Abstract
Chlamydiae are widely distributed pathogens of human populations, which can lead to serious reproductive and other health problems. In our search for novel antichlamydial metabolites from marine derived microorganisms, one new (1), along with two known (2, 3) dimeric indole derivatives were isolated from sponge derived actinomycete Rubrobacter radiotolerans. The chemical structures of these metabolites were elucidated by NMR spectroscopic data as well as CD calculations. All three metabolites suppressed chlamydial growth in a concentration-dependent manner. Among them, compound 1 exhibited the most effective antichlamydial activity with IC50 values of 46.6 ~ 96.4 μM in production of infectious progeny. Compounds appeared to target the mid-stage of the chlamydial developmental cycle by interfering with reticular body replication but not directly inactivating infectious elementary body.
Keywords: Chlamydia, dimeric indole, sponges, actinomycete, Rubrobacter radiotolerans, antichlamydial
Introduction
Chlamydiae are a group of obligate intracellular bacterial pathogens responsible for diseases in a range of hosts including humans [1, 2]. Chlamydia trachomatis is arguably the most common cause of sexually transmitted infections worldwide. Urogenital chlamydial infections frequently lead to serious complications including ectopic pregnancy, abortion, infertility, and pelvic inflammatory disease. In addition, C. trachomatis infection is the leading cause of eye disease as well, causing blindness in approximately 6 million and affecting the health of over 400 million worldwide [3, 4]. Chlamydia pneumoniae is an etiologic agent of respiratory diseases such as pneumonia and bronchitis and a possible risk factor for atherosclerosis, stroke and Alzheimer’s disease [5–7].
Chlamydiae have a unique biphasic life cycle consisting of two alternating cellular forms, the infectious but non-proliferative elementary body (EB) and the replicative but non-infectious reticulate body (RB). Chlamydial infection is initiated by EB adhering to and invading the host cell, and forming a specialized vacuole, the so-called inclusion. Within the inclusion, EB differentiates into RB. After round of replication, RBs reorganize back to EBs. Most chlamydial developmental cycles are complete in 40–72 hours when EBs are released from the host cell [1, 8–10].
Current therapies of chlamydial infections are broad-spectrum antibiotics, including azithromycin, doxycycline and fluoroquinolones [11–13]. Although chlamydiae are susceptible to a number of antibiotics, data suggest that incomplete antibiotic therapy can result in persistence and long-term infection [14]. In addition, antibiotic-resistant chlamydiae have been detected [11, 15–18]. During the past few decades, plenty of Chlamydia research efforts have been focused on vaccine development, but not resulting in an effective vaccine for human use [19, 20]. The merge of antibiotic-resistant chlamydiae and lack of approved vaccine necessitates identification of novel antichlamydials through chemical synthesis [21–23] or from natural resources [24–26].
Endozoic microorganisms inhabiting the inner tissues of animals produce of a huge diversity of secondary metabolites that deliver interesting pharmacological properties and are recognized as a prolific source of biologically active molecules [27]. In our continuous search for antichlamydial metabolites from marine derived microorganisms, we generated one new (1), along with two previously reported dimeric indole derivatives (2, 3) [28] from sponge derived actinomycete Rubrobacter radiotolerans. We found that all the three metabolites inhibited Chlamydia in a concentration-dependent manner, with compound 1 being most effective.
Results and Discussion
Compound 1 was obtained as a yellow, amorphous solid and was found to have a molecular formula of C21H22N2O3 by HRESIMS analysis. Absorption observed at 3445 cm−1 in its IR spectrum suggested the presence of hydroxyl functionality. Additionally, sixteen olefinic carbons at δC 138.3–108.5 and three oxygen-bearing carbons at δC 74.3–61.9 were observed. The remaining carbons were also categorized by the DEPT spectrum.
The NMR data of compound 1 was almost identical to that of compound 2 [28]. However, in HPLC analyses, two distinct peaks were generated. In combination with nuances of 13C NMR data of the side chain (C-8′–C-10′), compounds 1 and 2 were suggested to be epimer. This result was further confirmed by using optical rotation value and CD analysis, which showed a positive cotton effect (CE) around 223 nm and a negative CE around 216 nm of compound 1, suggested the absolute configuration of C-8′ was unambiguously as 8′R (Fig. 1).
Fig. 1.
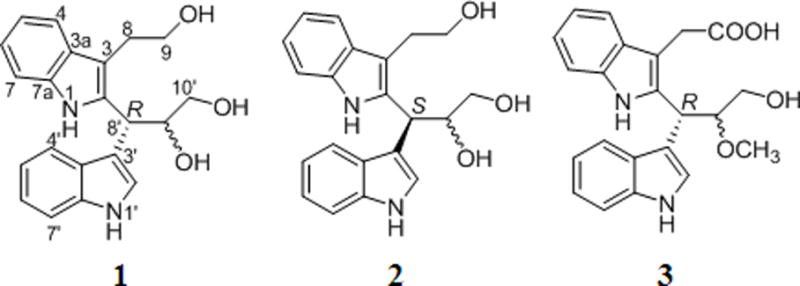
Chemical structures of compounds 1–3.
We hypothesized that all the three compounds were derived from condensation of two tryptophan molecules. The early steps of the biosynthetic process were proposed to involve the deamination of a tryptophan unit to yield 3-(1H-indol-3-yl)acrylic acid. The newly formed allyl part attacked another tryptophan moiety at position 2 to obtain intermediate material, resulting in the formation of both R and S isomers in our study. And then, the intermediate material was further underwent oxidation-reduction to give compounds 1 to 3 (Fig. 2).
Fig. 2.
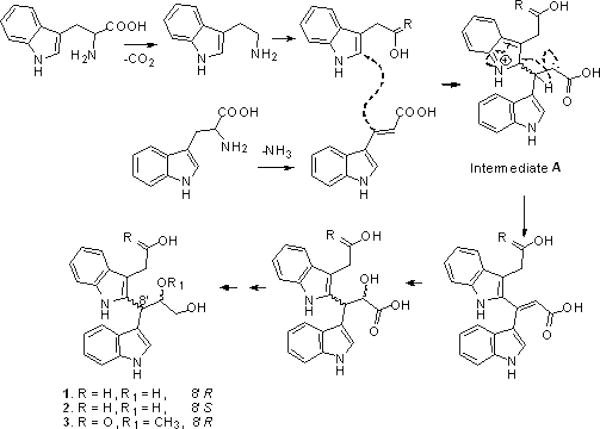
Plausible biosynthetic pathway of compounds 1–3.
Compound 1: yellow powder; [α]23D −8.9 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 203 (2.54), 225 (2.16), 278 (1.94) nm; IR vmax 3445, 2927, 2856, 1408, and 1383 cm−1; CD (c 1.4 × 10−3 M, MeOH) Δ∊ (nm) 4.92 (212); −9.46 (226); 1H-NMR (CD3OD): 7.51 (1H, dd, J = 7.8, 1.0, H-4′); 7.45 (1H, dd, J = 7.8, 1.0, H-7′); 7.44 (1H, s, H-2′); 7.30 (1H, dd, J = 8.1, 1.0, H-4); 7.26 (1H, dd, J = 8.0, 1.0, H-7); 7.03 (1H, ddd, J = 8.0, 7.8, 1.0, H-6′); 6.99(1H, ddd, J = 8.0, 7.8, 1.0, H-5′); 6.93 (1H, ddd, J = 8.1, 8.0, 1.0, H-5); 6.93(1H, ddd, J = 8.0, 8.0, 1.0, H-6); 4.78 (1H, d, J = 5.7, H-8'); 4.40 (1H, ddd, J = 10.5, 5.7, 4.0, H-9′); 3.70 (2H, t, J = 7.0, H-9); 3.57 (1H, dd, J = 10.5, 4.0, H-10′); 3.45 (1H, dd, J = 10.5, 5.7, H-10′); 3.08 (2H, t, J = 7.0, H-8); 138.3 (C-2); 137.7 (C-7a′); 137.3 (C-7a); 129.1 (C-3a′); 127.5 (C-3a); 122.8 (C-6′); 122.4 (C-2′); 121.1 (C-5′); 119.4 (C-4′); 119.4 (C-6); 119.1 (C-5); V118.0 (C-7′); 114.7 (C-3′); 110.7 (C-4); 110.4 (C-7); 108.5 (C-3); 74.3 (C-9′); 65.5 (C-10′); 61.9 (C-9); 36.9 (C-8′); 29.1 (C-8); (+)-ESIMS m/z 373 [M + Na]+.
The antichlamydial effect of compounds 1–3 was first determined using C. trachomatis L2, a sexually transmitted serotype responsible for lymphogranuloma venereum. Compared with negative control DMSO (final concentration, 0.5%), compounds 1–3 all inhibited both formation and growth of C. trachomatis L2 inclusions in a concentration-dependent manner. Compound 1 was the most effective, which completely blocked inclusion formation at 480 μM (Fig. 3A), analogous to the effect of positive control tetracycline (final concentration, 11.25 μM) [26]. Yields of infectious progeny EBs were further determined to evaluate the inhibitory effect of compounds 1–3. As expected, all three compounds exhibited similar concentration-dependent inhibition and 480 μM of compound 1 or positive control completely inhibited the production of progeny EBs (Fig. 3B). Using non-linear regression analysis by Graphpad Prism 5 software, the IC50 values, defined as the concentration at which the yield of infectious progeny EBs was reduced by 50% relative to negative control and presented as Mean (95% confidence interval), were calculated as 85.62 μM (77.45 ~ 94.65 μM), 227.0 μM (198.1 ~ 260.0 μM) and 238.7 μM (209.8 ~ 271.7 μM) for compounds 1–3, respectively.
Fig. 3.
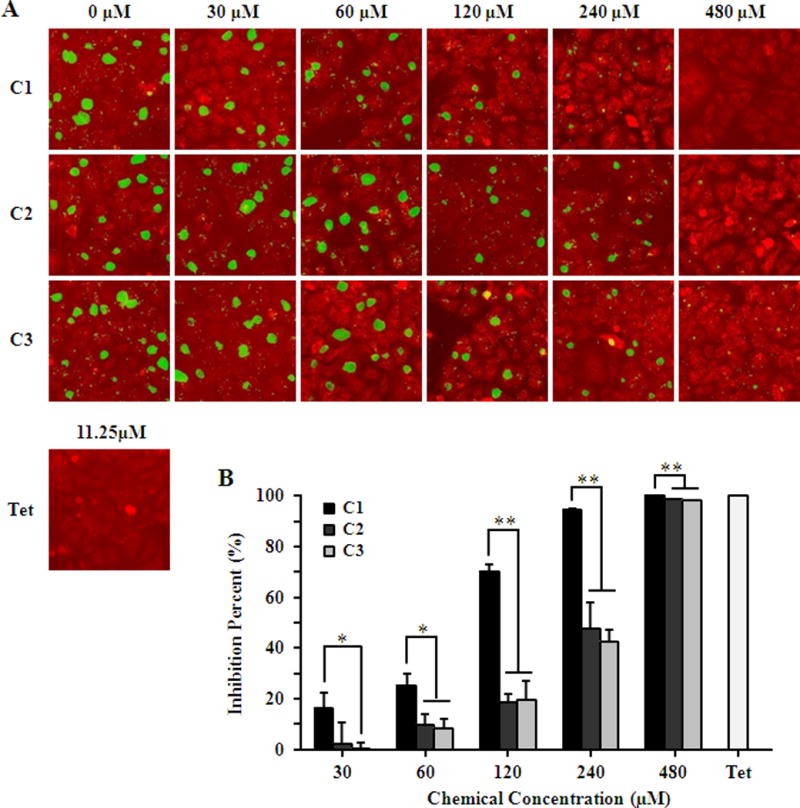
Inhibition effect of compounds on C. trachomatis L2. HeLa cells were infected with C. trachomatis L2 EBs at a MOI of 0.2 in the presence of 0.5% DMSO, 11.25 μM tetracycline (Tet) or various concentrations of compounds 1–3. (A) Cells were fixed 36 hours post-infection. The chlamydial inclusions were strained with a polyclonal anti-EB antibody (green) and cells were counterstained with Evans blue (red). (B) The inhibitory effect of compounds on the production of infectious progeny EBs was determined by counting recoverable IFUs in compound or Tet treated samples relative to untreated negative control. Asterisks indicate compound 1 inhibition levels were significantly different from compound 2 and 3 (*P<0.05, **P<0.01 by Student’s t test). (Color figure available online only)
As Chlamydia relied on the host cells for survival and replication, it was important to confirm whether the observed inhibitory effect was due to the direct action of the compounds on Chlamydia rather than an indirect cytotoxic effect on the host cells. Potential cytotoxic effect of compounds 1–3 on host cells was assessed by treatment of uninfected cells with 480 μM of individual compound. 48 hours later, no substantial cytotoxic effect on host cells was observed from cell proliferation by counting cell numbers, morphological feature by Evan’s blue staining and nuclear integrity by DAPI staining (Fig. 4), indicating that compounds 1–3 were well tolerated by HeLa cells and their inhibitory effect was due to the direct action on Chlamydia but not cytotoxicity.
Fig. 4.
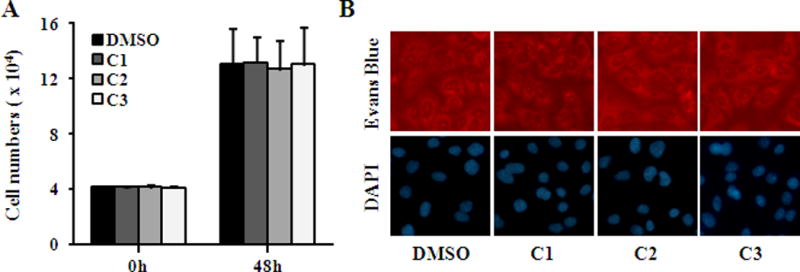
Lack of toxicity of compounds to host cells. Uninfected HeLa cells at 20% confluence were cultured with medium containing 480 μM individual compound or 0.5% DMSO and were enumerated (A) or stained with Evans blue or DAPI (B) 48 h later. (Color figure available online only)
The antichlamydial effect of compounds 1–3 was additionally confirmed using C. trachomatis C, a blinding ocular serotype, C. trachomatis D, one of the most common sexually transmitted serotype, and C. muridarum strain Nigg II, a mouse pathogen used to model human chlamydial infections in mice by evaluating the yields of infectious progeny EBs. All three Chlamydia strains were inhibited by compounds in similar concentration-dependent pattern as observed in C. trachomatis L2, only with slight differences of susceptibility (Fig. 5). As expected, compound 1 was the most active one, which fully abolished the production of progeny EBs at 480 μM, analogous to positive control tetracycline. The IC50 values of the three compounds on all Chlamydia strains were calculated based on concentration-response inhibition data by generating concentration-response curves (Figure 5S) and presented in Table 1.
Fig. 5.
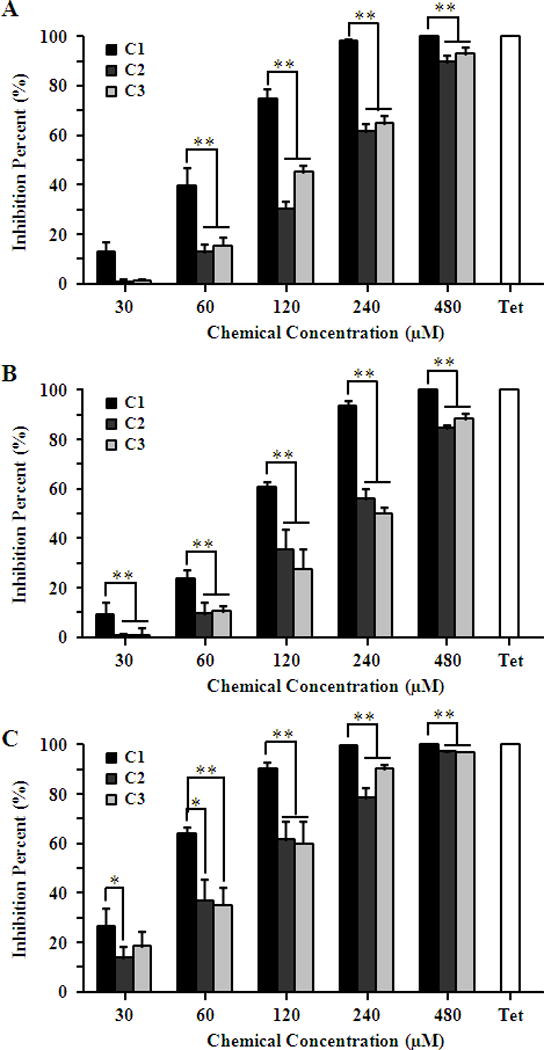
Inhibition effect of compounds on C. trachomatis C and D serotypes and C. muridarum. HeLa cells were infected with C. trachomatis C (A) or D (B) or C. muridarum (C) EBs at a MOI of 0.2 in the presence of 0.5% DMSO, 11.25 μM tetracycline (Tet) or various concentrations of compounds 1–3. The inhibitory effect of compounds on the production of infectious progeny EBs was determined by counting recoverable IFUs in compound or Tet treated samples relative to untreated negative control. Asterisks indicate compound 1 inhibition levels were significantly different from compound 2 and 3 (*P<0.05, **P<0.01 by Student’s t test).
Table 1.
IC50 values (μM) of compounds 1–3 for different Chlamydia. IC50 values were presented as Mean (95% confidence interval).
| C. trachomatis L2 | C. trachomatis C | C. trachomatis D | C. muridarum | |
|---|---|---|---|---|
| C1 | 85.62 (77.45 ~ 94.65) |
71.84 (66.76 ~ 77.30) |
96.24 (90.34 ~ 102.5) |
46.53 (43.97 ~ 49.24) |
| C2 | 227.0 (198.1 ~ 260.0) |
181.4 (170.9 ~ 192.6) |
191.7 (172.8 ~ 212.7) |
89.21 (78.97 ~ 100.8) |
| C3 | 238.7 (209.8 ~ 271.7) |
148.3 (133.6 ~ 164.7) |
212.2 (191.3 ~ 235.5) |
84.74 (74.63 ~ 96.22) |
Chlamydia is characterized by its unique developmental cycle of EB/RB alternation and this distinctive lifecycle offers multiple opportunities for therapeutic intervention. To understand whether compounds 1–3 exerted their antichlamydial effect by affecting the infectivity of EBs and/or the growth of RBs, C. trachomatis L2 EBs were pretreated with 480 μM of compounds 1–3 or 0.5% DMSO for 1 h at 4°C, and their titers were determined on HeLa cell monolayers after washing off the compounds. No detectable change of inclusion counts was observed (Fig. 6), suggesting that compounds 1–3 did not directly inactivate EBs. Next, we added individual compound to infected cultures at different times postinoculation (Fig. 7A), and then quantified infectious progenies that formed in these cultures. We found addition of 240 μM compound 1 at 0, 2 and 12 h postinoculation resulted in approximately 95% inhibition (Fig. 7B), whereas inhibition efficiency significantly dropped to 85% when the inhibitor was added at 24 h. Compounds 2 and 3 postinoculation revealed similar inhibition pattern (Fig. 7B). Since the EB differentiates into the RB around 6 h after cell entry, and RBs then replicate exponentially before they asynchronously reorganize into EBs [1], the time point inhibition data indicated that compounds 1–3 specifically targeted the mid-stage of the chlamydial developmental cycle by interfering with RB replication. Future studies are needed to identify detailed inhibition mechanism.
Fig. 6.
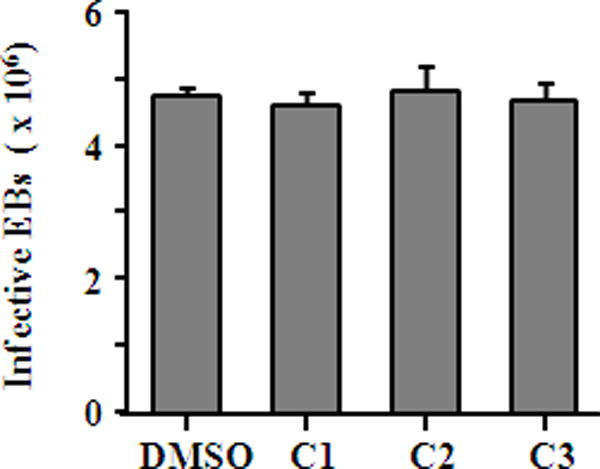
Lack of direct effect of compounds on Chlamydia EBs. C. trachomatis L2 EBs were incubated with 480 μM individual compound or 0.5% DMSO at 4°C for 1 h. The residual chemical was removed from EBs by two washes with SPG. Infective EBs counts were determined by measuring inclusion counts after inoculation of HeLa cells for 36 h.
Fig. 7.
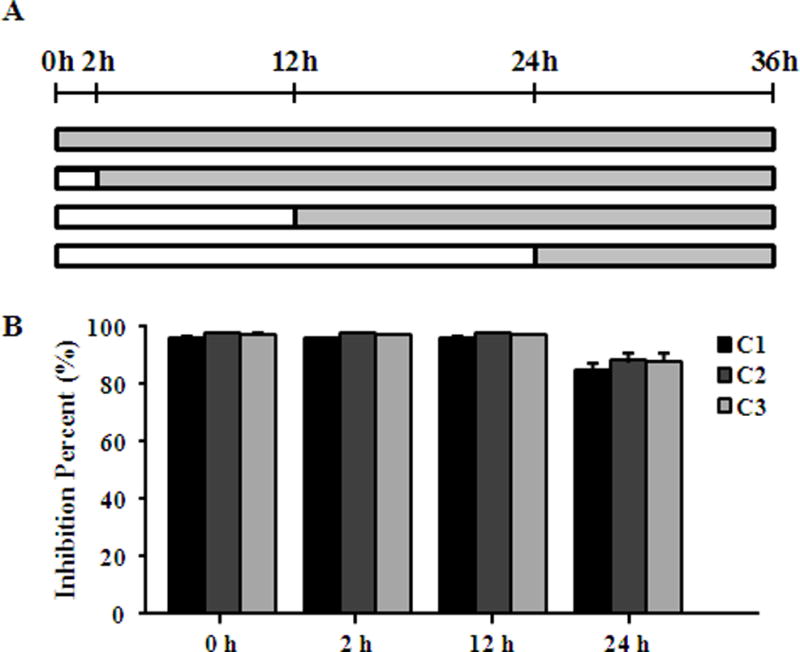
Impact of exposure times on antichlamydial effect of compounds. (A) HeLa cells infected with C. trachomatis L2 EBs at a MOI of 0.2 were treated with 240 μM compound 1 or 480 μM compound 2 or 3 at different time points as described. (B) The inhibitory effect of compounds on the production of infectious progeny EBs at different time points was determined by counting recoverable IFUs in compound treated samples relative to untreated negative control at 36 h.
In conclusion, one new dimeric indole metabolite, compound 1, along with the previously known metabolites (2, 3) were isolated and identified from the sponge’s endozoic actinomycete Rubrobacter radiotolerans. All three metabolites suppressed chlamydial growth in a concentration-dependent manner, which targeted the mid-stage of the chlamydial replication cycle. Among them, compound 1 was the most effective one. Our findings highlighted the potential of marine derived microorganisms as sources for screening promising lead molecules for development of antichlamydial agents.
Materials and Methods
Biological material isolation and identification
The actinomycete strain was isolated from a marine sponge Petrosia sp., which was collected by scuba (15 to 25 meters depth) in 2013, off the coast of Xisha Islands, China. Following a rinse with sterile sea water, small pieces of the surface and inner tissue of the sponge were homogenized and then inoculated. The pure actinomycete strain, designated as 05039, was identified as Rubrobacter radiotolerans by a morphological and biochemical analysis by Professor Guangtong Chen. A voucher sample (No PF2159) was deposited at the Department of Pharmacy, Nantong University.
Extraction, purification and structure elucidation
Fermentation was performed in 100 mL malt media in 300 mL Erlenmeyer flasks for subculture. For the massive culture, 100 mL of subculture was transferred into a 1 L Erlenmeyer flask containing 0.5 L culture media and fermentation was carried out on a rotary shaker (30°C, 150 rpm, 10 days). The cultured actinomycete (20 L) was extracted with 10 L of EtOAc, to afford the EtOAc extract, which was partitioned between n-hexane and 90% MeOH. The 90% MeOH layer was subjected to a stepped-gradient MPLC eluting with 30% to 100% MeOH to afford 16 fractions. Fraction 6 (50% MeOH fraction) was subjected to RP-HPLC (YMC packed J’sphere ODS-H80 column, 250 × 10 mm, 4 μm, 80 Å), eluted with 35% (1.0 mL/min, 220 nm) aqueous MeOH to afford compounds 2 (2.0 mg, Rt 90 mins) and 3 (1.4 mg, Rt 55 mins). Compound 1 was isolated from fraction 7 (55% MeOH fraction) with 35% (1.0 mL/min, 220 nm) aqueous MeOH (2.3 mg, Rt 62 mins) and further purified by sephadex LH-20 column, eluted with MeOH and CH2Cl2 (1:1).
Chlamydial strains and culture conditions
C. trachomatis C (strain TW3), D (strain UW-3/CX) and L2 (strain 434/Bu) and C. muridarum (strain Nigg II) were purchased from ATCC. HeLa cells purchased from ATCC were used for all cell culture experiments, and were routinely cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Sigma-Aldrich) and 10 μg/mL gentamicin at 37°C in a humidified atmosphere of 5% CO2. EB stocks were prepared in sucrose-phosphate-glutamate (SPG) and stored at −80°C as previously described [21].
Chlamydia infection and inhibition assay
The inhibition effect of compounds was evaluated as previously described, and quantified by IC50 [21, 26]. Cell monolayers were seeded onto coverslips in 24-well plate for immunofluorescence staining or directly in 48-well plate for determining the production of infectious progeny EBs. After overnight incubation, cells with 60~70% confluence were infected at multiplicity of infection (MOI) 0.2 inclusion-forming unit (IFU) per cell with different chlamydial strains. Individual compound, dissolved in DMSO, was added to final concentrations of 30, 60, 120, 240, or 480 μM simultaneously with chlamydial inoculation. DMSO (final concentration, 0.5%) or 11.25 μM tetracycline (Sigma Aldrich, purity 99%) [26] was used as negative or positive controls. Centrifugation (900 × g for 1 h at room temperature) was used to facilitate infection of the C. trachomatis C and D serotypes. Thirty-six (C. trachomatis L2 and C. muridarum) or 40 hours (C. trachomatis C and D) postinfection, cells were scraped or fixed with prechilled methanol at room temperature for 10 min. Methanol-fixed cells were kept in PBS at 4°C until subjected to sequential staining processed with a primary antibody and a FITC-conjugated secondary antibody. Evans blue was used as counterstain. Images were shot by Olympus IX51 fluorescence microscope. The scraped-off cells were disrupted by brief sonication to release infectious progeny EBs. The lysates were used to infect HeLa cell monolayers grown in 96-well plate following 1:10 serial dilution. The recoverable IFUs were quantified by immunofluorescence staining described as above. Percent inhibition was calculated based on recoverable IFUs in compound-treated samples relative to untreated negative control. IC50 values were calculated by non-linear regression from the concentration-response inhibition data using Graphpad Prism 5 software.
Cytotoxicity of compounds toward HeLa cells
HeLa cells were seeded onto 24-well plate at a density of 2 × 104 cells per well. After overnight growth, cells were switched into culture medium containing 480 μM of individual compound or 0.5% DMSO. Forty-eight hours later, cells were fixed with methanol and stained with Evans blue for morphology or DAPI for nuclear integrity. Otherwise, 48 hours later, cells were detached from the plastic by trypsinization and enumerated by hemacytometer.
Elementary body infectivity assay
L2 EBs were suspended into SPG at 4 × 105 IFU/mL and exposed to 480 μM of individual compound or 0.5% DMSO for 1 h at 4°C. After twice washing with SPG to eliminate the residual chemical, EBs were suspended into culture medium and immediately added to HeLa cell monolayers. Thirty-six hours later, infective EBs counts were determined by measuring inclusion counts after immunofluorescence staining as above.
Statistical analysis
All experiments were carried out as three replicates in minimum and results except IC50 were expressed as mean ± standard deviation. IC50 values, presented as Mean (95% confidence interval), were calculated by non-linear regression choosing “log(inhibitor) vs. normalized response – Variable slope” equation from the concentration-response inhibition data using Graphpad Prism 5 software. For statistical analysis, Student’s t-test (p < 0.05 or p < 0.01) was applied.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grants No. 31370209, No. 31400165 and No. 21402100), the United States National Institutes of Health (Grant No. AI122034), and Qing Lan Project.
Abbreviations
- EB
Elementary body
- RB
Reticular body
Footnotes
Supporting information
1H, 13C and 2D NMR data of compound 1 and concentration-response curves for generating IC50 values of three compounds on each Chlamydia strain are available as Supporting Information.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Abdelrahman YM, Belland RJ. The chlamydial developmental cycle. FEMS Microbiol Rev. 2005;29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Stephens RS, Myers G, Eppinger M, Bavoil PM. Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved. FEMS Immunol Med Microbiol. 2009;55:115–119. doi: 10.1111/j.1574-695X.2008.00516.x. [DOI] [PubMed] [Google Scholar]
- 3.Potroz MG, Cho NJ. Natural products for the treatment of trachoma and Chlamydia trachomatis. Molecules. 2015;20:4180–4203. doi: 10.3390/molecules20034180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan H. Blindness-causing trachomatous trichiasis biomarkers sighted. Invest Ophthalmol Vis Sci. 2012;53:2560. doi: 10.1167/iovs.12-9835. [DOI] [PubMed] [Google Scholar]
- 5.Belland RJ, Ouellette SP, Gieffers J, Byrne GI. Chlamydia pneumoniae and atherosclerosis. Cell Microbiol. 2004;6:117–127. doi: 10.1046/j.1462-5822.2003.00352.x. [DOI] [PubMed] [Google Scholar]
- 6.Grau AJ, Urbanek C, Palm F. Common infections and the risk of stroke. Nat Rev Neurol. 2010;6:681–694. doi: 10.1038/nrneurol.2010.163. [DOI] [PubMed] [Google Scholar]
- 7.Balin BJ, Little CS, Hammond CJ, Appelt DM, Whittum-Hudson JA, Gerard HC, Hudson AP. Chlamydophila pneumoniae and the etiology of late-onset Alzheimer’s disease. J Alzheimers Dis. 2008;13:371–380. doi: 10.3233/jad-2008-13403. [DOI] [PubMed] [Google Scholar]
- 8.Hybiske K, Stephens RS. Mechanisms of Chlamydia trachomatis entry into nonphagocytic cells. Infect Immun. 2007;75:3925–3934. doi: 10.1128/IAI.00106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci U S A. 2007;104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beeckman DS, De Puysseleyr L, De Puysseleyr K, Vanrompay D. Chlamydial biology and its associated virulence blockers. Crit Rev Microbiol. 2014;40:313–328. doi: 10.3109/1040841X.2012.726210. [DOI] [PubMed] [Google Scholar]
- 11.Sandoz KM, Rockey DD. Antibiotic resistance in Chlamydiae. Future Microbiol. 2010;5:1427–1442. doi: 10.2217/fmb.10.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Workowski KA, Levine WC, Wasserheit JN, Centers for Disease C, Prevention AG U.S. Centers for Disease Control and Prevention Guidelines for the treatment of sexually transmitted diseases: an opportunity to unify clinical and public health practice. Ann Intern Med. 2002;137:255–262. doi: 10.7326/0003-4819-137-4-200208200-00010. [DOI] [PubMed] [Google Scholar]
- 13.Workowski KA. Centers for Disease Control and Prevention Sexually Transmitted Diseases Treatment Guidelines. Clin Infect Dis. 2015;61(Suppl 8):S759–762. doi: 10.1093/cid/civ771. [DOI] [PubMed] [Google Scholar]
- 14.Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. Chlamydial persistence: beyond the biphasic paradigm. Infect Immun. 2004;72:1843–1855. doi: 10.1128/IAI.72.4.1843-1855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokoi S, Yasuda M, Ito S, Takahashi Y, Ishihara S, Deguchi T, Maeda S, Kubota Y, Tamaki M, Fukushi H. Uncommon occurrence of fluoroquinolone resistance-associated alterations in GyrA and ParC in clinical strains of Chlamydia trachomatis. J Infect Chemother. 2004;10:262–267. doi: 10.1007/s10156-004-0332-4. [DOI] [PubMed] [Google Scholar]
- 16.Khachatryan AR, Besser TE, Call DR. The streptomycin-sulfadiazine-tetracycline antimicrobial resistance element of calf-adapted Escherichia coli is widely distributed among isolates from Washington state cattle. Appl Environ Microbiol. 2008;74:391–395. doi: 10.1128/AEM.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Francesco A, Donati M, Rossi M, Pignanelli S, Shurdhi A, Baldelli R, Cevenini R. Tetracycline-resistant Chlamydia suis isolates in Italy. Vet Rec. 2008;163:251–252. doi: 10.1136/vr.163.8.251. [DOI] [PubMed] [Google Scholar]
- 18.Binet R, Bowlin AK, Maurelli AT, Rank RG. Impact of azithromycin resistance mutations on the virulence and fitness of Chlamydia caviae in guinea pigs. Antimicrob Agents Chemother. 2010;54:1094–1101. doi: 10.1128/AAC.01321-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Igietseme JU, Eko FO, Black CM. Chlamydia vaccines: recent developments and the role of adjuvants in future formulations. Expert Rev Vaccines. 2011;10:1585–1596. doi: 10.1586/erv.11.139. [DOI] [PubMed] [Google Scholar]
- 20.Mabey DC, Hu V, Bailey RL, Burton MJ, Holland MJ. Towards a safe and effective chlamydial vaccine: Lessons from the eye. Vaccine. 2014;32:1572–1578. doi: 10.1016/j.vaccine.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bao X, Gylfe A, Sturdevant GL, Gong Z, Xu S, Caldwell HD, Elofsson M, Fan H. Benzylidene acylhydrazides inhibit chlamydial growth in a type III secretion- and iron chelation-independent manner. J Bacteriol. 2014;196:2989–3001. doi: 10.1128/JB.01677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Good JA, Silver J, Nunez-Otero C, Bahnan W, Krishnan KS, Salin O, Engstrom P, Svensson R, Artursson P, Gylfe A, Bergstrom S, Almqvist F. Thiazolino 2-pyridone amide inhibitors of Chlamydia trachomatis infectivity. J Med Chem. 2016;59:2094–2108. doi: 10.1021/acs.jmedchem.5b01759. [DOI] [PubMed] [Google Scholar]
- 23.Hanski L, Ausbacher D, Tiirola TM, Strom MB, Vuorela PM. Amphipathic beta2,2-amino acid derivatives suppress infectivity and disrupt the intracellular replication cycle of Chlamydia pneumoniae. PLoS One. 2016;11:e0157306. doi: 10.1371/journal.pone.0157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horan AC, Shearer MC, Hedge V, Beyazova ML, Brodsky BC, King A, Berrie R, Cardaci K, Nimeck M. A family of novel macrocyclic lactones, the saccharocarcins produced by Saccharothrix aerocolonigenes subsp. antibiotica. I. Taxonomy, fermentation, isolation and biological properties. J Antibiot (Tokyo) 1997;50:119–125. doi: 10.7164/antibiotics.50.119. [DOI] [PubMed] [Google Scholar]
- 25.Salin O, Alakurtti S, Pohjala L, Siiskonen A, Maass V, Maass M, Yli-Kauhaluoma J, Vuorela P. Inhibitory effect of the natural product betulin and its derivatives against the intracellular bacterium Chlamydia pneumoniae. Biochem Pharmacol. 2010;80:1141–1151. doi: 10.1016/j.bcp.2010.06.051. [DOI] [PubMed] [Google Scholar]
- 26.Abdelmohsen UR, Cheng C, Reimer A, Kozjak-Pavlovic V, Ibrahim AK, Rudel T, Hentschel U, Edrada-Ebel R, Ahmed SA. Antichlamydial sterol from the red sea sponge Callyspongia aff. implexa. Planta Med. 2015 doi: 10.1055/s-0035-1545721. [DOI] [PubMed] [Google Scholar]
- 27.Mondol MA, Kim JH, Lee MA, Tareq FS, Lee HS, Lee YJ, Shin HJ. Ieodomycins A-D, antimicrobial fatty acids from a marine Bacillus sp. J Nat Prod. 2011;74:1606–1612. doi: 10.1021/np200223r. [DOI] [PubMed] [Google Scholar]
- 28.Li JL, Huang L, Liu J, Song Y, Gao J, Jung JH, Liu Y, Chen G. Acetylcholinesterase inhibitory dimeric indole derivatives from the marine actinomycetes Rubrobacter radiotolerans. Fitoterapia. 2015;102:203–207. doi: 10.1016/j.fitote.2015.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


