Abstract
Ecdysis (moulting) is the defining character of Ecdysoza (arthropods, nematodes and related phyla). Despite superficial similarities, the signalling cascade underlying moulting differs between Panarthropoda and the remaining ecdysozoans. Here, we reconstruct the evolution of major components of the ecdysis pathway. Its key elements evolved much earlier than previously thought and are present in non-moulting lophotrochozoans and deuterostomes. Eclosion hormone (EH) and bursicon originated prior to the cnidarian-bilaterian split, whereas ecdysis-triggering hormone (ETH) and crustacean cardioactive peptide (CCAP) evolved in the bilaterian last common ancestor (LCA). Identification of EH, CCAP and bursicon in Onychophora and EH, ETH and CCAP in Tardigrada suggests that the pathway was present in the panarthropod LCA. Trunk, an ancient extracellular signalling molecule and a well-established paralog of the insect peptide prothoracicotropic hormone (PTTH), is present in the non-bilaterian ctenophore Mnemiopsis leidyi. This constitutes the first case of a ctenophore signalling peptide with homology to a neuropeptide.
Research organism: P. dumerilii
eLife digest
Animals such as insects, crabs and spiders belong to one of the most species-rich animal groups, called the arthropods. These animals have exoskeletons, which are hard, external coverings that support their bodies. Arthropods shed their exoskeletons as they grow, a process called ecdysis or moulting, and this behaviour is controlled by a set of hormones and small protein-like molecules called neuropeptides that allow communication between neurons.
Other animals, such as roundworms, also moult; and together with arthropods they are classified into a group called the Ecdysozoa. Since moulting is a common behaviour in ecdysozoans, it was previously assumed that its signalling components had evolved in the common ancestor of roundworms and arthropods, although differences in the moulting machinery between both groups exist.
Here, De Oliveira et al. investigate the evolutionary origins of the arthropod moulting machinery and find that some of the hormones and neuropeptides involved appeared long before the arthropods themselves.
Database searches showed that important hormones and neuropeptides involved in arthropod moulting can be found in diverse animal groups, such as jellyfish, molluscs and starfish, confirming that these molecules evolved before the last common ancestor of roundworms and arthropods. These animals must therefore use the hormones and neuropeptides in many processes unrelated to moulting. De Oliveira et al. also found that roundworms have lost most of these molecules, and that moulting in these animals must be driven by a different complement of hormones and neuropeptides.
These results invite research into the role of moulting hormones and neuropeptides in animals outside the Ecdysozoa. They also show that signalling pathways and the processes they regulate are highly adaptable: two animals can use the same hormone in entirely different processes, but conversely, the same behaviour may be regulated by different molecules depending on the animal. This means that the evolution of a process and the evolution of its regulation can be decoupled, a finding that has important implications for the study of signalling pathways and their evolution.
Introduction
Ecdysis or moulting, which describes the process of shedding the outer integument, the cuticle, is a defining feature of Ecdysozoa (arthropods, tardigrades, onychophorans, nematodes and related phyla) (Aguinaldo et al., 1997; Schmidt-Rhaesa et al., 1998; de Rosa et al., 1999; Dunn et al., 2008; Telford et al., 2008). Despite superficial similarities of the ‘moulting behaviour’ within Ecdysozoa, the neuroendocrine components underlying this process remain elusive for the majority of the ecdysozoans outside of Arthropoda. This includes well-established model organisms such as the nematode Caenorhabditis elegans, for which the gene regulatory network responsible for ecdysis remains to be fully resolved (Frand et al., 2005; reviewed by Page et al., 2014 and Lažetić and Fay, 2017).
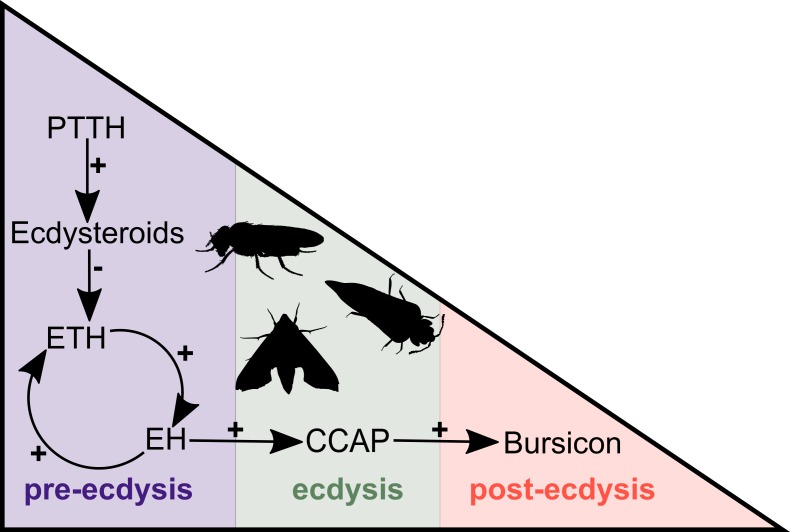
In arthropods, ecdysis can be divided into three distinct stages, pre-ecdysis, ecdysis and post-ecdysis. Each of these stages correlates with major behavioural, molecular and cellular changes and encompasses a series of specific muscular contractions controlled by a cascade of hormones and neuropeptides (Truman, 2005). Studies in insects have revealed that the major components of this peptidergic signalling pathway are ecdysis-triggering hormone (ETH), eclosion hormone (EH), crustacean cardioactive peptide (CCAP) and bursicon (Gammie and Truman, 1997a; Gammie and Truman, 1997b; Zitnan et al., 1999; Clark et al., 2004; Kim et al., 2006a; Kim et al., 2006b; Arakane et al., 2008; Lee et al., 2013). The process begins with the release of prothoracicotropic hormone (PTTH) from neurohemal organs. PTTH initiates a signalling cascade that results in the biosynthesis of ecdysteroids (i.e., steroid hormones synthesised from ingested cholesterol), including ecdysone (E) and 20-hydroxyecdysone (20E) (Figure 1). The decline of the ecdysone titre due to the ecdysone-inactivating enzyme cytochrome P450 protein Cyp18a1 (Guittard et al., 2011; reviewed by Rewitz et al., 2013) triggers the release of ETH that, in turn, causes the release of EH. These two hormones mutually enhance one another in a positive feedback loop to control and regulate pre-ecdysis behaviour (Figure 1). With the ensuing release of CCAP, caused by EH, pre-ecdysis ceases and the ecdysis motor program is initiated. Finally, bursicon responds to the increasing levels of CCAP and initiates post-ecdysis behaviour and cuticle tanning (Figure 1).
Figure 1. Simplified overview of the neuropeptide/hormone signalling pathway at moulting.
PTTH initiates a signalling cascade that results in the biosynthesis of ecdysone. The decline of the ecdysone titre triggers the release of ETH that, in turn, causes the release of EH. These two hormones mutually enhance one another in a positive feedback loop to control and regulate pre-ecdysis behaviour. With the ensuing release of CCAP, caused by EH, pre-ecdysis ceases and the ecdysis motor program is started. Finally, bursicon responds to the increasing levels of CCAP and initiates post-ecdysis behaviour and cuticle tanning. This figure is based on the studies of McNabb et al. (1997) and Clark et al. (2004). Animal silhouettes were obtained under Public Domain licence at phylopic (http://phylopic.org/), unless otherwise indicated. Beetle: T. Michael Keesey after Ponomarenko (available for reuse under https://creativecommons.org/publicdomain/zero/1.0/); moth: by Gareth Monger (available for reuse under https://creativecommons.org/licenses/by/3.0/); Drosophila: Thomas Hegna (available for reuse under https://creativecommons.org/publicdomain/zero/1.0/).
Comparative biochemical, genomic and transcriptomic analyses revealed that ecdysteroids and the required genes responsible for their biosynthesis are present outside of Ecdysozoa, showing that some key molecular players of moulting predate the origin of Ecdysozoa (Mendis et al., 1984; Nolte et al., 1986; Garcia et al., 1989; Barker et al., 1990; Schumann et al., 2018). Such integrative and comparative analyses have so far not been conducted on the major components of the peptidergic signalling system underlying moulting. To fill this gap in knowledge, we explored the distribution of PTTH, ETH, EH, CCAP and bursicon ligand-receptor pairs across Metazoa.
Results and discussion
PTTH is a neurohormone with a proposed origin at the base of Arthropoda that is believed to have evolved from the duplication of the ancient and widely distributed bilaterian signalling molecule-encoding gene trunk (Rewitz et al., 2009; Jékely, 2013). By screening 39 metazoan genomes and 57 transcriptomes (Supplementary file 1), we found that the PTTH peptide is present in Drosophila and Tribolium but absent in the house spider Parasteatoda tepidariorum and the crustacean Parhyale hawaiensis, suggesting that PTTH is an insect innovation (Figure 2A,B, Figure 2—figure supplement 1A, Figure 2—source data 1). The ablation of PTTH-producing neurons in Drosophila generates an imbalance in ecdysone biosynthesis, causing developmental delay, prolonged duration of feeding and larger individuals with reduced fecundity (McBrayer et al., 2007). These findings indicate that PTTH, at least in Drosophila, regulates developmental timing and body size, but is not essential for moulting. Trunk, the paralog of ptth, has previously been identified in arthropods, annelids, mollusks and the cephalochordate Branchiostoma floridae (Jékely, 2013). Our study expands the phyletic distribution of trunk to onychophorans, tardigrades, gastrotrichs, brachiopods, nemerteans, ectoprocts, phoronids and hemichordates (Figure 2A,B, Figure 2—figure supplement 1A, Figure 2—source data 1). Although we did not find a trunk ortholog in the genome of the sea anemone Nematostella vectensis, our similarity searches against the NCBI protein database led to the identification of this protein in other anthozoans, namely Stylophora pistillata (PFX31008.1) and Orbicella faveolata (XP_020630744.1 and XP_020630745.1). More importantly, our multi-species screen also recovered a trunk-like peptide in the ctenophore Mnemiopsis leidyi with high similarity (p-value < 1e-05) to lophotrochozoan, deuterostome and ecdysozoan trunk sequences (Figure 2—figure supplement 1A; Figure 3A,B). By similarity-based clustering we were able to demonstrate homology of the ctenophore trunk-like peptide with the insect trunk paralog, ptth (Figure 3A, Figure 3—source data 1; see also Rewitz et al., 2009; Jékely, 2013). This extends the phyletic distribution of trunk to the ctenophores (see, e.g., Halanych, 2004; Dunn et al., 2008; Moroz et al., 2014; Jékely et al., 2015; Pisani et al., 2015 for discussion).
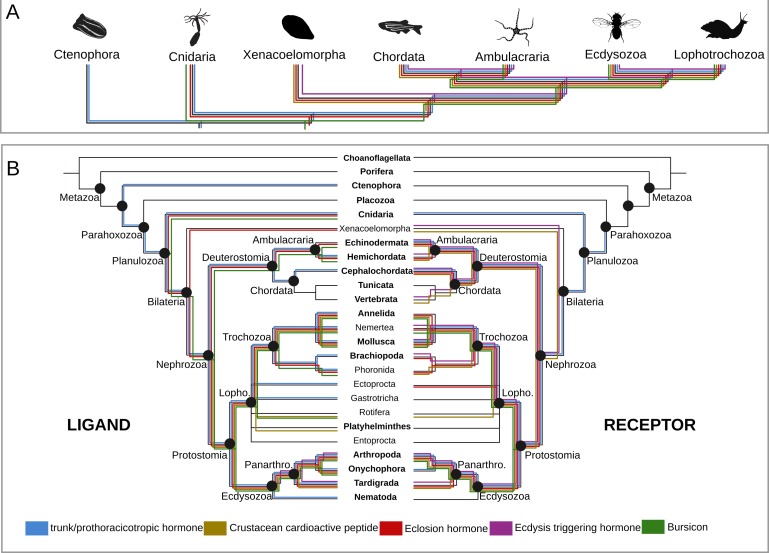
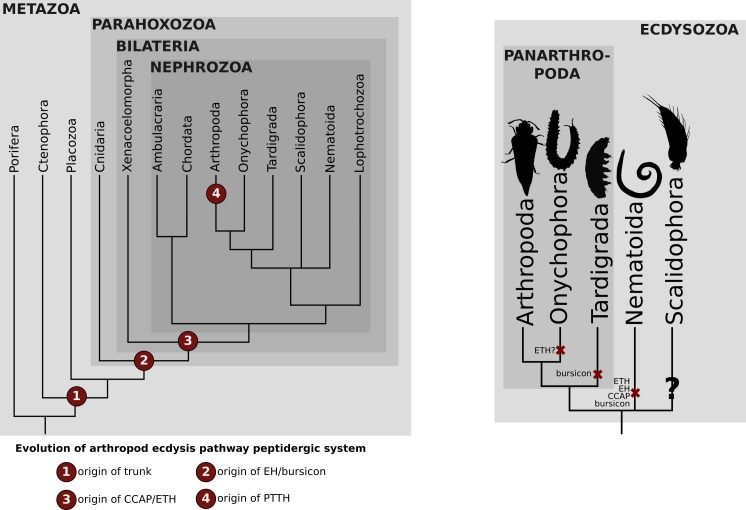
Figure 2. Origin and distribution of the key ligand-receptor components of the arthropod moulting signalling pathway across Metazoa.
(A) Simplified phylogeny (based on Dunn et al., 2014) of Metazoa showing the lineages in which the key components of the arthropod moulting signalling pathway are present. Note that Porifera and Placozoa, that lack the moulting pathway components investigated here, are omitted for clarity. Coloured lines indicate the presence of a given ligand and/or receptor in a given lineage. Eclosion hormone and bursicon peptidergic systems originated prior to the cnidarian-bilaterian split, whereas the ecdysis-triggering hormone and crustacean cardioactive peptide trace back to the last common ancestor of Bilateria. PTTH is an insect-specific neuropeptide. (B) Expanded phylogeny of Metazoa with Porifera as the earliest branching clade (adapted from Dunn et al., 2014). Coloured lines indicate the presence of a given ligand (right side) and receptor (left side) in a given lineage. Phylum name in bold indicates the availability of genomic data. Note that although the trunk ortholog was not retrieved from the genomes of Nematostella vectensis and Caenorhabditis elegans, similarity searches against publicly available protein databases identified this gene in other cnidarian and nematode species. Animal silhouettes were obtained under Public Domain licence at phylopic (http://phylopic.org/), unless otherwise indicated. Credited images: Ctenophora: Martini (available for reuse under https://creativecommons.org/publicdomain/zero/1.0/); Cnidaria: Jack Warner (available for reuse under https://creativecommons.org/publicdomain/zero/1.0/); Xenacoelomorpha: Andreas Hejnol (available for reuse under https://creativecommons.org/licenses/by-nc/3.0/); Chordata: Jake Warner (available for reuse under https://creativecommons.org/publicdomain/zero/1.0/); Ambulacraria: Noah Schlottman (photograph from Casey Dunn available for reuse under https://creativecommons.org/licenses/by-sa/3.0/); Ecdysozoa: Thomas Hegna based on picture by Nicolas Gompel (available for reuse under https://creativecommons.org/publicdomain/mark/1.0/); Lophotrochozoa: Fernando Carezzano (available for reuse under https://creativecommons.org/publicdomain/zero/1.0/).
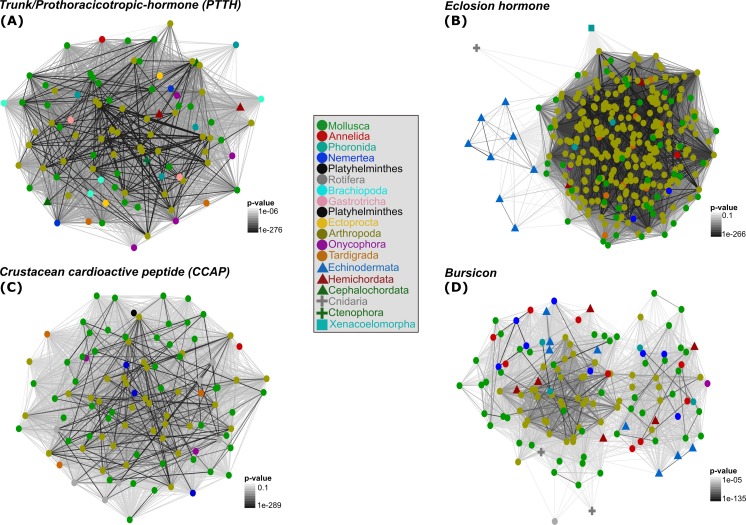
Figure 2—figure supplement 1. 2D cluster maps of trunk/PTTH, EH, CCAP and bursicon ligands reflecting the evolutionary relatedness of the key arthropod moulting components among metazoans.
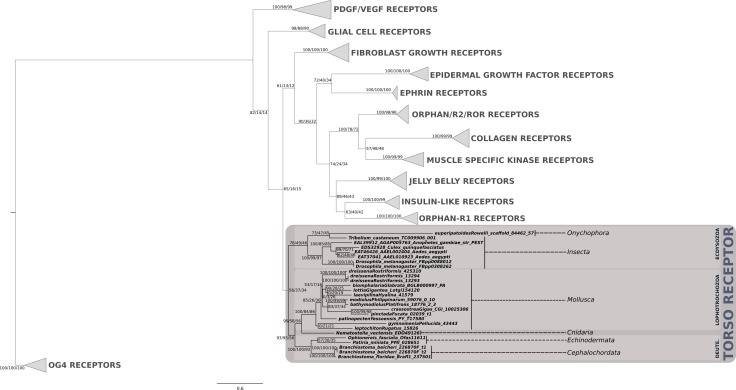
Figure 2—figure supplement 2. Phylogenetic analysis of the PTTH/trunk receptor tyrosine kinase torso showing the presence of torso receptor in cnidarians, lophotrochozoans, ecdysozoans and deuterostomes.
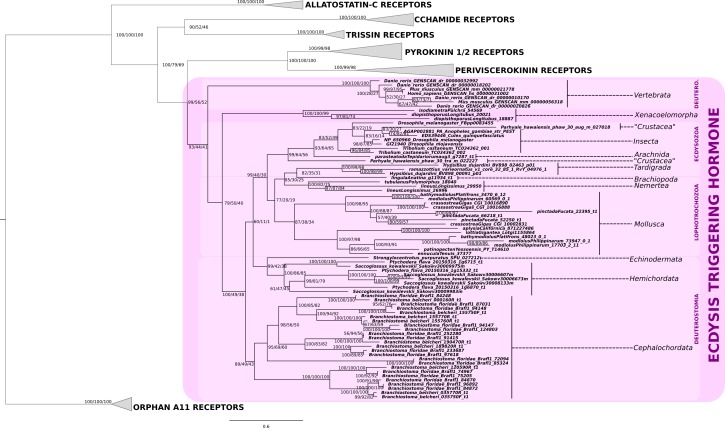
Figure 2—figure supplement 3. Phylogenetic analysis of the ecdysis-triggering hormone receptor showing the presence of ETH-receptor in bilaterians.
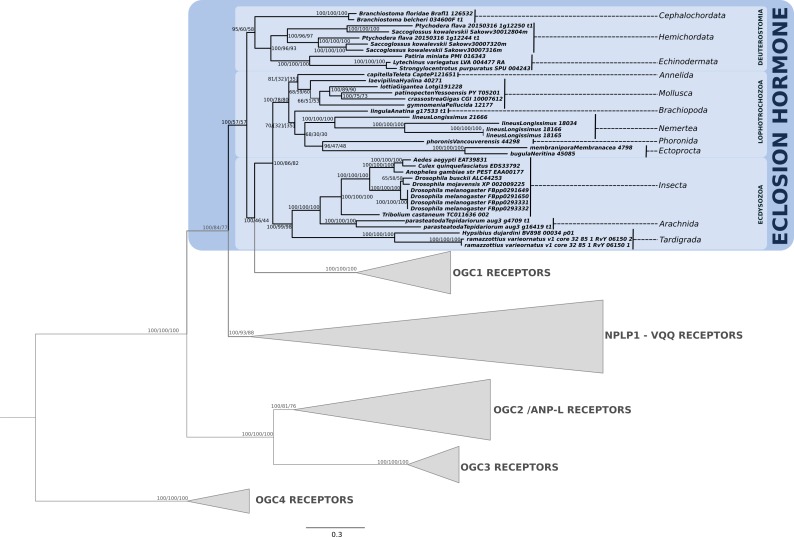
Figure 2—figure supplement 4. Phylogenetic analysis of the guanylyl cyclase eclosion hormone receptor showing the presence of EH-receptor in ecdysozoans, lophotrochozoans, ambulacrarians and cephalochordates.
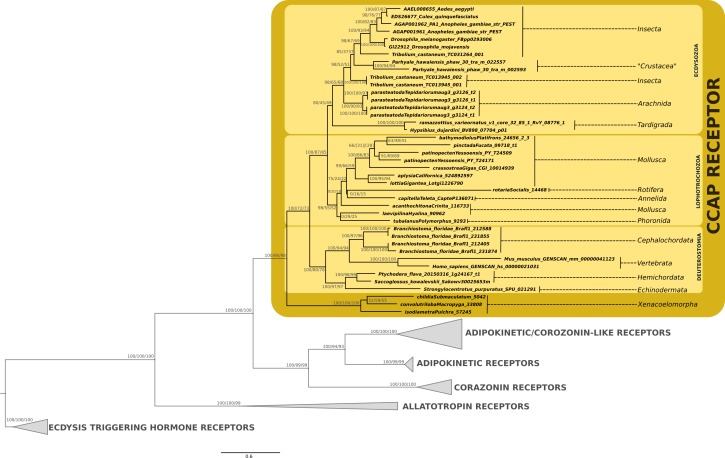
Figure 2—figure supplement 5. Phylogenetic analysis of the G protein-coupled CCAP receptor showing the presence of CCAP-receptor in ecdyzosoans, lophotrochozoans, deuterostomes (including vertebrates) and acoels.
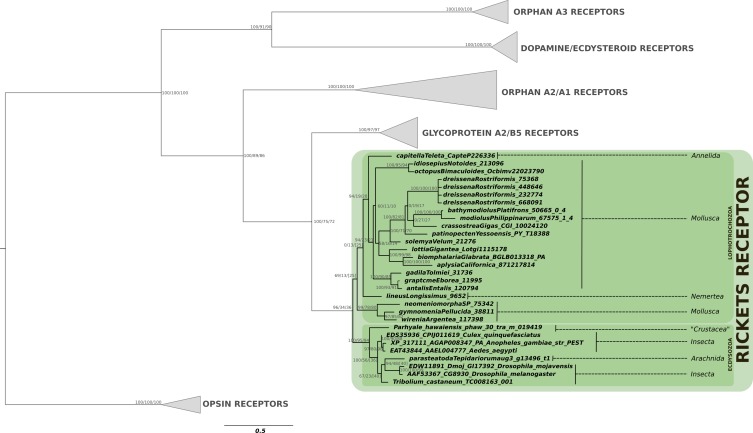
Figure 2—figure supplement 6. Phylogenetic analysis of the bursicon G protein-coupled receptor rickets showing the presence of rickets receptor in arthropods and lophotrochozoans.
Figure 3. Cluster analysis of prothoracicotropic hormone (ptth), trunk, noggin orthologs and multiple sequence alignment of the ctenophore trunk-like peptide and the metazoan ortholog sequences.
(A) 2D cluster map of ptth, trunk and noggin genes. Red triangles correspond to ptth homologs, green parallelograms correspond to noggin homologs and red circles correspond to trunk homologs. The ctenophore trunk gene sequence is represented by the pink star. Edges represent BLAST connections of P value > 1e-05. Note that the ctenophore trunk peptide is indirectly connected to insect PTTH sequences via transitive BLAST connections. (B) Multiple sequence alignment representation of ctenophore trunk sequence and its metazoan orthologs produced by Jalview 2 (Waterhouse et al., 2009). Only the sequences directly connected to the ctenophore sequence in the 2D cluster map are included in the multiple sequence alignment. The conservation histogram corresponds to the number of conserved amino acid physico-chemical properties for each column of the alignment.
PTTH and trunk share a common receptor, the tyrosine kinase torso (Rewitz et al., 2009). Similar to its ligands, torso (Rewitz et al., 2009) proved also to be much more ancient than commonly assumed. We identified torso sequences in deuterostomes, lophotrochozoans, cnidarians and ecdysozoans (Figure 2—figure supplement 2), indicating that the trunk-torso neuropeptide signalling pathway dates back at least as far as the last common ancestor of Cnidaria, Ctenophora and Bilateria and is thus not restricted to Bilateria as suggested previously (e.g., Jékely, 2013) (Figures 2A,B and 4A).
Figure 4. Distribution of the arthropod peptidergic system components throughout Metazoa.
(A) Simplified phylogeny of Metazoa with Porifera as the most basally branching clade (adapted from Dunn et al., 2014) showing the origin of the trunk/PTTH, eclosion-hormone (EH), bursicon, crustacean cardioactive peptide (CCAP) and ecdysis-triggering hormone (ETH) peptigergic systems. (B) Distribution of the arthropod peptigerdic system components within Panarthropoda. Secondary losses are depicted by the red crosses followed by the name of the peptide system absent in the lineage. Note that ETH and bursicon, two vital components underlying moulting in insects, were possibly secondarily lost in the Onychophora and Tardigrada (indicated by the red cross), respectively. Genomic and transcriptomic homology searches within the Kinorhyncha, Priapulida and Loricifera (condensed into the clade Scalidophora in Figure 1B) were not performed in this study (indicated by the question mark). Animal silhouettes were obtained under Public Domain licence at phylopic (http://phylopic.org/), unless otherwise indicated. Arthropoda: T. Michael Keesey after Ponomarenko (available for reuse under https://creativecommons.org/publicdomain/zero/1.0/); Onychophora: Noah Schlottman, photo by Adam G. Clause (available for reuse under https://creativecommons.org/licenses/by-sa/3.0/); Tardigrada: Fernando Carezzano (available for reuse under https://creativecommons.org/publicdomain/zero/1.0/); Nematoida: Mali'o Kodis, image from the Smithsonian Institution (available for reuse under https://creativecommons.org/licenses/by-nc-sa/3.0/); Scalidophora: Noah Schlottman, photo by Martin V. Sørensen (available for reuse under https://creativecommons.org/licenses/by-sa/3.0/).
In insects, the first hormone released in response to decreasing ecdysone levels is usually ETH (Zitnan et al., 1996; Zitnan et al., 1999) although in the lepidopteran Manduca sexta the neuropeptide corozanin acts as the trigger for the release of ETH from the epitracheal glands (Kim et al., 2004). Knockdown of the eth gene in Drosophila (Park et al., 2002) and of eth and its receptors in Tribolium and Schistocerca (Arakane et al., 2008; Lenaerts et al., 2017) lead to lethality at the expected onset of ecdysis, demonstrating the essential role of the ETH peptidergic signalling system in moulting (Park et al., 2002; Arakane et al., 2008; Lenaerts et al., 2017; Shi et al., 2017). Our screening and phylogenetic analyses confirmed the presence of ETH and its receptor in tardigrades and arthropods, thus corroborating previous studies (Figure 2B, Figure 2—figure supplement 3, Figure 2—source data 2) (Zitnan et al., 1996; Zitnan et al., 1999; Park et al., 2002; Arakane et al., 2008; Veenstra et al., 2012; Lenaerts et al., 2017; Koziol, 2018; Zhu et al., 2019). For Arthropoda, the ETH ligand was only found in insects (Drosophila and Tribolium), but was lacking in the crustacean Parhyale hawaiensis and the arachnid Parasteatoda tepidariorum. However, studies on the two mites Panonychus citri and Tetranychus urticae as well as several decapods have shown the presence of the ETH ligand in chelicerates and crustaceans (in which the homology was reconfirmed by our clustering analysis) (Veenstra et al., 2012; Veenstra, 2016; Zhu et al., 2019). Surprisingly, we did not find the entire ETH signalling pathway in the two onychophoran genomes analysed herein (Figures 2B and 4B, Figure 2—figure supplement 3, Figure 2—source data 2). Due to the fragmented nature of the onychophoran genomes a final statement whether this signalling system was indeed lost in this lineage cannot be made at present.
No eth ortholog was identified outside the panarthropods, including the nematode C. elegans, suggesting that this gene originated in the last common ancestor of Panarthropoda (Figure 2B). Our findings on the distribution of the ETH receptor are in agreement with the results of previous studies and demonstrate the presence of this receptor in arthropods (insects, crustaceans and arachnids), mollusks, nemerteans, brachiopods, echinoderms, cephalochordates (in which we found a substantial expansion of eth-receptor homologs in the genomes of Branchiostoma floridae and B. belcheri), vertebrates and acoels (Park et al., 2003; Roller et al., 2010; Veenstra et al., 2012; Mirabeau and Joly, 2013; Lenaerts et al., 2017; Thiel et al., 2018; Zhu et al., 2019) (Figures 2B and 4A, Figure 2—figure supplement 3, Figure 2—source data 2). This provides evidence for the presence of individual components of the ETH signalling system already at the base of Bilateria (Figures 2A and 4A).
Eclosion hormone (EH) was first identified as a blood-borne factor (Truman and Riddiford, 1970) in three lepidopteran species, Hyalophora cecropia, Antheraea polyphemus and Antheraea pernyi. A positive feedback loop between EH and ETH was found in Manduca and suggested in Tribolium (Ewer et al., 1997; Arakane et al., 2008), whereas in Drosophila, EH has been described either acting downstream of ETH (Kim et al., 2006a; Kim et al., 2006b) or in a positive endocrine feedback loop with ETH (Krüger et al., 2015). Despite EH being a key regulator of ecdysis in insects, eh knockout in Drosophila melanogaster did not abolish ecdysis, but instead produced flies with discrete behavioural deficits such as slow and uncoordinated eclosion. This shows that EH is involved, but does not play an essential role, in moulting in these insects (McNabb et al., 1997). This result, however, has been contested in a more recent study using eh null mutants in Drosophila (Krüger et al., 2015), showing that the lack of eh function is lethal during larval fruit fly ecdysis.
Traditionally considered to be confined to arthropods, recent studies showed the presence of EH and its receptor, a guanylyl cyclase, in echinoderms and tardigrades (Zandawala et al., 2017; Koziol, 2018). Our study corroborates these findings but considerably expands the presence of the EH ligand to cnidarians, acoels, hemichordates, lophotrochozoans (mollusks, annelids, nemerteans and phoronids) and onychophorans (Figure 2B, Figure 2—figure supplement 1B, Figure 2—source data 3). All EH ligand orthologs harbour the six cysteine conserved residues (Zitnan et al., 2007) except for Cnidaria, in which only five are present. The identification of the EH receptor in ambulacrarians, mollusks, annelids, nemerteans and phoronids suggests co-evolution of this ligand-receptor pair throughout Metazoa (Figure 2B, Figure 2—figure supplement 4, Figure 2—source data 3). Although the eh-receptor gene was not found in Cnidaria, Xenacoelomorpha and Onychophora, its distribution includes the Brachiopoda, Ectoprocta and Cephalochordata lineages. Consequently, our findings shift the ancestry of this peptidergic pathway back to the cnidarian-bilaterian split (Figures 2A and 4A).
First isolated from the shore crab Carcinus maenas, crustacean cardioactive peptide (CCAP) is a highly conserved amidated neuropeptide that increases heart rate in crustaceans and insects (Stangier et al., 1987; Cheung et al., 1992; Lehman et al., 1993; Suggs et al., 2016). CCAP has multiple functions in addition to its cardioacceleratory activity, such as accelerating the frequency and amplitude of oviduct contractions in the locust Locusta migratoria (Donini et al., 2001) and regulating the release of digestive enzymes in the cockroach Periplaneta americana (Sakai et al., 2006). CCAP is important for ecdysis in crustaceans and insects where it initiates the stereotyped sequence of behaviours that mark the end of the pre-ecdysis stage (Gammie and Truman, 1997a; Gammie and Truman, 1997b; Phlippen et al., 2000; Arakane et al., 2008; Lee et al., 2013). However, transgenic Drosophila larvae lacking CCAP neurons moult normally and only exhibit a prolonged pre-ecdysis behaviour (Clark et al., 2004).
Only three studies focusing on the CCAP signalling pathway components are available outside of Arthropoda. In the snail Lymnaea stagnalis (Vehovszky et al., 2005), immunostaining revealed a dense network of CCAP-positive fibres that likely function to regulate parts of the feeding behaviour. In the oyster Saccostrea glomerata (In et al., 2016) and in the cuttlefish Sepia officinalis (Endress et al., 2018), in vivo bioassays using synthesised neuropeptides and immunohistochemistry suggested that the CCAP signalling pathway is involved in reproduction (e.g., spawning, oocyte transport, egg-laying). Additionally, Sepia CCAP has been shown to increase the tonus of the vena cava, demonstrating its role in the regulation of hemolymph circulation (Endress et al., 2018). These results indicate that in both, mollusks and arthropods, CCAP functions in feeding, reproduction and regulation of hemolymph circulation, suggesting that these may have been its ancestral roles. In arthropods, co-option of CCAP into the ecdysis pathway expanded this set of functions to include moulting.
The CCAP receptor is a G protein-coupled receptor (GPCR) that was first described from the Drosophila genome and subsequently identified in many other insects (Cazzamali et al., 2003; Arakane et al., 2008; Vogel et al., 2013). We confirm here the presence of a CCAP ligand in mollusks, annelids, arthropods and tardigrades, as stated earlier (Veenstra, 2010; Jékely, 2013; Mirabeau and Joly, 2013; Conzelmann et al., 2013; Stewart et al., 2014; Ahn et al., 2017; Zhang et al., 2018; Koziol, 2018), and extend the distribution of the CCAP ligand to three additional lophotrochozoan phyla (Nemertea, Platyhelminthes and Rotifera) as well as to the remaining panarthropod phylum Onychophora (Figures 2B and 4B, Figure 2—figure supplement 1C, Figure 2—source data 4). Interestingly, the CCAP ligand is absent from all investigated deuterostome genomes analysed (Figure 2—figure supplement 1C, Figure 2B, Figure 2—source data 4). The ccap-receptor ortholog was found in acoels, lophotrochozoans and panarthropods except Onychophora (Figure 2—figure supplement 5). Surprisingly, we found the receptor also in all deuterostome phyla (Echinodermata, Hemichordata, Vertebrata, Cephalochordata) except Tunicata (Figure 2—figure supplement 5, Figure 2—source data 4). These results reinforce the suggested origin of the ligand-receptor pair at the base of Bilateria and points towards a possible loss of the CCAP ligand in the Deuterostomia lineage (Figures 2B and 4A, Figure 2—figure supplement 5, Figure 2—source data 4).
Bursicon was identified as a neurohormone responsible for cuticle sclerotization and melanisation (tanning) during post-ecdysis (Cottrell, 1962a; Cottrell, 1962b; Fraenkel and Hsiao, 1965). Recent studies have shown that bursicon also has a mild effect on the regulation of pre-ecdysis and is important for the proper execution of post-ecdysis in Manduca, Drosophila and Tribolium as well as for the development of wings and other integumentary structures (Baker and Truman, 2002; Dewey et al., 2004; Arakane et al., 2008; Bai and Palli, 2010). Together with the ecdydis-triggering hormone signalling system, bursicon is an indispensable component of the moulting behaviour in insects (Arakane et al., 2008). Previous studies show that bursicon is present outside Ecdysozoa, for example in the anthozoan Nematostella vectensis, the echinoderm Strongylocentrotus purpuratus as well as in annelids and mollusks (Jékely, 2013; Conzelmann et al., 2013; Stewart et al., 2014; Ahn et al., 2017; Zhang et al., 2018). Our work confirms the presence of the complete bursicon signalling system in all arthropod genomes analysed here and extends its distribution (receptor and/or ligand) to the hemichordate, nemertean, phoronid, rotifer and onychophoran phyla (Figure 2B, Figure 2—figure supplement 1D, Figure 2—source data 5). Interestingly, bursicon and its receptor rickets are absent in tardigrades, suggesting the loss of the bursicon peptidergic signalling in this lineage (Figures 2B and 4B, Figure 2—figure supplement 6, Figure 2—source data 5). The latter findings are corroborated by independent proneuropeptide and peptide prohormone surveys in the tardigrade genomic and EST data that also failed to detect this ligand-receptor pair in different tardigrade species (Christie et al., 2011; Koziol, 2018). We did not identify the receptor in any deuterostome or non-arthropod ecdysozoan lineage (Figure 2B).
The PTTH, ETH, EH, CCAP and bursicon peptide signalling systems are lacking in the nematode Caenorhabditis elegans (cf. our study and, e.g., Page et al., 2014; Lažetić and Fay, 2017; Figure 2B, Figure 2—figure supplement 1). Additionally, other key moulting components, such as the ecdysteroid ecdysone (E), 20-hydroxyecdysone (20E), and various halloween gene products have also been reported absent from the C. elegans genome (Frand et al., 2005; Schumann et al., 2018). An extensive body of research on moulting in C. elegans suggests an entirely different molecular machinery controlling this behaviour in this free-living nematode (Russel et al., 2011; for review Lažetić and Fay, 2017).
Interestingly, however, E and 20E were identified in parasitic nematodes (Cleator et al., 1987; Shea et al., 2004) and, outside Ecdysozoa, in the platyhelminth Monieza expansa, the gastropod mollusks Lymnaea stagnalis and Helix pomatia as well as in the hirudinean annelid Hirudo medicinalis (Mendis et al., 1984; Nolte et al., 1986; Garcia et al., 1989; Barker et al., 1990).
Conclusion
We show that key peptidergic components of the arthropod ecdysis pathway emerged prior to the protostome-deuterostome split, and thus considerably earlier than commonly assumed. EH, CCAP and the bursicon signalling systems are more widespread among non-moulting animals than previously appreciated. The presence of the eth-receptor ortholog in ecdysozoans, lophotrochozoans and deuterostomes, in combination with the restriction of its known ligand to insects, arachnids and tardigrades, suggests a scenario in which promiscuous ligand/receptor relationships can lead to novel signalling interactions that provide new opportunities for natural selection to generate novel functions (Figure 2B). The identification of the near complete suite of the peptidergic arthropod ecdysis pathway components in Onychophora and Tardigrada strongly suggests that the entire pathway was at least functional in the last common ancestor of Panarthropoda and maybe as early as in the ur-ecdysozoan (Figures 2B and 4B). However, considering the crucial role of the ETH and bursicon signalling systems in insect moulting, together with the apparent secondary loss of ETH in Onychophora and bursicon in Tardigrada (Figure 4B), the consequences of harbouring only the partial set of the ecdysis signalling genes should be the focus of future assessments. Independent recruitment of novel peptidergic components into insect ecdysis has been shown (cf. Kim et al., 2004; Kim et al., 2006a; Kim et al., 2006b; extensively reviewed by Zitnan et al., 1996), illustrating the evolutionary plasticity of this signalling pathway and calling for more detailed functional investigations into the role of individual components during moulting of the various ecdysozoan lineages.
Materials and methods
Data collection, filtering, sequence reconstruction and proteome prediction
To obtain a comprehensive sampling across Metazoa, ecdysozoan, deuterostome and non-bilaterian protein-coding sequence (CDS) databases were downloaded from publicly available sites and combined with previous lophotrochozoan transcriptomes (see De Oliveira et al., 2019). The acoel transcriptomic data were pre-processed and assembled as described in De Oliveira et al. (2019). The databases include representatives from the following phyla: Porifera, Ctenophora, Cnidaria, Placozoa, Xenacoelomorpha, Echinodermata, Hemichordata, Chordata, Annelida, Brachiopoda, Ectoprocta, Entoprocta, Gastrotricha, Mollusca, Nemertea, Phoronida, Platyhelminthes, Rotifera, Arthropoda, Tardigrada, Onychophora and Nematoda. The choanoflagellate Monosiga brevicollis was used as outgroup. Supplementary file 1 summarises the databases and the publicly available repositories from which they were obtained. Sequence read archive (SRA) accession numbers for xenacoelomorph databases are also shown.
Sensitive similarity searches with jackhmmer
Sensitive probabilistic iterative similarity searches based on profile hidden Markov models (HMMs) were performed with jackhmmer (Johnson et al., 2010) against the respective metazoan and choanoflagellate databases. Insect eh, eth ccap, ptth and bursicon orthologs were retrieved from NCBI (National Center for Biotechnology Information) and their respective receptors from Vogel et al. (2013). These sequences were used as queries in the similarity searches. The searches were performed under the default parameters using varying e-value thresholds (1 to 1e-06) controlled by the options –E and –domE, as defined in jackhmmer. The best hits found in the metazoan and choanoflagellate databases were stored in fasta format and used in the subsequent analyses.
Clustering and phylogenetic analyses
EH, ETH, CCAP, PTTH and bursicon ligand candidates retrieved from the metazoan and choanoflagellate databases were used as input, together with their respective insect orthologs, in the program clans (Frickey and Lupas, 2004) under different e-value thresholds (0.1 to 1e-06) and blast programs, that is blastp or psiblast (Camacho et al., 2009). Singleton sequences (isolated unconnected sequences) were excluded from the map. To further improve the orthology assessment, multiple sequence alignments were performed with mafft (Katoh and Standley, 2013) and the presence of shared conserved amino acid regions and residues were investigated with aliview (Larsson, 2014). The final 3D maps were collapsed into 2D after the clustering for easier visualisation.
Putative EH, ETH, CCAP, PTTH and bursicon receptor candidates retrieved from the metazoan and choanoflagellate databases were aligned with mafft together with their respective orthologs, when found, and subsequently trimmed with BMGE software under the following parameters: –h 1 –b 1 –m BLOSUM30 –t AA (Criscuolo and Gribaldo, 2010). Outgroups for the phylogenetic analyses were defined according to Vogel et al. (2013).
Phylogenetic analyses were performed using RAxML (Stamatakis, 2014), PhyML (Guindon et al., 2010) and mrbayes (Ronquist et al., 2012) softwares using the appropriate best-fit model of amino acid substitution. RaxML was executed under default parameters and rapid bootstrap. PhyML was executed under the default parameters and an optimised starting tree (-o tlr option). The number of bootstrap values was set to 1.000 in RaxML and PhyML and the number of generations used in mrbayes was determined using a convergence diagnostic. All runs in mrbayes were performed with the samplefreq and relative burn-in defined as 1000 and 25%, respectively. The three final phylogenetic trees obtained for each of the four different receptors were visualised and combined with TreeGraph2 (Stöver and Müller, 2010).
Data availability
All data generated in the course of this study are included in this article (Figure 2—source datas 1–5 and Figure 3—source data 1). The accession numbers for the publicly available datasets used in this work are available in Supplementary file 1. The 3D cluster peptide maps can be visualised and manipulated using the program clans (Frickey and Lupas, 2004); see ftp://ftp.tuebingen.mpg.de/pub/protevo/CLANS/). The multiple sequence alignment files can be viewed with aliview (Larsson, 2014). The phylogenetic tree files can be viewed using Figtree (http://tree.bio.ed.ac.uk/software/figtree/) or TreeGraph2 (Stöver and Müller, 2010).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Andreas Wanninger, Email: andreas.wanninger@univie.ac.at.
K VijayRaghavan, National Centre for Biological Sciences, Tata Institute of Fundamental Research, India.
K VijayRaghavan, National Centre for Biological Sciences, Tata Institute of Fundamental Research, India.
Funding Information
This paper was supported by the following grants:
Austrian Science Fund P29455-B29 to Andreas Wanninger.
Coordenação de Aperfeiçoamento de Pessoal de Nível Superior 6090/13-3 to André Luiz de Oliveira.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Resources, Data curation, Software, Formal analysis, Funding acquisition, Validation, Investigation, Visualization, Methodology, Writing—original draft, Writing—review and editing.
Conceptualization, Methodology, Writing—review and editing.
Conceptualization, Resources, Supervision, Funding acquisition, Investigation, Project administration, Writing—review and editing.
Additional files
Superphylum and/or phylum of the investigated species and the online repositories for each of the databases are also listed.
Data availability
All data generated or analysed during this study are included in the manuscript and supporting files. Source data files have also been provided. The molecular databases analysed in this study are publicly available and novel annotated sequences are included in the Source data 1-5. The list of investigated species and their respective links to a direct download are presented in the Supplementary File 1 (Table S1). The 3D proneuropeptide/prohormone maps as well as all the multiple sequence alignments and the phylogenetic trees generated in this study are available in the Source Data 1-5 enclosed in the original submission. The 3D maps in .rtf format can be visualised and inspected with the software clans (ftp://ftp.tuebingen.mpg.de/pub/protevo/CLANS/). The multiple sequence alignments used in the phylogenetic inferences can be graphically visualised using aliview (http://www.ormbunkar.se/aliview/#DOWNLOAD). The phylogenetic tree files can be viewed using an appropriate phylogetic tree viewer such as Figtree (http://tree.bio.ed.ac.uk/software/figtree/).
References
- Aguinaldo AM, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. Evidence for a clade of Nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- Ahn SJ, Martin R, Rao S, Choi MY. Neuropeptides predicted from the transcriptome analysis of the gray garden slug Deroceras reticulatum. Peptides. 2017;93:51–65. doi: 10.1016/j.peptides.2017.05.005. [DOI] [PubMed] [Google Scholar]
- Arakane Y, Li B, Muthukrishnan S, Beeman RW, Kramer KJ, Park Y. Functional analysis of four neuropeptides, EH, ETH, CCAP and Bursicon, and their receptors in adult ecdysis behavior of the red flour beetle, Tribolium castaneum. Mechanisms of Development. 2008;125:984–995. doi: 10.1016/j.mod.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Bai H, Palli SR. Functional characterization of bursicon receptor and genome-wide analysis for identification of genes affected by bursicon receptor RNAi. Developmental Biology. 2010;344:248–258. doi: 10.1016/j.ydbio.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JD, Truman JW. Mutations in the Drosophila glycoprotein hormone receptor, rickets, eliminate neuropeptide-induced tanning and selectively block a stereotyped behavioral program. The Journal of Experimental Biology. 2002;205:2555–2565. doi: 10.1242/jeb.205.17.2555. [DOI] [PubMed] [Google Scholar]
- Barker GC, Chitwood DJ, Rees HH. Ecdysteroids in helminths and annelids. Invertebrate Reproduction & Development. 1990;18:1–11. doi: 10.1080/07924259.1990.9672124. [DOI] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzamali G, Hauser F, Kobberup S, Williamson M, Grimmelikhuijzen CJ. Molecular identification of a Drosophila G protein-coupled receptor specific for crustacean cardioactive peptide. Biochemical and Biophysical Research Communications. 2003;303:146–152. doi: 10.1016/S0006-291X(03)00302-4. [DOI] [PubMed] [Google Scholar]
- Cheung CC, Loi PK, Sylwester AW, Lee TD, Tublitz NJ. Primary structure of a cardioactive neuropeptide from the tobacco hawkmoth, Manduca sexta. FEBS Letters. 1992;313:165–168. doi: 10.1016/0014-5793(92)81436-P. [DOI] [PubMed] [Google Scholar]
- Christie AE, Nolan DH, Garcia ZA, McCoole MD, Harmon SM, Congdon-Jones B, Ohno P, Hartline N, Congdon CB, Baer KN, Lenz PH. Bioinformatic prediction of arthropod/nematode-like peptides in non-arthropod, non-nematode members of the ecdysozoa. General and Comparative Endocrinology. 2011;170:480–486. doi: 10.1016/j.ygcen.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Clark AC, del Campo ML, Ewer J. Neuroendocrine control of larval ecdysis behavior in Drosophila: complex regulation by partially redundant neuropeptides. Journal of Neuroscience. 2004;24:4283–4292. doi: 10.1523/JNEUROSCI.4938-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleator M, Delves CJ, Howells RE, Rees HH. Identity and tissue localization of free and conjugated ecdysteroids in adults of Dirofilaria immitis and Ascaris suum. Molecular and Biochemical Parasitology. 1987;25:93–105. doi: 10.1016/0166-6851(87)90022-3. [DOI] [PubMed] [Google Scholar]
- Conzelmann M, Williams EA, Krug K, Franz-Wachtel M, Macek B, Jékely G. The neuropeptide complement of the marine annelid Platynereis dumerilii. BMC Genomics. 2013;14:906. doi: 10.1186/1471-2164-14-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell CB. The imaginal ecdysis of blowflies. The control of cuticuar hardening and darkening. The Journal of Experimental Biology. 1962a;39:395–412. [Google Scholar]
- Cottrell CB. The imaginal ecdysis of blowflies. Detection of the blood-borne darkening factor and determination of some of its properties. The Journal of Experimental Biology. 1962b;39:413–430. [Google Scholar]
- Criscuolo A, Gribaldo S. BMGE (Block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evolutionary Biology. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Oliveira AL, Calcino A, Wanninger A. Extensive conservation of the proneuropeptide and peptide prohormone complement in mollusks. Scientific Reports. 2019;9:4846. doi: 10.1038/s41598-019-40949-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rosa R, Grenier JK, Andreeva T, Cook CE, Adoutte A, Akam M, Carroll SB, Balavoine G. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- Dewey EM, McNabb SL, Ewer J, Kuo GR, Takanishi CL, Truman JW, Honegger HW. Identification of the gene encoding Bursicon, an insect neuropeptide responsible for cuticle sclerotization and wing spreading. Current Biology. 2004;14:1208–1213. doi: 10.1016/j.cub.2004.06.051. [DOI] [PubMed] [Google Scholar]
- Donini A, Agricola H, Lange AB. Crustacean cardioactive peptide is a modulator of oviduct contractions in Locusta migratoria. Journal of Insect Physiology. 2001;47:277–285. doi: 10.1016/S0022-1910(00)00112-8. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, Sørensen MV, Haddock SH, Schmidt-Rhaesa A, Okusu A, Kristensen RM, Wheeler WC, Martindale MQ, Giribet G. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Giribet G, Edgecombe GD, Hejnol A. Animal phylogeny and its evolutionary implications. Annual Review of Ecology, Evolution, and Systematics. 2014;45:371–395. doi: 10.1146/annurev-ecolsys-120213-091627. [DOI] [Google Scholar]
- Endress M, Zatylny-Gaudin C, Corre E, Le Corguillé G, Benoist L, Leprince J, Lefranc B, Bernay B, Leduc A, Rangama J, Lafont AG, Bondon A, Henry J. Crustacean cardioactive peptides: expression, localization, structure, and a possible involvement in regulation of egg-laying in the cuttlefish Sepia officinalis. General and Comparative Endocrinology. 2018;260:67–79. doi: 10.1016/j.ygcen.2017.12.009. [DOI] [PubMed] [Google Scholar]
- Ewer J, Gammie SC, Truman JW. Control of insect ecdysis by a positive-feedback endocrine system: roles of eclosion hormone and ecdysis triggering hormone. The Journal of Experimental Biology. 1997;200:869–881. doi: 10.1242/jeb.200.5.869. [DOI] [PubMed] [Google Scholar]
- Fraenkel G, Hsiao C. Bursicon, a hormone which mediates tanning of the cuticle in the adult fly and other insects. Journal of Insect Physiology. 1965;11:513–556. doi: 10.1016/0022-1910(65)90137-X. [DOI] [Google Scholar]
- Frand AR, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. PLOS Biology. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- Gammie SC, Truman JW. An endogenous elevation of cGMP increases the excitability of identified insect neurosecretory cells. Journal of Comparative Physiology A: Sensory, Neural, and Behavioral Physiology. 1997a;180:329–337. doi: 10.1007/s003590050052. [DOI] [PubMed] [Google Scholar]
- Gammie SC, Truman JW. Neuropeptide Hierarchies and the Activation of Sequential Motor Behaviors in the Hawkmoth, Manduca sexta. The Journal of Neuroscience. 1997b;17:4389–4397. doi: 10.1523/JNEUROSCI.17-11-04389.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Gharbi J, Girault JP, Hetru C, Lafont R. Ecdysteroid metabolism in leeches. Invertebrate Reproduction & Development. 1989;15:57–68. doi: 10.1080/07924259.1989.9672022. [DOI] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guittard E, Blais C, Maria A, Parvy JP, Pasricha S, Lumb C, Lafont R, Daborn PJ, Dauphin-Villemant C. CYP18A1, a key enzyme of Drosophila steroid hormone inactivation, is essential for metamorphosis. Developmental Biology. 2011;349:35–45. doi: 10.1016/j.ydbio.2010.09.023. [DOI] [PubMed] [Google Scholar]
- Halanych KM. The new view of animal phylogeny. Annual Review of Ecology, Evolution, and Systematics. 2004;35:229–256. doi: 10.1146/annurev.ecolsys.35.112202.130124. [DOI] [Google Scholar]
- In VV, Ntalamagka N, O'Connor W, Wang T, Powell D, Cummins SF, Elizur A. Reproductive neuropeptides that stimulate spawning in the Sydney rock oyster (Saccostrea glomerata) Peptides. 2016;82:109–119. doi: 10.1016/j.peptides.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Jékely G. Global view of the evolution and diversity of metazoan neuropeptide signaling. PNAS. 2013;110:8702–8707. doi: 10.1073/pnas.1221833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jékely G, Paps J, Nielsen C. The phylogenetic position of ctenophores and the origin(s) of nervous systems. EvoDevo. 2015;6:1. doi: 10.1186/2041-9139-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LS, Eddy SR, Portugaly E. Hidden markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Spalovská-Valachová I, Cho KH, Zitnanova I, Park Y, Adams ME, Zitnan D. Corazonin receptor signaling in ecdysis initiation. PNAS. 2004;101:6704–6709. doi: 10.1073/pnas.0305291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Zitnan D, Galizia CG, Cho KH, Adams ME. A command chemical triggers an innate behavior by sequential activation of multiple peptidergic ensembles. Current Biology. 2006a;16:1395–1407. doi: 10.1016/j.cub.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Zitnan D, Cho KH, Schooley DA, Mizoguchi A, Adams ME. Central peptidergic ensembles associated with organization of an innate behavior. PNAS. 2006b;103:14211–14216. doi: 10.1073/pnas.0603459103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol U. Precursors of neuropeptides and peptide hormones in the genomes of tardigrades. General and Comparative Endocrinology. 2018;267:116–127. doi: 10.1016/j.ygcen.2018.06.012. [DOI] [PubMed] [Google Scholar]
- Krüger E, Mena W, Lahr EC, Johnson EC, Ewer J. Genetic analysis of eclosion hormone action during Drosophila larval ecdysis. Development. 2015;142:4279–4287. doi: 10.1242/dev.126995. [DOI] [PubMed] [Google Scholar]
- Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lažetić V, Fay DS. Worm. Vol. 6. Taylor & Francis; 2017. Molting in C. elegans. [Google Scholar]
- Lee D, Orchard I, Lange AB. Evidence for a conserved CCAP-signaling pathway controlling ecdysis in a hemimetabolous insect, Rhodnius prolixus. Frontiers in Neuroscience. 2013;7:207. doi: 10.3389/fnins.2013.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman HK, Murgiuc CM, Miller TA, Lee TD, Hildebrand JG. Crustacean cardioactive peptide in the sphinx moth, Manduca sexta. Peptides. 1993;14:735–741. doi: 10.1016/0196-9781(93)90106-Q. [DOI] [PubMed] [Google Scholar]
- Lenaerts C, Cools D, Verdonck R, Verbakel L, Vanden Broeck J, Marchal E. The ecdysis triggering hormone system is essential for successful moulting of a major hemimetabolous pest insect, Schistocerca gregaria. Scientific Reports. 2017;7:46502. doi: 10.1038/srep46502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBrayer Z, Ono H, Shimell M, Parvy JP, Beckstead RB, Warren JT, Thummel CS, Dauphin-Villemant C, Gilbert LI, O'Connor MB. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Developmental Cell. 2007;13:857–871. doi: 10.1016/j.devcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb SL, Baker JD, Agapite J, Steller H, Riddiford LM, Truman JW. Disruption of a behavioral sequence by targeted death of peptidergic neurons in Drosophila. Neuron. 1997;19:813–823. doi: 10.1016/S0896-6273(00)80963-0. [DOI] [PubMed] [Google Scholar]
- Mendis AH, Rees HH, Goodwin TW. The occurrence of ecdysteroids in the cestode, Moniezia expansa. Molecular and Biochemical Parasitology. 1984;10:123–138. doi: 10.1016/0166-6851(84)90001-X. [DOI] [PubMed] [Google Scholar]
- Mirabeau O, Joly JS. Molecular evolution of peptidergic signaling systems in bilaterians. PNAS. 2013;110:E2028–E2037. doi: 10.1073/pnas.1219956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL, Kocot KM, Citarella MR, Dosung S, Norekian TP, Povolotskaya IS, Grigorenko AP, Dailey C, Berezikov E, Buckley KM, Ptitsyn A, Reshetov D, Mukherjee K, Moroz TP, Bobkova Y, Yu F, Kapitonov VV, Jurka J, Bobkov YV, Swore JJ, Girardo DO, Fodor A, Gusev F, Sanford R, Bruders R, Kittler E, Mills CE, Rast JP, Derelle R, Solovyev VV, Kondrashov FA, Swalla BJ, Sweedler JV, Rogaev EI, Halanych KM, Kohn AB. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510:109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte A, Koolman J, Dorlöchter M, Straub H. Ecdysteroids in the dorsal bodies of pulmonates (gastropoda): Synthesis and release of ecdysone. Comparative Biochemistry and Physiology Part A: Physiology. 1986;84:777–782. doi: 10.1016/0300-9629(86)90405-6. [DOI] [Google Scholar]
- Page AP, Stepek G, Winter AD, Pertab D. Enzymology of the nematode cuticle: a potential drug target? International Journal for Parasitology: Drugs and Drug Resistance. 2014;4:133–141. doi: 10.1016/j.ijpddr.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Filippov V, Gill SS, Adams ME. Deletion of the ecdysis-triggering hormone gene leads to lethal ecdysis deficiency. Development. 2002;129:493–503. doi: 10.1242/dev.129.2.493. [DOI] [PubMed] [Google Scholar]
- Park Y, Kim YJ, Dupriez V, Adams ME. Two subtypes of ecdysis triggering hormone receptor in Drosophila Melanogaster. The Journal of Biological Chemistry. 2003 doi: 10.1074/jbc.M301119200. [DOI] [PubMed] [Google Scholar]
- Phlippen MK, Webster SG, Chung JS, Dircksen H. Ecdysis of decapod crustaceans is associated with a dramatic release of crustacean cardioactive peptide into the haemolymph. The Journal of Experimental Biology. 2000;203:521–536. doi: 10.1242/jeb.203.3.521. [DOI] [PubMed] [Google Scholar]
- Pisani D, Pett W, Dohrmann M, Feuda R, Rota-Stabelli O, Philippe H, Lartillot N, Wörheide G. Genomic data do not support comb jellies as the sister group to all other animals. PNAS. 2015;112:15402–15407. doi: 10.1073/pnas.1518127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz KF, Yamanaka N, Gilbert LI, O'Connor MB. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science. 2009;326:1403–1405. doi: 10.1126/science.1176450. [DOI] [PubMed] [Google Scholar]
- Rewitz KF, Yamanaka N, O'Connor MB. Developmental checkpoints and feedback circuits time insect maturation. Current Topics in Developmental Biology. 2013;103:1–33. doi: 10.1016/B978-0-12-385979-2.00001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller L, Zitnanová I, Dai L, Simo L, Park Y, Satake H, Tanaka Y, Adams ME, Zitnan D. Ecdysis triggering hormone signaling in arthropods. Peptides. 2010;31:429–441. doi: 10.1016/j.peptides.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel S, Frand AR, Ruvkun G. Regulation of the C. elegans molt by pqn-47. Developmental Biology. 2011;360:297–309. doi: 10.1016/j.ydbio.2011.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Satake H, Takeda M. Nutrient-induced alpha-amylase and protease activity is regulated by crustacean cardioactive peptide (CCAP) in the cockroach midgut. Peptides. 2006;27:2157–2164. doi: 10.1016/j.peptides.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Schmidt-Rhaesa A, Bartolomaeus T, Lemburg C, Ehlers U, Garey JR. The position of the arthropoda in the phylogenetic system. Journal of Morphology. 1998;238:263–285. doi: 10.1002/(SICI)1097-4687(199812)238:3<263::AID-JMOR1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Schumann I, Kenny N, Hui J, Hering L, Mayer G. Halloween genes in Panarthropods and the evolution of the early moulting pathway in ecdysozoa. Royal Society Open Science. 2018;5:180888. doi: 10.1098/rsos.180888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea C, Hough D, Xiao J, Tzertzinis G, Maina CV. An rxr/usp homolog from the parasitic nematode, Dirofilaria immitis. Gene. 2004;324:171–182. doi: 10.1016/j.gene.2003.09.032. [DOI] [PubMed] [Google Scholar]
- Shi Y, Jiang HB, Gui SH, Liu XQ, Pei YX, Xu L, Smagghe G, Wang JJ. Ecdysis triggering hormone signaling (ETH/ETHR-A) Is required for the Larva-Larva ecdysis in Bactrocera dorsalis (Diptera: Tephritidae) Frontiers in Physiology. 2017;8:587. doi: 10.3389/fphys.2017.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangier J, Hilbich C, Beyreuther K, Keller R. Unusual cardioactive peptide (CCAP) from pericardial organs of the shore crab Carcinus maenas. PNAS. 1987;84:575–579. doi: 10.1073/pnas.84.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MJ, Favrel P, Rotgans BA, Wang T, Zhao M, Sohail M, O'Connor WA, Elizur A, Henry J, Cummins SF. Neuropeptides encoded by the genomes of the akoya pearl oyster Pinctata fucata and Pacific oyster Crassostrea gigas: a bioinformatic and peptidomic survey. BMC Genomics. 2014;15:840. doi: 10.1186/1471-2164-15-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöver BC, Müller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suggs JM, Jones TH, Murphree SC, Hillyer JF. CCAP and FMRFamide-like peptides accelerate the contraction rate of the antennal accessory pulsatile organs (auxiliary hearts) of mosquitoes. The Journal of Experimental Biology. 2016;219:2388–2395. doi: 10.1242/jeb.141655. [DOI] [PubMed] [Google Scholar]
- Telford MJ, Bourlat SJ, Economou A, Papillon D, Rota-Stabelli O. The evolution of the ecdysozoa. Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363:1529–1537. doi: 10.1098/rstb.2007.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel D, Franz-Wachtel M, Aguilera F, Hejnol A. Xenacoelomorph neuropeptidomes reveal a major expansion of neuropeptide systems during early bilaterian evolution. Molecular Biology and Evolution. 2018;35:2528–2543. doi: 10.1093/molbev/msy160. [DOI] [Google Scholar]
- Truman JW. Hormonal control of insect ecdysis: endocrine cascades for coordinating behavior with physiology. Vitamins & Hormones. 2005;73:1–30. doi: 10.1016/S0083-6729(05)73001-6. [DOI] [PubMed] [Google Scholar]
- Truman JW, Riddiford LM. Neuroendocrine control of ecdysis in silkmoths. Science. 1970;167:1624–1626. doi: 10.1126/science.167.3925.1624. [DOI] [PubMed] [Google Scholar]
- Veenstra JA. Neurohormones and neuropeptides encoded by the genome of Lottia gigantea, with reference to other mollusks and insects. General and Comparative Endocrinology. 2010;167:86–103. doi: 10.1016/j.ygcen.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Veenstra JA, Rombauts S, Grbić M. In silico cloning of genes encoding neuropeptides, neurohormones and their putative G-protein coupled receptors in a spider mite. Insect Biochemistry and Molecular Biology. 2012;42:277–295. doi: 10.1016/j.ibmb.2011.12.009. [DOI] [PubMed] [Google Scholar]
- Veenstra JA. Similarities between decapod and insect neuropeptidomes. PeerJ. 2016;4:e2043. doi: 10.7717/peerj.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vehovszky A, Agricola HJ, Elliott CJ, Ohtani M, Kárpáti L, Hernádi L. Crustacean cardioactive peptide (CCAP)-related molluscan peptides (M-CCAPs) are potential extrinsic modulators of the buccal feeding network in the pond snail Lymnaea stagnalis. Neuroscience Letters. 2005;373:200–205. doi: 10.1016/j.neulet.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Vogel KJ, Brown MR, Strand MR. Phylogenetic investigation of peptide hormone and growth factor receptors in five dipteran genomes. Frontiers in Endocrinology. 2013;4:193. doi: 10.3389/fendo.2013.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandawala M, Moghul I, Yañez Guerra LA, Delroisse J, Abylkassimova N, Hugall AF, O'Hara TD, Elphick MR. Discovery of novel representatives of bilaterian neuropeptide families and reconstruction of neuropeptide precursor evolution in ophiuroid echinoderms. Open Biology. 2017;7:170129. doi: 10.1098/rsob.170129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang Y, Li Y, Li W, Li R, Xie X, Wang S, Hu X, Zhang L, Bao Z. Identification and characterization of neuropeptides by transcriptome and proteome analyses in a bivalve mollusc Patinopecten yessoensis. Frontiers in Genetics. 2018;9 doi: 10.3389/fgene.2018.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Zhang W, Li G, Sun Q-Z, Wang J-J, Smagghe G, Jiang H-B. Molecular characterization of ecdysis triggering hormone and its receptor in Citrus red mite (Panonychus citri) Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2019;230:100–105. doi: 10.1016/j.cbpa.2019.01.003. [DOI] [PubMed] [Google Scholar]
- Zitnan D, Kingan TG, Hermesman JL, Adams ME. Identification of ecdysis-triggering hormone from an epitracheal endocrine system. Science. 1996;271:88–91. doi: 10.1126/science.271.5245.88. [DOI] [PubMed] [Google Scholar]
- Zitnan D, Ross LS, Zitnanova I, Hermesman JL, Gill SS, Adams ME. Steroid induction of a peptide hormone gene leads to orchestration of a defined behavioral sequence. Neuron. 1999;23:523–535. doi: 10.1016/S0896-6273(00)80805-3. [DOI] [PubMed] [Google Scholar]
- Zitnan D, Kim YJ, Zitnanová I, Roller L, Adams ME. Complex steroid-peptide-receptor cascade controls insect ecdysis. General and Comparative Endocrinology. 2007;153:88–96. doi: 10.1016/j.ygcen.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žitňan D, Adams ME. Insect Endocrinology. Academic Press; 2012. Neuroendocrine regulation of ecdysis; pp. 253–309. [Google Scholar]