Abstract
While the advent of combination antiretroviral therapy (cART) has dramatically increased the lifespan of people living with HIV-1 paradoxically, the prevalence of NeuroHIV in people treated with cART is on the rise. It has been well documented that despite the effectiveness of cART in suppressing viremia, CNS continues to harbor viral reservoirs with persistent low-level virus replication. This, in turn, leads to the presence and accumulation of early viral protein - HIV-1 Tat, that is a well-established cytotoxic agent. In the current study, we demonstrated that exposure of mouse microglia to HIV-1 Tat resulted both in a dose- and time-dependent upregulation of miRNA-34a, with concomitant downregulation of NLRC5 (a negative regulator of NFκB signaling) expression. Using bioinformatics analyses and Argonaute immunoprecipitation assay NLRC5 was identified as a novel target of miRNA-34a. Transfection of mouse primary microglia with miRNA-34a mimic significantly downregulated NLRC5 expression, resulting in increased expression of NFκB p65. In contrast, transfection of cells with miRNA-34a inhibitor upregulated NLRC5 levels. Using pharmacological approaches, our findings showed that HIV-1 Tat-mediated microglial activation involved miRNA-34a-mediated downregulation of NLRC5 with concomitant activation of NFκB signaling. Reciprocally, inhibition of miRNA-34a blocked HIV-1 Tat-mediated microglial activation. In summary, our findings identify yet another novel mechanism of HIV-1 Tat-mediated activation of microglia involving the miRNA-34a-NLRC5-NFκB axis. These in vitro findings were also validated in the medial prefrontal cortices of HIV-1 transgenic rats as well as in SIV-infected rhesus macaques. Overall, these findings reveal the involvement of miRNA-34a-NLRC5-NFκB signaling axis in HIV-1 Tat-mediated microglial inflammation.
Keywords: HIV-1 Tat, inflammasome, microglial activation, miRNA-34a, neuroinflammation, NLRC5, NFκB
1. INTRODUCTION
Currently, over 40 million HIV infected people are living worldwide, out of which nearly fifty percent of them receive combination antiretroviral therapy (cART). Despite the ability of cART to control viremia, HIV-infected individuals continue to experience central nervous system (CNS) complications, collectively termed as NeuroHIV. Though the exact molecular pathways underlying NeuroHIV are not clearly understood, the role of exacerbated cellular oxidative stress, dysregulated energy metabolism, immune activation, inflammation, and neuronal damage have been implicated in the process (Saylor et al., 2016). It has been proposed that impaired penetration of antiretrovirals in the CNS, neurotoxicity allied with antiretrovirals, persistent low-level viral replication, release of viral proteins and secretion of proinflammatory cytokines by infected or infiltered cells, collectively involve in the pathogenesis of HIV-1-mediated neurodegeneration (Carroll and Brew, 2017; Jayadev and Garden, 2009; Vassallo et al., 2013).
HIV-1 transactivator of transcription (Tat) is one among the nine HIV-1-encoded viral proteins that have received considerable attention owing to its toxicity on many CNS cells (Bagashev and Sawaya, 2013; Nath et al., 1999). There are many reports that confirmed the existence of both protein and mRNA expression of HIV-1 Tat in the brains of HIV-1-infected individuals (Hudson et al., 2000; Wiley et al., 1996). HIV-1 Tat also elicits neuroinflammation by activating glial cells and by escalating viral replication in latently infected cells (Ensoli et al., 1993; Frankel and Pabo, 1988). Extensive reports also suggest the mechanism(s) by which HIV-1 Tat impacts microglial functioning, via modulation of numerous signaling pathways (Bagashev and Sawaya, 2013; Chivero et al., 2017; Minghetti et al., 2004; Periyasamy et al., 2018b; Thangaraj et al., 2018). Additionally, HIV-1 Tat has also been shown to elicit other detrimental effects on the CNS, such as neurotoxicity, cellular activation, and endothelial dysfunction (Maubert et al., 2015).
MicroRNAs (miRs) are a class of small noncoding RNAs and function as critical regulators of several genes by modulating their post-transcriptional expression. Extensive studies also signify the impact of several brain-enriched microRNAs in regulating gene expression, microglial quiescence, and neuronal activities (Davis et al., 2015; Ponomarev et al., 2011; Yu et al., 2015). A growing body of evidence has shown that miRNA-34a is one of the brain-enriched microRNAs that is upregulated in various neurological as well as aging-related diseases (Bavamian et al., 2015; Jauhari et al., 2018; Kou et al., 2016; Li et al., 2011; Reynolds et al., 2018). In mammalians, the miRNA-34 family includes three processed miRs that are encoded by two different genes: miRNA-34a is encoded by its transcript, while miRNA-34b and miRNA-34c share a common primary transcript (Agostini and Knight, 2014; Rokavec et al., 2014). In mice, miRNA-34a is ubiquitously expressed with the highest expression in the brain, while miRNA-34b/c is mainly expressed in lung tissues (Bommer et al., 2007). The two miRNA-34 genes thus have tissue-specific functions. There are extensive studies documenting miRNA-34a mediated modulation of proteins that regulate synaptic targets and neuronal morphology and function (Agostini et al., 2011a; Agostini et al., 2011b; Bavamian et al., 2015; Morgado et al., 2015). Recently, it has also been reported that microRNA-34a along with microRNA-138 negatively regulates the expression of astrocytic SIRT1 in HIV-1 Tat exposed astrocytes, thereby resulting in astrogliosis – a hallmark feature of aging in the CNS (Hu et al., 2017). Although HIV-1 proteins have been shown to dysregulate gene expression profiles of various CNS cells (thereby contributing to HIV-1 neuropathology), there exists a gap in knowledge on the exact epigenetic mechanism(s) underlying HIV-1 Tat-mediated activation of microglia, specifically in the context of dysregulated microRNAs. In the current study, we found a novel molecular mechanism involved in HIV-1 Tat-mediated activation of microglia. Our findings suggest HIV-1 Tat-mediated upregulation of microRNA-34a targets NLRC5, which ultimately activates the NFκB signaling resulting ultimately into activation of microglia.
2. MATERIALS AND METHODS
2.1. Reagents
Antibodies have been acquired from the following sources: NLRC5 (sc-515668; 1:1000 dilution) from Santa Cruz Biotechnology, NFκB p65 (ab16502; 1:2000 dilution) from Abcam, P-NFκB p65 (3031; 1:500 dilution), IKKα (2682; 1:1000 dilution), and P-IKKα/β (2697; 1:500 dilution) from Cell Signaling Technology, IKKγ (559675; 1:1000 dilution), P-IKKγ (562590; 1:500 dilution) from BD Biosciences, CD11b (NB110–89474; 1:200 dilution) from Novus Biological Company, Iba-1 (019–19741; 1:200 dilution) from Wako Pure Chemical Industries, Ltd. Peroxidase-AffiniPure Goat Anti-Rabbit IgG (H+L) (111035-003; 1:10000 dilution) and Peroxidase-conjugated AffiniPure Goat Anti-Mouse IgG (H+L) (115-035-003; 1:1000 dilution) from Jackson ImmunoResearch Inc. Endotoxin-free, HIV-1 recombinant Tat (1032–10) was bought from ImmunoDX. TaqMan® miRNA Reverse Transcription Kit (4366596), TaqMan® miRNA assays for miRNA-34a (000426), TaqMan™ miRNA Control Assay - U6 snRNA (4427975), and TaqMan® Universal PCR Master Mix, no AmpErase® UNG (4324018) were bought from Applied Biosystems. miRIDIAN miRNA-34–3p mimic (C-311258–00), miRIDIAN miRNA-34–3p hairpin inhibitor (IH-311258–01), miRIDIAN miRNA mimic negative control (CN-001000), and miRIDIAN miRNA hairpin negative control (IN-001005) were purchased from Dharmacon. Lipofectamine™ RNAiMAX transfection reagent (13778150), and Opti-MEM® I Reduced Serum Media (31985070) were purchased from Life Technologies. ProLong® Gold Antifade Mountant with DAPI (P36935) was obtained from Molecular Probes.
2.2. Animals
HIV-1 transgenic rats were generated using a noninfectious provirus expressing 7 of the 9 HIV-1 viral proteins, such as Tat, Env, Rev, Nef, Vif, Vpr, and Vpu (Peng et al., 2010). Male Sprague Dawley (5-months old) HIV-1 transgenic rats (HIV-1, F344) with age, sex, and background-matched controls were used in this study. The animals were housed in clean polypropylene cages under conditions of constant temperature and humidity, with a 12–12 h day-night cycle, during which time they had free access to food and water ad libitum. Animal experiments were conducted according to the protocols approved by the University of Nebraska Medical Center and the National Institutes of Health (Thangaraj et al., 2018). HIV-1 transgenic rats were killed by isoflurane overdose followed by transcardial perfusion with ice-cold PBS for brain removal. Medial prefrontal cortical regions of the brain were separated and used for extraction of protein and total RNA.
2.3. Rhesus macaques and Simian immunodeficiency virus (SIV) infection
Briefly, 2- to 3-year-old Indian rhesus macaques (Macaca mulatta) of both the sexes were randomly divided into two groups (saline and SIV group). The SIV group was chronically infected with the SIVR71/17E virus for ~52 weeks. The detailed methodology of SIV infection and information on viral load and other parameters related disease pathogenesis has been described in our previous publications (Bokhari et al., 2011; Chivero et al., 2017; Hu et al., 2012; Pendyala et al., 2015; Periyasamy et al., 2018b). Briefly, rhesus macaques were deeply anesthetized using an intramuscular injection of ketamine (3 mg/kg) and medetomidine (0.15 mg/kg) following which laparotomies were performed. The animals were then exsanguinated from the descending aorta and were perfused transcardially with saline. Brains were dissected and portions of the all the brain regions were collected and stored frozen at −80°C for later protein and RNA analyses. In this study, we used the archival frozen medial prefrontal cortices from saline and SIV groups of rhesus macaques.
2.4. Mouse primary microglial culture
Mouse primary microglial cells were prepared from 1–3 days-old newborn pups of either sex bred from C57B1/6 as specified, under standard conditions as described (Skaper et al., 2012) with minor modifications (Periyasamy et al., 2018b; Thangaraj et al., 2018). For subsequent experiments, we used the microglia purity if >95% pure which was evaluated by immunocytochemistry using the antibody specific for Iba-1 (1:200 dilution).
2.5. BV-2 cells
BV-2-immortalized cell line was obtained from Dr. Sanjay Maggirwar (University of Rochester Medical Center, Rochester, NY, USA) and was routinely maintained in DMEM (Invitrogen, 11995–065) with 10% heat-inactivated fetal bovine serum (FBS, Invitrogen, 16000–044) at 37 °C and 5% CO2 and used up to 20 passages.
2.6. TaqMan® miRNA assays for miRNA-34a
The expression of miRNA-34a was quantified using TaqMan® miRNA assays as described (Periyasamy et al., 2018b). Briefly, total RNA was extracted using Quick-RNA™ MiniPrep Plus (Zymo Research, R1058) as per the manufacturer’s protocol and quantified using Thermo Scientific NanoDrop™ 8000 spectrophotometer. Total RNA thus isolated was reverse transcribed to synthesize cDNA for individual miRNA using specific miRNA primers from the TaqMan® miRNA assays and the TaqMan® miRNA Reverse Transcription kit. The reverse transcription product was then diluted 1:10 for the following PCR reaction. Each PCR reaction was carried out in triplicate, and six independent experiments were run. TaqMan® miRNA assays were performed using an Applied Biosystems® QuantStudio™ 3 Real-Time PCR System (Applied Biosystems, Grand Island, NY). The expression level of miRNA-34a was calculated by normalizing to U6 snRNA.
2.7. Transient transfection of miRNA-34a mimic and inhibitor
Mouse primary microglial cells were seeded into 6-well plates (3×105 cells per well) and were transiently transfected with 30 pmol of miRNA-34a mimic, miRNA-34a inhibitor, and miRNA control using Lipofectamine™ RNAiMAX as described (Periyasamy et al., 2018b). Following transfection, cells were exposed to HIV-1 Tat (50 ng/mL) for another 24 h, and total RNA and proteins were extracted for further investigation as indicated.
2.8. Pharmacological inhibition of NFκB complex using BMS345541
Mouse primary microglial cells were seeded into 6-well (3×105 cells per well) for 24 h at 37°C in a humidified, 5% CO2 incubator. Overnight serum-starved mouse primary microglial cells were pretreated with BMS345541 (10 μM; a selective allosteric inhibitor of IKK that blocks NFκB-dependent transcription) for 1 h followed by exposure of cells to HIV-1 Tat (50 ng/mL) for additional 24 h, followed by extraction of total RNA and protein for further investigations as indicated.
2.9. Quantitative polymerase chain reaction (qPCR)
qPCR experiments were performed according to the protocol described (Guo et al., 2016; Periyasamy et al., 2016; Periyasamy et al., 2018a). Briefly, total RNA was extracted using Quick-RNA™ MiniPrep Plus (Zymo Research, R1058) as per the manufacturer’s protocol and quantified using Thermo Scientific NanoDrop™ 8000 spectrophotometer. Reverse transcription reactions were performed using iScript™ Reverse Transcription Supermix for RT-qPCR (Bio-Rad, 1708841), as per manufacturer’s instructions. qPCRs were completed using TaqMan® Universal PCR Master Mix, no AmpErase® UNG in an Applied Biosystems® QuantStudio™ 3 Real-Time PCR System. Each reaction was carried out in triplicate, and six independent experiments were run. Gapdh was used as a housekeeping control for the normalization and the fold change in expression was obtained by the 2−ΔΔCT method.
2.10. Western blotting
Western blotting was performed using standard procedures as described previously (Guo et al., 2016; Periyasamy et al., 2016; Periyasamy et al., 2018a). Briefly, the control and treated microglial cells were harvested and lysed using the 200 μL of RIPA buffer (Cell Signaling Technology, 9806). Lysates were centrifuged at 12000 g for 10 min at 4°C, and the protein content of the supernatant was determined by a BCA assay using Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, 23227) as per the manufacturer’s instructions. 10 μg of soluble proteins were resolved in a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by blotting onto a polyvinylidene fluoride membrane (Millipore, IPVH00010). Then, the membranes were blocked with 5% nonfat dry milk (in 1X TTBS buffer) for 1 h at room temperature followed by overnight incubation with the indicated primary antibodies at 4°C. After 3 times washing, the membranes were incubated with a secondary antibody for 1 h at room temperature. Next, the protein signals were visualized using Super Signal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, 34078). Each band intensity was normalized to the internal control, β-actin (Sigma-Aldrich; A5316; 1:5,000 dilution), and the data were presented as a relative fold change by using ImageJ analysis software (Schneider et al., 2012).
2.11. Immunocytochemistry
Immunocytochemistry was performed using standard procedures as described previously (Guo et al., 2016; Periyasamy et al., 2016). Briefly, mouse primary microglial cells were seeded in a 24-well plate containing sterile glass coverslips (11 mm) at a density of 5×104 cells per well at 37°C in a humidified, 5% CO2 incubator for 24 h. Overnight serum-starved mouse primary microglial cells were then treated with the respective agents for the indicated time. Cells were then rinsed 2 times with 1× PBS at room temperature and fixed with 4% paraformaldehyde in PBS for 20 min at room temperature, followed by permeabilization with 0.3% Triton X-100 (Fisher Scientific, BP151–500) in PBS. Then, the permeabilized cells were incubated in a blocking buffer for 1 h at room temperature followed by overnight incubation of NLRC5 and CD11b primary antibodies at 4°C. After that, goat anti-rabbit IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 488 (ThermoFisher Scientific, A-11008; 1:500 dilution) and goat anti-mouse IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 594 (ThermoFisher Scientific, A-11005; 1:500 dilution) were added for 2 h to detect the expression of indicated proteins. Coverslips were then mounted on glass slides with ProLong® Gold Antifade Reagent with DAPI (Molecular Probes, P36935). Fluorescence images were taken on a Zeiss Observer using a Z1 inverted microscope (Carl Zeiss, Thornwood, NY, USA) and the acquired images were analyzed using the AxioVs 40 Version 4.8.0.0 software (Carl Zeiss MicroImaging GmbH).
2.12. miRNA-34a target validation
miRNA-34a target validation was performed using miRNA Target Immunoprecipitation (IP) Kit (Active Motif, 25500) as described in the manufacturer’s protocol. Briefly, mouse primary microglial cells were plated onto 150 mm plate and transfected with either miRNA control or miRNA-34a mimic for 24 h. An equal number of cells were used for the IP to minimize variability. Cells were then washed in the cold 1x PBS, scraped, and then lysed with a lysis buffer. IP uses G-coupled magnetic beads and a pan-Ago antibody to precipitate the miRNA/mRNA complex. An isotype antibody control was also performed in parallel. The precipitated complex was collected, and the RNA purified from the complex by phenol:chloroform:isoamyl alcohol (25:24:1) reagent. Purified RNA was then reverse transcribed into cDNA with iScript™ Reverse Transcription Supermix for RT-qPCR, and specific primers for NLRC5 were used for qPCR. Gapdh was used as a housekeeping control for the normalization. Data were analyzed by comparing the cells transfected with miRNA-34a mimic, or negative control and the fold enrichment of NLRC5 was determined as described by the manufacturer.
2.13. Statistical analysis
All the data were expressed as mean ± SEM, and statistical significance was determined using GraphPad Prism version 6.01 (San Diego, CA, USA). The detailed statistical analysis used is shown in each figure caption for all studies. For the in vivo experiments, an unpaired Student t test was used to comparing between the two groups. However, non-parametric Kruskal-Wallis One-way ANOVA followed by Dunn’s post hoc test was used to find the statistical significance between multiple groups. Values were statistically significant when p < 0.05.
3. RESULTS
3.1. Upregulation of miRNA-34a in the medial prefrontal cortices of SIV-infected rhesus macaques and HIV-1 transgenic rats
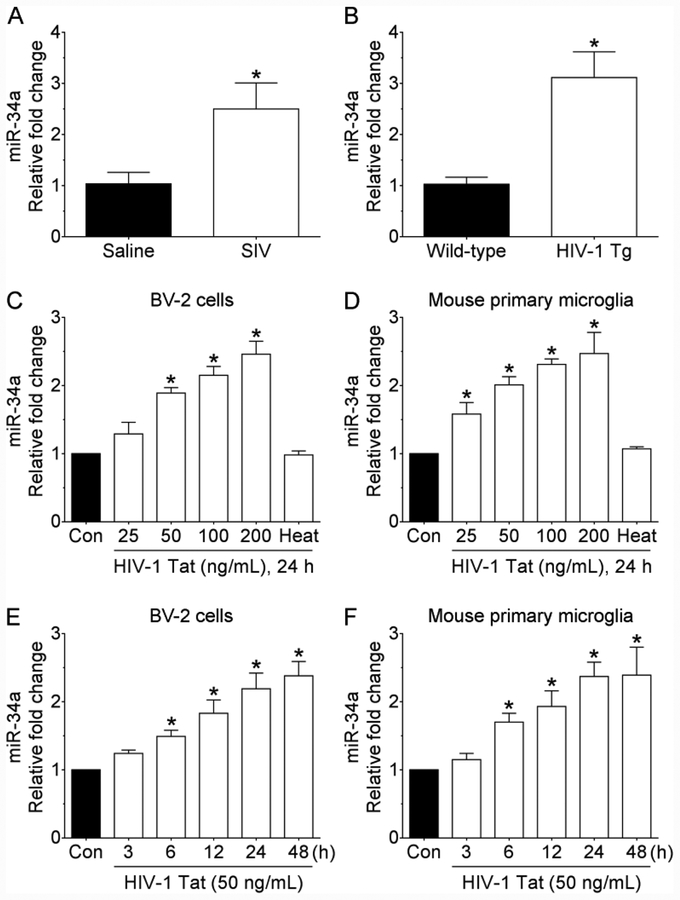
Based on the fact that miRNA-34a is significantly elevated in the brain as well as in PBMCs of HIV-1 infected individuals (Chang et al., 2011; Duskova et al., 2013; Witwer et al., 2012), we first sought to assess the expression profile of miRNA-34a in the medial prefrontal cortices of saline and SIV-infected rhesus macaques using qPCR. As represented in Figure 1A, expression of mature miRNA-34a was significantly increased in the medial prefrontal cortices of SIV-infected rhesus macaques compared with the saline group. Next, we wanted to confirm this observation in the medial prefrontal cortices of 5-month old male wild-type and age- and sex-matched HIV-1 transgenic rats. Interestingly, as shown in Figure 1B, the expression of mature miRNA-34a was also significantly increased in the medial prefrontal cortices of HIV-1 transgenic rats compared with that of the wild-type rats.
FIGURE 1.
Increased expression of miRNA-34a both in vivo and in vitro. (A) qPCR analysis showing increased expression of miRNA-34a in the medial prefrontal cortices of SIV-infected rhesus macaques compared with the saline group. Data are mean ± SEM. An unpaired Student’s t test was used to determine the statistical significance. *p < 0.05 versus saline. (B) qPCR analysis showing increased expression of miRNA-34a in the medial prefrontal cortices of HIV-1 transgenic rats compared with the wild-type rats. Data are mean ± SEM. An unpaired Student’s t test was used to determine the statistical significance. *p < 0.05 versus wild-type. qPCR analysis showing dose-dependent upregulation of miRNA-34a expression in HIV-1 Tat exposed BV-2 cells (C) as well as in mouse primary microglial cells (D). qPCR analysis showing time-dependent upregulation of miRNA-34a expression in HIV-1 Tat (50 ng/mL) exposed BV-2 cells (E) as well as in mouse primary microglial cells (F). Data are mean±SEM from six independent experiments. Nonparametric Kruskal–Wallis one-way ANOVA followed by Dunn’s post hoc test was used to determine the statistical significance of multiple groups. *p < 0.05 versus control. Con, Control.
3.2. Upregulation of miRNA-34a in HIV-1 Tat exposed mouse microglial cells
Based on the findings that the medial prefrontal cortices of both SIV-infected rhesus macaques as well as HIV-1 transgenic rats exhibited significant upregulation of miRNA-34a, we next sought to validate these observations in the murine BV-2 cells as well as in the purified cultures of mouse primary microglial cells exposed to HIV-1 Tat protein (as a surrogate of HIV-1/SIV infection). Both BV-2 cells and mouse primary microglial cells were exposed to varying doses of HIV-1 Tat (25, 50, 100, and 200 ng/ml; for 24 h) and assessed for the expression of mature miRNA-34a. As shown in Figure 1C and 1D, the expression of mature miRNA-34a was dose-dependently upregulated in both BV-2 cells and mouse primary microglial cells exposed to HIV-1 Tat. As expected, heat-inactivated HIV-1 Tat did not have any effect on mature miRNA-34a expression (Figure 1C and 1D). Based on these findings, the concentration of 50 ng/ml of HIV-1 Tat was selected for all further experiments and is in keeping with the circulating levels of HIV-1 Tat found in the serum and cerebrospinal fluid of HIV-1-infected individuals (range, 1–40 ng/ml) (Westendorp et al., 1995; Xiao et al., 2000). It has also been suggested that the local extracellular concentrations of HIV-1 Tat in the CNS could be even higher, especially in the vicinity of HIV-1-infected perivascular cells (Hayashi et al., 2006). Next, we performed time-course experiments to determine the optimal time of HIV-1 Tat-mediated upregulation of miRNA-34a in both the cell types. As shown in Figure 1E and 1F, exposure of both BV-2 cells and mouse primary microglial cells to HIV-1 Tat significantly upregulated the expression levels of mature miRNA-34a starting at 6 h post exposure.
3.3. Upregulation of miRNA-34a targets 3′-UTR of NLRC5 in microglial cells
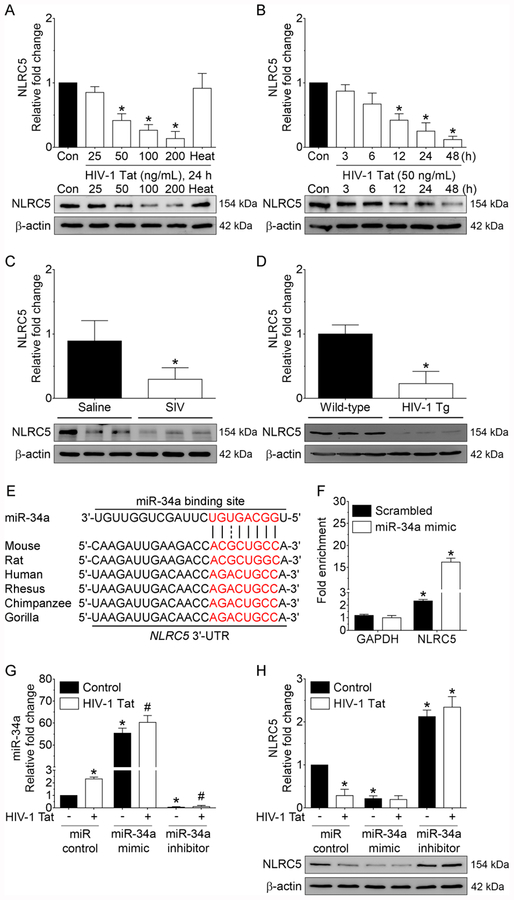
Since miRs primarily function through inhibiting their target mRNA by binding to the 3′-UTR region of the mRNA, we next sought to identify a novel miRNA-34a target that could be critical for microglial activation using online miRNA target prediction database such as TargetScan (http://www.targetscan.org). Using these analyses, we found the NOD-like receptor family CARD domain containing 5 (NLRC5), as a potential target of miRNA-34a. Next, we sought to investigate the expression levels of NLRC5 in HIV-1 Tat exposed mouse microglial cells. Mouse primary microglial cells were exposed to varying doses of HIV-1 Tat (25, 50, 100, and 200 ng/ml; for 24 h) and assessed for the expression of the NLRC5 protein. As shown in Figure 2A, expression of the NLRC5 protein was dose-dependently downregulated in mouse primary microglial cells exposed to HIV-1 Tat for 24 h, and as expected, heat-inactivated HIV-1 Tat did not affect NLRC5 expression (Figure 2A). We then performed a time-course experiment to determine time-dependent downregulation of NLRC5 protein in HIV-1 Tat (50 ng/mL) exposed mouse primary microglial cells. As shown in Figure 2B, exposure of mouse primary microglial cells to HIV-1 Tat significantly downregulated NLRC5 expression levels starting at 12 h. Next, we wanted to validate these in vitro findings in the medial prefrontal cortices of saline and SIV-infected rhesus macaques as well as in the medial prefrontal cortices of both wild-type and HIV-1 transgenic rats. As shown in Figure 2C and 2D, the expression level of NLRC5 protein was significantly decreased in the medial prefrontal cortices of SIV-infected rhesus macaques as well as in HIV-1 transgenic rats compared to their respective controls. Figure 2E demonstrates a conserved miRNA-34a binding site within the NLRC5 3′-UTR of most species. Based on this, we next examined the binding of miRNA-34a with its target, NLRC5, using the miRNA Target IP Kit. Briefly, mouse primary microglial cells were transfected with either miRNA-34a mimic or miRNA control for 24 h, followed by pulldown of mRNAs using anti-Ago antibody (Supplementary Figure 1A and 1B) and assessed for the enrichment of NLRC5 using qPCR. As shown in Figure 2F, miRNA Target IP confirmed NLRC5 as a direct target of miRNA-34a, based on significant enrichment of NLRC5 mRNA bound to Ago protein in cells overexpressing miRNA-34a.
FIGURE 2.
Downregulation of NLRC5 both in vitro and in vivo. Representative western blotting analysis showing the dose-dependent (A) and time-dependent (B) downregulation of NLRC5 expression in HIV-1 Tat exposed mouse primary microglial cells. Data are mean±SEM from six independent experiments. Nonparametric Kruskal–Wallis one-way ANOVA followed by Dunn’s post hoc test was used to determine the statistical significance of multiple groups. *p < 0.05 versus control. Con, Control. (C) Representative western blotting analysis showing decreased expression of the NLRC5 protein in the medial prefrontal cortices of SIV-infected rhesus macaques compared with the saline group. Data are mean ± SEM. An unpaired Student’s t test was used to determine the statistical significance. *p < 0.05 versus saline. (D) Representative western blotting analysis showing decreased expression of the NLRC5 protein in the medial prefrontal cortices of HIV-1 transgenic rats compared with the wild-type rats. Data are mean ± SEM. An unpaired Student’s t test was used to determine the statistical significance. *p < 0.05 versus wild-type. (E) Putative miRNA-34a binding sites in the 3′-UTR of NLRC5 gene. (F) miRNA target validation assay confirmed enrichment of the miRNA-34a target mRNA, NLRC5 in Ago IP compared with total RNA isolated from mouse primary microglial cells transfected with miRNA-34a mimic or miRNA control. GAPDH was used as a housekeeping control. (G) qPCR analysis showing expression levels of miRNA-34a in mouse primary microglial cells transfected with miRNA control, miRNA-34a mimic or miRNA-34a inhibitor following exposure to HIV-1 Tat (50 ng/ml) for 24 h. (H) Representative western blotting analysis showing expression of the NLRC5 protein in mouse primary microglial cells transfected with control, miRNA-34a mimic or miRNA-34a inhibitor following exposure to HIV-1 Tat (50 ng/ml) for 24 h. β-Actin was probed as an internal control for all the experiments. Nonparametric Kruskal–Wallis one-way ANOVA followed by Dunn’s post hoc test was used to determine the statistical significance of multiple groups. *p < 0.05 versus control; #p < 0.05 versus HIV-1 Tat.
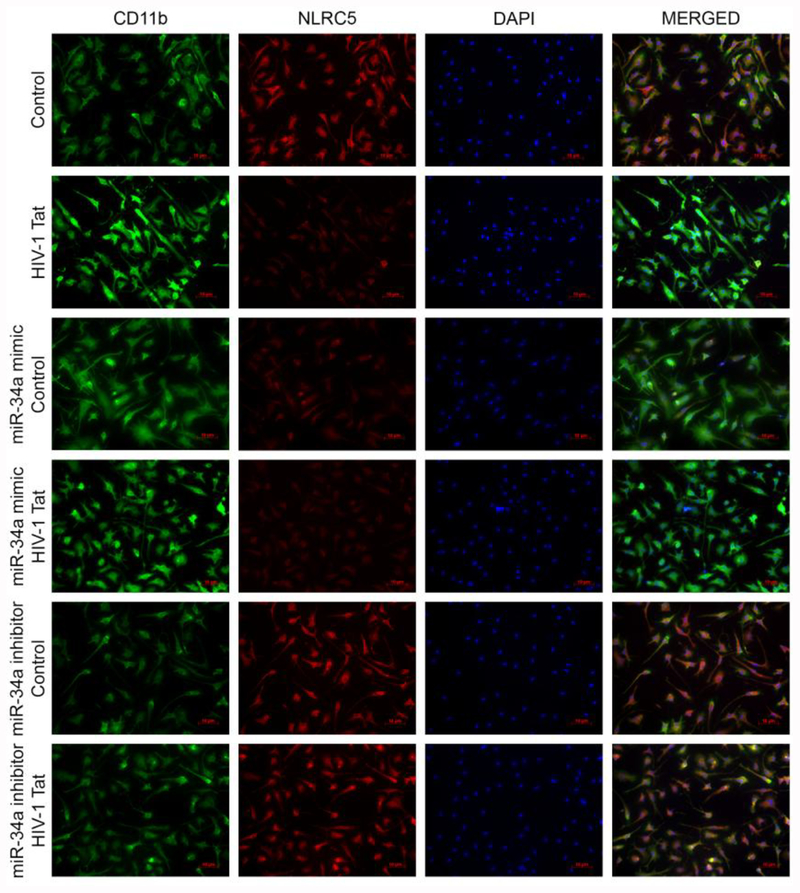
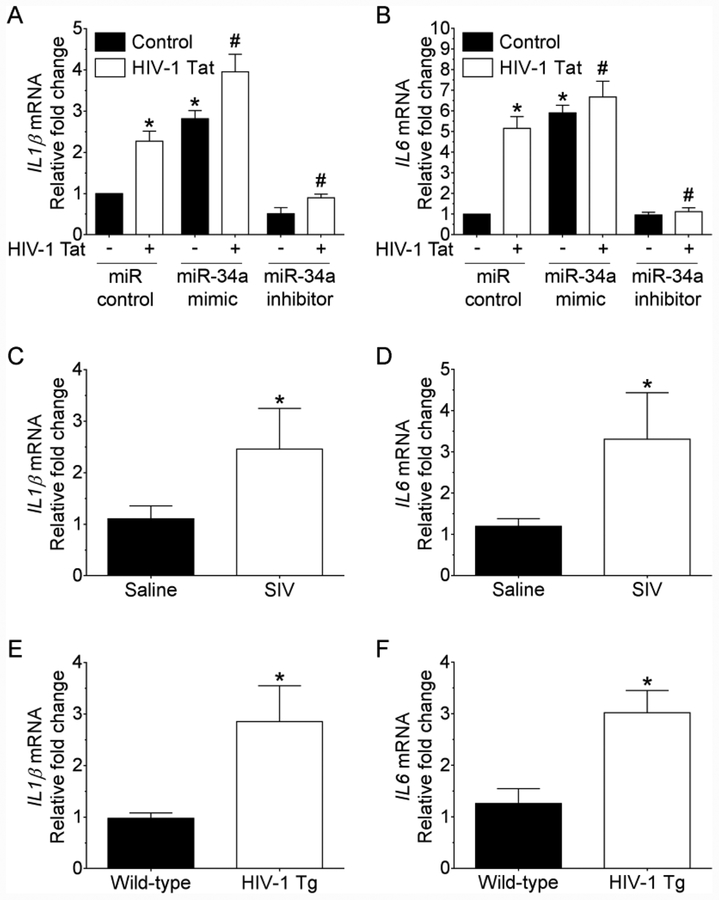
Next, we wanted to demonstrate the effects of miRNA-34a on NLRC5 expression levels in mouse primary microglial cells transiently transfected with miRNA-34a mimic, miRNA-34a inhibitor or miRNA control followed by HIV-1 Tat exposure for 24 h. The transfection efficiency of miRNA-34a is shown in Figure 2G. Interestingly, mouse primary microglial cells transfected with miRNA-34a mimic demonstrated a further downregulation of NLRC5 in the presence of HIV-1 Tat compared with cells transfected miRNA control (Figure 2H). As expected, and in contrast, cells transfected with miRNA-34a inhibitor failed to demonstrate any downregulation of NLRC5 in the presence of HIV-1 Tat. Further validation of these findings was done by double immunostaining of mouse primary microglial cells transfected with miRNA control, miRNA-34a mimic or miRNA-34a inhibitor that was exposed to HIV-1 Tat (Figure 3).
FIGURE 3.
miRNA-34a-mediated downregulation of NLRC5 activates microglia. Representative immunocytochemistry staining showing expression levels of NLRC5 (red) and microglial activation marker, CD11b (green) in mouse primary microglial cells transiently transfected with miRNA-34a mimic or miRNA-34a inhibitor followed by exposure of cells to HIV-1 Tat (50 ng/mL for 24 h). Scalebar: 10 μM.
3.4. NLRC5 negatively regulates the IKK-mediated activation of NFκB in microglia
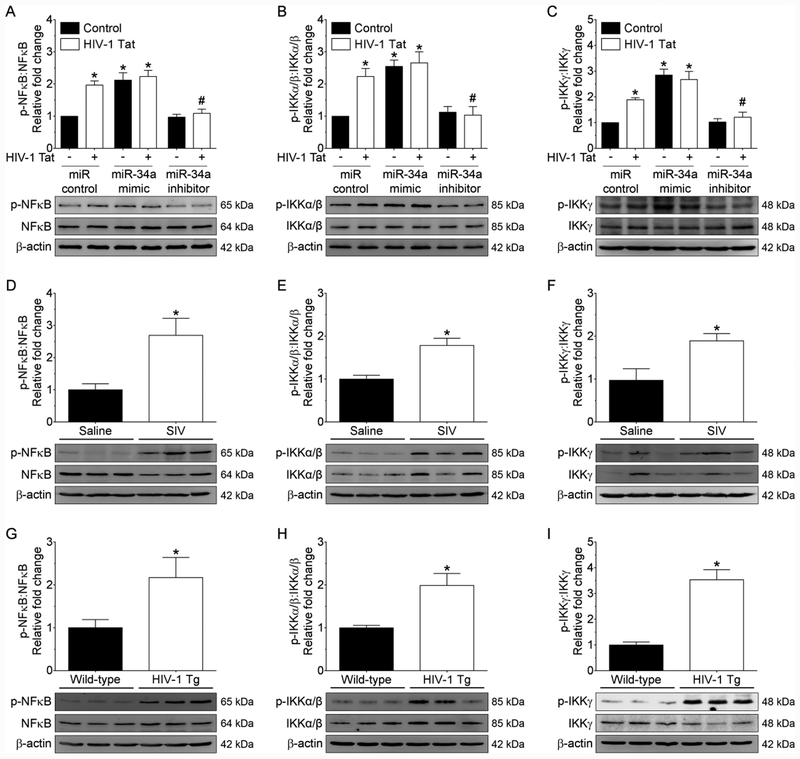
Recent studies have provided evidence of a negative association between NLRC5 and NFκB-mediated activation of proinflammatory cytokines in macrophages, with the involvement of inhibition of IKK complex (Cui et al., 2010a; Meng et al., 2015). NLRC5 is known to inhibit NFκB-dependent responses by interacting with IKKα and IKKβ and blocking their phosphorylation (Cui et al., 2010a). The next step then was to assess the role of miRNA-34a in downregulation of NLRC5 with subsequent activation of NFκB signaling. Mouse primary microglial cells were transfected with either miRNA-34a mimic or miRNA-34a inhibitor followed by exposure of cells to HIV-1 Tat for 24 h. As shown in Figure 4A, transfection of microglial cells with miRNA-34a mimic notably increased the expression of NFκB p65 compared with cells transfected with miRNA control. On the other hand, miRNA-34a inhibitor transfected cells failed to demonstrate upregulation of NFκB p65 subunit phosphorylation in the presence of HIV-1 Tat (Figure 4A). Interestingly and as expected, phosphorylation of IKK complex subunits such as IKKα/β and IKKγ was dramatically increased in miRNA-34a mimic transfected microglial cells and was significantly blocked in miRNA-34a inhibitor transfected microglial cells (Figure 4B and 4C). Further, we sought to determine the expression levels of both total and phosphorylated forms of NFκB p65, IKKα/β, and IKKγ in the medial prefrontal cortices of saline and SIV-infected rhesus macaques as well as wild-type and HIV-1 transgenic rats. As shown in Figure 4D–4F, there was significant upregulation of phosphorylated NFκB p65, IKKα/β, and IKKγ proteins in the medial prefrontal cortices of SIV-infected rhesus macaques compared with saline administered rhesus macaques. Similarly, there was a significant increase in the phosphorylated forms of NFκB p65, IKKα/β, and IKKγ proteins in the medial prefrontal cortices of HIV-1 transgenic rats compared with wild-type rats (Figure 4G–4I).
FIGURE 4.
NLCR5 negatively regulates IKK-mediated microglial NFκB signaling. Representative western blotting analysis showing phosphorylated and total forms of NFκB p65 (A), IKKα/β (B), and IKKγ (C) in mouse primary microglial cells transfected with control, miRNA-34a mimic or miRNA-34a inhibitor, following exposure of cells to HIV-1 Tat (50 ng/ml; 24 h). Nonparametric Kruskal–Wallis one-way ANOVA followed by Dunn’s post hoc test was used to determine the statistical significance of multiple groups. *p < 0.05 versus control; #p < 0.05 versus HIV-1 Tat. Representative western blotting analysis showing the phosphorylated and total forms of NFκB p65 (D), IKKα/β (E), and IKKγ (F) in the medial prefrontal cortices of SIV-infected rhesus macaques and saline controls. Data are mean ± SEM. An unpaired Student’s t test was used to determine the statistical significance. *p < 0.05 versus wild-type. Representative western blotting analysis showing the phosphorylated and total forms of NFκB p65 (G), IKKα/β (H), and IKKγ (I) in the medial prefrontal cortices of HIV-1 transgenic and wild-type rats. Data are mean ± SEM. An unpaired Student’s t test was used to determine the statistical significance. *p < 0.05 versus saline. β-Actin was probed as an internal control for all the experiments.
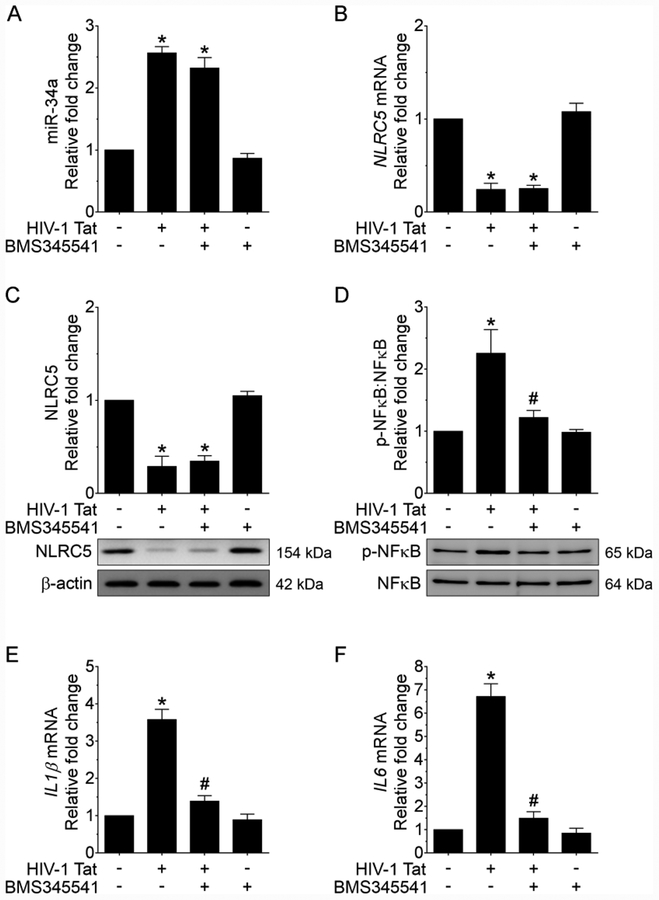
Next, we sought to determine whether blocking NFκB signaling by inhibiting phosphorylation of IKK complex using the pharmacological inhibitor could prevent HIV-1 Tat-mediated downregulation of NLRC5. Mouse primary microglial cells were pretreated with the pharmacological inhibitor, 10 μM BMS345541 (a selective allosteric inhibitor of IKK that blocks NFκB-dependent transcription) for 1 h, followed by exposure of cells to HIV-1 Tat for additional 24 h. Cells were then assessed for the expression levels of miRNA-34a. As shown in Figure 5A, pretreatment of microglial cells with the inhibitor failed to improve HIV-1 Tat-mediated upregulation of miRNA-34a thereby suggesting that activation of miRNA-34a lies upstream of NFκB activation. Additionally, we also determined the expression levels of both NLRC5 mRNA and protein in cells pretreated with this inhibitor followed by exposure of cells to HIV-1 Tat. As shown in Figure 5B and 5C and similar to the findings presented above, cells pretreated with the inhibitor failed to improve HIV-1 Tat-mediated downregulation of NLRC5. In contrast, however, in microglial cells pretreated with BMS345541 HIV-1 Tat failed to upregulate the phosphorylation of NFκB p62 (Figure 5D) as well as the expression of downstream proinflammatory cytokines such as IL1β, and IL6 (Figure 5E and 5F).
FIGURE 5.
Pharmacological inhibition of NFκB signaling alleviates HIV-1 Tat-mediated microglial inflammation. qPCR analysis showing expression of miRNA-34a (A) and NLRC5 mRNA (B) in mouse primary microglial cells pretreated with IKK complex inhibitor - BMS345541 for 1 h followed by exposure of cells to HIV-1 Tat (50 ng/ml; 24 h). Representative western blotting analysis showing expression levels of NLRC5 (C) and NFκB p65 (D) in mouse primary microglial cells pretreated with IKK complex inhibitor - BMS345541 (10 μM) for 1 h followed by exposure of cells to HIV-1 Tat (50 ng/ml; 24 h). qPCR analysis demonstrating mRNA expression of IL1β (E) and IL6 (F) in mouse primary microglial cells pretreated with IKK complex inhibitor - BMS345541 for 1 h followed by exposure of cells to HIV-1 Tat (50 ng/ml; 24 h). Data are mean±SEM of six independent experiments. β-Actin was probed as an internal control for all the experiments. Nonparametric Kruskal–Wallis one-way ANOVA followed by Dunn’s post hoc test was used to determine the statistical significance of multiple groups. *p < 0.05 versus control; #p < 0.05 versus HIV-1 Tat.
3.5. NLRC5-mediated negative regulation of NFκB signaling activates microglial proinflammatory cytokines
Next, we sought to examine the expression levels of proinflammatory cytokines such as IL1β and IL6 in mouse primary microglial cells transfected with either miRNA-34a mimic or miRNA-34a inhibitor for 24 h followed by exposure of cells to HIV-1 Tat. As shown in Figure 6A and 6B, mRNA expression levels of IL1β and IL6 were significantly elevated in miRNA-34a mimic transfected microglial cells in the presence of HIV-1 Tat. In contrast, in cells transfected with miRNA-34a inhibitor, HIV-1 Tat failed to upregulate the mRNA expression of IL1β and IL6. Additionally, we also determined the expression levels of IL1β and IL6 mRNA in the medial prefrontal cortices of saline and SIV-infected rhesus macaques. As shown in Figure 6C and 6D, expression levels of IL1β and IL6 were significantly increased in the medial prefrontal cortices of SIV-infected rhesus macaques compared with saline controls. Similarly, expression levels of IL1β and IL6 were significantly increased in the medial prefrontal cortices of HIV-1 transgenic rats compared with the wild-type controls (Figure 6E and 6F). Overall, HIV-1 Tat-mediated upregulation of miRNA-34a resulted in downregulation of NLRC5 which, in turn, negatively regulated NFκB p65 signaling, ultimately leading to increased production of proinflammatory cytokines and ensuing neuroinflammation.
FIGURE 6.
miRNA-34a upregulates the expression of proinflammatory cytokines. qPCR analysis showing mRNA expression levels of IL1β (A), and IL6 (B) in mouse primary microglial cells transfected with control, miRNA-34a mimic or miRNA-34a inhibitor followed by exposure of cells to HIV-1 Tat (50 ng/ml;24 h). Data are mean±SEM of six independent experiments. β-Actin was probed as an internal control for all the experiments. Nonparametric Kruskal–Wallis one-way ANOVA followed by Dunn’s post hoc test was used to determine the statistical significance of multiple groups. *p < 0.05 versus control; #p < 0.05 versus HIV-1 Tat. qPCR analysis showing increased mRNA expression of IL1β (C) and IL6 (D) in the medial prefrontal cortices of SIV-infected and saline-injected rhesus macaques. qPCR analysis showing increased mRNA expression of IL1β (E) and IL6 (F) in the medial prefrontal cortices of HIV-1 transgenic and wild-type rats. Data are mean ± SEM. An unpaired Student’s t test was used to determine the statistical significance. *p < 0.05 versus saline.
4. DISCUSSION
This study provides new mechanistic insights into HIV-1 Tat-mediated upregulation of miRNA-34a in regulating microglial activation via targeting the NLRC5 inflammasome signaling axis. Herein, we have demonstrated that HIV-1 Tat exposure significantly elevated the expression of miRNA-34a resulting in targeting of the 3′-UTR of NLRC5 inflammasome protein, in microglial cells. HIV-1 Tat-mediated reduced expression of NLRC5 resulted in activation of microglia involving negative regulation of NFκB p65 signaling.
We first determined the expression levels of miRNA-34a since this miRNA has been reported to play a role in premature aging associated with various diseases (Bavamian et al., 2015; Chang et al., 2011; Duskova et al., 2013; Hu et al., 2017; Jauhari et al., 2018; Kou et al., 2016; Li et al., 2011; Reynolds et al., 2018; Witwer et al., 2012). It is well known that numerous inducible miRNAs like miRNA-34a are under transcriptional control by the proinflammatory transcription factor NFκB in the CNS and other tissues. Upregulated NFκB could thus play a vital role in the upregulation of various proinflammatory cytokines and ensuing neurodegeneration and cancer (Bhattacharjee et al., 2016; Cui et al., 2010b; Li et al., 2012; Lukiw, 2012; Lukiw et al., 2008). Several studies have demonstrated that increased expression of miRNA-34a-signalling is associated with various neurological, neuroimmune, neuroinflammatory or neurodegenerative pathologies (Agostini et al., 2011b; Bhattacharjee et al., 2016; Boon et al., 2013; Gao et al., 2013; Junker et al., 2009; Kofman et al., 2013; Li et al., 2011; Piccio et al., 2007; Wang et al., 2009). The involvement of miRNA-34a in HIV-1 Tat-mediated microglial activation, however, has never been explored before and is particularly interesting because of the participation of NLRC5 and NFκB p65 signaling axis in cellular activation.
Based on the premise that microglial activation associated with the release of cytotoxic mediators such as proinflammatory cytokines, chemokines, and ROS contributes to the pathogenesis of HIV-1-associated neurocognitive disorders (Chen et al., 2017), we focused our study on the involvement of miRNA-34a in microglial activation during SIV infection as well as following HIV-1 Tat exposure (as a surrogate for HIV infection; see below). Upregulation of miRNA-34a in the medial prefrontal cortices of both HIV-1 transgenic rats as well as SIV-infected rhesus macaques indicated an association of upregulated miRNA-34a with neuroinflammation. Next, we validated these findings in vitro. The rationale for using HIV-1 Tat in cell culture was based on the fact that despite effective suppression of viremia by cART, HIV-1 Tat protein continues to be present in the CNS and lymph nodes of treated patients (Hudson et al., 2000; Wiley et al., 1996), leading to underlying inflammation both in the brain and periphery. Furthermore, several reports are confirming the occurrence of low-level virus replication in the CNS of cART-treated patients (Hudson et al., 2000; Wiley et al., 1996), thereby providing credence to the use of HIV-1 Tat in our in vitro studies. Our study demonstrates that exposure of mouse primary microglial cells to HIV-1 Tat demonstrated a dose- and time-dependent upregulation of miRNA-34a.
NLRC5 is a new member of the Nod-like receptor (NLR) family that is abundantly expressed in mucosal epithelial cells and brain (Kuenzel et al., 2010). Similar to other classic NLR members, NLRC5 is emerging as a regulator of immune responses to combat intruding microbes (Lamkanfi and Kanneganti, 2012). One of the most conspicuous features of NLRC5 is that its expression closely correlates with inflammatory responses (Benko et al., 2010). Recent studies have demonstrated that NLRC5 negatively regulates NFκB signaling via direct binding to the NFκB regulators IKKα/IKKβ, thereby preventing both the recruitment of IKKγ as well as nuclear translocation of NFκB (Benko et al., 2010; Cui et al., 2010a). Additionally, NLRC5 has also been speculated to modulate the transactivation potential of nuclear NFκB leading to increased production of the anti-inflammatory cytokine, IL10 (Benko et al., 2010; Cui et al., 2010a). Typically, NFκB serves as a critical point of convergence for multiple signaling pathways that are relevant for the antiviral response and subsequent activation of the adaptive immune responses (Hacker and Karin, 2006). Consequently, downregulation of NLRC5 could represent a unique mechanism by which HIV-1 Tat interferes with the host defense system, leading to persistent inflammation and cellular activation. In our study, exposure of microglial cells to HIV-1 Tat resulted in significant downregulation of NLRC5 protein with concomitant activation of NFκB signaling, thereby suggesting a possible association of NLRC5-mediated activation of NFκB signaling axis in microglia. Additionally, this study, for the first time, reports a novel 3′-UTR target of miRNA-34a, such as NLRC5. Using multiple prediction algorithms, we found that a putative miRNA-34a targeting site was located within the 3′-UTR of NLRC5, which was also potentially related to HIV-1 Tat-mediated microglial inflammation. In the present study, we also demonstrated that HIV-1 Tat-mediated upregulation of miRNA-34a decreased the expression of the 3′-UTR target protein NLRC5, in microglial cells. As shown earlier, reduced expression of NLRC5 can activate the NFκB signaling axis via negative regulation (Benko et al., 2010; Cui et al., 2010a). It is well-established that proinflammatory cytokines secreted by activated glia or damaged cells play a fundamental role in triggering neuroinflammation. Accordingly, it is likely that HIV-1 Tat-mediated microglial activation leads to overexpression of proinflammatory cytokines, further underscoring the detrimental role of HIV-1 Tat in mediating microglial activation. This study was mainly focused on HIV-1 Tat-mediated microglial inflammation in vitro. These findings were also validated in vivo using medial prefrontal cortices of wild-type and HIV-1 transgenic rats as well as in archival medial prefrontal cortices of SIV-infected rhesus macaques. Future studies are, however, warranted to explore other host and viral components responsible for NLRC5 downregulation and microglial activation.
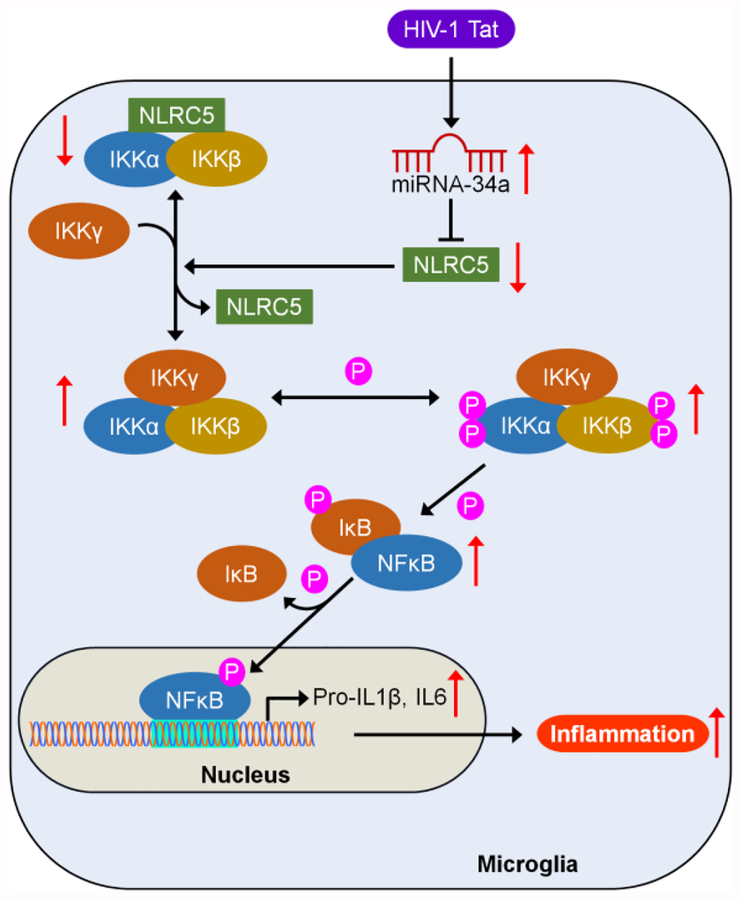
In conclusion, this study demonstrated that HIV-1 Tat-mediated upregulation of miRNA-34a targets the 3′-UTR of NLRC5 in microglial cells and that downregulated NLRC5 inflammasome negatively regulates NFκB p65 signaling axis via modulation of phosphorylated IKK complex, leading, in turn, to increased expression of proinflammatory cytokines such as IL1β, and IL6 and ensuing microglial activation (Figure 7). These novel results provide functional insights into the interaction of HIV-1 Tat with miRNA-34a-NLRC5-NFκB signaling axis and implicate that silencing of miRNA-34a, as well as inhibition of phosphorylation of IKK complex, could be developed as an alternative approach for the treatment of HIV-1 Tat-mediated microglial inflammation.
FIGURE 7.
Schematic diagram demonstrating HIV-1 Tat-mediated upregulation of miRNA-34a, leading to downregulation of its target - NLRC5, which in turn, negatively regulates the NFκB signaling axis, resulting in increased expression of proinflammatory cytokines such IL1β and IL6 in microglia.
Supplementary Material
Highlights:
miRNA-34a expression is increased in the frontal cortices of both SIV-infected rhesus macaques and in the HIV-1 transgenic rats.
miRNA-34a targets the inflammasome protein, NLRC5.
NLRC5 negatively regulates the NFκB signaling axis.
HIV-1 Tat-mediated microglial activation involves miRNA-34a-NLRC5-NFκB signaling axis.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health Grants P30MH062261, R01MH106425, R01DA043138, and R01DA035203 to S.B., and R03DA044087 to P.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare no competing financial interests.
REFERENCES
- Agostini M, Knight RA, 2014. miR-34: from bench to bedside. Oncotarget 5, 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Killick R, Candi E, Sayan BS, Rivetti di Val Cervo P, Nicotera P, McKeon F, Knight RA, Mak TW, Melino G, 2011a. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci U S A 108, 21093–21098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, Forsythe ID, Young KW, Ventura A, Concepcion CP, Han YC, Candi E, Knight RA, Mak TW, Melino G, 2011b. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci U S A 108, 21099–21104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagashev A, Sawaya BE, 2013. Roles and functions of HIV-1 Tat protein in the CNS: an overview. Virol J 10, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavamian S, Mellios N, Lalonde J, Fass DM, Wang J, Sheridan SD, Madison JM, Zhou F, Rueckert EH, Barker D, Perlis RH, Sur M, Haggarty SJ, 2015. Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Mol Psychiatry 20, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benko S, Magalhaes JG, Philpott DJ, Girardin SE, 2010. NLRC5 limits the activation of inflammatory pathways. J Immunol 185, 1681–1691. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S, Zhao Y, Dua P, Rogaev EI, Lukiw WJ, 2016. microRNA-34a-Mediated Down-Regulation of the Microglial-Enriched Triggering Receptor and Phagocytosis-Sensor TREM2 in Age-Related Macular Degeneration. PLoS One 11, e0150211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokhari SM, Hegde R, Callen S, Yao H, Adany I, Li Q, Li Z, Pinson D, Yeh HW, Cheney PD, Buch S, 2011. Morphine potentiates neuropathogenesis of SIV infection in rhesus macaques. J Neuroimmune Pharmacol 6, 626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER, 2007. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol 17, 1298–1307. [DOI] [PubMed] [Google Scholar]
- Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Muller OJ, Potente M, Zeiher AM, Hermeking H, Dimmeler S, 2013. MicroRNA-34a regulates cardiac ageing and function. Nature 495, 107–110. [DOI] [PubMed] [Google Scholar]
- Carroll A, Brew B, 2017. HIV-associated neurocognitive disorders: recent advances in pathogenesis, biomarkers, and treatment. F1000Res 6, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JR, Mukerjee R, Bagashev A, Del Valle L, Chabrashvili T, Hawkins BJ, He JJ, Sawaya BE, 2011. HIV-1 Tat protein promotes neuronal dysfunction through disruption of microRNAs. J Biol Chem 286, 41125–41134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NC, Partridge AT, Sell C, Torres C, Martin-Garcia J, 2017. Fate of microglia during HIV-1 infection: From activation to senescence? Glia 65, 431–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivero ET, Guo ML, Periyasamy P, Liao K, Callen SE, Buch S, 2017. HIV-1 Tat Primes and Activates Microglial NLRP3 Inflammasome-Mediated Neuroinflammation. J Neurosci 37, 3599–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen ZJ, Wang RF, 2010a. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell 141, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JG, Li YY, Zhao Y, Bhattacharjee S, Lukiw WJ, 2010b. Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by microRNA-146a and NF-kappaB in stressed human astroglial cells and in Alzheimer disease. J Biol Chem 285, 38951–38960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GM, Haas MA, Pocock R, 2015. MicroRNAs: Not “Fine-Tuners” but Key Regulators of Neuronal Development and Function. Front Neurol 6, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duskova K, Nagilla P, Le HS, Iyer P, Thalamuthu A, Martinson J, Bar-Joseph Z, Buchanan W, Rinaldo C, Ayyavoo V, 2013. MicroRNA regulation and its effects on cellular transcriptome in human immunodeficiency virus-1 (HIV-1) infected individuals with distinct viral load and CD4 cell counts. BMC Infect Dis 13, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensoli B, Buonaguro L, Barillari G, Fiorelli V, Gendelman R, Morgan RA, Wingfield P, Gallo RC, 1993. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J Virol 67, 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel AD, Pabo CO, 1988. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 55, 1189–1193. [DOI] [PubMed] [Google Scholar]
- Gao H, Zhao H, Xiang W, 2013. Expression level of human miR-34a correlates with glioma grade and prognosis. J Neurooncol 113, 221–228. [DOI] [PubMed] [Google Scholar]
- Guo ML, Periyasamy P, Liao K, Kook YH, Niu F, Callen SE, Buch S, 2016. Cocaine-mediated downregulation of microglial miR-124 expression involves promoter DNA methylation. Epigenetics 11, 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker H, Karin M, 2006. Regulation and function of IKK and IKK-related kinases. Sci STKE 2006, re13. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Pu H, Andras IE, Eum SY, Yamauchi A, Hennig B, Toborek M, 2006. HIV-TAT protein upregulates expression of multidrug resistance protein 1 in the blood-brain barrier. J Cereb Blood Flow Metab 26, 1052–1065. [DOI] [PubMed] [Google Scholar]
- Hu G, Liao K, Yang L, Pendyala G, Kook Y, Fox HS, Buch S, 2017. Tat-Mediated Induction of miRs-34a & −138 Promotes Astrocytic Activation via Downregulation of SIRT1: Implications for Aging in HAND. J Neuroimmune Pharmacol 12, 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Yao H, Chaudhuri AD, Duan M, Yelamanchili SV, Wen H, Cheney PD, Fox HS, Buch S, 2012. Exosome-mediated shuttling of microRNA-29 regulates HIV Tat and morphine-mediated neuronal dysfunction. Cell Death Dis 3, e381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson L, Liu J, Nath A, Jones M, Raghavan R, Narayan O, Male D, Everall I, 2000. Detection of the human immunodeficiency virus regulatory protein tat in CNS tissues. J Neurovirol 6, 145–155. [DOI] [PubMed] [Google Scholar]
- Jauhari A, Singh T, Singh P, Parmar D, Yadav S, 2018. Regulation of miR-34 Family in Neuronal Development. Mol Neurobiol 55, 936–945. [DOI] [PubMed] [Google Scholar]
- Jayadev S, Garden GA, 2009. Host and viral factors influencing the pathogenesis of HIV-associated neurocognitive disorders. J Neuroimmune Pharmacol 4, 175–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, Bittner R, Lassmann H, Wekerle H, Hohlfeld R, Meinl E, 2009. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 132, 3342–3352. [DOI] [PubMed] [Google Scholar]
- Kofman AV, Kim J, Park SY, Dupart E, Letson C, Bao Y, Ding K, Chen Q, Schiff D, Larner J, Abounader R, 2013. microRNA-34a promotes DNA damage and mitotic catastrophe. Cell Cycle 12, 3500–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou X, Liu X, Chen X, Li J, Yang X, Fan J, Yang Y, Chen N, 2016. Ampelopsin attenuates brain aging of D-gal-induced rats through miR-34a-mediated SIRT1/mTOR signal pathway. Oncotarget 7, 74484–74495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenzel S, Till A, Winkler M, Hasler R, Lipinski S, Jung S, Grotzinger J, Fickenscher H, Schreiber S, Rosenstiel P, 2010. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J Immunol 184, 1990–2000. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Kanneganti TD, 2012. Regulation of immune pathways by the NOD-like receptor NLRC5. Immunobiology 217, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang K, Chen X, Meng H, Song M, Wang Y, Xu X, Bai Y, 2012. Transcriptional activation of microRNA-34a by NF-kappa B in human esophageal cancer cells. BMC Mol Biol 13, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Khanna A, Li N, Wang E, 2011. Circulatory miR34a as an RNAbased, noninvasive biomarker for brain aging. Aging (Albany NY) 3, 985–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ, 2012. NF-small ka, CyrillicB-regulated micro RNAs (miRNAs) in primary human brain cells. Exp Neurol 235, 484–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ, Zhao Y, Cui JG, 2008. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem 283, 31315–31322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maubert ME, Pirrone V, Rivera NT, Wigdahl B, Nonnemacher MR, 2015. Interaction between Tat and Drugs of Abuse during HIV-1 Infection and Central Nervous System Disease. Front Microbiol 6, 1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Cai C, Sun T, Wang Q, Xie W, Wang R, Cui J, 2015. Reversible ubiquitination shapes NLRC5 function and modulates NF-kappaB activation switch. J Cell Biol 211, 1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minghetti L, Visentin S, Patrizio M, Franchini L, Ajmone-Cat MA, Levi G, 2004. Multiple actions of the human immunodeficiency virus type-1 Tat protein on microglial cell functions. Neurochem Res 29, 965–978. [DOI] [PubMed] [Google Scholar]
- Morgado AL, Xavier JM, Dionisio PA, Ribeiro MF, Dias RB, Sebastiao AM, Sola S, Rodrigues CM, 2015. MicroRNA-34a Modulates Neural Stem Cell Differentiation by Regulating Expression of Synaptic and Autophagic Proteins. Mol Neurobiol 51, 1168–1183. [DOI] [PubMed] [Google Scholar]
- Nath A, Conant K, Chen P, Scott C, Major EO, 1999. Transient exposure to HIV-1 Tat protein results in cytokine production in macrophages and astrocytes. A hit and run phenomenon. J Biol Chem 274, 17098–17102. [DOI] [PubMed] [Google Scholar]
- Pendyala G, Periyasamy P, Callen S, Fox HS, Lisco SJ, Buch SJ, 2015. Chronic SIV and morphine treatment increases heat shock protein 5 expression at the synapse. J Neurovirol 21, 592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Vigorito M, Liu X, Zhou D, Wu X, Chang SL, 2010. The HIV-1 transgenic rat as a model for HIV-1 infected individuals on HAART. J Neuroimmunol 218, 94–101. [DOI] [PubMed] [Google Scholar]
- Periyasamy P, Guo ML, Buch S, 2016. Cocaine induces astrocytosis through ER stress-mediated activation of autophagy. Autophagy 12, 1310–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periyasamy P, Liao K, Kook YH, Niu F, Callen SE, Guo ML, Buch S, 2018a. Cocaine-Mediated Downregulation of miR-124 Activates Microglia by Targeting KLF4 and TLR4 Signaling. Mol Neurobiol 55, 3196–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periyasamy P, Thangaraj A, Guo ML, Hu G, Callen S, Buch S, 2018b. Epigenetic Promoter DNA Methylation of miR-124 Promotes HIV-1 Tat-Mediated Microglial Activation via MECP2-STAT3 Axis. J Neurosci 38, 5367–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccio L, Buonsanti C, Mariani M, Cella M, Gilfillan S, Cross AH, Colonna M, Panina-Bordignon P, 2007. Blockade of TREM-2 exacerbates experimental autoimmune encephalomyelitis. Eur J Immunol 37, 1290–1301. [DOI] [PubMed] [Google Scholar]
- Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL, 2011. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med 17, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds RH, Petersen MH, Willert CW, Heinrich M, Nymann N, Dall M, Treebak JT, Bjorkqvist M, Silahtaroglu A, Hasholt L, Norremolle A, 2018. Perturbations in the p53/miR-34a/SIRT1 pathway in the R6/2 Huntington’s disease model. Mol Cell Neurosci 88, 118–129. [DOI] [PubMed] [Google Scholar]
- Rokavec M, Li H, Jiang L, Hermeking H, 2014. The p53/miR-34 axis in development and disease. J Mol Cell Biol 6, 214–230. [DOI] [PubMed] [Google Scholar]
- Saylor D, Dickens AM, Sacktor N, Haughey N, Slusher B, Pletnikov M, Mankowski JL, Brown A, Volsky DJ, McArthur JC, 2016. HIV-associated neurocognitive disorder--pathogenesis and prospects for treatment. Nat Rev Neurol 12, 234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW, 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaper SD, Argentini C, Barbierato M, 2012. Culture of neonatal rodent microglia, astrocytes, and oligodendrocytes from cortex and spinal cord. Methods Mol Biol 846, 67–77. [DOI] [PubMed] [Google Scholar]
- Thangaraj A, Periyasamy P, Liao K, Bendi VS, Callen S, Pendyala G, Buch S, 2018. HIV-1 TAT-mediated microglial activation: role of mitochondrial dysfunction and defective mitophagy. Autophagy 14, 1596–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassallo M, Dunais B, Durant J, Carsenti-Dellamonica H, Harvey-Langton A, Cottalorda J, Ticchioni M, Laffon M, Lebrun-Frenay C, Dellamonica P, Pradier C, 2013. Relevance of lipopolysaccharide levels in HIV-associated neurocognitive impairment: the Neuradapt study. J Neurovirol 19, 376–382. [DOI] [PubMed] [Google Scholar]
- Wang X, Liu P, Zhu H, Xu Y, Ma C, Dai X, Huang L, Liu Y, Zhang L, Qin C, 2009. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer’s disease, inhibits bcl2 translation. Brain Res Bull 80, 268–273. [DOI] [PubMed] [Google Scholar]
- Westendorp MO, Frank R, Ochsenbauer C, Stricker K, Dhein J, Walczak H, Debatin KM, Krammer PH, 1995. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature 375, 497–500. [DOI] [PubMed] [Google Scholar]
- Wiley CA, Baldwin M, Achim CL, 1996. Expression of HIV regulatory and structural mRNA in the central nervous system. AIDS 10, 843–847. [DOI] [PubMed] [Google Scholar]
- Witwer KW, Watson AK, Blankson JN, Clements JE, 2012. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology 9, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Neuveut C, Tiffany HL, Benkirane M, Rich EA, Murphy PM, Jeang KT, 2000. Selective CXCR4 antagonism by Tat: implications for in vivo expansion of coreceptor use by HIV-1. Proc Natl Acad Sci U S A 97, 11466–11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Zhou S, Yi S, Gu X, 2015. The regulatory roles of non-coding RNAs in nerve injury and regeneration. Prog Neurobiol 134, 122–139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.