Abstract
Background:
We investigated the serum proteome of hormone-sensitive prostate cancer patients to determine candidate biomarkers associated with androgen deprivation therapy (ADT) efficacy.
Methods:
Serum proteomes generated using isobaric mass tags for relative and absolute quantitation (iTRAQ) were analyzed by reverse-phase liquid chromatography coupled to tandem mass spectrometry. The advanced hormone-sensitive prostate cancer cohorts studied were 1) untreated “paired” pre-ADT and 4-month post-ADT hormone-sensitive patients (n=15); 2) “early ADT failure” patients (n=10) failing ADT with-in a short period of time; 3) “late ADT failure” patients (n=10) failing ADT after a prolonged response time. Differential abundance was assessed, and Ingenuity Pathway Analysis (IPA) was used to identify interaction networks in selected candidates from these comparisons.
Results:
Between “post-ADT” and combined “early” and “late” ADT failure 149 differentially detected candidates were observed, and between “early” and “late” ADT failure 98 candidates were observed; 47 candidates were common in both comparisons. IPA network enrichment analysis of the 47 candidates identified three interaction networks (p<0.01) including 17-β-estradiol (E1), NfKB complex P38-MAPK as pathways with potential markers of response to ADT.
Conclusions:
A global proteomic analysis identified pathways with markers of ADT response, which will need validation in independent datasets.
Keywords: Proteomics, Androgen deprivation therapy, prostate cancer, predictive, biomarkers
Microabstract:
A global serum proteomic analysis was analyzed using liquid chromatography coupled with tandem mass spectrometry to identify predictive biomarkers in hormone sensitive prostate cancer patients undergoing androgen deprivation therapy. We identified 47 candidates and then performed network enrichment analysis which implicated beta estradiol, NfKB complex and P38-MAPK complex pathways as candidate pathways with potential predictive biomarkers of androgen deprivation.
Introduction
Prostate cancer is a leading cause of cancer related morbidity and mortality in US males(1). Androgen deprivation therapy (ADT) is a standard systemic hormonal therapy used for treating advanced prostate cancer and also for non-metastatic stage patients with poor prognosis in the presence of high risk features for early clinical progression. ADT slows disease progression but the duration of ADT response can range from a few months to several years (2–6) before a state of castration resistance emerges. Currently there are no validated serum based predictive markers available for application in this clinical setting (7). Therapeutic advances in the treatment of hormone sensitive prostate cancer have led to upfront combinations with ADT for increasing the overall efficacy of primary ADT (8, 9). These combinations work best for patients with clinical poor risk features or with high volume disease; biomarkers are not yet used to decide whether these patients receive ADT alone or these combinations. The most well-known biomarker in prostate cancer, serum prostatic specific antigen (PSA), lacks evidence as a predictive factor for defining ADT response duration (10–12). PSA does not adequately reflect underlying heterogeneity in tumor biology. Beyond PSA, other clinical and tumor factors (such as Gleason score), androgen levels, germline markers, that have been evaluated as predictors of hormonal therapy response are yet to be validated (13–17).
We hypothesized that serum based candidates of ADT response could be identified by comparing global serum proteomic signatures in advanced prostate cancer patients initiating ADT and failing the treatment after variable durations of response. We profiled serum samples taken from multiple advanced stage hormone sensitive prostate cancer patient cohorts to test this hypothesis.
Methods
Study population
Whole blood was collected from patients visiting a tertiary level hospital after obtaining written consent for participation in an advanced prostate cancer biomarker registry at the University of Rochester, NY. Institutional Review Board approved written consents were used and IRB approval was received for research on specimens used. Specimens from three distinct patient cohorts of non-localized stage prostate cancer were identified for serum proteomic profiling. The first cohort consisted of fifteen patients with untreated advanced hormone sensitive prostate cancer with “paired” “pre-ADT” initiation and a 4-month “post-ADT” serum specimen (a total of 30 serum specimens) between 2007 and 2008. The second cohort included serum collected from ten patients progressing on ADT for non-localized hormone sensitive prostate cancer with-in a short (<2 years) period of time (“early ADT failure”). The third cohort consisted of ten additional patients progressing on ADT after a long (>3 years) response duration (“late ADT failure”).
The definition of ADT failure for metastatic prostate cancer and progression to castrate resistance stage was defined as development of new metastatic lesions on imaging or biochemical progression (two serially rising PSAs at least one week apart) during continuous androgen deprivation. Serum was collected using a uniform processing protocol for all specimens as described under Supplementary Methods.
Serum proteome generation and mass spectrometry methods
Removal of high abundance proteins in serum was performed using a Hu-14 Multiple Affinity Removal Column, 4.6 × 100mm (Product Number 5188–6558; Agilent Technologies, Newcastle, DE USA), attached to an Agilent 1100 binary gradient HPLC system (Agilent Technologies, Inc. Santa Clara, CA). Subsequently, a total of 50 μg protein of each sample was used for iTRAQ labeling. This was followed by high pH reverse phase fractionation. Following fractionation of serum samples we performed tandem mass spectrometry with nanoLC-MS/MS analysis using a LTQ-Orbitrap Velos (Thermo-Fisher Scientific, San Jose, CA) mass spectrometer equipped with a “Plug and Play” nano ion source (CorSolutions, LLC, Ithaca, NY) using high energy collision dissociation (HCD). Data processing following mass spectrometry and identification of proteins was performed using Proteome Discoverer 1.1 software. Detailed methods are included in Supplementary Methods.
Methods.
Statistical Methods
Samples were randomly allocated to a mass spectrometry experiment and iTRAQ label via randomized block methods (18). Two samples were replicated for quality control purposes. Thus, a total of 52 samples from 35 subjects were assayed in 13 mass spectrometry experiments with the 4-plex iTRAQ system (Supplementary Figure S1). In addition to the laboratory quality-control assessments, global quality and bias were assessed graphically (19, 20).
The natural log peptide abundance values were normalized using a linear model to remove global experimental effects (21). The residual values from the model fit were used as normalized values in further analyses. Per-protein linear mixed effects models with contrast statements were used to assess differential abundance.
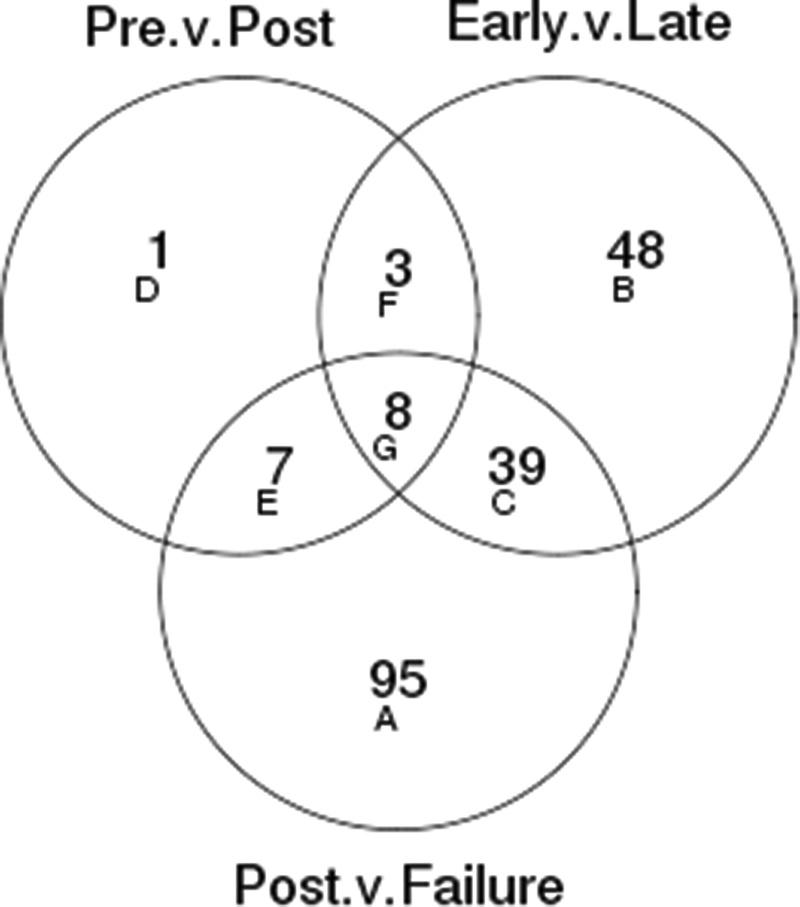
Three comparisons were conducted: pre-ADT vs. post-ADT (15 paired samples), early vs. late failure (ten samples each), and post-ADT vs. any failure (early plus late failure cohorts). A random effect for subject was used in order to account for correlation between paired samples for the pre-ADT vs. post-ADT analysis. Only the first replicates from the two duplicated samples were included in differential abundance analyses. Statistical significance was assessed using a False Discovery Rate (FDR) of 0.20 for each comparison (22). Volcano plots for each comparison were created, with fold changes being reported on the log2 scale for ease of interpretation. Differentially abundant proteins in each of these comparisons are presented in Venn diagrams illustrating the overlap of candidates in these comparisons (at an FDR of 0.20 or less). Experimentally observed candidates with differential abundance in these comparisons were then grouped using Ingenuity Pathway Analysis (IPA) as described in the Supplementary Methods.
Results
Study Population description
Three of fifteen subjects were diagnosed initially in metastatic stage and proceeded to receive ADT as first treatment without undergoing any primary treatment to the prostate. None of the three initially metastatic patients had high volume metastatic disease as in greater than 3 metastatic sites. All fifteen patients received ADT alone as initial treatment for hormone sensitive disease. The early and late ADT failure cohorts (n=10, each) included subjects selected after failure of ADT. For the castration resistant states all patients met the criteria of PSA progression and did not have any new metastases. Patient demographics including age, initial treatments at diagnosis, time between initial diagnosis and start of ADT for metastatic hormone sensitive disease, and wherever appropriate PSA and time on ADT are summarized in Table 1. Description of proteomic analyses
Table 1.
| early (N=10) |
late (N=10) |
pair (N=15) |
Total (N=35) |
|
|---|---|---|---|---|
| Age | ||||
| N | 10 | 10 | 15 | 35 |
| Mean (SD) | 73.6 (9.2) | 76.6 (8.0) | 65.7 (9.2) | 71.1 (9.9) |
| Median | 71.5 | 75.5 | 65.0 | 70.0 |
| Q1,Q3 | 68.0,81.0 | 71.0,83.0 | 61.0,71.0 | 65.0, 80.0 |
| Range | (59.0–87.0) | (67.0–90.0) | (49.0–87.0) | (49.0–90.0) |
| Gleason score at initial diagnosis | ||||
| N | 10 | 7 | 14 | 31 |
| Mean (SD) | 8.3 (1.2) | 6.7 (2.1) | 7.1 (0.9) | 7.4 (1.4) |
| Median | 8.0 | 7.0 | 7.0 | 7.0 |
| Q1, Q3 | 7.0, 9.0 | 5.0, 9.0 | 7.0, 8.0 | 7.0, 8.0 |
| Range | (7.0–10.0) | (3.0–9.0) | (6.0–9.0) | (3.0–10.0) |
| Initial prostate treatments | ||||
| ADT | 6 (60.0%) | 0 (0.0%) | 1 (6.7%) | 7 (20.0%) |
| ADTC | 1 (10.0%) | 0 (0.0%) | 1 (6.7%) | 2 (5.7%) |
| ADTX | 1 (10.0%) | 0 (0.0%) | 1 (6.7%) | 2 (5.7%) |
| S | 0 (0.0%) | 3 (30.0%) | 8 (53.3%) | 11 (31.4%) |
| SADTX | 1 (10.0%) | 0 (0.0%) | 0 (0.0%) | 1 (2.9%) |
| SX | 0 (0.0%) | 0 (0.0%) | 2 (13.3%) | 2 (5.7%) |
| SXADT | 0 (0.0%) | 1 (10.0%) | 0 (0.0%) | 1 (2.9%) |
| X | 1 (10.0%) | 6 (60.0%) | 2 (13.3%) | 9 (25.7%) |
| Time (months) between diagnosis and start of ADT treatments | ||||
| N | 10 | 10 | 15 | 35 |
| Mean (SD) | 8.5 (23.8) | 54.0 (40.2) | 43.2 (60.8) | 36.4 (49.4) |
| Median | 1.0 | 54.0 | 22.3 | 21.7 |
| Q1, Q3 | 0.0, 1.0 | 24.0,73.1 | 6.8, 45.1 | 1.0,48.0 |
| Range | (0.0–76.0) | (−1.9–132.5) | (0.0–200.1) | (−1.9–200.1) |
| Median time (months) on ADT for hormones sensitive metastatic disease before failure (Range) | 16.5 (1,23) | 52 (39, 246) | NA | |
| PSA (ng/ml) at the start of ADT for paired samples | ||||
| N | NA | NA | 15 | 15 |
| Mean (SD) | 27.4 (47.1) | 27.4 (47.1) | ||
| Median | 3.2 | 3.2 | ||
| Q1,Q3 | 1.0,35.0 | 1.0,35.0 | ||
| Range | (0.1–152.0) | (0.1–152.0) | ||
| 4-month mean PSA (ng/ml) after ADT initiation | ||||
| N | NA | NA | 15 | 15 |
| Mean (SD) | 1.0 (1.7) | 1.0 (1.7) | ||
| Median | 0.3 | 0.3 | ||
| Q1, Q3 | 0.0, 1.1 | 0.0, 1.1 | ||
| Range | (0.0–6.2) | (0.0–6.2) | ||
| PSA (ng/ml) at the time of ADT failure | ||||
| N | 10 | 10 | NA | 20 |
| Mean (SD) | 39.9 (48.8) | 16.7 (24.4) | 28.3 (39.4) | |
| Median | 27.4 | 4.3 | 9.6 | |
| Q1,Q3 | 2.9,53.1 | 1.0,26.9 | 1.4,42.0 | |
| Range | (0.7–152.1) | (0.5–76.6) | (0.5–152.1) |
NA: Not Applicable; PSA: Prostate Specific Antigen; ADT: Androgen Deprivation Therapy; SD: Standard Deviation; Q1: 25th Percentile; Q3: 75th Percentile; S-Surgery/Radical Prostatectomy; X-Primary radiation therapy to prostate
A total of 586 proteins/peptides were detected after restricting to the non-replicate experiment set (N=50), with 153 detected in all 13 mass spectrometry experiments and 208 detected in only 1 mass spectrometry experiment (Supplementary Figure S2: “Proteins and Peptides Coverage plots”). In order to identify predictors of ADT response we were interested in candidate proteins/peptides that changed significantly during ADT in the comparisons between the 4 month post ADT initiation cohort proteome and any failure, as well as between the proteomes of “early” and “late” ADT failure cohorts. This was based on the assumption that to be considered as circulatory predictive markers of response to ADT, at the very least, candidates would have to demonstrate change in abundance with ADT initiation and again at the time of ADT failure. Volcano plots for each comparison are presented in Supplementary figure S5. Venn diagrams illustrating the overlap of proteins/peptides with differential abundance in these comparisons by FDR of 0.20 or less are presented in Figure 1. At a FDR level of 0.20, 19 proteins/peptides were found to be differentially abundant in the “pre” versus “post” ADT initiation comparison in the first cohort; 98 in the “early-ADT failure” versus “late-ADT” comparison, and 149 in the “post ADT initiation” versus all failure comparisons. There were 47 proteins (regions C+G in Figure 1) that were differentially abundant in both the early versus late and post versus failure comparisons (Supplementary .xls file, “Final List of 47 proteins normdata.venn”, provides identity of various proteins based on these comparisons). These candidates were analyzed further in network analyses.
Figure 1.
Venn diagrams showing the number of proteins significant with FDR ≤ 0.2 and the number in common between each of the three comparisons performed. “Pre” refers to patients in cohort 1 before initiation of ADT. “Post” refers to the paired 4 month sample after initiating ADT. “Early” indicates the “Early-ADT” failure cohort and “Late” indicates the “Late-ADT failure cohort.
Description of pathway analysis
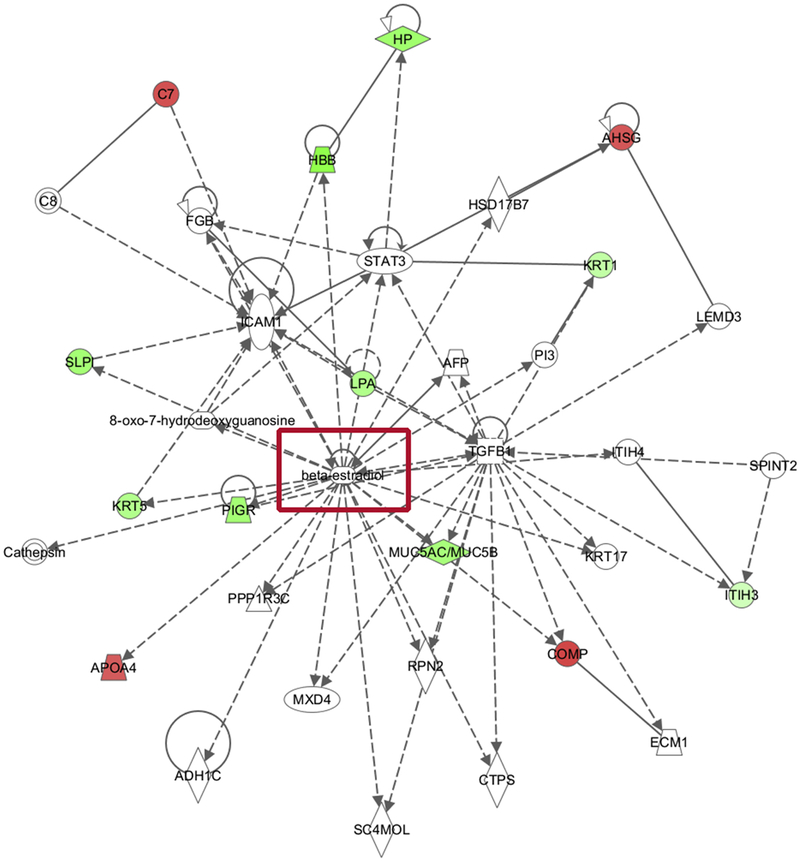
In total, 47 candidates were considered to be significantly altered for pathway level enrichment analysis. The genes corresponding to these proteins were overlaid onto the molecular network collected by the IPA database. Only experimentally observed interactions were included in this analysis, assuring the high confidence of network knowledge. A representative gene network identified in the candidate proteins was provided in Figure 2. This network was centered on Beta-estradiol and the genes within the network correspond to organismal injury and abnormalities, respiratory disease and cellular compromise (significance score is 29, corresponding to p-value less than 1E-29). A network legend, and the corresponding gene lists are provided in the supplementary data. Two other networks were also identified as significant hits in the analysis. One was centered on NfKB complex (Supplementary Figure S6), and the other was centered on P38-MAPK complex (Supplementary Figure S7).
Figure 2.
Pathway Enrichment Network Results of 47 proteins after comparing Post-ADT proteome with failure proteome (from the above Venn diagram (areas G + C) showing beta estradiol (red box) as a key gene implicated in the pathway network analysis (P value less than 10E-30 considered significant).
Green: signifies decrease in protein abundance in the comparison
Red: signifies Increase in protein abundance
Discussion
Since no serum based predictive biomarkers for ADT response are known we performed a serial global analysis of the serum proteome in patients starting ADT for advanced stage disease. To generate serum proteomic profiles we used iTRAQ analyzed by reverse-phase liquid chromatography coupled to tandem mass spectrometry (LC/MS/MS) and undertook a strategy in multiple “discovery level” cohorts and then followed by conducting a network analysis of 47 candidate biomarkers. Of the three networks implicated based on biological relevance in this hormonally driven tumor we focused on one, a hormonal based network in an independent cohort of patients. Variation in sex steroids may in fact have some relevance in predicting response to hormonal ablation (23) but this observation will need further evaluation in independent cohorts. Post ADT initiation measurements of PSA and testosterone as prognostic and predictive biomarkers have been investigated in advanced prostate cancer (24, 25) although are not validated yet for clinical practice. A serially rising serum PSA is a sensitive measure of progressive disease, but its pre-treatment level as a predictive marker has not been established and is more likely to correlate with tumor burden. A seven-month post ADT treatment serum PSA decline has been observed to have a level of association with long-term response, but has not been validated as a predictive biomarker. Eisenberger et al found that 76% of patients with metastatic prostate cancer treated with ADT had a PSA decrease to less than 4.0 ng/mL. In general, a nadir level is reached 2.5 to 3 months after starting ADT. The same investigators found that the median time to PSA less than 4 ng/mL was less than 82 days, whereas others have found 99% of patients reaching nadir levels by 3 to 4 months. Normalization of a rising PSA and PSA kinetics (PSA doubling time; PSA velocity) have been evaluated in clinical studies and appear to have some prognostic significance, but there is no consensus on either absolute PSA measurements or PSA kinetics to demonstrate predictive value to ADT.
By using emerging proteomic based technologies it has become possible to perform this type of large-scale study of protein abundance, in which thousands of protein candidates can be identified in a single experiment as candidates for prospective evaluation. Our study is the first of its kind to perform a global proteome analysis using an iTRAQ approach and we were able to identify 47 potential candidates and at least two other networks that might serve as biomarkers of response. The need to investigate these in future is relevant as currently no serum based proteomic biomarkers are available to identify ADT response. Comparative proteomic approaches employing two-dimensional gel electrophoresis (2-DE) in conjunction with mass spectrometry (MS) have provided valuable information for human plasma proteome projects (26–28). However, 2-DE has well-recognized limitations such as its limited applicability to hydrophobic proteins and those of extreme molecular weight or isoelectric point. Furthermore, 2-DE suffers from a limited dynamic range (29). For these reasons, solution-based “shotgun” techniques based on two-dimensional liquid chromatography (2D-LC) are widely used as an alternative approach, in which low pH reverse phase (RP) LC is always used as the second chromatographic dimension, coupled directly with tandem MS (MS/MS) detection (30, 31). Importantly, 2D-LC is highly compatible with a stable isotope label-based quantitative technique employing iTRAQ, allowing multiplexed (high throughput) quantitative analysis (32) across multiple biological samples. We used this technique for identifying serum based candidates of ADT response and then used pathway enrichment tools from a manually curated repository containing thousands of biological interactions and functional annotations extracted from published literature to identify clinically relevant candidates for prospective testing.
Despite the sensitive and global proteome detection techniques used in this hypothesis generating study, there are limitations. Our study population is from a single institution and limited to 35 advanced prostate cancer patients. The identified candidates are observed to have potential to be pursued based on a liberal statistical cut off value of FDR=0.2. This may be a result of a limited cohort size and event rate, and will need further follow up and evaluation in larger cohorts with careful and uniform collection and processing protocols to avoid pre-analytic bias. A serum proteome based signature for ADT response based on other networks identified in our analysis (NfKB complex and P38-MAPK complex) or the individual 47 candidates may also need to be included beyond hormonal pathways. An independent validation set of pre and post ADT initiation serum estrogen levels will be needed to determine whether estrogen levels can serve as predictive biomarkers of ADT efficacy in advanced prostate cancer.
Supplementary Material
Funding support
This work was supported by the National Institutes of Health [5R21CA133536] (MK and SZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement:
The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Eisenberger MA, Blumenstein BA, Crawford ED, Miller G, McLeod DG, Loehrer PJ, et al. Bilateral orchiectomy with or without flutamide for metastatic prostate cancer. N Engl J Med. 1998;339(15):1036–42. [DOI] [PubMed] [Google Scholar]
- 3.Crawford ED, Eisenberger MA, McLeod DG, Spaulding JT, Benson R, Dorr FA, et al. A controlled trial of leuprolide with and without flutamide in prostatic carcinoma. N Engl J Med. 1989;321(7):419–24. [DOI] [PubMed] [Google Scholar]
- 4.Denis LJ, Keuppens F, Smith PH, Whelan P, de Moura JL, Newling D, et al. Maximal androgen blockade: final analysis of EORTC phase III trial 30853. EORTC Genito-Urinary Tract Cancer Cooperative Group and the EORTC Data Center. European urology. 1998;33(2):144–51. [DOI] [PubMed] [Google Scholar]
- 5.Newling DW, Denis L, Vermeylen K. Orchiectomy versus goserelin and flutamide in the treatment of newly diagnosed metastatic prostate cancer. Analysis of the criteria of evaluation used in the European Organization for Research and Treatment of Cancer--Genitourinary Group Study 30853. Cancer. 1993;72(12 Suppl):3793–8. [DOI] [PubMed] [Google Scholar]
- 6.Prostate Cancer Trialists Collaborative Group. Maximum androgen blockade in advanced prostate cancer: an overview of the randomised trials. Prostate Cancer Trialists’ Collaborative Group. Lancet. 2000;355(9214):1491–8. [PubMed] [Google Scholar]
- 7.Grivas PD, Robins DM, Hussain M. Predicting response to hormonal therapy and survival in men with hormone sensitive metastatic prostate cancer. Crit Rev Oncol Hematol. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fizazi K, Tran N, Fein L, Matsubara N, Rodriguez-Antolin A, Alekseev BY, et al. Abiraterone plus Prednisone in Metastatic, Castration-Sensitive Prostate Cancer. N Engl J Med. 2017. [DOI] [PubMed] [Google Scholar]
- 9.Sweeney CJ, Chen YH, Carducci M, Liu G, Jarrard DF, Eisenberger M, et al. Chemohormonal Therapy in Metastatic Hormone-Sensitive Prostate Cancer. N Engl J Med. 2015;373(8):737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vicini FA, Vargas C, Abner A, Kestin L, Horwitz E, Martinez A. Limitations in the use of serum prostate specific antigen levels to monitor patients after treatment for prostate cancer. J Urol. 2005;173(5):1456–62. [DOI] [PubMed] [Google Scholar]
- 11.Smith JA Jr., Lange PH, Janknegt RA, Abbou CC, deGery A. Serum markers as a predictor of response duration and patient survival after hormonal therapy for metastatic carcinoma of the prostate. J Urol. 1997;157(4):1329–34. [PubMed] [Google Scholar]
- 12.Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Waldron W, et al. SEER Cancer Statistics Review, 1976–2008, National Cancer Institute; Bethesda, MD2011: Available from: http://seer.cancer.gov/csr/1975-2008/, based on November 2010 SEER data submission, posted to the SEER website, 2011. [Google Scholar]
- 13.Ross RW, Xie W, Regan MM, Pomerantz M, Nakabayashi M, Daskivich TJ, et al. Efficacy of androgen deprivation therapy (ADT) in patients with advanced prostate cancer: association between Gleason score, prostate-specific antigen level, and prior ADT exposure with duration of ADT effect. Cancer. 2008;112(6):1247–53. [DOI] [PubMed] [Google Scholar]
- 14.Quinn DI, Henshall SM, Sutherland RL. Molecular markers of prostate cancer outcome. Eur J Cancer. 2005;41(6):858–87. [DOI] [PubMed] [Google Scholar]
- 15.Kohli M, Riska SM, Mahoney DW, Chai HS, Hillman DW, Rider DN, et al. Germline predictors of androgen deprivation therapy response in advanced prostate cancer. Mayo Clin Proc. 2012;87(3):240–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geller J, Albert JD, Nachtsheim DA, Loza D. Comparison of prostatic cancer tissue dihydrotestosterone levels at the time of relapse following orchiectomy or estrogen therapy. J Urol. 1984;132(4):693–6. [DOI] [PubMed] [Google Scholar]
- 17.Perachino M, Cavalli V, Bravi F. Testosterone levels in patients with metastatic prostate cancer treated with luteinizing hormone-releasing hormone therapy: prognostic significance? BJU Int. 2010;105(5):648–51. [DOI] [PubMed] [Google Scholar]
- 18.Oberg AL, Vitek O. Statistical design of quantitative mass spectrometry-based proteomic experiments. J Proteome Res. 2009;8(5):2144–56. [DOI] [PubMed] [Google Scholar]
- 19.Cunningham JM, Oberg AL, Borralho PM, Kren BT, French AJ, Wang L, et al. Evaluation of a new high-dimensional miRNA profiling platform. BMC Med Genomics. 2009;2:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oberg AL, Mahoney DW. Statistical methods for quantitative mass spectrometry proteomic experiments with labeling. BMC Bioinformatics. 2012;13 Suppl 16:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberg AL, Mahoney DW, Eckel-Passow JE, Malone CJ, Wolfinger RD, Hill EG, et al. Statistical analysis of relative labeled mass spectrometry data from complex samples using ANOVA. J Proteome Res. 2008;7(1):225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin X, Zhang H, Ye D, Yao X, Zhang S, Dai B. Variations in circulating sex steroid levels in metastatic prostate cancer patients with combined androgen blockade: observation and implication. Andrology. 2013;1(3):512–6. [DOI] [PubMed] [Google Scholar]
- 24.Harshman LC, Chen YH, Liu G, Carducci MA, Jarrard D, Dreicer R, et al. Seven-Month Prostate-Specific Antigen Is Prognostic in Metastatic Hormone-Sensitive Prostate Cancer Treated With Androgen Deprivation With or Without Docetaxel. J Clin Oncol. 2018;36(4):376–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrylak DP, Ankerst DP, Jiang CS, Tangen CM, Hussain MH, Lara PN Jr., et al. Evaluation of prostate-specific antigen declines for surrogacy in patients treated on SWOG 99–16. J Natl Cancer Inst. 2006;98(8):516–21. [DOI] [PubMed] [Google Scholar]
- 26.Choi KS, Song L, Park YM, Marshall J, Lund AL, Shion H, et al. Analysis of human plasma proteome by 2DE- and 2D nanoLC-based mass spectrometry. Prep Biochem Biotechnol. 2006;36(1):3–17. [DOI] [PubMed] [Google Scholar]
- 27.Rai AJ, Gelfand CA, Haywood BC, Warunek DJ, Yi J, Schuchard MD, et al. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005;5(13):3262–77. [DOI] [PubMed] [Google Scholar]
- 28.Echan LA, Tang HY, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5(13):3292–303. [DOI] [PubMed] [Google Scholar]
- 29.Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci U S A. 2000;97(17):9390–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2(1):43–50. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Qiang X, Owsiany K, Zhang S, Thannhauser TW, Li L. Evaluation of different multidimensional LC-MS/MS pipelines for isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomic analysis of potato tubers in response to cold storage. J Proteome Res. 2011;10(10):4647–60. [DOI] [PubMed] [Google Scholar]
- 32.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3(12):1154–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.