/ MS data sets applied to a population study analysing urine samples from two cohorts, Shanxi (People’s Republic of China) and Honolulu (USA). This comprises approaches to optimize the information extraction from metabonomic data bases in an organized and consistent fashion. In particular, the presented strategy is aimed at coping with analytical variation and at reducing the number of variables, by bi-linear compression, to a smaller subset of information-rich variables prior to multivariate data analysis. The presented results show that the evaluation of the “information optimized” data by means of multivariate projections provided robust, predictive and highly interpretable (transparent) models for screening of large metabonomic LC/MS data sets. The advantages with the presented method compared to already existing methods for handling metabolic LC/MS data are three-fold. (i) The model transparency: it is easy and straightforward to trace the model results back to the informative parts of the raw data (time points and important m/z values). This is also a feature obtained using the peak detection or the curve resolution approaches described above. (ii) All samples are treated equally throughout the whole data set, meaning that each sample is described by the same number of variables. This is only achieved by the summing data approach mentioned above. (iii) This is the first method, to our knowledge, that has shown that the model used for information extraction from LC/MS data is also valid for extraction of the relevant information in new independent samples (here represented by separate well plates). This has important implications for the building and evaluation of large metabonomic LC/MS data bases, where there is a requirement for the comparison of samples measured over time and/or at different laboratories (instruments) by means of a robust and reliable modelling framework based on an organised and information dense data format.
In summary the presented method takes advantage of and combines the positive features from the common existing approaches for handling and analysing metabolic LC/MS data. In addition, this approach is unique in the sense that it is a method dealing with the problem of coping with analytical variations or drifts while still maintaining the important biological variation in the data, which in turn will be highlighted by the multivariate modelling.
The presented population data is a part of INTERMAP (international study of macro- and micro-nutrients and blood pressure), which is a large epidemiological study estabilished in 1996 by Northwestern University, Chicago, USA. Information was collected from people within USA, Japan, UK, and the People’s Republic of China, with the aim to investigate whether there is a correlation between dietary intake and changes in blood pressure. It has already been demonstrated through the use of 1D 1H NMR that profiles of inter- and intra-population differences can be produced, with variations being noted from people with health conditions and unusual dietary intakes.18,19 Being a more sensitive technique than NMR, LC/MS has the potential to become a powerful tool in metabolic profiling. However, it is of great importance that a strong and organised statistical strategy is applied to the complex data sets that are being generated.
Materials and methods
Chemicals
All aqueous solutions were prepared using purified distilled water (18.2 MΩ) from a Millipore MilliQ system (MA, USA). HPLC grade acetonitrile was purchased from JT Baker (NJ, USA). Formic acid, extra pure grade (98–100%), was purchased from Fluka (WI, USA). All other materials were purchased from Sigma-Aldrich (MO, USA).
Sample preparation
Aliquots of the urine samples stored at −40 °C were allowed to completely thaw at room temperature. 50 μL of urine from each sample were placed in a 96 well plate, and diluted with 150 μL of distilled water (1 : 4 sample dilutions).
LC/MS analysis
Chromatography was performed on a Waters Metabonomics System comprising of an Alliance® 2795XC, equipped with a column oven and a 2996 PDA detector, coupled to a Micromass® Q-Tof micro™ mass spectrometer equipped with an electrospray source operating in positive ion mode and a Lockspray™ interface for accurate mass measurements. The source temperature was set at 120 °C with a cone gas flow of 50 L h−1, a desolvation gas temperature of 250 °C and a nebulization gas flow of 450 L h−1. The capillary voltage was set at 3.2 kV for positive ion mode with a cone voltage of 40 V, a scan time of 1.0 s and an interscan delay of 0.10 s. A collision energy of 10 V was employed with a collision gas pressure of 5.3 × 10−5 Torr. Leucine enkephalin was employed as the lockmass at a concentration of 50 fmol μL−1 (in 50 : 50 ACN : H2O, 0.1% formic acid) at a flow rate of 30 μL via a lock spray interface. All mass spectral data were collected in centroid mode.
A 10 μL aliquot of diluted urine was injected onto a 2.1 × 100 mm Symmetry® C18 3.5 μm column. The column was eluted with a linear gradient of 0–20% of 0.1% formic acid in acetonitrile (B) over 0.5–4 min, and 20–95% of B over 4–8 min; the composition was held at 95% of B for 0.1 minute then returned to 100% of 0.1% formic acid (aq) (A) at 9.1 min at an eluent flow rate of 600 μL min−1. “Purge–wash–purge” cycle was employed on the autosampler, with 75% aqueous methanol used as the wash solvent and 0.1% aqueous formic acid used as the purge solvent; this ensured that the carry-over between injections was minimized. The mass spectrometric data were collected in full scan mode 100 to 850 m/z from 0–12 min.
Peak detection and alignment
Prior to any data analysis, the mass resolution was reduced to unit resolution. In addition linear interpolation was used to make each scan number correspond to the same time point across all samples. To define the number of chromatographic peaks an average ion chromatogram from all samples was calculated for each m/z channel and then each averaged m/z channel was searched individually. The criteria used was that a peak had to have an intensity above a certain specified “noise threshold level” and additionally, the first derivative had to be positive before and negative after a peak. A search for peaks was then carried out on all sample chromatograms. The search was limited to regions near the peaks detected in the average ion chromatogram. Detected peaks were represented with the maximum peak intensity and were also aligned to the corresponding peak found in the average ion chromatogram. This procedure was repeated for all m/z channels. The use of the maximum intensity for peak representation may not be the optimal way of using the information, but alignment of the chromatograms becomes easier since the peak shape does not need to be considered. Recent results presented by Torgrip et al.20 suggest that this approach works well for the alignment of metabolic GC and NMR data.
Data reduction and compression
The next step in the proposed strategy was data reduction. This was done by dividing the chromatographic dimension into a number of narrow, equally sized time windows (2 scans), (step a in Fig. 1). The use of a narrow window size was allowed due to the fact that the data were already aligned in the previous step. The data in each time window hence held a three dimensional data structure defined by the sample dimension, the chromatographic time dimension and the mass spectral dimension. Summing of these three dimensional “data cubes” in the chromatographic dimension produced a two dimensional data table (XSUM) defined by the sample dimension and the summed spectral dimension. Compression of the information in the two dimensional data table down to a small number of latent variables was then performed for each sample by means of alternating regression (AR;21 step b in Fig. 1). The choice of AR as the preferred method for data compression, instead of e.g. PCA, was based on the fact that AR does not have an orthogonality constraint, which is reasonable when the mass spectra for the different compounds do not have to be orthogonal to each other. The same also applies to the concentrations of the metabolites. Ideally the AR results will be easier to relate back to the raw data since the spectral profiles need not to be mixed in the AR components. Compression of the data provided a data table with fewer variables and also aided in the identification of metabolites since the m/z values correlated with each other between the samples, which probably originated from the same metabolite. Thus, correlated m/z values will end up in the same spectral profile. AR is an iterative method that alternates between two operations until conversion (see eqns. (1) and (2)). Both the “concentration” (intensity vectors) (C) and the mass spectrum (S) were given a non-negative constraint, meaning that neither of the two could ever include negative values. This constraint was applied after each operation in the AR algorithm.
| (1) |
where C = intensity vector, S = mass spectral profile, and the superscript T denotes transposition and −1 matrix inversion.
| (2) |
Fig. 1.
An overview of the data pre-processing steps. The data in the three dimensional data cube are represented by the peak height in the position of the detected peaks and the peaks are aligned towards an average sample. The data cube is then divided into time windows (segments) and each segment is summed in the time direction to form a matrix (XSUM), (step a). XSUM is compressed into intensity vectors and mass profiles using alternating regression (AR) (step b). The intensity vectors from all time windows are then combined to form the final matrix (X) which is subjected to multivariate analysis (step c).
The number of AR components to extract for each time window was decided by means of a PCA prior to the AR procedure. The chosen number of AR components was equal to the number of principal components needed to describe 95% of the variation in a specific time window. The AR procedure was performed for each time window individually, with the samples used for model validation (test set) excluded during the calculations. The spectral profiles (S) were then used to predict the intensity vectors (C) for the test set samples (see eqns. (1) and (2)).
By combining the concentration profiles from all the time windows the final “reduced” data table (X) was obtained, which was then subjected to further multivariate analysis (MVA; step c in Fig. 1).
Multivariate data analysis
Partial least square discriminant analysis (PLS-DA)22 was used as the classification method for modelling the discrimination between the urine samples collected from the two different populations (Shanxi (People’s Republic of China) and Honolulu (USA)). The number of significant components for the PLS-DA classification models was estimated by seven-fold, component-wise cross-validation.23 For validation purposes an independent test set was selected including 253 samples (3 separate well plates) out of the total 600 samples (7 well plates). The remaining 347 samples (4 separate well plates) were used for the model building. The peak detection and data compression processes were carried out using the 347 model samples and the 253 test samples were then treated accordingly. A PLS-DA model was calculated using the 347 model samples and the predictive ability of independent samples (well plates) was evaluated applying the model to the selected 253 test set samples. For all the multivariate analyses the variables in the data (X) were log transformed (X = log (X + 1)), mean centered and scaled to unit variance.
Non-processed LC/MS files were exported to MATLAB software 6.5 (Mathworks, Natick, MA), where all data pre-treatment procedures, such as peak detection, alignment, data reduction and compression were carried out.
Multivariate analysis was performed using the SIMCA-P + 10.5 software (Umetrics AB, Umeå, Sweden).
Results
The peak detection process applied to the 347 model samples rendered in 5680 detected chromatographic peaks. After the AR compression step the number of variables in the final data table (X) was reduced to 597 latent variables summarizing the metabolite concentrations in the selected model samples.
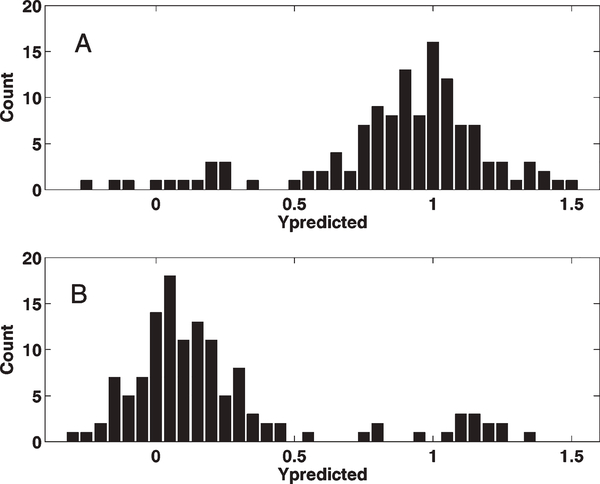
The calculated PLS-DA model based on the 347 model samples gave six significant components, according to crossvalidation, describing 46.4% of the variation in X (R2X = 0.464), 90.5% of the variation in the response Y (class) (R2Y = 0.905), and predicting 81.9% of the variation in the response Y (class), according to cross-validation (Q2Y = 0.819). 343 out of the 347 model samples (98.8%) were correctly classified with regards to population, as visualized in the PLS score plot for the two first components (Fig. 2). Using the model to predict the class identity of the 253 independent test samples gave a correct classification in 221 out of the 253 cases (87.4%), and by examining the distributions of the prediction results a clear picture of the high predictive ability of the model was obtained (Figs. 3a and b).
Fig. 2.
PLS-DA score plot for the first two components (T2/T1) showing the separation between the two cohorts based on the 347 model samples. Shanxi population (★) and Honolulu population (+).
Fig. 3.
Histograms for the prediction of the 253 test set samples. x-axis: predicted values, y-axis: count in each interval (interval = 0.05). (A) Prediction results for class 1 samples (Honolulu). (B) Prediction results for class 2 samples (Shanxi). Samples predicted to belong to the Honolulu class should have a value close to 1, while samples predicted to belong to the Shanxi class should have a value close to 0. 221 of 253 samples (87.4%) were correctly classified.
Lately, the understanding of the “mechanisms” of PLS, including PLS-DA, estimation has increased substantially. We now know that PLS (and similar methods) are affected by the presence of systematic variation in X that is not related to Y. Examples of such variation are the variation of baseline, unknown constituents, or effects of changing instrumental equipment. This results in an increase in the number of PLS components and complicates the model interpretation. It is now known that there exists only one Y-related component for a single Y-variable and that the interpretation of PLS models, in the single Y case, should be based on the first loading vector w1.24 Interpretation of the PLS loadings (component 1 versus component 2) (Fig. 4), identified the variable “P250_C04” (time window 250, AR component 4) to be positively correlated to the Honolulu population and the variable “P284_C01” (time window 284, AR component 1) to be positively correlated to the Shanxi population in the first loading vector w1. The variable “P250_C04” refers to the 5.72 min time point where the most intense m/z in the corresponding m/z profile was m/z 288 (Fig. 5a). Inspection of the ion chromatogram for m/z 288 around the time point 5.72 min revealed generally higher intensities for the Honolulu population compared to the Shanxi population (Fig. 5b and c). Similarly, for the variable “P284_C01”, referring to time point 6.50 min, where the most intense m/z is m/z 355 (Fig. 6a). The corresponding ion chromatogram for m/z 355 around time point 6.50 min clearly showed higher intensities for the majority of the Shanxi population in comparison to the Honolulu population (Fig. 6b and c). The same interpretation can be carried out for all the variables contributing to the separation between the two populations, giving a simple and straight forward explanation of the differences between the cohorts.
Fig. 4.
PLS-DA variable loadings plot for the two first components (w*c2/w*c1) explaining the separation between the two cohorts. Variable P250_C04 (time window: 250, AR component: 4) was found to be positively correlated to the Honolulu population. Variable P284_C01 (time window: 284, AR component: 1) was found to be positively correlated to the Shanxi population.
Fig. 5.
(A) The mass spectrum for time window 250 (5.72 min) with the most intense m/z = 288. (B) Ion chromatogram for m/z = 288 in time window 250 for the Honolulu samples. (C) Ion chromatogram for m/z = 288 in time window 250 for the Shanxi samples.
Fig. 6.
(A) The mass spectrum for time window 284 (6.50 min) with the most intense m/z = 355. (B) Ion chromatogram for m/z = 355 in time window 284 for the Honolulu samples. (C) Ion chromatogram for m/z = 355 in time window 284 for the Shanxi samples.
Discussion
LC/MS has the potential to become a powerful tool in metabolic research (metabolomics/metabonomics). However in order to accomplish this there are still a few hurdles to overcome. A lot of work still remains to be done on the analytical side to develop procedures that give robust and reproducible results between samples and studies for these extremely complex investigations. Another crucial issue lies in the interface between the instrumental analysis and the statistical modelling and evaluation. In order to analyse and interpret these extremely complex data, organised statistical approaches that can handle many and correlated variables are a requirement. Multivariate projection methods (e.g. PCA and PLS) have proved to be suitable for these purposes. However, to be able to create robust and interpretable (transparent) models for comparison of many samples, the data produced by the analytical technique (e.g. LC/MS) must fulfil certain criteria. Such criteria are that each sample (observation) is characterized by the same number of variables and each of these variables is represented across all observations. In addition, it is vital that a variable in one sample is the same for all the other samples (i.e. has the same biological meaning or represents the same metabolite). If these criteria are fulfilled the odds for obtaining accurate and reliable statistical models are considerably lowered, enabling facilitated interpretation and improved understanding of complex relationships deciding various biological events. Nevertheless, if these criteria are not perfectly matched, which can often be the case for these types of samples/studies, it must still be possible to create organised strategies for the analysis and interpretation of the generated data. With the presented method a representative data set is obtained where the relevant metabolic variation is retained for further analysis e.g. multivariate statistical analysis. This organised approach allows the comparison of a large number of samples based on their LC/MS metabolic profiles, which has implications for data base construction and comparisons, but also, importantly, offers a robust and transparent framework for the interpretation of the investigated biological system. This includes finding relevant systematic patterns among samples, identifying influential variables, verifying the findings by tracing the results back to the raw data (both mass spectral and chromatographic), and finally using the models for prediction of new independent samples.
As for any model describing any biological system the main focus has to be directed towards the interpretation of the relevant systematic changes associated with specific physiological or pathological events, and thereby create a means for improved understanding of the complex mechanisms causing these events to take place. Hence, it is of utmost importance to create what we here define as transparent models where the relevant variation can be extracted, visualised and then verified going back to the parts of the original data highlighted by the model. This transparency will have at least two important implications. Firstly, it will provide a platform for experienced expertise within the biological/medical community to base their interpretations on by highlighting e.g metabolites or correlations among metabolites that are responsible for the detected patterns. This will work to establish a connection between statistical and biological significance (meaning), which is crucial for these types of applications. Secondly, this transparency also allows the user to investigate and interpret the variation on which the prediction of new samples is based, to make sure that prediction results are not obtained from spurious changes or irrelevant systematic changes within the data. Another important topic addressed in this work is the importance of creating models giving accurate predictions of independent samples and how to validate this predictive ability. It is obvious that a model describing a biological system in relation to e.g. diseases or other metabolic events must be able to provide reliable predictions of future samples, to be considered as a potential tool for diagnosis or prognosis. Here this was investigated by constructing the data table (X) and building the model from a set of four well plates, all including samples from both populations. An independent set consisting of three well plates was then used for validating the models predictive ability, and hence described according to the model sample parameters. Proofs of the presented methods ability were the accurate predictions of independent samples together with an appropriate interpretation of the urinary metabolic signatures associated with the difference between the cohorts.
Conclusion
The presented approach shows how it is possible to optimize the information extraction from metabonomic LC/MS data sets in an organized and consistent fashion, and hence accomplish a means for the efficient analysis and evaluation of these complex data. In particular, the strategy focuses on coping with analytical variation and at reducing the number of variables, by bi-linear compression, to a smaller subset of information-rich variables prior to multivariate data analysis. The presented results showed that evaluation of the “information optimized” data by multivariate projection methods (PLS-DA) provided robust, predictive and highly interpretable (transparent) models for screening of large metabonomic LC/MS data sets, here exemplified by a metabonomic population study. It was clearly shown that the classification of the two sample populations (Shanxi (People’s Republic of China) and Honolulu (USA)) based on urine LC/MS data was successful with a high predictive ability. Furthermore, the high transparency of the obtained model allowed a total interpretation of the metabolic patterns associated with the population differences, which could be traced back to the raw data for verification.
The suggested method is a general approach for the data handling, analysis, and evaluation of large metabolic LC/MS data sets and should be regarded as a contribution to the analysis of biosystems in terms of improving information extraction and understanding of complex interactions between metabolites.
Acknowledgements
PJ and HA acknowledge funding from EU strategic funds and SSF/UPSC.
The INTERMAP project was supported by NIH Research Grant R01 HL50490 funded by the Office of Dietary Supplements, NIH and the National Heart, Lung and Blood Institute, by the Chicago Health Research Foundation, and by national agencies in Japan (the Ministry of Education, Science, Sports, and Culture, Grant-in-Aid for Scientific Research [A], No. 090357003), People’s Republic of China and the UK. The funders had no role in the design and conduct of the study, collection, management, analysis, and interpretation of the data, and preparation, review, or approval of the manuscript. The authors would like to thank Paul Elliot and Queenie Chan (Department of Epidemiology and Public Health, Faculty of Medicine, St Mary’s Campus, Imperial College London, UK), Jeremiah Stamler (Department of Preventive Medicine, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA), Hugo Kesteloot (Central Laboratory, Akademisch Ziekenhuis St. Rafael, Leuven, Belgium), Hirotsugu Ueshima (Department of Health Science, Shiga University of Medical Science, Otsu, Shiga, Japan), and Beifan Zhou (Department of Epidemiology, Fu Wai Hospital and Cardiovascular Institute, Chinese Academy of Medical Sciences, Beijing, People’s Republic of China) for their coordination and direction of the INTERMAP project and constructive discussions.
References
- 1.van der Greet J, Stroobant P and van der Heijden R, Curr. Opin. Chem. Biol, 2004, 8, 559–565. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson JK, Holmes E, Lindon JC and Wilson ID, Nat. Biotechnol, 2004, 22, 10, 1268–1274. [DOI] [PubMed] [Google Scholar]
- 3.Kell DB, Curr. Opin. Microbiol, 2004, 7, 3, 296–307. [DOI] [PubMed] [Google Scholar]
- 4.Lindon JC, Nicholson JK, Holmes E and Everett JR, Concepts Magn. Reson, 2000, 12, 289–320. [Google Scholar]
- 5.Nicholson JK, Lindon JC and Holmes E, Xenobiotica, 1999, 29, 11, 1181–1189. [DOI] [PubMed] [Google Scholar]
- 6.Griffin JL, Curr. Opin. Chem. Biol, 2003, 7, 5, 648–654. [DOI] [PubMed] [Google Scholar]
- 7.Jonsson P, Gullberg J, Nordström A, Kusano M, Kowalczyk M, Sjöström M and Moritz T, Anal. Chem, 2004, 76, 1738–1745. [DOI] [PubMed] [Google Scholar]
- 8.Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN and Willmitzer L, Nat. Biotechnol, 2000, 18, 1157–1161. [DOI] [PubMed] [Google Scholar]
- 9.Plumb RS, Stumpf CL, Grainger JH, Castro-Perez J, Haselden JN and Dear GJ, Rapid Commun. Mass Spectrom, 2003, 17, 2632–2638. [DOI] [PubMed] [Google Scholar]
- 10.Idborg-Björkman H, Edlund PO, Kvalheim OM, Schuppe-Koistinen I and Jacobsson SP, Anal. Chem, 2003, 75, 4784–4792. [DOI] [PubMed] [Google Scholar]
- 11.Plumb RS, Stumpf CL, Gorenstein MV, Castro-Perez J, Dear GJ, Sweatman AM, Connor SC and Haselden JN, Rapid Commun. Mass Spectrom, 2002, 16, 1991–1996. [DOI] [PubMed] [Google Scholar]
- 12.Wold S, Esbensen K and Geladi P, Chemom. Intell. Lab. Syst, 1987, 2, 37–52. [Google Scholar]
- 13.Jackson JE, A Users’ Guide to Principal Components, Wiley, New York, 1991. [Google Scholar]
- 14.Wold S, Ruhe A, Wold H and Dunn WJ III, SIAM J. Sci. Stat. Comput, 1984, 5, 735–743. [Google Scholar]
- 15.Wold S, Trygg J, Berglund A and Antti H, Chemom. Intell. Lab. Syst, 2001, 58, 131–150. [Google Scholar]
- 16.Holmes E and Antti H, Analyst, 2002, 127, 12, 1549–1557. [DOI] [PubMed] [Google Scholar]
- 17.Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, Bethell HWL, Clarke S, Schofield PM, McKilligin E, Mosedale DE and Grainger DJ, Nat. Med. (N.Y.), 2002, 8, 12, 1439–1444. [DOI] [PubMed] [Google Scholar]
- 18.Lenz EM, Bright J, Wilson ID, Hughes A, Morrisson J, Lindberg H and Lockton A, J. Pharm. Biomed. Anal, 2004, 36, 4, 841–849. [DOI] [PubMed] [Google Scholar]
- 19.Maibaum E et al. , 2004, to be published. [Google Scholar]
- 20.Torgrip RJO, Åberg M, Karlberg B and Jacobsson SP, J. Chemom, 2003, 17, 11, 573–582. [Google Scholar]
- 21.Karjalainen EJ, Chemom. Intell. Lab. Syst, 1989, 7, 31–38. [Google Scholar]
- 22.Sjöström M, Wold S and Söderström B, PLS discriminant plots in Pattern Recognition in Practice II, Elsevier Science Publ; Holland BV, 1986, pp. 461–470. [Google Scholar]
- 23.Wold S, Technometrics, 1978, 20, 397–405. [Google Scholar]
- 24.Trygg J and Wold S, J. Chemom, 2002, 16, 119–128. [Google Scholar]